Abstract
Synthetic biology efforts began in simple single-cell systems, which were relatively easy to manipulate genetically1. The field grew exponentially in the last two decades, and one of the latest frontiers are synthetic developmental programs for multicellular mammalian systems2,3 to genetically control features such as patterning or morphogenesis. These programs rely on engineered cell-cell communications, multicellular gene regulatory networks and effector genes. Here, we contextualize the first of these synthetic developmental programs, examine molecular and computational tools that can be used to generate next generation versions, and present the general logic that underpins these approaches. These advances are exciting as they represent a novel way to address both control and understanding in the field of developmental biology and tissue development4–7. This field is just at the beginning and it promises to be of major interest in the upcoming years of biomedical research.
Keywords: Synthetic biology, tissue engineering, organoids, self-organization, morphogenesis, patterning, synthetic receptors, synthetic development, multicellular mammalian systems, computational modeling
Graphical Abstract
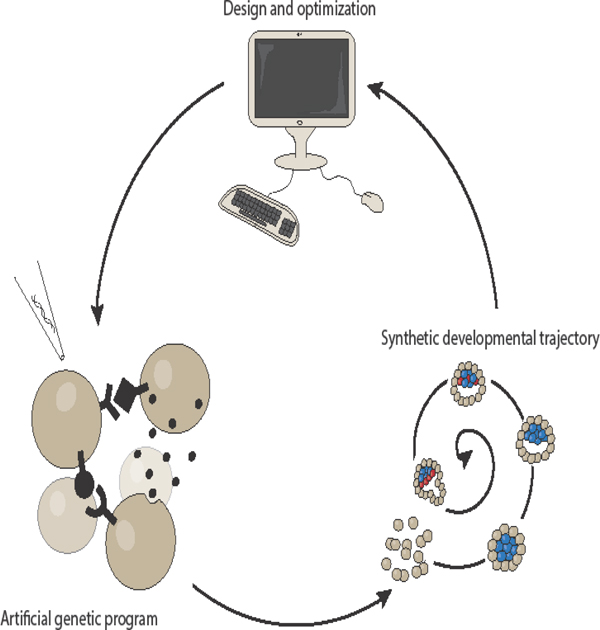
Introduction
The early days of synthetic biology were devoted to engineering simple genetic circuits in simple single-cell systems, which were relatively easy to manipulate genetically. As the field has progressed, our understanding and tools have become more sophisticated, first extending to simple eukaryotic cells, and eventually to mammalian cells1. More recently, synthetic developmental programs have been developed for multicellular mammalian systems2,3 to genetically control features such as patterning or morphogenesis. To achieve this, these genetic programs take usually inspiration from natural genetic circuits that pattern and build the embryo. A general logic of both natural and synthetic developmental program is the linking of cell-cell communication channels, patterning through multicellular gene regulatory networks, and morphogenesis through changes in cell biophysical parameters via effector genes (Fig. 1).
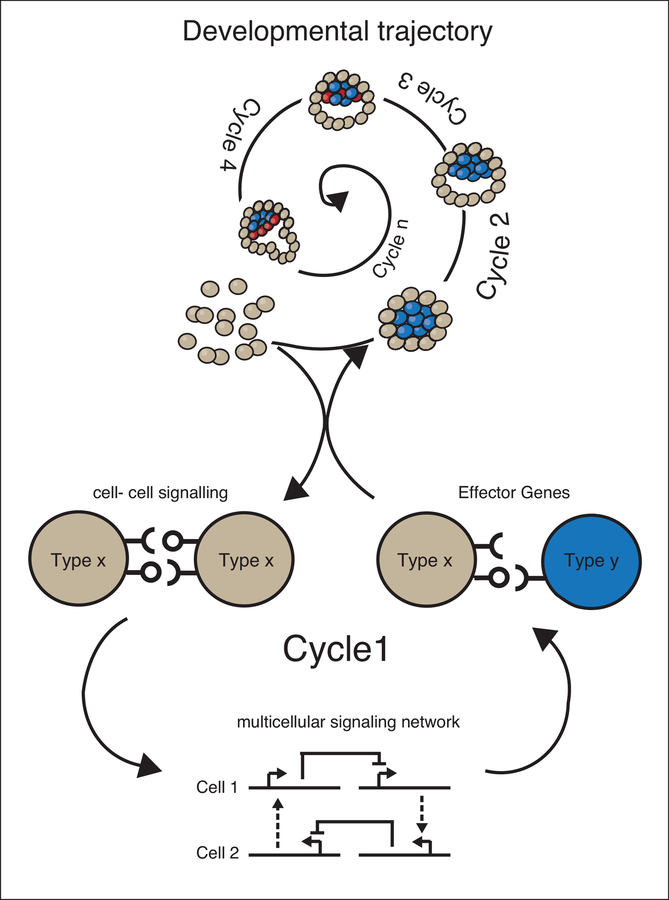
Figure 1 – Deconstruction of developmental trajectory into cycles and molecular implementations.
The spiral represents the developmental trajectory unfolding over time through a series of cycles (Cycle 1, Cycle 2, etc.). Each cycle is driven by the underlying genetic program made of: 1) cell-cell signaling, 2) multicellular networks that generate patterns, 3) and changes in cells’ physical and/or biological properties by changing expression of one or more effector genes. The new signaling, biological, and physical status of the multicellular system defines the new starting conditions for a new cycle.
This new field has the potential to contribute to different areas of research:
Study of developmental principles. By using an understanding-by-building framework4, this approach can attempt to recapitulate endogenous morphogenetic networks and/or transitions to see what it takes to build them from the bottom-up, thereby illuminating defining design principles.
Controlling tissue development in a dish. Current approaches, such as organoids and tissue engineering, display shortcomings, especially at the level of single cell control. Synthetic development can try to correct features of in vitro tissues that are not recapitulating actual organs due to growth in isolation5. Additionally, this approach could deliver a new generation of organs or tissues with enhanced synthetic capacities.
Expression of creativity. This field could benefit from a certain degree of “free-form” exploration, where the only motivation is the curiosity and ingenuity of the researcher. This spirit has been a strong defining feature of the early days of synthetic biology, and likely underlies its current popularity.
While this is an exciting time in the field, it is also in its early days, and it is important to continue developing the tools and conceptual frameworks necessary to realize its full potential.
In this review, we first introduce an abstraction logic for developmental programs in multiple components (cell-cell signaling, multicellular networks and effector genes); for each of these components, we present the expanding array of relevant synthetic biology tools, including ones that could be generated through a combination of existing tools; then we describe the first examples of how the first synthetic developmental programs have been achieved; finally we give an overview of the parallel computational efforts that have been used for modeling endogenous developmental system, and that may be used in the future to guide design of synthetic developmental systems.
Abstraction of development
The goal of synthetic development is to guide the formation of multicellular mammalian structures by engineering genetic programs in cells. This is conceptually similar to what happened during evolutionary times to generate the instructions in the DNA code to instruct embryonic development. During embryonic development, genetically encoded, evolutionarily selected programs guide cells from an amorphous aggregate (and before that, a single cell) to a multicellular structure with integrated functions. For example, during early mammalian development, the equipotent cells of the morula differentiate such that cells on the outside of the morula become placental precursors, while those on the interior become embryonic stem cells. The compacted morula then forms an inner cavity containing an inner cell mass, which undergoes a subsequent cycle of morphogenesis to differentiate into epiblast stem cells and primitive endoderm cells. These transitions produce the nascent blastocyst6. We propose an abstraction that deconstructs developmental trajectories like this one into various cycles. This abstraction moves from the characterization of cells as having a dual nature as both information processors and material7. For each such cycle, we deconstruct the molecular and cellular logics that propel the transitions as: cell-cell communication systems, multicellular genetic networks, and physical or biological cell changes.
Cell-cell communication describes the ways in which cells send and receive signals to and from each other and their environment. Cell-cell communication pathways can be linked in multicellular networks, when their output changes the communication itself. Networks contain feedback and non-linearity that generate different cellular states in a population of cells (i.e. patterning). Finally, physical and biological cellular changes occur when cells acquire different physical features or differentiation routes—including changes to cell adhesiveness, shape, identity, etc. Cell-cell communication, multicellular networks, and physical changes create a highly dynamic system since all the components affect each-other: cell-cell signaling pathways generate patterning networks, and different parts of the pattern execute different functional programs that in turn generate new states and new communication networks. In this way, a fluid yet very robust process of computation and morphogenesis unfolds over time until, from an amorphous beginning, the cell aggregate develops into a complex tissue (Fig. 1).
We think that in order to implement synthetic versions of these types of complex programs it is important to abstract their logic. Abstractions in synthetic biology have been very valuable as they can work as mission statements guiding the kinds of genetic programs or synthetic proteins that need to be made, which kinds of control are helpful and what kind of collective behaviors we want to implement. The framework presented here does not represents the only possible definition or abstraction, but is our own interpretation based on our knowledge and experience in the field; not every facet of it is well-defined and many grey areas still exist for exploration.
Synthetic cell communication pathways
Mammalian multicellular development uses a handful cell-cell communication pathways8, which can be distinguished by input features such as contact-dependence, short-versus long-range diffusibility, cell versus extracellular matrix (ECM), and physical versus chemical signals. The input itself is either produced by other cells or present in the extracellular environment and allows a community of cells to communicate with each other to coordinate their behaviors.
The logic of these communication pathways revolves around protein sensors that connect inputs from the cell’s exterior to intracellular changes (Fig. 2). At the molecular level, this is achieved with protein sensor domains for the inputs (A) linked to transducers (B) activating an effector domain for the output (C) that effect changes in the cell. To build synthetic receptors, these domains have been linked to form chimeric proteins9–11,3,12,13. In Fig. 2 we map existing receptors based on categorical features, such as sensor, transduction and effector domains.
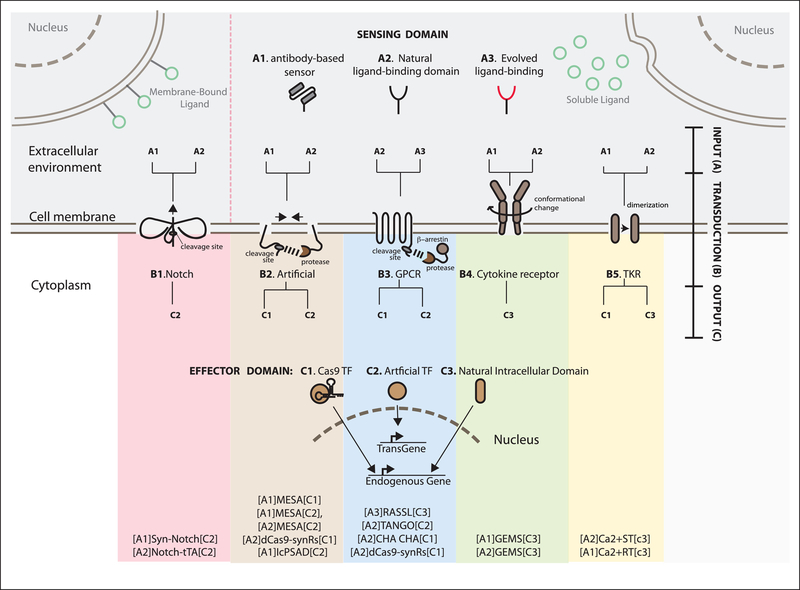
Figure 2 – Mapping existing synthetic receptor systems based on their input, transduction and output characteristics.
A schematic of various synthetic receptors that process information from the extracellular space (top), to inside the cell (bottom).
Sensing domains are classified as antibody-based (A1), based on natural ligand binding domains (A2), or generated via directed evolution (A3). Effector domains are natural (C3) if endogenous domains are used. For transcriptional outputs, we distinguish artificial transcription factors like Gal4 or tTA that activate exogenous expression cassettes (transgene) (C2), or dCas9-based transcription factors that activate endogenous targets (C3). Each sensor and effector domain can be mounted onto a part of the receptor protein that executes the transduction (B). B1 is contact dependent, i.e. is activated only via membrane-bound ligand (green circles presented on a neighboring cell in the upper left corner). B2–5 all recognize ligands that are soluble in the extracellular environment (green circles secreted from other cells in the upper right corner).
At the far bottom, we list the synthetic receptor systems that have been implemented to date using alphanumeric codes according to the notation introduced in the text of [input]transduction[output]. For example, [A1]synNotch[C2] represents the synthetic receptor that uses an antibody-based sensor as input (A1), and activates an artificial transcription factor as output (C2), and so on. The works that describe in the details those receptors are in the references below and in the text.
SynNotch = Synthetic Notch24; MESA = Modular Extracellular Sensor Architecture15,64,65; IcPSAD = Intracellular protein sensor actuator device66; GEMS = generalized extracellular molecule sensor14; RASSL = Receptor activated solely by synthetic ligand17; Tango19; ChaCha20; dCas9 synRs = dCas9 synthetic receptor21; Ca2+ST = calcium sensing-rewiring tool67; Ca2+RT = calcium rewiring tool18,68; GPCR = G-protein coupled receptor; TKR= tyrosine kinase receptor.
Different kinds of transduction logic result in differences in signaling features. Contact-dependent synthetic receptors (synNotch) have been developed and used for multicellular patterning (see below), based on the framework of the native contact-dependent receptor Notch (Fig. 2B.1). Several versions of synthetic soluble-signal dependent pathways have been developed, either by using endogenous transduction frameworks (GEMS, TANGO, CHA-CHA and calcium-signaling based) or by using dimerization-based transduction (MESA and dCas9-synR) (Fig. 2B.2–5). How receptors with different molecular logics affect communication and determine their suitability to particular applications or network types is an area of current investigation.
The modularity of signaling pathways allows for the generation of different combinations of input/transduction/output. For this reason, we propose the following nomenclature for synthetic receptors: [Input]Transduction[Output]. For example, GEMS receptors that recognize rapamycin and activate JACK/STAT pathways would be described as [rapamycin]GEMS[JACK/STAT]; anti-CD19 synNotch that activates the orthogonal transcription factor tTA, which in turn activates an mCherry reporter, would be [CD19]synNotch[tTA→mCherry] and so on.
To design synthetic signaling pathways there is a spectrum of possibilities framed by two extremes: using completely orthogonal communication pathways, or rewiring endogenous ones. When synthetic control of a native cellular behavior is desired, mixed synthetic/endogenous receptors are useful. For example, endogenous pathways can be rendered activatable by synthetic ligands using programmable sensor domains linked by a transducers to an endogenous effector domain or CRISPR-Cas9 based transcription factor, as in [rapamycin]GEMS[JAK/STAT]14 and similar [VEGF]MESA[Cas9-TF→IL-2]15,16, [CNO]RASSL[GPCR signaling]17, and [sCD14]Ca2+RT[CaRQ→migration]18.
In other cases, endogenous sensing can be rewired to drive a synthetic response, as in [vasopressin]TANGO[tTA→luciferase]19, [NMB]ChaCha[Crispr-VPR→IFN-gamma]20, and [VEGF]dCas9-synR[Crispr-VP64→TNFalfa/TSP-1]21. Another way to achieve similar rewiring is to act at the transcriptional level: a synthetic cassette with an endogenous promoter responsive to endogenous pathways can contain a user-defined effector gene22. In this way, TNF pathway activation was rewired to produce anti-TNF proteins at the transcriptional level23. Using endogenous sensors or outputs can be beneficial for detecting endogenous ligands, or using the endogenous response of a particular pathway.
In other cases, it’s more fitting to be orthogonal to endogenous pathways. To design a program that is completely orthogonal to endogenous ones, synthetic control on both input and output is needed, as in [GFP]synNotch[tTA→mCherry]24,25, and [rapamycin]MESA[Gal4]15. Such programs can be useful in designing complex patterning in multicellular or other systems with minimal cross-talk with endogenous cellular activity.
In the future, we anticipate the field of synthetic receptors to provide an array of different molecular controls. For example, new sensors can enable receptors to be designed to sense other inputs, including mechanical inputs and ones presented on the extracellular matrix. One important point for the field will be to be able to provide guidance for users of these technologies by systematically comparing the different synthetic signaling pathways.
Patterning through synthetic multicellular networks
Synthetic cell communication pathways can be used to connect multiple cells to form multicellular communication networks. Networks like these are important for generating spatial and temporal patterns of differential gene expression (Fig. 3a).
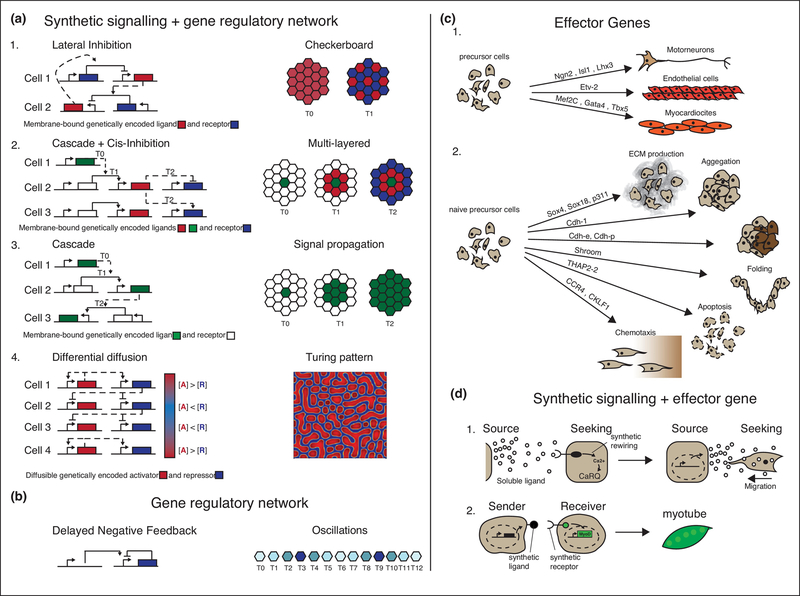
Figure 3 – Multicellular signaling networks and effector genes can drive patterning of different cell states and biophysical changes.
(a-b) Examples of spatial and temporal patterning guided by multicellular synthetic gene regulatory networks implemented in mammalian cells. The gene regulatory networks are presented on the left-hand side; on the right hand side are the corresponding cellular patterns. The gene regulatory networks are represented as blocks-and-arrows schemes: the solid arrows are the regulatory links of the intracellular network, acting in the same cell; the dashed arrows are the intercellularly regulatory links between two cells. (a.1) Representation of network scheme and resulting checkerboard pattern generated using a single cell population expressing NOTCH receptor (blue), repressing its own ligand DELTA (red); this network was implemented in 2D in CHO cells26. (a.2) Representation of network scheme and resulting multi-layered pattern generated using two distinct cell lines, where cell type 1 expresses a constitutively active green fluorescent protein (GFP) ligand (green) and cells 2–3 express a [GFP]synNotch[tTA→CD19 ligand], a [CD19]synNotch[tTA→BlueFluorescentProtein] (blue) and an mCherry reporter (red). When co-cultured, cell type 1 acts as a nucleation center for a signal cascade where cells 2 that are in contact with cells 1 fluoresce red and create a secondary ligand, which then signals to neighboring cells 3 to fluoresce blue. This network was implemented in 2D in MDCK cells24. (a.3) Representation of network scheme and resulting signal propagation pattern generated using two distinct cell lines, where cell 1 constitutively expresses the NOTCH ligand DELTA (green) and cells 2–3 express the synthetic modular receptor [DELTA]NOTCH[tTA→DELTA]. When co-cultured, cell 1 triggers signal propagation in neighboring cells 2–3. This network was implemented using MDCK cells as cell 1 and CHO cells as cells 2–327. (a.4) Network scheme and resulting Turing pattern obtained by using two different genetically encoded signaling molecules characterized by differential diffusion properties: Nodal the short range network activator (red) and Lefty the long range network inhibitor (blue). The network was implemented in 2D in 293AD Cells30. (b) In the oscillation program a desynchronized temporal pattering was generated in a cell population using the transcriptional repressor Hes1 (blue) that is able to repress its own expression after a delay encoded by its introns. This network was implemented in 2D in CHO cells and 3T3 mouse fibroblasts33,69.
(c) Examples of effector genes driving physical (1.) or biological (2.) differentiation when overexpressed. (c1.) Examples of effector genes that, when exogenously overexpressed, induce cell differentiation into motoneurons from human induced pluripotent stem cells, endothelial cells from a lung fibroblast cell line, and cardiomyocytes from a fibroblast cell line70–72. (c.2) Examples of effector genes changing selected cells physical properties: production of type II collagen proteins was achieved in adipose stem cells by overexpression of the Sox trio (Sox5, Sox6 and Sox9)73. The rest of the figure c2 is an adaptation from37,74.
(d) Examples of effector genes induced by synthetic signaling pathways. (d.1) Soluble ligand CD14, released by source cells (HEK293), activates [sCD14]Ca2+RT[CaRQ→migration] on the membrane of seeking cells (HEK293) causing intracellular Ca2+ release from the endoplasmic reticulum. In the receiver cells, Calcium signaling is rewired via CaRQ to trigger migration towards the source cell18. (d.2) CD19 ligand exposed on the membrane of a sender cell activates a [CD19]synNotch[tTA→myoD]receptor that drives Myo-D expression: overexpression of myoD in turn drives myoblast differentiation from embryonic fibroblasts24.
Contact-dependent receptors have been used to generate precise single-cell patterns in 2D. The Notch pathway has been reconstituted in epithelial cells (CHO) to regenerate the checkerboard pattern displayed during the development of the inner ear. To do so, Notch-responsive promoters are linked to the repression of the endogenous Notch ligand Delta in a circuit known as lateral-inhibition. Starting from a uniform population of cells expressing the same program, this system generates two cellular states (receptor high or ligand high)26 (Fig. 3a.1). In other examples, signaling cascades have been implemented with Notch/tTA receptors or synNotch. In these experiments, the core network is a positive feedback network where receptor activation leads to the induction of ligands for subsequent communications. This can lead to signal propagation in epithelial cells27, or to alternating activation of different responses in epithelial monolayers24, depending on the number of pathways and ligands (Fig. 3a.2–3).
Endogenous soluble ligand-dependent pathways have been used to generate longer-range patterning. In one example, a reconstituted SHH (sonic hedgehog) pathway was used to generate a morphogen-like expression gradient of reporter genes from a signal source in fibroblasts in 2D28. The signal (Shh) was produced by sender cells restricted to one side of the cell culture, and the response in receiver cells closer to the signal source decreased as distance from the source increased. In another recent example, Turing-like patterns were generated with a reconstituted Nodal/Lefty pathway in epithelial cells. To reconstitute the local activation (positive feedback) and long-range inhibition (negative feedback) typical of Turing systems, the responses to Nodal were rewritten to include the induction of both Nodal itself (positive feedback) and of its diffusible inhibitor Lefty (negative feedback). The result was the formation of multicellular patterns of spots reminiscent of some Turing patterns29,30 (Fig. 3a.4).
Gene regulatory networks that achieve purely temporal patterning are also possible to implement in mammalian cell with synthetic biology tools. For example, negative feedback with response delay gives rise to oscillations31–33 (Fig. 3b). Other temporal controls, such as memory and timed pulses34, have also been implemented in mammalian cells. These intracellular networks are not usually coordinated at the cell population level. We anticipate progress in this area to connect intracellular temporal networks with spatial control and coordination, thanks to links between the cell-cell communication pathways and the intracellular gene regulatory networks.
Physical and biological cellular changes through effector genes
In order for a group of cells to change their shape to form structurally complex tissues, cells need to take on different physical and/or biological features. In this section, we focus on cellular changes underlying morphogenetic and developmental processes. Genes that underlie these physical and biological events are called here effector genes (Fig. 3c). A long series of effector genes cassettes are known to reprogram cell behaviors, either through differentiation into distinct cell fates via reprogramming factors35,36, or by changes in cells’ physical features, such as adhesion or shape37. Changes of cell fate affect both physical and biological features, making them a major motor of multicellular development and morphogenesis. Processes such as proliferation, apoptosis, and cell fusion can also be powerful, as they control the number of cells. Additionally, changing isolated physical properties of a single cell can be responsible for morphogenetic transitions at the multicellular level. Cytoskeletal players can induce cell shape changes that can underlie behaviors such as apical constriction and folding, and cell adhesion molecules can control multicellular aggregation status and sorting38,39.
It will be interesting to see if synthetic versions of effector genes can be generated for mammalian cells, and how much they can drive orthogonal programs of morphogenesis in the presence of endogenous effectors. Recently, synthetic versions of adhesion proteins have been generated in bacterial cells40. Additionally, other cell behaviors have been identified as drivers of multicellular morphogenesis: (i) the “fluidity” status of multicellular tissues was shown to be important for tissue elongation in vivo41, although driver genes have not yet been clearly identified; (ii) cytoskeletal dynamics, for example supracellular contractions of multicellular actomyosin cables, were recently shown to drive neural crest cells chemotaxis42.
Effector genes induced by synthetic cell-cell communication pathways are another building block of morphogenesis (Fig. 3d). Synthetic versions of this concept have been developed: synthetic signaling pathways that can sense chemical compounds or chemotactic agents via RASSL or Ca2+RT are wired to respond with a chemotactic behavior18,43 (Fig. 3d.1). In other examples, synNotch pathways turn on (i) the transcription factor Snail to change cell behavior from epithelial to mesenchymal/fibroblastoid; or (ii) the master regulator MyoD that turns fibroblasts into myotubes24 (Fig. 3d.2).
It will be interesting to see which synthetic pathways can drive effector genes efficiently enough to guide robust changes in cell behavior that can control multicellular dynamics.
Synthetic developmental trajectories
To generate an architecturally complex tissue starting from a uniform cell population, signaling, patterning, and effectors are linked into a developmental trajectory (Fig. 1). This allows cell behavior to be changed, through effector genes and multicellular networks, in a precise spatial and temporal fashion in order to generate functional units. So far, the full generation of a functional tissue via a synthetic artificial network has not been achieved.
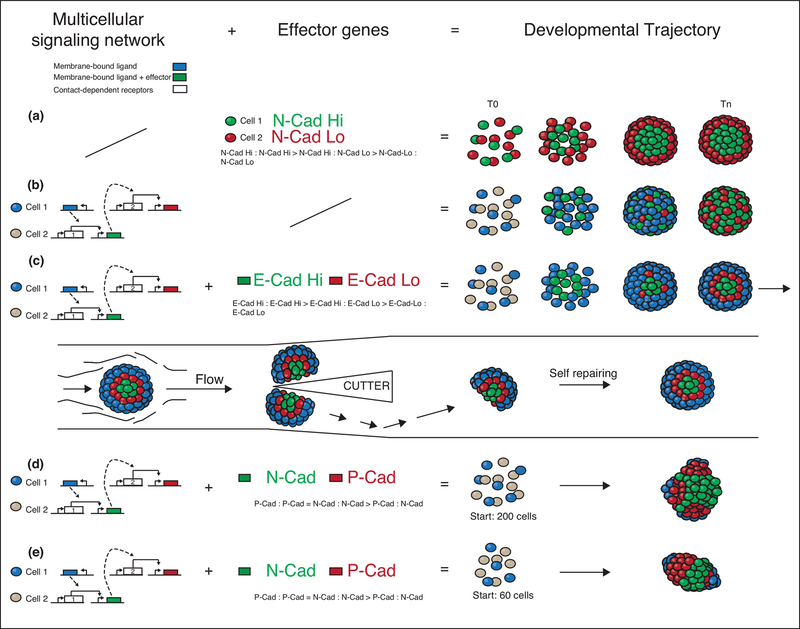
However, Toda et al. recently combined a synthetic signaling-based multicellular network with simple morphogenetic effectors of the adhesion family to generate multi-step synthetic developmental trajectories44 for generating structural units (Fig. 4). Two channels of orthogonal contact-dependent synNotch pathways were used as the synthetic cell signalling component; the network was back-and-forth communication between two different cell populations; the effectors were adhesion molecules of the cadherin family. In one example (Fig. 4c), the communication from cell 1 induced E-cadherin in cell 2, and a new signal that communicated back to induce lower levels of E-cadherin in subset of the cell 1 population. The resulting structure was a radially symmetric three-layered spheroid, which was more ordered and complex than when they used only effectors or only networks (Fig. 4a,b). Structures generated in this way displayed robustness against perturbations such as cutting in half (Fig. 4 c, second row). Moreover, similar networks with different effectors (different members of the cadherin family) generated polarized structures (Fig. 4d,e), showing that changing one parameter of the system could generate multiple variants—a feature we might call “synthetic evolvability.”
Figure 4 – Engineered synthetic developmental trajectories in multicellular fibroblast spheroids.
The combination of multicellular signaling network driving effector genes are shown on the left hand side; the resulting developmental trajectories are shown on the right.
The multicellular regulatory network (dashed arrows represent the intercellular signaling, solid arrows the intracellular signaling) (left most column), downstream effector genes (central column) and the corresponding developmental trajectory (right column) for each genetic program are shown. (a) No regulatory network is present, N-cad is constitutively expressed at high level in the green cells and at low levels in the red cell line39, generating a 2-layered structure. (b) The regulatory network based on synNotch signaling drives the expression of fluorescent proteins (no effector genes). The blue cell line activates the gray cell line turning it green, the green cell line then activates the blue cell line turning it red. (c) The regulatory network is the same as in (b), but it now drives expression of effector genes as well: high levels of E-cadherin (E-cad) in green cells and low levels of E-cad in red cells. The differences in the expression levels of E-Cad (null in blue cells, low in red cells, and high in green cells) generated in a temporally controlled fashion cause spatial rearrangements and cell sorting into a three-layered structure. The three-layer structure is able to self-repair after cleavage (c, second row). (d-e) The same regulatory network as in (b) drives the expression of N-Cadherin (N-cad) (green) and P-Cadherin (P-cad) (red), generating (d) multiple poles (initial co-culture conditions of 200 cells) or (e) a single pole (initial co-culture conditions of 60 cells) 75.
Continued progress in this area could pave the way for a field devoted to synthetic development where synthetic multicellular networks can execute a variety of effector functions in cells, including more complex morphogenesis. In the future, we expect more complex circuits, coupled with effector genes, will generate more complex morphogenesis and even functional structures.
Computational approaches can facilitate synthetic development
The design of developmental trajectories in vitro for synthetic developmental systems (as well as organoids) currently proceeds through trial and error. A more deterministic method involving computational models could provide powerful tools for synthetic development5. Below, we review how, in recent works, the aforementioned developmental trajectory components of multicellular signaling networks, and physical changes are modeled separately to yield patterning, and how they can be integrated to form comprehensive morphogenesis models.
Multicellular signaling networks have been described via sets of differential equations modeling the level of each reactive species/protein on some kind of lattice that describes the cells. Usually ordinary differential equations (ODES) are used for temporal patterning and partial differential equations (PDES) for spatial patterning. For contact-dependent signaling, Shaya et al.45 provide an innovative example. Utilizing a fixed heterogeneous cell size lattice, the authors described extracellular membrane-bound ligand/receptor levels via ODES on each inter-cell boundary, as opposed to the homogenous cell method in prior works46–48. By doing so, they generated a patterning model with high spatial resolution, which was able to match in vitro observations that small cells are biased towards one fate and large cells towards another. For diffusible signals, Li et al.28 provided an outstanding example by developing a minimalist mathematical framework to investigate how a specific diffusible signal can affect gradient patterning. By describing the diffusible ligand levels via reaction-diffusion PDES, and other species’ (receptor, repressor, etc.) levels via coupled ODES per discretized space, the authors recapitulated and predicted key patterning results.
The morphological effect of effector genes is usually modeled via mechanical modeling. For example, cell-cell or cell-ECM adhesion is typically modeled as energy, which is then converted into a probability of cells sticking together49–51 or converted into equations of motion52,53. Growth can also lead to patterning, such as brain folding, and can be modeled by mechanical deformation equations54. In the discretized cell case, it can be modeled by adding a volume constraint to a cell, thus designating a cell’s “ideal” volume. Increasing the “ideal” volume over time leads to growth55 while proliferation can be modeled by simple division at a specified volume55–58. Apoptosis can be considered the opposite of growth, and therefore can be implemented by decreasing the “ideal” volume over time until the cell disappears58. For more complex/different formulations of growth, proliferation, and apoptosis, we refer the readers to56–59, 60, and56, respectively.
Integrating models of multicellular signaling networks with mechanical models makes it possible to describe complex native developmental trajectories. For example, secreted diffusible signals directing motion can capture chicken feather primordia striping and spotting61. Secreted diffusible ligands directing mitosis instead recapitulate a different biological process, ex vivo kidney explant branching60. Adding differential adhesion allows modeling of tooth germ formation59. Joining multiple signals with multiple outputs achieves even higher complexity, as Hester et al.62 demonstrate by mixing lateral inhibition synchronization with secreted diffusible signals to dynamically describe somitogenesis. Indeed, utilizing multiple inputs, such as multiple diffusible ligands, allows for the description of other complex processes, such as palate fusion56.
Computational models have been helpful for modeling native developmental trajectories. The ability to model cell communication networks and effector genes in silico parallels the abstraction we described for synthetic development, making it especially well-suited for this application. It would be interesting to adjust and link existing models for various signals and responses in order to generate a variety of in silico synthetic developmental trajectories. For example, they can be used to screen different possible structures, establish parameter dependencies, study different forms of computation, and extract algorithms that can be used in other biologically inspired computational contexts. Ideally, a closed loop with modeling and implementation would allow the most progress in this area.
Conclusions
We have described how we can deconstruct developmental morphogenetic programs in order to start reconstructing them in novel ways. We discussed how cell-cell signaling, multicellular networks, and effector genes form a core motif for programming developmental trajectories. Many controllable cell-cell communication channels are currently available through synthetic receptor and synthetic pathway engineering, and some of them have already been used to either build patterning networks or drive cellular physical and biological changes.
Challenges ahead include expanding the toolkit for cell-cell communication, alongside its characterization in various cellular contexts. Much of the focus has been on model cell lines, and we will want to continue expanding towards either therapeutically useful cells, such as immune cells or stem cells, or embryonic cells for modeling development. Moreover, advances in developmental biology research will help identify genetic, biochemical or logic controllers for mesoscale phenomena63, ushering in the next generation of synthetic developmental systems. The ultimate challenge will be to design genetic program at the single cell level to control behaviors at the multicellular scale. One important step in that direction will be the generation of closed loop computational/experimental frameworks for guiding and exploring network/morphogenesis relationships.
For the first time, we find ourselves equipped with the tools and basic knowledge necessary for engineering self-organization in multicellular systems. This multi-disciplinary effort could enhance our control of the development and functional behavior of complex multicellular systems and serve as a valuable testing ground for cell signaling in multicellular contexts. We expect this effort to uncover basic biological principles and create the next generation of therapies for regenerative medicine.
Acknowledgements
The authors want to thank Cristy Lytal for revising the text, Marion Johnson and the other members of the Morsut Lab for the stimulating discussions and for revising the text. This work was supported by an R00 grant from the National Institute of Biomedical Imaging and Bioengineering to LM (4R00EB021030-03), and from a Startup Fund from the Department of Stem Cell Biology and Regenerative Medicine at USC.
L.M. invented intellectual property related to the synNotch receptor that has been licensed to Cell Design Labs and receives royalty payments for this through UCSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cameron DE, Bashor CJ & Collins JJ A brief history of synthetic biology. Nature Reviews Microbiology 12, 381–390 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Black JB, Perez-Pinera P & Gersbach CA Mammalian Synthetic Biology: Engineering Biological Systems. Annu. Rev. Biomed. Eng (2017). doi: 10.1146/annurev-bioeng-071516-044649 [DOI] [PubMed]
- 3.Wieland M & Fussenegger M Engineering Molecular Circuits Using Synthetic Biology in Mammalian Cells. Annu. Rev. Chem. Biomol. Eng 3, 209–234 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Elowitz M & Lim WA Build life to understand it. Nature (2010). doi: 10.1038/468889a [DOI] [PMC free article] [PubMed]
- 5.Velazquez JJ, Su E, Cahan P & Ebrahimkhani MR Programming Morphogenesis through Systems and Synthetic Biology. Trends Biotechnol 36, 415–429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White MD, Zenker J, Bissiere S & Plachta N Instructions for Assembling the Early Mammalian Embryo. Dev. Cell 45, 667–679 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Morsut L Programming cells to build tissues with synthetic biology: a new pathway towards engineering development and regeneration. In “Regenerative Engineering and Developmental Biology: Principles and Applications,” edited by Gardiner David M. CRC Press; (September 1, 2017) [Google Scholar]
- 8.Barolo S & Posakony JW Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes and Development (2002). doi: 10.1101/gad.976502 [DOI] [PubMed]
- 9.Lim WA Designing customized cell signalling circuits. Nature Reviews Molecular Cell Biology (2010). doi: 10.1038/nrm2904 [DOI] [PMC free article] [PubMed]
- 10.Gordley RM, Bugaj LJ & Lim WA Modular engineering of cellular signaling proteins and networks. Current Opinion in Structural Biology (2016). doi: 10.1016/j.sbi.2016.06.012 [DOI] [PMC free article] [PubMed]
- 11.Sprinzak D & Elowitz MB Reconstruction of genetic circuits. Nature (2005). doi: 10.1038/nature04335 [DOI] [PubMed]
- 12.Arber C, Young M & Barth P Reprogramming cellular functions with engineered membrane proteins. Current Opinion in Biotechnology (2017). doi: 10.1016/j.copbio.2017.06.009 [DOI] [PMC free article] [PubMed]
- 13.Brenner M, Cho JH & Wong WW Synthetic biology: Sensing with modular receptors. Nature Chemical Biology (2017). doi: 10.1038/nchembio.2290 [DOI] [PubMed]
- **14.Scheller L, Strittmatter T, Fuchs D, Bojar D & Fussenegger M Generalized extracellular molecule sensor platform for programming cellular behavior Leo. Nat. Chem. Biol (2018). doi: 10.1038/s41589-018-0046-zDescription of the GEMS synthetic receptors for engineering input-output cellular controls. The transduction logic is based on an elegant use of conformational change of the EPO-R tyrosin-kinase receptor platform. It has been used to link a number of synthetic or endogenous inputs to endogenous pathways activation.
- 15.Hartfield RM, Schwarz KA, Muldoon JJ, Bagheri N & Leonard JN Multiplexing Engineered Receptors for Multiparametric Evaluation of Environmental Ligands. ACS Synth. Biol (2017). doi: 10.1021/acssynbio.6b00279 [DOI] [PMC free article] [PubMed]
- 16.Schwarz KA, Daringer NM, Dolberg TB & Leonard JN Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol (2017). doi: 10.1038/nchembio.2253 [DOI] [PMC free article] [PubMed]
- 17.Conklin BR et al. Engineering GPCR signaling pathways with RASSLs. Nat. Methods (2008). doi: 10.1038/nmeth.1232 [DOI] [PMC free article] [PubMed]
- 18.Qudrat A & Truong K Antibody-based fusion proteins allow Ca2 + rewiring to most extracellular ligands (2017). doi: 10.1021/acssynbio.7b00323 [DOI] [PubMed]
- 19.Barnea G et al. The genetic design of signaling cascades to record receptor activation 105, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Kipniss NH et al. Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nat. Commun (2017). doi: 10.1038/s41467-017-02075-1Description of the development of ChaCha receptor system that couples CRISPR-dCas9 genome regulation to diverse natural and synthetic extracellular signals via GPCRs. CRISPR ChaCha is dose-dependent, reversible, and can activate multiple endogenous genes simultaneously in response to extracellular ligands that can be either endogenous or synthetic.
- *21.Baeumler TA et al. Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors Article Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. CellReports 20, 2639–2653 (2017).Description of the step-wise development and optimization of the dCas9-synR class of synthetic receptors that integrate the highly programmable and portable dCas9 signal transduction module. Both receptor-tyrosin kinases and G-protein-coupled receptor scaffold has been used to rewire endogenous or artificial inputs into activation of user-defined endogenous genes.
- 22.Krawczyk K, Scheller L, Kim H & Fussenegger M Rewiring of endogenous signaling pathways to genomic targets for therapeutic cell reprogramming [DOI] [PMC free article] [PubMed]
- 23.Brunger JM, Zutshi A, Willard VP, Gersbach CA & Guilak F Genome Engineering of Stem Cells for Autonomously Regulated, Closed-Loop Delivery of Biologic Drugs. Stem Cell Reports (2017). doi: 10.1016/j.stemcr.2017.03.022 [DOI] [PMC free article] [PubMed]
- *24.Morsut L et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell (2016). doi: 10.1016/j.cell.2016.01.012Description of the synNotch synthetic receptors class. The framework is the Notch contact-dependent signaling. It has been rewired to accept both endogenous and synthetic inputs, and activate reporter genes as well as biological changes in the receiving cells, including diffferentiation. Simple logic gates and multicellular patterning networks are demonstrated as well.
- 25.Roybal KT et al. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell 167, 419–432.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Matsuda M, Koga M, Woltjen K, Nishida E & Ebisuya M Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat. Commun 6, 6195 (2015).The Notch pathway has been reconstituted in epithelial cells (CHO) to regenerate the cell-type bifurcation pattern displayed during development of, for example, the inner ear. Notch-responsive promoters are linked to repression of endogenous Notch ligand Delta in a circuit known as lateral-inhibition.
- 27.Matsuda M, Koga M, Nishida E & Ebisuya M Synthetic Signal Propagation Through Direct Cell-Cell Interaction. Sci. Signal 5, ra31–ra31 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Li P et al. Morphogen gradient reconstitution reveals Hedgehog pathway design principles. Science (80-. ) (2018). [DOI] [PMC free article] [PubMed]
- 29.Sekine R, Shibata T & Ebisuya M Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. bioRxiv (2018). doi: 10.1101/372490 [DOI] [PMC free article] [PubMed]
- 30.Sekine R, Shibata T & Ebisuya M Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tigges M, Marquez-Lago TT, Stelling J & Fussenegger M A tunable synthetic mammalian oscillator. Nature (2009). doi: 10.1038/nature07616 [DOI] [PubMed]
- 32.Stricker J et al. A fast, robust and tunable synthetic gene oscillator. Nature (2008). doi: 10.1038/nature07389 [DOI] [PMC free article] [PubMed]
- 33.Santorelli M et al. Reconstitution of an Ultradian Oscillator in Mammalian Cells by a Synthetic Biology Approach. ACS Synth. Biol 7, 1447–1455 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Saxena P, Bojar D, Zulewski H & Fussenegger M Generation of glucose-sensitive insulin-secreting beta-like cells from human embryonic stem cells by incorporating a synthetic lineage-control network. J. Biotechnol (2017). doi: 10.1016/j.jbiotec.2017.07.018 [DOI] [PubMed]
- 35.Cherry ABC & Daley GQ Reprogramming cellular identity for regenerative medicine. Cell (2012). doi: 10.1016/j.cell.2012.02.031 [DOI] [PMC free article] [PubMed]
- 36.Black JB & Gersbach CA Synthetic transcription factors for cell fate reprogramming. Current Opinion in Genetics and Development (2018). doi: 10.1016/j.gde.2018.05.001 [DOI] [PubMed]
- 37.Davies JA Synthetic morphology: Prospects for engineered, self-constructing anatomies. J. Anat 212, 707–719 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zartman JJ & Shvartsman SY Unit Operations of Tissue Development: Epithelial Folding. Annu. Rev. Chem. Biomol. Eng (2010). doi: 10.1146/annurev-chembioeng-073009-100919 [DOI] [PMC free article] [PubMed]
- 39.Foty RA & Steinberg MS The differential adhesion hypothesis: a direct evaluation. Dev. Biol 278, 255–263 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Glass DS & Riedel-Kruse IH A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell (2018). doi: 10.1016/j.cell.2018.06.041 [DOI] [PubMed]
- 41.Mongera A et al. A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature (2018). doi: 10.1038/s41586-018-0479-2 [DOI] [PMC free article] [PubMed]
- 42.Shellard A, Trepat X & Mayor R Supracellular contraction at the rear of cell groups drives collective chemotaxis 343, 339–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JS et al. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc. Natl. Acad. Sci (2014). doi: 10.1073/pnas.1402087111 [DOI] [PMC free article] [PubMed]
- **44.Toda S, Blauch LR, Tang SKY, Morsut L & Lim WA Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science (80-. ) 162, eaat0271 (2018).Description of the implementation of a multistep synthetic developmental trajectory in naïve fibroblast spheroid. The developmental trajectory is programmed via synNotch-based multicellular networks that drive changes in cell-cell adhesions using Cadherin effector genes. These simple programs generate synthetic developmental trajectory that display robust self-organization into multidomain structures, well-choreographed sequential assembly, cell type divergence, symmetry breaking, and the capacity for regeneration upon injury.
- 45.Shaya O et al. Cell-Cell Contact Area Affects Notch Signaling and Notch-Dependent Patterning. Dev. Cell 40, 505–511.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collier JR, Monk NAM, Maini PK & Lewis JH Pattern Formation by Lateral Inhibition with Feedback: a Mathematical Model of Delta-Notch Intercellular Signalling. J. Theor. Biol 183, 429–446 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J & Elowitz MB Mutual Inactivation of Notch Receptors and Ligands Facilitates Developmental Patterning. PLOS Comput. Biol 7, e1002069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owen MR, Sherratt JA & Wearing HJ Lateral Induction by Juxtacrine Signaling Is a New Mechanism for Pattern Formation. Dev. Biol 217, 54–61 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Wang Y et al. Spheroid Formation of Hepatocarcinoma Cells in Microwells: Experiments and Monte Carlo Simulations. PLoS One 11, e0161915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Thomas GL, Swat M, Shirinifard A & Glazier JA Computer Simulations of Cell Sorting Due to Differential Adhesion. PLoS One 6, e24999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glazier JA & Graner F Simulation of the differential adhesion driven rearrangement of biological cells. Phys. Rev. E 47, 2128–2154 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Flenner E et al. Kinetic Monte Carlo and cellular particle dynamics simulations of multicellular systems. Phys. Rev. E 85, 31907 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Farhadifar R, Röper J-C, Aigouy B, Eaton S & Jülicher F The Influence of Cell Mechanics, Cell-Cell Interactions, and Proliferation on Epithelial Packing. Curr. Biol 17, 2095–2104 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Budday S, Steinmann P, Goriely A & Kuhl E Size and curvature regulate pattern selection in the mammalian brain. Extrem. Mech. Lett 4, 193–198 (2015). [Google Scholar]
- 55.Izaguirre JA et al. CompuCell, a multi-model framework for simulation of morphogenesis. Bioinformatics 20, 1129–1137 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Hutson MS, Leung MCK, Baker NC, Spencer RM & Knudsen TB Computational Model of Secondary Palate Fusion and Disruption. Chem. Res. Toxicol 30, 965–979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belmonte JM et al. Virtual-tissue computer simulations define the roles of cell adhesion and proliferation in the onset of kidney cystic disease. Mol. Biol. Cell 27, 3673–3685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swat MH, Thomas GL, Shirinifard A, Clendenon SG & Glazier JA Emergent Stratification in Solid Tumors Selects for Reduced Cohesion of Tumor Cells: A Multi-Cell, Virtual-Tissue Model of Tumor Evolution Using CompuCell3D. PLoS One 10, e0127972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marin-Riera M, Moustakas-Verho J, Savriama Y, Jernvall J & Salazar-Ciudad I Differential tissue growth and cell adhesion alone drive early tooth morphogenesis: An ex vivo and in silico study. PLOS Comput. Biol 14, e1005981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lambert B et al. Bayesian inference of agent-based models: a tool for studying kidney branching morphogenesis. J. Math. Biol 76, 1673–1697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin C-M et al. Spots and stripes: Pleomorphic patterning of stem cells via p-ERK-dependent cell chemotaxis shown by feather morphogenesis and mathematical simulation. Dev. Biol 334, 369–382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hester SD, Belmonte JM, Gens JS, Clendenon SG & Glazier JA A Multi-cell, Multi-scale Model of Vertebrate Segmentation and Somite Formation. PLOS Comput. Biol 7, e1002155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blanchard GB, Fletcher AG & Schumacher LJ The devil is in the mesoscale: Mechanical and behavioural heterogeneity in collective cell movement. Semin. Cell Dev. Biol (2018). doi: 10.1016/J.SEMCDB.2018.06.003 [DOI] [PubMed]
- 64.Daringer NM, Dudek RM, Schwarz KA & Leonard JN Modular Extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol (2014). doi: 10.1021/sb400128g [DOI] [PMC free article] [PubMed]
- 65.Schwarz KA, Daringer NM, Dolberg TB & Leonard JN Rewiring human cellular input–output using modular extracellular sensors. Nat. Publ. Gr 13, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siciliano V et al. Engineering modular intracellular protein sensor-actuator devices. Nat. Commun 1–7 (2018). doi: 10.1038/s41467-018-03984-5 [DOI] [PMC free article] [PubMed]
- *67.Qudrat A & Truong K Engineering Synthetic Proteins to Generate Ca 2+ Signals in Mammalian Cells (2016). doi: 10.1021/acssynbio.6b00310Series of papers describing rewiring of Ca2+-signaling into user-defined responses. This Ca2+ rewiring systems that consists of chimeric receptor that generates a Ca2+ signal upon ligand binding; this Ca2+ signal is then transduced via a chimeric RhoA protein that activates cell migration in response to a Ca2+ signal.
- 68.Mosabbir AA, Qudrat A & Truong K A monoclonal antibody acts as a migratory cue via Ca2+ rewiring. Integr. Biol (2018). doi: 10.1039/c7ib00203c [DOI] [PubMed]
- 69.Swinburne I. a, Miguez DG, Landgraf D & Silver P. a. Intron length increases oscillatory periods of gene expression in animal cells Email alerting service Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev 2342–2346 (2008). doi: 10.1101/gad.1696108 [DOI] [PMC free article] [PubMed]
- 70.Hester ME et al. Rapid and Efficient Generation of Functional Motor Neurons From Human Pluripotent Stem Cells Using Gene Delivered Transcription Factor Codes. Mol. Ther 19, 1905–1912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita R et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. PNAS (2014). doi: 10.1073/pnas.1413234112 [DOI] [PMC free article] [PubMed]
- 72.Wang L et al. Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming. 237–244 (2014). doi: 10.1161/CIRCRESAHA.116.305547 [DOI] [PMC free article] [PubMed]
- 73.Im G & Kim H Electroporation-mediated gene transfer of SOX trio to enhance chondrogenesis in adipose stem cells. OARSI 19, 449–457 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Cachat E, Liu W, Hohenstein P & Davies JA A library of mammalian effector modules for synthetic morphology 1–11 (2014). [DOI] [PMC free article] [PubMed]
- 75.Toda S, Blauch LR, Tang SKY, Morsut L & Lim WA Programming selforganizing multicellular structures with synthetic cell-cell signaling. Science (80-. ) (2018). [DOI] [PMC free article] [PubMed]