Abstract
The innate immune system forms an evolutionarily ancient line of defense against invading pathogens and endogenous danger signals. Within certain cells of innate immunity, including epithelial cells and macrophages, intricate molecular machineries named inflammasomes sense a wide array of stimuli to mount inflammatory responses. Dysregulation in inflammasome signaling leads to a wide range of immune disorders such as gout, Crohn’s disease, and sepsis. Recent technological advances in cryo-electron microscopy (cryo-EM) have enabled the structural determination of several key signaling molecules in inflammasome pathways, from which macromolecular assembly emerges as a common mechanistic theme. Through the assembly of helical filaments, symmetric disks, and transmembrane pores, inflammasome pathways employ highly dynamic yet ordered processes to relay and amplify signals. These unprecedentedly detailed views of inflammasome signaling not only revolutionize our understanding of inflammation, but also pave the way for the development of therapeutics against inflammatory diseases.
Introduction
Innate immunity is mediated by a series of germline-encoded pattern recognition receptors (PRRs) that respond to pathogen-associated and damage-associated molecular patterns (PAMPs and DAMPs). The already identified PRR families include transmembrane proteins such as Toll-like receptors (TLRs) and C-type lectin receptors (CLRs), as well as cytosolic proteins such as RIG-I like receptors (RLRs), AIM2-like receptors (ALRs), nucleotide-binding domain (NBD) and leucinerich repeat-containing (LRR) proteins (NLRs), and cyclic GMP-AMP synthase (cGAS) (Lamkanfi and Dixit 2014; Kesavardhana and Kanneganti 2017; Kagan, Magupalli, and Wu 2014).
When triggered by PAMPs and DAMPs, many PRRs stimulate the downstream transcription of inflammation-related genes encoding pro-inflammatory cytokines, interferons, and antimicrobial proteins. By contrast, certain NLRs and ALRs induce inflammatory signaling through the assembly of cytosolic multimeric protein complexes known as canonical inflammasomes, which comprise the NLR and ALR sensors, adaptors such as apoptosis-associated, speck-like protein containing a caspase recruitment domain (ASC), and effectors such as caspase-1 (Schroder and Tschopp 2010; Yin et al. 2015; Lu and Wu 2015) (Figure 1a). In addition to canonical inflammasomes, cytosolic lipopolysaccharides (LPS) from Gram-negative bacteria and oxidized phospholipids can directly engage caspase-11 to form the non-canonical inflammasome, which then activates caspase-11 (Kayagaki et al. 2011; Kayagaki et al. 2013; Zanoni et al. 2016). Inflammasome-activated caspases promote the proteolytic maturation of cytokines interleukin-1β (IL-1 β) and IL-18, and cleave effector molecule gasdermin D (GSDMD) to free its active N-terminal fragment (GSDMD-NT) from its auto-inhibitory C-terminal fragment (GSDMD-CT) (Kayagaki et al. 2015; Shi et al. 2015) (Figure 1a). Membrane pore formation by GSDMD-NT regulates cytokine release and results in pyroptosis, a highly inflammatory form of programmed cell death (Liu et al. 2016; Qi 2016; Shi, Gao, and Shao 2017; Wright and Bryant 2016).
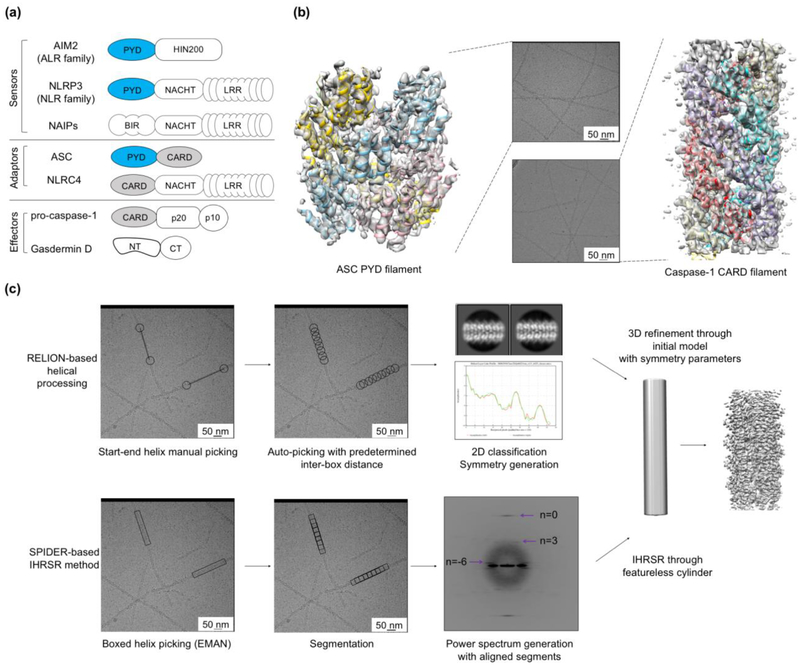
Figure 1. Molecular components and filamentous structures in inflammasome signaling.
(a) Domain organization of proteins involved in inflammasome pathways. (b) Raw cryo-EM images and structures of ASCPYD (PDB ID: 3J63) and caspase-1CARD (PDB ID: 5FNA) filaments overlaid with their cryo-EM maps. (c) General procedures of helical processing. For data processing in RELION, in the manual picking step, black circles indicate the start-end coordinates, and black lines indicate the length of the picked filaments. In the auto-picking step, the distance between different black circles is defined as the inter-box distance, which equals the number of asymmetric units multiplied by the helical rise. Good 2D class averages are selected to generate layer line information (He and Scheres 2017). For the IHRSR method in SPIDER, helical filaments are picked in box manually with EMAN (Ludtke, Baldwin, and Chiu 1999). In the segmentation step, the rectangle box length is determined empirically (Egelman 2010). The image source of the powerspectrum (Shen et al. 2019).
In this review, we discuss recent structural studies of inflammasome signaling using the cutting edge cryo-EM. A characteristic of inflammasome pathways is the assembly of large macromolecular complexes, often heterogeneous and intractable to X-ray crystallography. Cryo-EM structures of these complexes have provided detailed mechanistic insights into inflammasome signaling and a new paradigm for signal transduction.
Inflammasome filaments reconstructed by cryo-EM
Widely distributed among inflammasome sensors, adaptors, and effectors, the death domain (DD) superfamily comprises protein-protein interaction domains crucial in inflammatory signaling, and consists of death domain (DD), death effector domain (DED), caspase recruitment domain (CARD), and Pyrin domain (PYD). Structurally, all family members share an antiparallel six-helix bundle architecture despite differences in their primary sequence (Ferrao et al. 2012; Park et al. 2007). For ALR and NLR PYD-containing (NLRP) inflammasomes, the PYD of ALR and NLRP sensors is responsible for recruiting the adaptor protein ASC, which has an N-terminal PYD and a C-terminal CARD, through homotypic PYD-PYD interactions. The C-terminal CARD of ASC then serves as a scaffold for the recruitment of downstream effector caspase-1 through CARD-CARD interactions. On the other hand, the assembly of NLR CARD-containing (NLRC) inflammasomes is ASC-independent, as NLRCs directly engage caspase-1 via CARD-CARD interactions.
Prior to the cryo-EM revolution, nuclear magnetic resonance (NMR) and X-ray crystallography were used to determine DD superfamily structures in inflammasomes (PDB ID: NLRP3, 2NAQ, 3QF2; NLRP1, 1PN5; NLRP4, 4EWI; NLRP7, 2KM6; NLRP10, 2DO9; and NLRP12, 2l6A), and often an acidic environment was used in these two methods to keep the proteins in a monomeric or dimeric form. Under physiological pH, however, DD superfamily proteins often assemble into filamentous polymers. The recently resolved cryo-EM structures of ASCPYD and caspase-1CARD filaments reveal the molecular basis for the helical assembly in these polymers (Lu et al. 2014; Lu et al. 2016), thereby providing an explanation for signal amplification and threshold kinetics during inflammasome signaling (Figure 1b). The ASCPYD filament displays a C3 point symmetry with each helical strand possessing a right-handed 53° twist and a 13.9 Å rise, whereas the caspase-1CARD filament contains a single helical strand with a left-handed 100.2° twist and a 5.1 Å rise. Common to these DD family filaments is that they are stabilized by three types of DD interactions mediated by six complementary surfaces (Lu et al. 2014; Lu et al. 2016; Li et al. 2018; Fu et al. 2016; Shen et al. 2019).
Successful determination of these DD filament structures mainly relied on helical processing in RELION (He and Scheres 2017) and iterative helical real-space reconstruction (IHRSR) in SPIDER (Egelman 2000) (Figure 1c). Instead of single particles, helical filaments were picked from the micrographs and segmented with an appropriate inter-box distance. In RELION, the extracted segments were 2D classified to generate averages with optimal features. Power spectra were calculated to obtain the layer lines, which were used to deduce the twist and rise of the helical object. With an initial 3D model, which can be a featureless cylinder or a simulated helical lattice with the deduced helical parameter, RELION can perform 3D classification or direct 3D refinement with local refinement of the suggested symmetry. For IHRSR, the box length for segmentation was tested empirically with consideration for the different radii of curvature of filaments in different cases (Egelman 2010). The 2D classification step is dispensable, as the symmetry could be estimated by indexing the power spectra from aligned segments and then optimized iteratively from a starting cylinder model.
The discovery of filament structures formed by the DD superfamily has far-reaching biological implications. A classical picture of cell signaling is that ligand-induced conformational changes at receptors, often dimerization or trimerization, lead to the recruitment and activation of downstream molecules such as secondary messengers and enzymes. In innate immunity, the formation of higher-order complexes via homotypic protein-protein interactions suggests a novel and more mechanistically complex mode of signal transduction. Consistent with this higher-order signaling hypothesis, biomolecules can form punctate aggregates, exhibit cooperativity during oligomerization, and display threshold behaviors in cellular response (Wu 2013). It is possible that higher-order assembly represents a general signaling mechanism employed in biological processes beyond inflammasome activation and innate immunity.
Cryo-EM structure of the NAIP-NLRC4 inflammasome
An important characteristic of inflammasome signaling is the switch from auto-inhibition to activation upon ligand engagement. With clearly identified ligands like flagellin (such as Salmonella typhimurium FliC or Legionella pneumophila FlaA) and rod proteins from the Type III secretion system (T3SS) (such as PrgJ), the NAIP-NLRC4 inflammasomes have been ideal targets for structural and mechanistic dissection (Vance 2015). Reconstituted PrgJ-NAIP2-NLRC4 inflammasome using CARD-deleted NLRC4 (NLRC4ΔCARD) revealed disk-like structures with 11 or 12 subunits (Figure 2a) (Hu et al. 2015; Zhang et al. 2015). Biochemical characterization and gold labeling surprisingly showed that only one subunit in each disk belongs to the PrgJ-NAIP2 complex, while the remaining subunits are NLRC4ΔCARD (Hu et al. 2015; Zhang et al. 2015). Because NAIP2 also has a similar domain organization to NLRC4, cryo-EM reconstruction was performed by applying the 11- or 12-fold symmetry assuming that all subunits are NLRC4ΔCARD and uncovered the active conformation of NLRC4ΔCARD (Figure 2b) (Hu et al. 2015; Zhang et al. 2015).
Figure 2. Nucleated polymerization of the NAIP-NLRC4 inflammasome.
(a) A cryo-EM micrograph shows disk-like PrgJ-NAIP2-NLRC4ΔCARD inflammasome particles boxed and 11- and 12-mer 2D class averages (Zhang et al. 2015). (b) Auto-inhibited (PDB ID: 4KXF) and active conformations of NLRC4ΔCARD. The latter is a single subunit from the cryo-EM structure of the disk-like PrgJ-NAIP2-NLRC4ΔCARD inflammasome complex (PDB ID: 6B4B, 3JBL). (c) Mechanism of FlaA recognition within a partial FlaA-NAIP5-NLRC4ΔCARD inflammasome disk (PDB ID: 6B5B). (d) The whole process of NLRC4ΔCARD inflammasome activation from a single FlaA-bound NAIP5 to a final 11-subunit disk (PDB ID: 6B4B, 3JBL, 6B5B). (e) Cryo-ET map of the FliC-D0L-NAIP5-NLRC4 inflammasome (EMDB ID: 2901), showing a spiral architecture with a central CARD filament.
In comparison with the crystal structure of NLRC4ΔCARD in complex with ADP in a closed, auto-inhibited conformation (Hu et al. 2013), the active conformation featured a large domain rotation (~90°) at the junction between the NBD-helical domain 1 (HD1) module and the winged helix domain (WHD)-helical domain 2 (HD2)-LRR module, leading to an open conformation of NLRC4ΔCARD that is likely facilitated by nucleotide exchange (Figure 2b). Recent cryo-EM structures of the FlaA-NAIP5-NLRC4ΔCARD inflammasome resolved a NAIP from the NLRC4ΔCARD subunits (Tenthorey et al. 2017; Yang et al. 2018) (Figure 2c). Here, the active NAIP5 is in complex with FlaA, revealing the molecular basis for ligand recognition by NAIP5. Through ligand-receptor binding analysis, the authors proposed multi-surface innate immune recognition as an efficient way to overcome single-point mutation-induced pathogen invasion.
The fact that only one subunit belongs to the ligand-sensor complex in these inflammasome structures suggests an elegant mechanism of ligand-induced inflammasome activation (Figure 2d). Upon ligand engagement, NAIP undergoes conformational changes to bind to an NLRC4ΔCARD protomer, and induces conformational changes of NLRC4ΔCARD to expose the oligomerization or “catalytic” interface for the recruitment of another NLRC4ΔCARD molecule. The self-propagation proceeds until a whole disk-like inflammasome is assembled (Figure 2d). The conformational change from auto-inhibited to active NLRC4ΔCARD releases the positively charged “catalytic” surface to recruit the negatively charged NBD surface of another molecule to initiate and propagate NLRC4ΔCARD polymerization (Hu et al. 2015; Zhang et al. 2015).
These studies provide important insights and raised two intriguing questions that require further structural studies of activated inflammasomes: 1. Do other NLRs form homo-oligomers or heterooligomers with the help of other NLRs, just like the NAIP-NLRC4 inflammasome? 2. How do the scaffold domains of NLRs - namely CARD and PYD - integrate into the disk-like inflammasome structure? Clues to the second question come from cryo-electron tomography (cryo-ET), which revealed that CARD may form the central helical filament of the NAIP-NLRC4 inflammasome surrounded by the disk-like structure in a spiral manner (Diebolder et al. 2015) (Figure 2e). Indeed, the dimensions and architecture of the NLRC4CARD filament support its integration into the core region (Li et al. 2018). Taken together, progress in NAIP-NLRC4 inflammasome research through the development of cryo-EM and cryo-ET methods provides valuable guidance for further studies of NLR inflammasomes.
Mechanistic elucidation of gasdermin pore formation by cryo-EM
Both canonical and non-canonical inflammasomes can induce inflammatory and lytic cell death commonly referred to as pyroptosis. Two independent genetic screens identified GSDMD, which belongs to the GSDM family, as the executioner of pyroptosis downstream of caspase-1 and murine caspase-11 (human homologs caspase-4 and −5) (Kayagaki et al. 2015; Shi et al. 2015). These inflammasome-activated caspases, as well as a caspase-8-dependent pathway during Yersinia infection (Orning et al. 2018; Sarhan et al. 2018), proteolytically activate GSDMD. GSDMD-NTs promote pyroptosis by directly binding membrane lipids and forming oligomeric membrane pores, while GSDMD-CT has an auto-inhibitory function that is removed by caspase cleavage (Ding et al. 2016; Liu et al. 2016). Adding to the complex signaling network involving GSDMD, recent studies have highlighted a non-pyroptotic role of GSDMD and placed GSDMD upstream of the NLRP3 inflammasome (Evavold et al. 2018; Orning et al. 2018; Platnich et al. 2018).
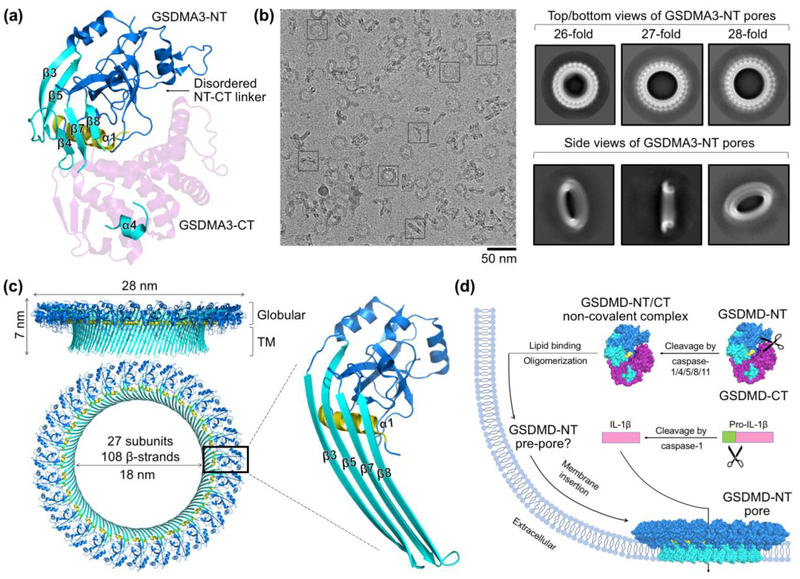
The X-ray crystal structure of mouse GSDMA3 provided valuable visualization of the auto-inhibited, inactive conformation of GSDM family proteins (Figure 3a) (Ding et al. 2016). However, a thorough structural understanding of GSDMs is hindered by the instability and heterogeneity of GSDM pores that render crystallization unfeasible. By testing GSDMs of different species, Ruan et al. reconstituted GSDMA3 pores on cardiolipin-containing liposomes and used cholate to solubilize the pores for cryo-EM structural determination (Ruan et al. 2018). RELION 2D classification showed 26-, 27-, and 28-fold symmetric top views with 27-fold as the major class, suggesting oligomerization heterogeneity of GSDM pores (Figure 3b). Top-view 27-fold 2D classes, and all side-view classes of which the symmetry is unclear, were selected for 3D classification without assumption of any symmetry, upon which the predominant 3D class was refined in RELION with C27 symmetry.
Figure 3. Molecular mechanism of GSDM pore formation.
(a) Crystal structure of mouse GSDMA3 (PDB ID: 5B5R) with key secondary structure elements labeled, GSDMA3-NT colored in blue, cyan, and yellow, and GSDMA3-CT colored in magenta. (b) A cryo-EM micrograph of detergent-solubilized GSDMA3 pores and 2D class averages of the pores generated in RELION (Ruan et al. 2018; Scheres 2012). (c) Cryo-EM structure of the 27-fold symmetric GSDMA3 membrane pore (PDB ID: 6CB8) with dimensions indicated and a magnified view of a pore-form GSDMA3-NT subunit. Color schemes follow those of the crystal structure in (a). (d) A model for pore formation by the GSDM family, where GSDM-NTs might oligomerize into a membrane-associated pre-pore before insertion of the β-barrel to form a transmembrane pore that allows the passage of cytoplasmic contents such as IL-1β.
The final pore structure at 3.8 Å resolution features a 108-stranded antiparallel β-barrel as the transmembrane region, capped by a soluble rim formed by the globular domain of each of the 27 subunits (Figure 3c). The inner diameter of the pore is approximately 18 nm, large enough for the passage of IL-1β but not all cytosolic components, suggesting a potential size-exclusion mechanism. In comparison with the auto-inhibited crystal structure, GSDMA3-NT undergoes large conformational changes, particularly at the β3-β4 and the β7-α4-β8 regions, for membrane insertion (Figure 3a, c). Adjacent to the basic α1 helix, a cryo-EM density is visible and likely represents the negatively charged head group of the acidic lipid cardiolipin, which indicates the crucial role of lipid binding in membrane pore formation. Interestingly, lining the membrane-inserted pore is a soluble ring without extended β-strands, which could be an intermediate prepore conformation of GSDMs (Ruan et al. 2018). It merits further study whether GSDM pore formation is a smooth continuum of structural transitions or comprises discrete steps from the auto-inhibited conformation, to a non-covalent complex of GSDM-NT and GSDM-CT (Ding et al. 2016), to an oligomerized but not membrane-inserted pre-pore, and finally to a membrane pore (Figure 3d).
Outlook and challenges
Cryo-EM has offered invaluable mechanistic insights into inflammasome signaling (Figure 4). Especially regarding the NLRC4 inflammasome, high-resolution structures have revealed molecular details of ligand-induced conformational change, release from auto-inhibition, selfpropagation through NBD and LRR interactions, and nucleation-induced polymerization of ASC and caspase-1 by DD-fold assembly. These structures also shed light on the activation mechanisms of other NLRs given their highly conserved domain architecture.
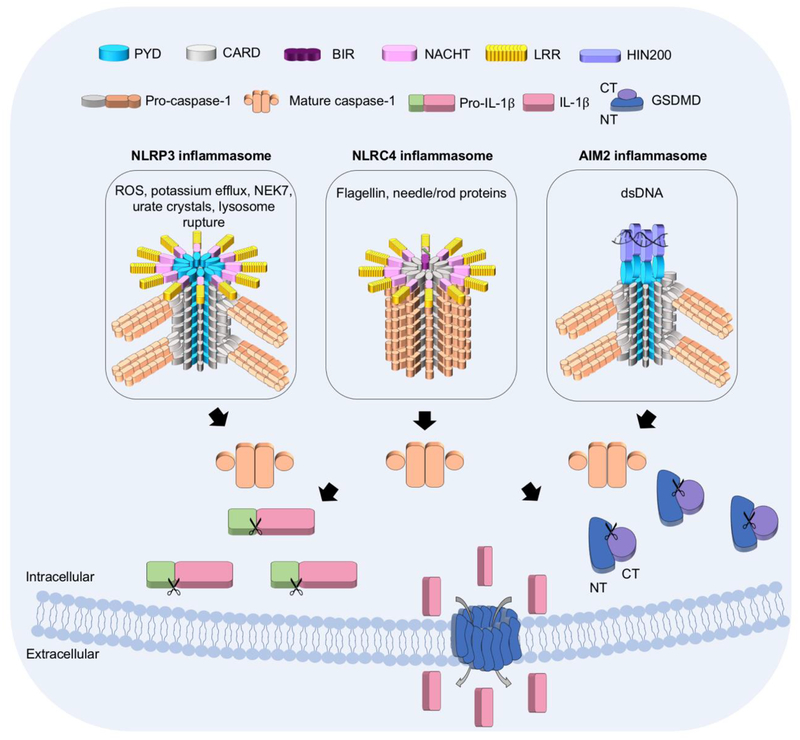
Figure 4. Structural biology of inflammasome signaling.
Overview of the inflammasome activation processes for NLRP3, NLRC4, and AIM2, with domains and proteins indicated above, and the stimuli and cartoon models of NLRP3, NLRC4, and AIM2 inflammasomes shown in boxes. NLRP3 and AIM2 require the adaptor protein ASC for proximity-induced activation of caspase-1. By contrast, NLRC4 can directly recruit and activate caspase-1. Active caspase-1 cleaves and releases the auto-inhibition of GSDMD. GSDMD-NT then forms membrane pores to induce pyroptosis. Active caspase-1 also cleaves pro-IL-1β into its mature form, which is released through GSDMD pores.
One future direction towards the understanding of inflammasome activation lies in ligand-receptor interactions. NEK7 kinase and lipoteichoic acid (LTA) from Gram-positive bacteria have been identified as activators of NLRP3 and NLRP6, respectively (He et al. 2016; Hara et al. 2018), but the structural basis for the recognition of these ligands remain elusive. Likewise, the interaction between LPS and caspase-11 in the non-canonical inflammasome pathway remains an intriguing topic (Hagar et al. 2013; Kayagaki et al. 2013; Shi et al. 2014). In addition to ligand-receptor interactions, the cooperativity of sensor molecules in inflammasome signaling merits further study. For example, NLRP3 and NLRC4 may together orchestrate inflammasome activation in macrophages (Qu et al. 2016), which suggests that the structural scaffolds of inflammasomes may be co-formed by different NLRs. A thorough depiction of inflammasome signaling by cryo-EM, from a translational perspective, will aid structure-guided drug development against inflammatory diseases.
Despite these interesting directions, structural study of ligand-receptor complexes and higher-order protein assemblies faces many challenges. Not only are inflammasome components difficult to express and purify, instability, heterogeneity in size and shape, and orientational preference also deter the structural determination of these molecules and their complexes by cryo-EM. For example, the effector protein GSDMD employs a higher-order assembly strategy to rupture cell membranes for cytokine release and pyroptosis (Figure 4). A previous attempt at reconstituting homogeneous human GSDMD pores in vitro was unsuccessful due to their aggregation and deformation after being detergent-solubilized from liposomes (Ruan et al. 2018). In addition, the cryo-EM densities at the globular domain of the GSDMA3 pore are only at a modest resolution, likely due to conformational dynamics. The field is awaiting a method to validate sequence registration into cryo-EM maps with relatively poor densities, possibly by selective labeling of reactive amino acid residues.
Highlights.
Helical assembly of PYD and CARD is revealed as one universal mechanism for inflammasome signaling by cryo-EM.
Cryo-EM facilitates the understanding of disk-like, self-propagating assembly of the NLRC4 inflammasome.
The cryo-EM structure of a gasdermin (GSDM) pore uncovers the molecular details of membrane insertion, providing a novel mechanism for cytokine secretion and pyroptosis execution.
Acknowledgements
This work was supported by National Institutes of Health grants (1DP1 HD087988, 1R01 AI139914 and 1R01 Al124491 to H.W.), and the Irvington Postdoctoral Fellowship from the Cancer Research Institute (to C.S.). We thank Liman Zhang and Jianbin Ruan for discussions and their cryo-EM images.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest.
References
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- •. Diebolder CA, Halff EF, Koster AJ, Huizinga EG, and Koning RI. 2015. ‘Cryoelectron Tomography of the NAIP5/NLRC4 Inflammasome: Implications for NLR Activation’, Structure, 23: 2349–57. [DOI] [PubMed] [Google Scholar]
- •. Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, and Shao F. 2016. ‘Pore-forming activity and structural autoinhibition of the gasdermin family’, Nature, 535: 111–6. [DOI] [PubMed] [Google Scholar]
- ••. Egelman EH 2000. ‘A robust algorithm for the reconstruction of helical filaments using single-particle methods’, Ultramicroscopy, 85: 225–34. [DOI] [PubMed] [Google Scholar]
- Egelman EH 2010. ‘Reconstruction of helical filaments and tubes’, Methods Enzymol, 482: 167–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, and Kagan JC. 2018. ‘The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages0027, Immunity, 48: 35–44 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Li J, Bergamin E, and Wu H. 2012. ‘Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily’, Sci Signal, 5: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu TM, Li Y, Lu A, Li Z, Vajjhala PR, Cruz AC, Srivastava DB, DiMaio F, Penczek PA, Siegel RM, Stacey KJ, Egelman EH, and Wu H. 2016. ‘Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex’, Mol Cell, 64: 236–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, and Miao EA. 2013. ‘Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock’, Science, 341: 1250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, and Nunez G. 2018. ‘The NLRP6 Inflammasome Recognizes Lipoteichoic Acid and Regulates Gram-Positive Pathogen Infection’, Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. He S, and Scheres SHW. 2017. ‘Helical reconstruction in RELION’, J Struct Biol, 198: 163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zeng MY, Yang D, Motro B, and Nunez G. 2016. ‘NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux’, Nature, 530: 354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, Deng H, Wang J, Chang J, and Chai J. 2013. ‘Crystal structure of NLRC4 reveals its autoinhibition mechanism’, Science, 341: 172–5. [DOI] [PubMed] [Google Scholar]
- ••. Hu Z, Zhou Q, Zhang C, Fan S, Cheng W, Zhao Y, Shao F, Wang HW, Sui SF, and Chai J. 2015. ‘Structural and biochemical basis for induced self-propagation of NLRC4’, Science, 350: 399–404. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Magupalli VG, and Wu H. 2014. ‘SMOCs: supramolecular organizing centres that control innate immunity’, Nat Rev Immunol, 14: 821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, and Dixit VM. 2015. ‘Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling’, Nature, 526: 666–71. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, and Dixit VM. 2011. ‘Non-canonical inflammasome activation targets caspase-11’, Nature, 479: 117–21. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, and Dixit VM. 2013. ‘Noncanonical inflammasome activation by intracellular LPS independent of TLR4’, Science, 341: 1246–9. [DOI] [PubMed] [Google Scholar]
- Kesavardhana S, and Kanneganti TD. 2017. ‘Mechanisms governing inflammasome activation, assembly and pyroptosis induction’, Int Immunol, 29: 201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, and Dixit VM. 2014. ‘Mechanisms and functions of inflammasomes’, Cell, 157: 1013–22. [DOI] [PubMed] [Google Scholar]
- Li Y, Fu TM, Lu A, Witt K, Ruan J, Shen C, and Wu H. 2018. ‘Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1’, Proc Natl Acad Sci U S A, 115: 10845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J. 2016. ‘Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores’, Nature, 535: 153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Lu A, Li Y, Schmidt FI, Yin Q, Chen S, Fu TM, Tong AB, Ploegh HL, Mao Y, and Wu H. 2016. ‘Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism’, Nat Struct Mol Biol, 23: 416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, and Egelman EH. 2014. ‘Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes’, Cell, 156: 1193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Lu A, and Wu H. 2015. ‘Structural mechanisms of inflammasome assembly’, FEBS J, 282: 435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, and Chiu W. 1999. ‘EMAN: semiautomated software for high-resolution single-particle reconstructions’, J Struct Biol, 128: 82–97. [DOI] [PubMed] [Google Scholar]
- Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, Berger SB, Gough PJ, Bertin J, Proulx MM, Goguen JD, Kayagaki N, Fitzgerald KA, and Lien E. 2018. ‘Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death’, Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, and Wu H. 2007. ‘The death domain superfamily in intracellular signaling of apoptosis and inflammation’, Annu Rev Immunol, 25: 561–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnich JM, Chung H, Lau A, Sandall CF, Bondzi-Simpson A, Chen HM, Komada T, Trotman-Grant AC, Brandelli JR, Chun J, Beck PL, Philpott DJ, Girardin SE, Ho M, Johnson RP, MacDonald JA, Armstrong GD, and Muruve DA. 2018. ‘Shiga Toxin/Lipopolysaccharide Activates Caspase-4 and Gasdermin D to Trigger Mitochondrial Reactive Oxygen Species Upstream of the NLRP3 Inflammasome’, Cell Rep, 25: 1525–36 e7. [DOI] [PubMed] [Google Scholar]
- Qi X 2016. ‘Formation of membrane pores by gasdermin-N causes pyroptosis’, Sci China Life Sci, 59: 1071–73. [DOI] [PubMed] [Google Scholar]
- Qu Y, Misaghi S, Newton K, Maltzman A, Izrael-Tomasevic A, Arnott D, and Dixit VM. 2016. ‘NLRP3 recruitment by NLRC4 during Salmonella infection’, J Exp Med, 213: 877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. Ruan J, Xia S, Liu X, Lieberman J, and Wu H. 2018. ‘Cryo-EM structure of the gasdermin A3 membrane pore’, Nature, 557: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, and Poltorak A. 2018. ‘Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection’, Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH 2012. ‘RELION: implementation of a Bayesian approach to cryo-EM structure determination’, J Struct Biol, 180: 519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, and Tschopp J. 2010. ‘The inflammasomes’, Cell, 140: 821–32. [DOI] [PubMed] [Google Scholar]
- Shen C, Lu A, Xie WJ, Ruan J, Negro R, Egelman EH, Fu TM, and Wu H. 2019. ‘Molecular mechanism for NLRP6 inflammasome assembly and activation’, Proc Natl Acad Sci U S A, 116: 2052–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Gao W, and Shao F. 2017. ‘Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death’, Trends Biochem Sci, 42: 245–54. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F. 2015. ‘Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death’, Nature, 526: 660–5. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, and Shao F. 2014. ‘Inflammatory caspases are innate immune receptors for intracellular LPS’, Nature, 514: 187–92. [DOI] [PubMed] [Google Scholar]
- ••. Tenthorey JL, Haloupek N, Lopez-Blanco JR, Grob P, Adamson E, Hartenian E, Lind NA, Bourgeois NM, Chacon P, Nogales E, and Vance RE. 2017. ‘The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion’, Science, 358: 888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE 2015. ‘The NAIP/NLRC4 inflammasomes’, Curr Opin Immunol, 32: 84–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JA, and Bryant CE. 2016. ‘The killer protein Gasdermin D’, Cell Death Differ, 23: 1897–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H 2013. ‘Higher-order assemblies in a new paradigm of signal transduction’, Cell, 153: 287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Yang X, Yang F, Wang W, Lin G, Hu Z, Han Z, Qi Y, Zhang L, Wang J, Sui SF, and Chai J. 2018. ‘Structural basis for specific flagellin recognition by the NLR protein NAIP5’, Cell Res, 28: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •. Yin Q, Fu TM, Li J, and Wu H. 2015. ‘Structural biology of innate immunity’, Annu Rev Immunol, 33: 393–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, and Kagan JC. 2016. ‘An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells’, Science, 352: 1232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••. Zhang L, Chen S, Ruan J, Wu J, Tong AB, Yin Q, Li Y, David L, Lu A, Wang WL, Marks C, Ouyang Q, Zhang X, Mao Y, and Wu H. 2015. ‘Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization’, Science, 350: 404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]