Abstract
Advances in electron microscopes, detectors and data processing algorithms have greatly facilitated the structural determination of many challenging integral membrane proteins that have been evasive to crystallization. These breakthroughs facilitate the application and development of various membrane protein solubilization approaches for structural studies, including reconstitution into lipid nanoparticles. In this review we discuss various approaches for preparing transmembrane proteins for structural determination with single-particle electron cryo microscopy (cryo-EM).
Keywords: transmembrane proteins, single-particle cryoelectron microscopy, detergents, amphipols, nanodiscs, SMA lipid particles, native nanodiscs
Introduction
Structural characterization of integral membrane proteins remains a major challenge, largely owing to difficulties in extracting them from their native lipid environment while maintaining structural and functional integrity. Structural characterization has conventionally been achieved using detergents, which are amphipathic molecules that keep membrane proteins in solution by forming micelles around the hydrophobic domains. However, detergent micelles fall short of mimicking the complexity of the native membrane environment. As such, discrepancies between detergent micelles and the cell membrane may destabilize or irreversibly inactivate detergent-solubilized membrane proteins.
In the wake of what has been coined the ‘resolution revolution’, single-particle cryoEM enables high-resolution structural determination of integral membrane proteins without the need for crystallization. Such capability facilitates the use of alternative, non-detergent solubilizing mediums for structural studies. Amphipols, which are short hydrophilic polymers that carry numerous hydrophobic side chains, allow membrane proteins to stay in solution in the absence of excess surfactant, due to their high affinity for the transmembrane surface of integral membrane proteins [1–3]. Soluble lipid-bilayer nanoparticle systems, stabilized by either membrane scaffold proteins (MSPs) [4] or saposin [5], facilitate protein-lipid interactions in a more ‘native’ environment and have thus become an increasingly popular method to stabilize membrane proteins for single-particle cryoEM studies. More recently, a nanodisc system that utilizes styrene–maleic acid (SMA) copolymers to solubilize membrane proteins directly from cell membranes show potential as the next generation nanodiscs [6–8].
The progress of single-particle cryoEM is facilitating rapid expansion and further optimization of all these alternative membrane protein preparation approaches for functional and structural studies of integral membrane proteins. Here we discuss both well-established and novel approaches for preparing membrane proteins for structural determination with single-particle cryoEM (Figure 1), and consider future development of this rapidly evolving field.
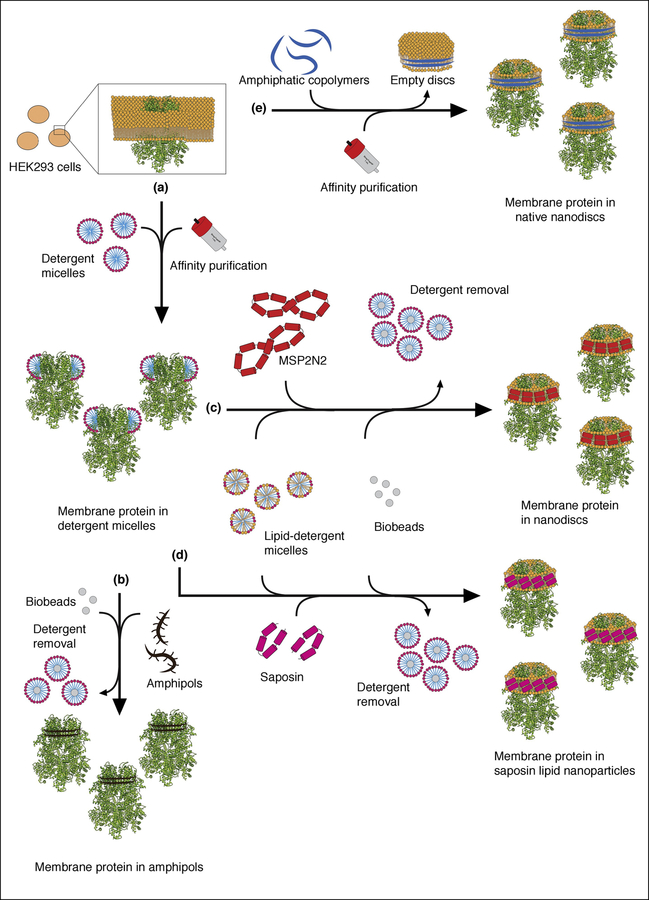
Figure 1. Schematic overview of the typical approaches for solubilization and purification of membrane proteins for single-particle cryo-EM studies.
Here, we used TRPM4 as an example to illustrate various approaches of solubilizing and purifying membrane proteins for single-particle cryo-EM structure determination. (a) The most common starting point is the extraction and solubilization of membrane proteins from cell membranes typically using detergents. (b) The detergent-solubilized membrane protein can be exchanged into amphipols using biobeads to remove the detergent. (c) The detergent-solubilized membrane protein can be reconstituted into MSP lipid nanodiscs by addition of detergent-solubilized lipids, MSP and biobeads to remove the detergent. (d) Alternatively, the detergent-solubilized membrane protein can be reconstituted into the Salipro system, adding detergent-solubilized lipids, saposin-A and biobeads. (e) Rather than breaking the cells and extracting the membrane protein with detergent, addition of SMA copolymers will extract it as native nanodiscs forming SMALPs.
General considerations
Detergents are amphipathic molecules with defined hydrophilic and hydrophobic domains. Currently, solubilization of whole cells or isolated membranes with detergent is the typical starting point for extracting and purifying endogenous or expressed, recombinant membrane proteins (Figure 1A). The choice of detergent in each step of protein purification has significant effects on the yield and quality of the protein. Originally developed for sample optimization prior to crystallization, fluorescent-based size-exclusion chromatography (FSEC) remains an efficient way to screen solubilization conditions that provide the highest yield of homogeneous protein [9]. Additional considerations are the inclusion of additives during solubilization or later in the purification protocol. For example, the soluble cholesterol mimic, cholesteryl hemisuccinate (CHS), has been used in a considerable number of high-resolution ion channel structures. CHS seems to stabilize a broad range of membrane proteins (irrespective of whether cholesterol is required for function), although one must also consider the possibility that CHS locks the protein in a specific conformation [10–12]. Addition of lipid extracts or synthetic lipid-detergent mixtures can also facilitate membrane protein purification and protein stability, as well as the addition of cofactors and natural or pharmacologic ligands. Moreover, structures of proteins purified in such ways can reveal important lipid-protein interactions [13].
Negative-stain EM is an efficient method to evaluate each step of a purification protocol and assess whether the sample is progressively purified and is eventually homogeneous and monodisperse. Unfortunately, aggregation or shearing during plunge freezing is not evident from negative-stain EM and can only be revealed by screening cryoEM grids. If there is suspicion that these problems are due to the chosen detergent, one may attempt to lower the detergent concentration or exchange the detergent-solubilized membrane protein into a different detergent or a detergent-free system [14].
Detergents
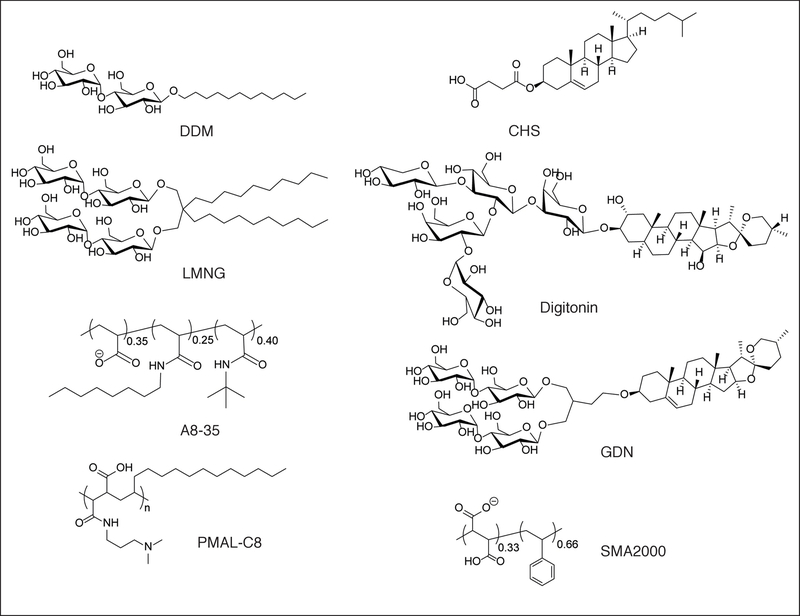
Detergents have limited solubility in water and exist in an equilibrium between free detergent monomers, free detergent micelles and micelles associated with the transmembrane domains of membrane proteins. To prevent dissociation of detergent from the membrane protein and thereby causing protein aggregation, the detergent concentration must remain sufficiently greater than its critical micelle concentration (CMC). However, excessive detergent poses a challenge for single-particle cryoEM because it influences the surface tension of water, thickness of the vitrified ice in which the particles are embedded and the particle distribution on the EM grid during plunge-freezing. Free detergent micelles in solution also increase the background noise in cryoEM images [15]. Nevertheless, modern single-particle cryoEM technology makes it generally feasible to determine high-resolution structures of membrane proteins in detergents. Typically, detergents with very small CMCs are more favorable for single-particle cryoEM analysis. Particularly, n-Dodecyl-β-D-Maltopyranoside (DDM) and maltose-neopentyl glycols (MNGs) with hydrophilic groups derived from maltose [16] have been used extensively in single-particle cryoEM of integral membrane proteins (Figure 2). Other detergents successfully applied to mammalian membrane proteins in single-particle cryoEM is the steroidal saponin digitonin [17], which can be extracted from the purple foxglove plant Digitalis purpurea (Figure 2). Due to batch-to-batch variations of Digitonin, the glyco-diosgenin (GDN) has started to appear in a number of studies as a synthetic substitute for Digitonin [18–20] (Figure 2).
Figure 2. Overview of popular detergents and amphipols used in single-particle cryoEM.
Figure includes the detergents n-Dodecyl-β-D-Maltopyranoside (DDM), Lauryl Maltose Neopentyl Glycol (LMNG), cholesteryl hemisuccinate (CHS), Digitonin and glyco-diosgenin (GDN), the amphipols A8–35 and PMAL-C8 and the styrene-maleic acid copolymer SMA2000.
Amphipols
Amhipols are amphipathic polymers that have a hydrophilic backbone decorated with multiple hydrophobic groups. Similar to detergents, amphipols self-assemble into well-defined particles that surround transmembrane domains [1–3]. A main difference between amphipols and detergents is that the affinity for the hydrophobic transmembrane domain is higher for the former. Thus, amphipols remain tightly associated with a membrane protein even when their concentration is sufficiently low such that there are no free amphipol molecules in solution [1–3]. For single-particle cryoEM, this avoids the free micelle problem associated with detergents and their negative influence on the background and ice thickness. In practice, membrane proteins are first solubilized with detergent and then exchanged into amphipols following affinity purification (Figure 1B). In some cases, the tight association with amphipols also stabilizes the conformation of membrane proteins [21–24]. The amphipols A8–35 and PMAL-C8 are most commonly used for single-particle cryoEM, although PMAL-C12 and -C16 are also commercially available (Figure 2). We hope that their successful use will stimulate the development and commercial production of additional amphipols having a wider range of physical-chemical properties [25].
MSP Nanodiscs
As the structure, function and stability of membrane proteins are all modulated by the surrounding membrane, it is beneficial to provide them with as native a lipid bilayer environment as possible. In the past, this could be accomplished with electron crystallography, in which membrane proteins are reconstituted into lipid bilayers as flat or tubular two-dimensional crystals [26–30]. For single-particle cryoEM, the most obvious approach is to use lipid nanodiscs (Figure 1C). Originally developed by Sligar and colleagues, nanodiscs are discoidal lipid bilayers stabilized by two encircling, amphipathic, helical protein belts, the so-called membrane scaffold proteins (MSPs) [4,31], which are constructed by one or several serially connected domains of apolipoprotein A1 [32].
Lipid nanodiscs have facilitated substantial progress in the structural and functional characterization of membrane proteins in the context of artificial lipid bilayers, resembling a more native-like bilayer environment [33]. Reconstitution of membrane proteins into nanodiscs is typically accomplished by incubating the purified and detergent-solubilized membrane protein with phospholipids and the MSP, followed by detergent removal with biobeads and/or dialysis and size exclusion chromatography (SEC). The MSP is selected to form a nanodisc that will accommodate the membrane protein. However, the final size of the nanodisc is also influenced by the lipid-protein ratio. Single-particle image analysis may also be impeded if the nanodisc is too large [1]. The reader is referred to recent reviews on lipid nanodiscs in cryoEM [34,35].
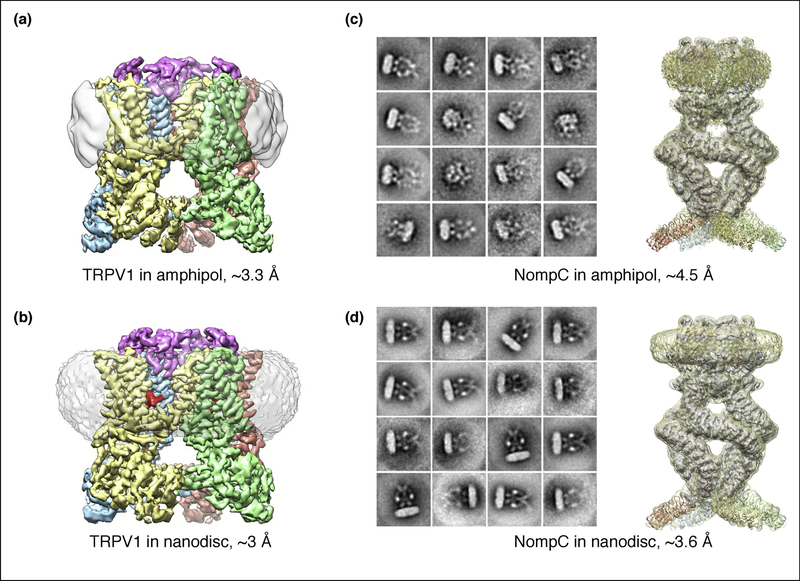
Nanodiscs may also be helpful for stabilizing conformational states of a protein that are less well resolved, or even unattainable, in detergent and amphipol-based preparations [10,36–38]. For instance, structures of the transient receptor potential (TRP) cation channel V1 (TRPV1) in a complex with DkTx spider toxin are better resolved in a nanodisc environment (Figure 3A and B), which enhanced the stability of a tripartite complex between the TRP channel, DkTx and bilayer lipids [18,22]. Another example is the TRPN1 channel (NompC) from Drosophila whose soluble and transmembrane domains are significantly better resolved in lipid nanodiscs compared to detergent and amphipols (Figure 3C and D) [39].
Figure 3. TRPV 1 and NompC in amphipols and lipid nanodiscs.
(a) and (b) TRPV1 with DkTx reconstituted in amphipol (a) and nanodiscs (b). Each subunit in the tetramer is displayed in a different color. The DkTx spider toxin is shown in red. Densities for the amphipol and nanodisc are shown in gray. (c) and (d) 2D class averages and 3D reconstructions of NompC in amphipol (c) and nanodiscs (d). The resolution improvement in nanodiscs is obvious for the two proteins.
Nevertheless, MSP nanodiscs have a number of drawbacks. The conventional protocol for reconstituting membrane proteins in nanodiscs with MSP starts from the detergent-solubilized sample. As discussed in the previous section, this initial solubilization may be harmful for the stability of the protein, disrupt specific conformational states and alter the activity. Also, it is almost impossible to reconstitute membrane proteins solubilized and purified in certain detergents, such as MNGs, which have a very small CMC. Importantly, detergent solubilization may strip away native cofactors, a potential problem that is not remedied by reconstitution into MSP nanodiscs. Nanodisc reconstitution also adds additional time to the sample preparation, which may be harmful for time-sensitive targets. Another approach is to add MSP and biobeads immediately after detergent solubilization and prior to affinity purification [40]. This approach may significantly shorten the exposure to detergent, but requires a considerable amount of MSP for reconstitution. On a practical level, finding the best lipid-to-MSP ratio is an important optimization step that can be a costly challenge, particularly for mammalian membrane proteins with low expression levels. In our own experience with TRP channels, a single type of MSP can pack the protein into a range of different disc sizes. Using the shortest MSP that can accommodate a target protein of a given size may generate more homogeneous nanodisc-protein particles, but with the potential risk that a too tightly bounded MSP may constrain functionally interesting conformational states.
Salipro
Another lipid nanoparticle option is the saposin-lipoprotein (Salipro) system (Figure 1D). This approach is similar to lipid nanodiscs but utilizes saposin A as the scaffolding protein [5]. While detailed cellular functions of saposin proteins are only partly understood, they serve as modulators of lipid degrading enzymes, and lipid-binding is inherent to their function [41–45]. The saposin protein family is composed of four (A–D) small (~10 kDa), amphipathic, α-helical glycoproteins. Crystal structures of saposins A-D have revealed the dynamic range of the four-helix, amphipathic bundle and shown that it can form discoidal lipoprotein particles when incubated with unilamellar liposomes [41,42,45]. By exploiting these properties, Frauenfeld and colleagues demonstrated that saposin A can function as a scaffold to reconstitute a number of different membrane proteins within a lipid environment [5,46].
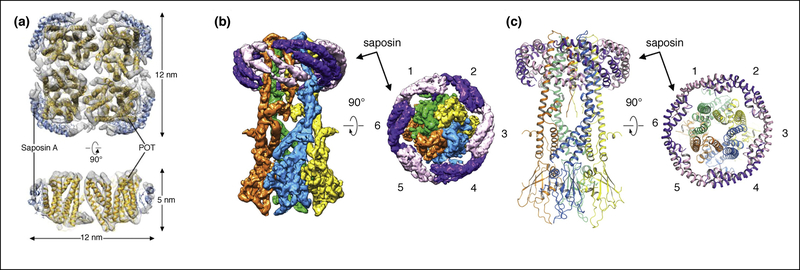
Similar to MSPs, saposin A can be expressed and purified readily from Escherichia coli [5]. Similar to nanodiscs, reconstitution requires optimization to find a suitable protein-to-lipid ratio. However, with only a single protein construct to choose from, the complexity of screening is significantly reduced [5,46]. With its relatively small size of ~10 kDa, saposin A is more adaptive towards membrane proteins of different sizes and shapes (Figure 4A). Thus, the Salipro system combines the advantages of adapting to the size of the incorporated membrane protein, similar to detergents and amphipols, while including a lipid environment similar to nanodiscs [47,48]. A recent high-resolution cryoEM structure of the mitochondrial calcium uniporter (MCU) was solved using saposin A. Nanodiscs were initially tested, but proved to be too large and flexible for the rather small transmembrane domain [49]. The cryoEM density map shows a clear picture of how the diagonally packed helices of saposin A facilitates the formation of a lipid nanoparticle around MCU (Figure 4B and C).
Figure 4. Single-particle cryo-EM of POT and MCU in saposin-lipid particles.
(a) Cryo-EM map of the homotetrameric bacterial peptide transporter PeptTSo2 (gold) with four Saposin A molecules (blue) [5]. (b) and (c). Cryo-EM map (b) and atomic model (c) of the homotetrameric MCU from Neosartorya fischeri stabilized by Saposin A shown as alternating purple and pink densities [45]. There are six Saposin A molecules wrapping around MCU.
The flip side of the adaptivity of saposin A is that, unlike nanodiscs supported by MSPs, the saposin A nanoparticle may provide a suitable environment for lipid-protein interactions, but without recapitulating the mechanical properties of a lipid bilayer. Furthermore, the 3D structure of saposin A may restrain the overall shape and conformation of the target protein.
Native nanodiscs
As noted in the previous section, reconstitution of membrane proteins into either MSP nanodiscs or saposin A nanoparticles requires solubilization and purification of the membrane protein with detergents. The detergent may have deleterious effects on the membrane protein, eliminate its interactions with important cellular cofactors, or affect its conformational state. Furthermore, reconstitution of any type fails to restore the original membrane environment. SMA co-polymers can directly solubilize cellular membranes into nanosized lipid particles (termed SMALPs), thus obviating the need for detergent solubilization. These nanodiscs, which are encircled by an SMA copolymer belt rather than an MSP protein, have been termed native nanodiscs, because they carry native lipids from the membrane in which the target protein resides (Figure 1E). This means that certain unique characteristics of the native membrane, such as lipid composition and asymmetry of the bilayer, may be preserved. Therefore, the native nanodisc approach has great potential as a method of extracting membrane proteins in their immediate native bilayer environment [6,7].
The SMA copolymer has been used for many years in the plastics and pharmaceutical industries and is cheap and highly accessible. Unfortunately, while commercially available SMA can readily extract membrane proteins, their use has a number of limitations. The highly heterogeneous composition of the copolymers likely influences sample homogeneity and extraction efficiency. Indeed, in the case of SMA2000 (2:1 ratio of styrene and maleic acid), our own experience shows that only a subset of the added polymers ends up in the lipid discs. Similarly, Pardo et al. showed that while commercial copolymers of all sizes form nanodiscs, the high molecular weight copolymers are more stable [50]. Furthermore, the negatively charged maleic acid makes the SMA copolymers pH sensitive, with the risk of “dynamic” crosslinking (i.e. non-covalent) from interactions with divalent cations and other positively charged cellular constituents. As such, the majority of available SMA copolymers pose a problem for enzymatic characterization of proteins that depend on cations and, on a more practical level, affinity purification that will be hindered by charge-charge repulsion with cellulose-based column resins.
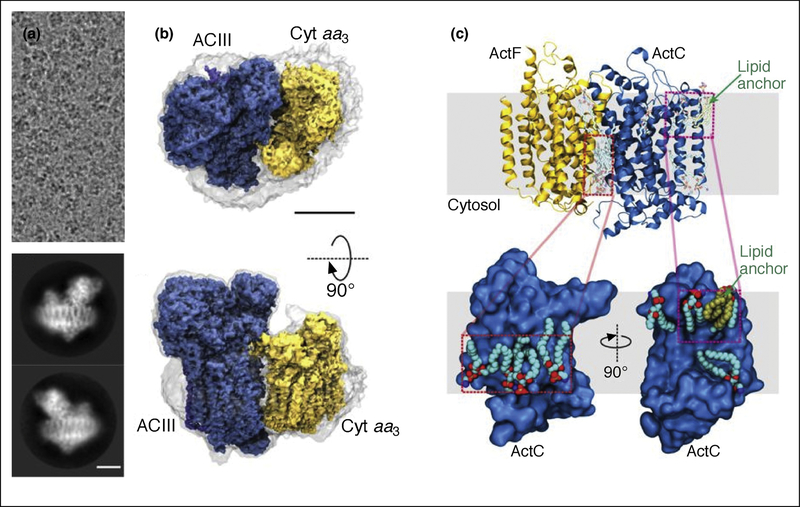
To-date, there are only a few single-particle cryoEM structures of membrane proteins in native nanodiscs, and all have been expressed in bacteria [48,51,52]. The first reported, high-resolution cryoEM structure of a membrane protein in native nanodiscs is that of the alternative complex III, which presumably captures the native lipid–protein interactions (Figure 5) [48]. This structure, however, shows only a very thin layer of density around the protein, suggesting that SMA encircles the protein tightly, with few lipids included in the nanodisc. If so, the system preserves the native lipid-protein interactions, but does not necessarily mimic a bilayer environment.
Figure 5. The alternative complex III in SMA native nanodiscs.
(a). Electron cryomicrograph and representative 2D class averages. (b). Side and top views of the cryoEM map of the complex. The gray surface indicates the boundary of the nanodisc. Scale bars, 50 Å. (c). Resolved lipids near the transmembrane α-helices [43].
Clearly, more effort is needed to better understand and exploit the native nanodisc system. For example, the isolation conditions with SMA copolymers are too harsh for mammalian targets, and the sample loss in the preparation process is too severe for membrane proteins with limited expression. Currently, typical approaches for using SMA co-polymers include high ionic strength buffers, such as 500 mM NaCl or L-arginine and/or 12–24 hours of incubation with affinity resins [48,51]. Such conditions may not be compatible with the typical fragility of membrane proteins, particularly of mammalian origin. Fortunately, a number of novel strategies for synthesizing SMA and similar amphipathic copolymers are emerging. These new polymers challenge some of the deficiencies of the polymers currently available and have facilitated characterization of the biophysical properties of this relatively novel system. A specifically interesting approach is the alternating diisobutylene maleic acid (DIBMA) copolymer, which is aliphatic and can therefore readily be quantified by UV absorbance, as opposed to the aromatic SMA copolymers. Furthermore, because of steric hindrance, the DIBMA copolymer is not prone to dynamic crosslinking in the presence of divalent cations and has high solubility [53]. In addition, the effect on the lipid acyl-chain order is not as significant as for the aromatic SMA copolymer. It was shown that the size of the DIBMA nanodisc increases with increasing lipid acyl-chain length and order, while it decreases with increasing ionic strength [54]. The poly(styrene-co-maleimide) (SMI) formed by modified SMA represents another, newly developed polymer engineered to be polycationic, hindering dynamic crosslinks with divalent cations [55].
Smith et al. used reversible addition fragmentation chain transfer (RAFT) to precisely control the SMA architecture, creating more homogeneous copolymers [56]. By this approach, FSEC showed that polymers of different compositions and lengths can produce different disc sizes for the same membrane protein. One may speculate that proteins of different transmembrane sizes might prefer polymers of certain compositions and lengths. Therefore, customizable copolymers may be a future strategy for achieving well-behaved native nanodisc systems.
Common for the majority of new copolymers for native nanodiscs is that their architecture is centered on the maleic acid footprint. Therefore, merely making minor adjustments to a system might not be the best long-term strategy. However, these initial polymers are helping us to better understand the native nanodisc system and its potential applications. Positively charged polymethracrylates have recently been shown to form nanodiscs, showcasing novel designs beyond SMA systems [57]. Readers are referred to extensive summaries on the types of polymers and the applications of native nanodisc [6,58].
Perspectives
Single-particle cryo-EM eliminates the need for crystallization in structural biology facilitating various approaches for studying membrane proteins in lipid bilayer environments. Although one may argue that it is always beneficial to study membrane proteins in a reconstituted lipid bilayer environment, in our own experience, reconstitution of certain membrane proteins may be unsuccessful. Such examples can be found in proteins solubilized and purified using detergents with very small CMCs, such as MNGs, which are almost impossible to remove using established protocols for nanodisc reconstitution [24]. For the purpose of determining high-resolution membrane protein structures, detergents or amphipols are still valid approaches and should not be ignored. Therefore, the choice of which system is most appropriate depends on the individual proteins being studied and what questions are being addressed. Nevertheless, we think that detergent-free isolation of membrane proteins embedded in soluble lipid bilayer systems will be the future framework for their isolation and characterization in a more native context. We envision that polymer screening can follow the same strategies as currently implemented for detergents using FSEC [9].
In addition to the reconstitution approaches discussed above, there are other methods, which although less popular, are very important for answering specific biological questions. Reconstituting membrane proteins into liposomes (proteoliposomes) is one example [59]. This approach potentially enables the analysis of ion channels at non-zero membrane voltages, which so far cannot be generated in any other lipid nanoparticle system, reconstituted or native.
Highlights.
Single-particle electron cryomicroscopy facilitates structural determination of membrane proteins without the need of crystallization.
Advances in membrane protein sample preparation are facilitating structural studies of membrane proteins.
Amphipols have a high affinity for the hydrophobic transmembrane domain and are able to keep membrane proteins soluble in the absence of free surfactant.
Soluble lipid nanoparticle systems offer a means to maintain a lipid-like environment for membrane proteins, while facilitating their structural studies by single-particle cryoEM.
Styrene-maleic acid copolymers facilitate extraction of membrane proteins in discs consisting of native membrane.
Acknowledgements
We thank all members of laboratories of Julius and Cheng for discussions, and thank Dr. Anton A.A. Smith for valuable discussions about native nanodiscs. H.E.A. is supported by a fellowship from Lundbeck Foundation. This work is supported partly by grants from National Institute of Health (R01NS047723 to D.J, and R01GM098672, P01GM111126, S10OD020054, and S10OD021741to Y.C.). Y.C. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1. **.Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. : Amphipols from A to Z. Annu Rev Biophys 2011, 40:379–408.This is an extensive review of amphipols. For those interested in using amphipols, it is worth to read this review.
- 2.Popot JL, Berry EA, Charvolin D, Creuzenet C, Ebel C, Engelman DM, Flotenmeyer M, Giusti F, Gohon Y, Hong Q, et al. : Amphipols: polymeric surfactants for membrane biology research. Cell Mol Life Sci 2003, 60:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tribet C, Audebert R, Popot JL: Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci U S A 1996, 93:15047–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuler MA, Denisov IG, Sligar SG: Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol Biol 2013, 974:415–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. **.Frauenfeld J, Loving R, Armache JP, Sonnen AF, Guettou F, Moberg P, Zhu L, Jegerschold C, Flayhan A, Briggs JA, et al. : A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 2016, 13:345–351.This was the first paper to describe and apply the Salipro system as a tool to study integral membrane proteins with single-particle cryo-EM.
- 6. **.Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA: The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur Biophys J 2016, 45:3–21.This is one of the first reviews describing the potential of the native nanodisc approach for studying integral membrane proteins.
- 7.Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas OR, Overduin M, et al. : Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem Soc Trans 2011, 39:813–818. [DOI] [PubMed] [Google Scholar]
- 8.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M: Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc 2009, 131:7484–7485. [DOI] [PubMed] [Google Scholar]
- 9.Hattori M, Hibbs RE, Gouaux E: A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 2012, 20:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, Cheng Y: Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 2018, 359:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, She J, Zeng W, Guo J, Xu H, Bai XC, Jiang Y: Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature 2017, 550:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschi M, Herzik MA Jr., Wie J, Suo Y, Borschel WF, Ren D, Lander GC, Lee SY: Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature 2017, 550:411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.She J, Guo J, Chen Q, Zeng W, Jiang Y, Bai XC: Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 2018, 556:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauer F, Gerle C, Fischer N, Oshima A, Shinzawa-Itoh K, Shimada S, Yokoyama K, Fujiyoshi Y, Stark H: GraDeR: Membrane Protein Complex Preparation for Single-Particle Cryo-EM. Structure 2015, 23:1769–1775. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt-Krey I, Rubinstein JL: Electron cryomicroscopy of membrane proteins: specimen preparation for two-dimensional crystals and single particles. Micron 2011, 42:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, et al. : Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods 2010, 7:1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Chen J: Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator. Cell 2016, 167:1586–1597 e1589. [DOI] [PubMed] [Google Scholar]
- 18. **.Gao Y, Cao E, Julius D, Cheng Y: TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016, 534:347–351.This is the first high resolution single-particle cryo-EM structure of a membrane protein using lipid nanodiscs as the stabilizing medium. The work demonstrated the feasibility of using lipid nanodiscs as a general platform for high-resolution structure determination by single-particle cryo-EM.
- 19.Pan X, Li Z, Zhou Q, Shen H, Wu K, Huang X, Chen J, Zhang J, Zhu X, Lei J, et al. : Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1. Science 2018, 362. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, Tarrasch JT, Li S, Sun Kobilka T, Kobilka BK, et al. : Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 2017, 546:248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SHW, Shi Y: An atomic structure of human gamma-secretase. Nature 2015, 525:212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. **.Cao E, Liao M, Cheng Y, Julius D: TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 2013, 504:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. **.Liao M, Cao E, Julius D, Cheng Y: Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504:107–112.These two papers described the first single-particle cryo-EM structure of an integral membrane protein, TRPV1, solved to near-atomic resolution. The structures were solved using amphipols as the stabilizing medium.
- 24.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D: Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015, 520:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popot JL: Membrane Proteins in Aqueous Solutions 2018.
- 26.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T: Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 2005, 438:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R: Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol 1996, 259:393–421. [DOI] [PubMed] [Google Scholar]
- 28.Henderson R, Unwin PN: Three-dimensional model of purple membrane obtained by electron microscopy. Nature 1975, 257:28–32. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y: The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J Mol Biol 1999, 286:861–882. [DOI] [PubMed] [Google Scholar]
- 30.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y: Structural determinants of water permeation through aquaporin-1. Nature 2000, 407:599–605. [DOI] [PubMed] [Google Scholar]
- 31.Bayburt TH, Grinkova YV, Sligar SG: Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters 2002, 2:853–856. [Google Scholar]
- 32.Bayburt TH, Carlson JW, Sligar SG: Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol 1998, 123:37–44. [DOI] [PubMed] [Google Scholar]
- 33. *.Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, et al. : Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 2011, 18:614–621.This was likely the first example of using lipid nanodisc for single-particle cryo-EM studies of integral membrane proteins. In this study, the SecYE complex reconstituted into lipid nanodiscs was attached to large ribosome particle, driving the image alignment.
- 34.Efremov RG, Gatsogiannis C, Raunser S: Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. Methods Enzymol 2017, 594:1–30. [DOI] [PubMed] [Google Scholar]
- 35.Mio K, Sato C: Lipid environment of membrane proteins in cryo-EM based structural analysis. Biophys Rev 2018, 10:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbuhler K, Ye W, Qi L, et al. : Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 2017, 552:426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi W, Li Y, Yoon SH, Ernst RK, Walz T, Liao M: Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 2017, 549:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava AP, Luo M, Zhou W, Symersky J, Bai D, Chambers MG, Faraldo-Gomez JD, Liao M, Mueller DM: High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science 2018, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin P, Bulkley D, Guo Y, Zhang W, Guo Z, Huynh W, Wu S, Meltzer S, Cheng T, Jan LY, et al. : Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 2017, 547:118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Wilkens S: Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H(+) -ATPase. Protein Sci 2017, 26:1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Prive GG: Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci U S A 2003, 100:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn VE, Leyko P, Alattia JR, Chen L, Prive GG: Crystal structures of saposins A and C. Protein Sci 2006, 15:1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruhn H: A short guided tour through functional and structural features of saposin-like proteins. Biochem J 2005, 389:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popovic K, Holyoake J, Pomes R, Prive GG: Structure of saposin A lipoprotein discs. Proc Natl Acad Sci U S A 2012, 109:2908–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossmann M, Schultz-Heienbrok R, Behlke J, Remmel N, Alings C, Sandhoff K, Saenger W, Maier T: Crystal structures of human saposins C andD: implications for lipid recognition and membrane interactions. Structure 2008, 16:809–817. [DOI] [PubMed] [Google Scholar]
- 46.Lyons JA, Boggild A, Nissen P, Frauenfeld J: Saposin-Lipoprotein Scaffolds for Structure Determination of Membrane Transporters. Methods Enzymol 2017, 594:85–99. [DOI] [PubMed] [Google Scholar]
- 47.Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, et al. : Structural basis for activation of voltage sensor domains in an ion channel TPC1. Proc Natl Acad Sci U S A 2018, 115:E9095–E9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. *.Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB: Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 2018, 557:123–126.This was the first high-resolution structure of an integral membrane protein complex using the native lipid nanodisc approach.
- 49. *.Nguyen NX, Armache JP, Lee C, Yang Y, Zeng W, Mootha VK, Cheng Y, Bai XC, Jiang Y: Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature 2018, 559:570–574.This study used the Salipro approach, and the final reconstruction reveals high-resolution densities contributed by saposin proteins. It is probably the first atomic structure determination of an integral membrane protein using Salipro.
- 50.Dominguez Pardo JJ, Koorengevel MC, Uwugiaren N, Weijers J, Kopf AH, Jahn H, van Walree CA, van Steenbergen MJ, Killian JA: Membrane Solubilization by Styrene-Maleic Acid Copolymers: Delineating the Role of Polymer Length. Biophys J 2018, 115:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parmar M, Rawson S, Scarff CA, Goldman A, Dafforn TR, Muench SP, Postis VLG: Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim Biophys Acta 2018, 1860:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu W, Fu Z, Xu GG, Grassucci RA, Zhang Y, Frank J, Hendrickson WA, Guo Y: Structure and activity of lipid bilayer within a membrane-protein transporter. Proc Natl Acad Sci U S A 2018, 115:12985–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oluwole AO, Klingler J, Danielczak B, Babalola JO, Vargas C, Pabst G, Keller S: Formation of Lipid-Bilayer Nanodiscs by Diisobutylene/Maleic Acid (DIBMA) Copolymer. Langmuir 2017, 33:14378–14388. [DOI] [PubMed] [Google Scholar]
- 54.Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S: Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew Chem Int Ed Engl 2017, 56:1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall SCL, Tognoloni C, Charlton J, Bragginton EC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ, et al. : An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale 2018, 10:10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith AAA, Autzen HE, Laursen T, Wu V, Yen M, Hall A, Hansen SD, Cheng Y, Xu T: Controlling Styrene Maleic Acid Lipid Particles through RAFT. Biomacromolecules 2017, 18:3706–3713. [DOI] [PubMed] [Google Scholar]
- 57.Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi JI, Ramamoorthy A: Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers. J Am Chem Soc 2017, 139:18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stroud Z, Hall SCL, Dafforn TR: Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights. Methods 2018. [DOI] [PubMed]
- 59. *.Wang L, Sigworth FJ: Liposomes on a streptavidin crystal: a system to study membrane proteins by cryo-EM. Methods Enzymol 2010, 481:147–164.This is a very interesting approach that attempts to determine the high resolution structure of membrane protein reconstituted into liposomes. This approach could potentially generate a voltage across the liposome membrane, thereby enabling determination of the atomic structures of membrane proteins, particularly voltage gated ion channels, under certain membrane potentials.