Abstract
The mechanisms by which common risk variants of small effect interact to contribute to complex genetic disorders remain unclear. Here, we apply a genetic approach, using isogenic human induced pluripotent stem cells (hiPSCs), to evaluate the effects of schizophrenia-associated common variants predicted to function as brain expression quantitative trait loci (SZ-eQTLs). By integrating CRISPR-mediated gene editing, activation and repression technologies to study one putative SZ-eQTL (FURIN rs4702) and four top-ranked SZ-eQTL genes (FURIN, SNAP91, TSNARE1, CLCN3), our platform resolves pre- and post-synaptic neuronal deficits, recapitulates genotype-dependent gene expression differences, and identifies convergence downstream of SZ-eQTL gene perturbations. Our observations highlight the cell-type-specific effects of common variants and demonstrate a synergistic effect between SZ-eQTL genes that converges on synaptic function. We propose that the links between rare and common variants implicated in psychiatric disease risk constitute a potentially generalizable phenomenon occurring more widely in complex genetic disorders.
INTRODUCTION
Genome-wide association studies (GWAS) of single-nucleotide polymorphisms (SNPs) continue to identify loci (143 and growing1,2) that are significantly associated with risk for schizophrenia (SZ). These common variant risk loci are enriched for genes expressed in pyramidal excitatory neurons (and a subset of GABAergic interneurons) 3,4, particularly in synaptic pathways 5,6.
By integrating GWAS and post-mortem brain expression quantitative trait loci (c/s-eQTL) studies, common variants that affect disease risk through regulation of gene expression can be identified. Recent estimates calculate that ~45.5% of SZ GWAS loci have brain eQTLs 5. While not necessarily true across all complex genetic disorders (e.g. inflammatory bowel disease 7), extensive evidence suggests that in SZ at least, the eQTL signal drives the GWAS signal 8,9. What remains unclear is the precise neuronal cell type(s) in which common variants act, and how they impact neuronal function. Because current case/control hiPSC designs are underpowered to discover the impact of common variants 10,11, it is necessary to utilize isogenic strategies to evaluate the impact of these genetic variants, alone and in combination, on gene expression and synaptic function across a variety of human neural cell types.
Here we apply a functional validation pipeline for common variants that incorporates leading genomic, hiPSC- and CRISPR-based approaches. For prioritized variants 12, we applied CRISPR editing to achieve allelic conversion when only one putative causal SNP was predicted at a SZ-associated locus, or CRISPR activation/inhibition (CRISPRa/i) to manipulate endogenous gene expression at loci containing several linked SNPs. We demonstrate that single and combinatorial isogenic comparisons can reveal the molecular and functional effects of common variants associated with SZ risk in a cell-type-specific manner. This work identifies a synergy between SZ-eQTL genes, supporting the hypothesis that common and rare SZ-associated variants occur in the same genes and/or converge on the same pathways - an unexpected finding that may apply more broadly across complex genetic disorders.
RESULTS
Prioritization of FURIN rs4702 for CRISPR editing and SNAP91/TSNARE1 for CRISPR activation/inhibition
Common variant SNPs and genes were prioritized from the expanding list of SZ-associated loci based on: i) strong evidence of genetic regulation of expression 12, ii) fine-mapping quantification of the number of putative causal SNPs at each locus 12, iii) RNA levels in hiPSC-derived neural cells (neural progenitor cells (NPCs), neurons and astrocytes), and iv) capacity for CRISPRa/i-mediated manipulation (Fig. 1; Supplementary Fig. 1).
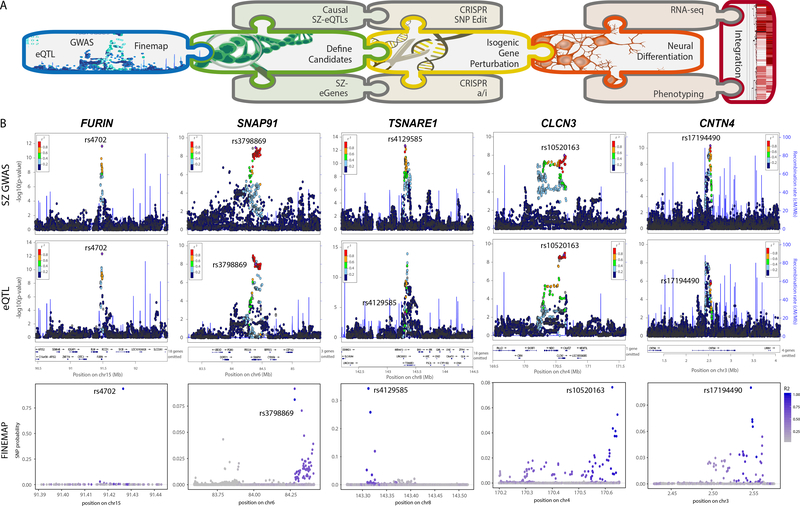
Figure 1. Prioritization of SZ-SNPs and SZ-genes for functional validation.
A. Schematic summary of analysis pipeline. B. Integration of GWAS (top), eQTL (middle) and fine-mapping (bottom) analysis for FURIN, SNAP91, TSNARE1, CLCN3 and CNTN4. Only FURIN shows evidence of having a single, high probability (0.94) putative causal SNP-eQTL. GWAS results derived from 34,241 cases, 45,604 controls and 1,235 trios 1 and association tested using additive logistic regression model and meta-analysis using an inverse-variance weighted fixed effects model. eQTL statistics generated from 467 DLPFC samples 12 using an additive linear model implemented, and significance assessed using t test.
Of the 108 SZ-GWAS loci identified by the Psychiatric Genomics Consortium (PGC2)-SZ GWAS 12, 19 harbored co-localized SZ-GWAS and c/s-eQTL (SNP-gene pairs within 1 Mb of a gene) signals in the CommonMind Consortium (CMC) post-mortem brain RNA-seq analysis 12. Of these, five were predicted to involve only a single protein-coding gene: furin (FURIN, down-regulated in risk allele individuals), t-SNARE domain containing 1 (TSNARE1, up-regulated), contactin 4 (CNTN4, up-regulated), voltage-sensitive chloride channel 3 (CLCN3, up-regulated), synaptosomal-associated protein of 91 kDa (SNAP91, up-regulated) 12. For FURIN only, the most significant GWAS-SNP (rs4702, NC_000015.10:g.90883330G>A) was also the most significant eQTL-SNP (Fig. 1B) and fine mapping analysis identified a single putative causal c/s-eQTL (rs4702, probability = 0.94) (Fig. 1B). Independent evidence suggested that FURIN rs4702 is an active c/s-eQTL in NGN2-excitatory neurons 13.
We considered post-mortem and hiPSC-based neural expression patterns of these five SZ-eQTL genes. Post-mortem brain findings were inconsistent: bulk immuno-panned adult post-mortem expression of FURIN was neuronal but enriched in astrocytes, SNAP91 and CNTN4 were neuron-specific, TSNARE1 was very low and CLCN3 was pan-neural 14, whereas cortical expression in GTEX 15 was robust for all but CNTN4 (Supplementary Fig. 1A), and single cell RNA-seq from PsychENCODE 16 showed comparable expression of CNTN4 in all neural cell types (Supplementary Fig. 1B). Therefore, we further prioritized these five genes based on own RNA-seq analysis of control hiPSC-derived NPCs, 6-week forebrain neurons, 21-day NGN2-excitatory neurons and primary astrocytes from three independent donors, which found reliable expression of four: SNAP91, TSNARE1, CLCN3, and FURIN (Supplementary Fig. 1C). To facilitate prioritization of other neuropsychiatric GWAS genes in hiPSC-based platforms, we make available a resource by which neural cell-type-specific expression of any gene can be easily cross-referenced in our hiPSC CRISPRa/i datasets as well as hiPSC-NPC & hiPSC-neuron RNA-seq datasets (schroden.shinyapps.io/BrennandLab-ExpressionApp-limited).
CRISPR activation (CRISPRa) and interference (CRISPRi) 17,18 was applied to up- and/or down-regulate endogenous gene expression in NGN2-excitatory neurons from two donors (Supplementary Table 1) 19,20. Three candidate guide RNAs (gRNAs) for each gene were screened in 8-day NGN2-excitatory neurons 21; the single-best was validated in hiPSC-NPCs, 8- and 21-day-old NGN2-excitatory neurons from three control donors, with efficacy generally decreasing over neuronal maturation (Supplementary Fig. 1D–F). CRISPRa/i resulted in smaller perturbations in neurons relative to NPCs, which were significant for three SZ-GWAS genes: SNAP91 (2.8-fold up, P < 1 × 10−4 and 2.4-fold down, P < 1 × 10−4), TSNARE1 (3.3-fold up, P < 1 × 10−4 and 4.2-fold down, P < 1 × 10−4) and CLCN3 (1.58-fold up, P <1 × 10−4; 1.2-fold down, P <1 × 10−3) (Supplementary Fig. 1F). FURIN showed decreased efficacy specifically with CRISPRi, consistent with the strong “active” neuron-specific H3K27ac peaks across the entire gene (Supplementary Fig. 1G).
For most SZ-GWAS genes, the number and/or distance between eQTLs in linkage disequilibrium was impractical for allelic conversion. By considering SZ GWAS, fine-mapping, brain cis-eQTL, cell-type-specific hiPSC-neural expression and CRISPRa/i efficacies in NGN2-excitatory neurons, we selected FURIN rs4702 for CRISPR editing and SNAP91/ TSNARE1/CLCN3 for CRISPRa/i-based functional evaluation.
Impact of CRISPR-mediated allelic conversion of FURIN rs4702
CRISPR-based allelic conversion was used to test the impact of FURIN rs4702 through isogenic comparisons (Fig. 2; Supplementary Fig. 2). gRNAs were designed to the target region that minimized potential off target effects 22 and to favor homozygous genomic editing 23. hiPSCs were nucleofected with a ssDNA repair template and Cas9 vector (Addgene #62988) with gRNA, followed by a 36-hour puromycin selection. To isolate low frequency edits, a pooled Taqman ddPCR screen (Fig. 2A, Supplementary Fig. 2A) was performed to enrich for cells with the desired allele; edited cells were identified via restriction fragment length polymorphism screening and validated by Sanger sequencing (Supplementary Fig. 2B). Although our targeting strategy incorporated a single point mutation in the Protospacer Adjacent Motif (PAM) to avoid recursive cutting through Cas9, unexpectedly, the final edited lines did not show evidence of PAM edits. We achieved seamless editing of FURIN rs4702 from AA to GG in four clonal lines from one control donor (NSB3182–3, Supplementary Table 1) (Supplementary Fig. 2B). The top three predicted on- and off-target cutting sites were confirmed for all hiPSCs by Sanger sequencing (Supplementary Fig. 2C).
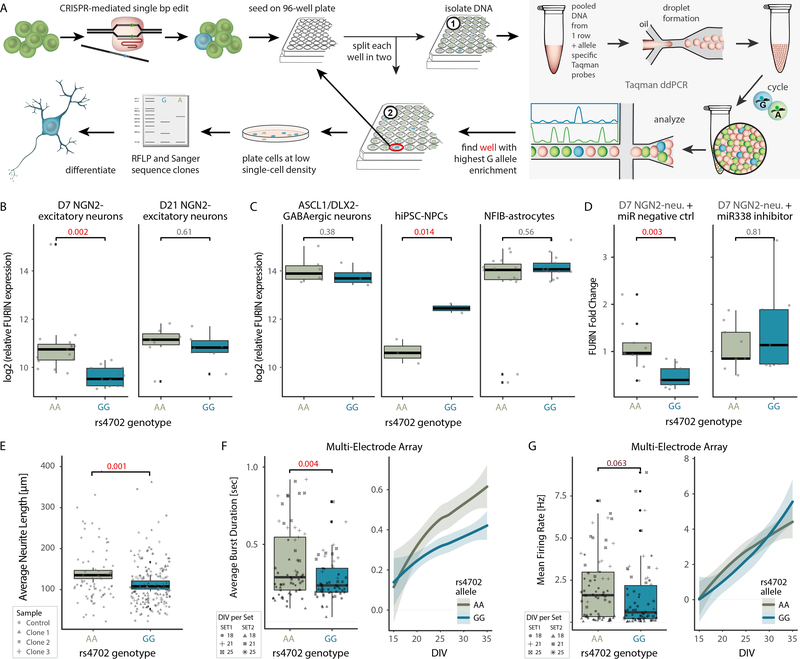
Figure 2. CRISPR editing demonstrates the cell-type-specific impact of rs4702 on FURIN expression and resulting neural phenotypes.
A. Schematic for CRISPR-mediated single base pair (bp) edit, pooled screening and validation strategy. B. log2 (FURIN expression) in rs4702 AA and GG D7 (left, n = 14) and D21 (right, n = 13) NGN2-excitatory neurons. qPCR was performed on four edited clones and two unedited clones of the same donor line in four independent experiments. C. log2 (FURIN expression) in rs4702 AA and GG hiPSC-derived inhibitory ASCL1/DLX2-GABAergic neurons (left, n = 6 ea), hiPSC-NPCs (center, n = 2 ea) and hiPSC-derived NFIB-astrocytes (right, n = 14 ea) through qPCR. D. log2 (FURIN expression) in rs4702 AA and GG D7 NGN2-excitatory neurons, following transfection with either a control miRNA (left, n = 8 ea) or miR338 (right, n = 8 ea). P values calculated using one-sided t test, a linear mixed-effects model controlled for variation between qPCR runs, n refers to biologically independent samples. E. Average neurite length of rs4702 AA and GG D7 NGN2-excitatory neurons, measured (semi-automated) from confocal images of GFP-labeled neurons. N = 40 biologically independent samples, totaling 379 neurons F. Average burst duration and G. Mean firing rate in rs4702 AA and GG D18-D25 NGN2-excitatory neurons (left) and at various time points from 15 to 35 days in vitro (DIV) (right). N = 44 biologically independent samples from 2 independent experiments. P values calculated using two-sided t test; line fitted through locally weighted smoothing (loess). Shaded areas (95% confidence interval), boxplots (quartiles), whiskers (largest/smallest observation within hinge ±1.5× inter-quartile range), and overlaid scatter plots (means of replicates from individual clones) indicated.
Allelic conversion from rs4702 AA to GG resulted in decreased FURIN mRNA levels in 7-day NGN2-excitatory neurons by qPCR (GG, P = 1.7 × 10−3 in D7 and P = 0.61 in D21 neurons) (Fig. 2B). rs4702 is predicted to be an eQTL for several genes beyond FURIN, both by the CMC and GTEX. In our edited NGN2-excitatory neurons, SV2B showed significantly increased expression in rs4702 GG cells, while PRC1 and RCCD1 were repressed with moderate significance and FES was unchanged (Supplementary Fig. 2D).
To evaluate cell-type-specific eQTL effects, rs4702 AA and GG hiPSCs were induced or differentiated into ASCL1/DLX2-GABAergic neurons 24, NFIB-astrocytes 25, and hiPSC-NPCs 26. Although we observed no significant differences in FURIN expression between AA and GG ASCL1/DLX2-GABAergic neurons (P = 0.38) and NFIB-astrocytes (P = 0.56), unexpectedly, we observed increased FURIN in GG hiPSC-NPCs (GG, P = 1.4 × 10−2). Moreover, consistent with reduced neural migration following repression of FURIN in hiPSC-NPCs 12, rs4702 GG hiPSC-NPCs (which showed increased FURIN expression) showed increased neural migration relative to isogenic AA controls (Supplementary Fig. 2E). These results highlight the complex and cell-type-specific nature of eQTL biology (Fig. 2C).
Cell-type-specific eQTL-regulated gene expression may differ in a more physiologically relevant environment, whereby heterogeneous cell types engage in both cell and non-cell autonomous gene regulation. To test this, we generated both 3D cortical spheroids (hCSs) comprising predominantly glutamatergic (and some GABAergic) neurons and astrocytes 27, and subpallium spheroids (hSSs) comprising largely GABAergic neurons and astrocytes 28. (Supplementary Fig. 2F). FURIN expression in hCSs was somewhat repressed (trending towards significance, P = 0.12) but was unchanged in hSSs (P = 0.74) (Supplementary Fig. 2F, G). Although generally consistent with our induced neurons, heterogeneous 3D cultures may less sensitively resolve cell-type-specific eQTL effects.
One mechanism by which cell-type specific eQTL-regulated gene expression can occur is post-transcriptional regulation. rs4702 is located in the FURIN 3’-UTR and the G allele occurs within a putative binding site for miR338 29. Treatment of NGN2-excitatory neurons with a miR338 inhibitor, but not a negative control, ameliorated genotype-dependent differences in FURIN expression (Fig. 2D), suggesting that regulatory activity of the rs4702 GG allele is dependent on miR338 expression levels.
Finally, we tested if the observed eQTL-dependent changes in FURIN expression were sufficient to achieve downstream biochemical and functional changes in neurons. FURIN proteolytically activates pro-nerve growth factor (NGF) 30, which in turn regulates neurite outgrowth 31. Imaging-based analysis of sparsely labeled AA and GG NGN2-excitatory neurons revealed significantly reduced neurite length in rs4702 GG neurons (P = 1 × 10−3, Fig. 2E). Moreover, population-based analysis of neuronal activity patterns by Axion multi-electrode array (MEA) further revealed significantly decreased average burst duration (P = 3.7 × 10−3, Fig. 2F) and, potentially, decreased mean firing rates (P = 6.3 × 10−2, Fig. 2G) in rs4702 GG neurons compared to isogenic controls. Additionally, a kinase array platform queried activity levels of serine/threonine protein kinases (>100) 32, finding altered kinase activity in seven protein kinase families, including AKT and MAPK, between rs4702 AA and GG NGN2-excitatory neurons (Supplementary Fig. 2H).
Our results indicate that isogenic comparisons following allelic conversion of a noncoding SNP can validate eQTLs predicted from postmortem brain analyses and yield phenotypic effects, while also underlining the great complexity and potential cell-type-specificity of these SNPs.
Altered SNAP91 and TSNARE1 expression impacts synaptic development
Synaptic defects 5,6 in excitatory neurons 3,4 are increasingly associated with SZ risk. Of the five SZ-eQTL genes identified by the CommonMind Consortium, three were efficiently and specifically modulated in NGN2-excitatory neurons using CRISPRa/i, and two, TSNARE1 and SNAP91, are linked to synaptic vesicle recycling. CRISPRa/i manipulation of endogenous gene expression was used to resolve the synaptic impact of SNAP91 and TSNARE1 through isogenic comparisons (Fig. 3; Supplementary Fig. 3). We conducted RNA-seq of 21-day NGN2-excitatory neurons with SNAP91 and TSNARE1 CRISPRa/i (Supplementary Fig. 3A–D, Supplementary Dataset 1, Supplementary Table 2) to elucidate impacts on the global transcriptome and explore the possibility of convergent downstream effects. Gene set enrichment analysis was performed across a collection of 698 manually curated gene sets with a neural theme (subdivided into eight categories) (Supplementary Dataset 2). We applied a competitive gene set test to evaluate enrichment of genes that were not necessarily genome-wide significant, allowing us to capture even subtle changes in expression, and identified sets of genes for which the distribution of t statistics differed from expectation. The results were clustered hierarchically by significance and regression coefficient of the enriched genes (Fig. 3A). Word cloud analysis of enriched gene sets (FDR < 10%) for SNAP91 and TSNARE1 revealed similar motifs (often containing the words “abnormal”, “neuron”, “morphology”, “brain”, “development”), suggesting that perturbation of these genes might impact various neural processes (Fig. 3B).
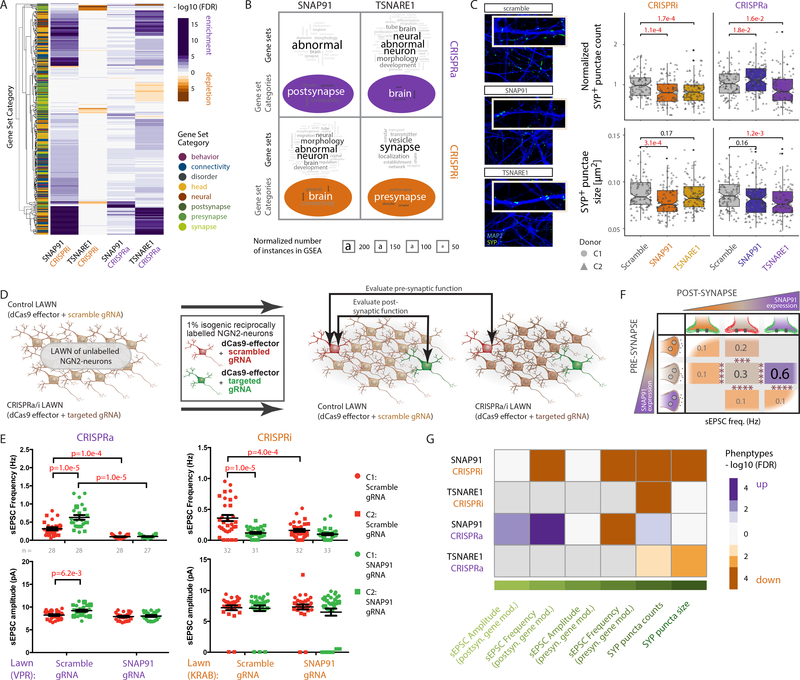
Figure 3. CRISPRa/i perturbations of the synaptic genes SNAP91 and TSNARE1 leads to transcriptomic changes and synaptic phenotypes.
A. Heat map of SNAP91 and TSNARE1 CRISPRa/i clustering in NGN2-excitatory neurons based on −log10 (P value) and regression coefficient of gene set enrichment analysis. Gene set enrichment tests were performed using a competitive test (accounting for inter-gene correlation) across 698 curated neural gene sets, summarized in 8 categories (denoted as y-axis color annotations). B. Word cloud analysis of enriched gene sets and summary categories from (A) for SNAP91 and TSNARE1 shows most frequently occurring gene set/category words. Font size denotes frequency, which was corrected by subtraction of the respective total word frequency in all used gene sets. C. Representative confocal microscopic images of NGN2-excitatory neurons with altered SNAP91 and TSNARE1 expression, immunostained against presynaptic SYP (SYNAPTOPHYSIN1; green) and dendritic MAP2 (blue) at day 24. Scale bar = 5 pm (left). Normalized SYP+ puncta counts (top) or size (bottom) of neurons with control (scramble gRNA, CRISPRa/i, n = 104 ea), or altered expression of SNAP91 (CRISPRa/i, n = 102 ea) or TSNARE1 (CRISPRa/i, n = 106 ea) from two cell lines (C1, C2). P values calculated using two-sided t tests. N refers to independent images from three independent experiments. Boxplots (quartiles), whiskers (largest/smallest observation within hinge ± 1.5 × inter-quartile range), and overlaid scatter plots (means of replicates from individual clones) indicated. D. Schematic of electrophysiological strategy to evaluate presynaptic and postsynaptic effects of manipulating gene expression. E. sEPSC frequency and amplitude of fluorescently labeled day 28–31 NGN2-excitatory neurons with SNAP91 CRISPRa (left, n = 111 individual neurons) or SNAP91 CRISPRi (right, n = 128 individual neurons). Data collected across three independent experiments. P values determined by multiple comparison test using two-way ANOVA. Plots show scatter graphs with mean and standard error for each group. F. Summary schematic of sEPSC frequency and amplitude of fluorescently labeled NGN2-excitatory neurons from (E). orange: SNAP91 CRISPRi, brown: control, purple: SNAP91 CRISPRa neurons. G. Heat map representation of observed synaptic phenotypes for SNAP91 and TSNARE1 CRISPRa/i based on −log10 (P value) and regression coefficient from (C) and (E).
Rodent studies suggest a role for SNAP91 in presynaptic function 33; similarly, TSNARE1 is predicted to function in presynaptic vesicle exocytosis 34. We evaluated isogenic pairs of CRISPRa/i-manipulated NGN2-excitatory neurons, co-cultured with human fetal astrocytes to improve synaptic maturation (Fig. 3C–G, Supplementary Fig. 3E–H). Through immunocytochemistry, CRISPRi of SNAP91 (1.2-fold down, P = 1.1 × 10−4) and TSNARE1 (1.2-fold down, P = 1.7 × 10−4) showed significant reduction in SYNAPTOPHYSIN1 (SYP)+ puncta number across two control donors, whereas CRISPRa increased SYP+ puncta for SNAP91 (1.11-fold up, P = 1.8× 10−2) and decreased for TSNARE1 (1.13-fold down, P = 1.6 × 10−2) (Fig. 3C top). All CRISPRa/i perturbations of SNAP91 and TSNARE1 reduced SYP+ puncta size (CRISPRi, SNAP91 1.12-fold down, P = 3.1 × 10−4, TSNARE1 1.04-fold down, P = 0.17; CRISPRa SNAP91 1.04-fold down, P = 0.16, TSNARE1 1.09-fold down, P = 1.2 × 10−3) (Fig. 3C bottom). To further test the potential of this approach for high throughput analyses, we employed an automated image analysis approach, which pointed to a reduction in synaptic puncta number and size following SNAP91 CRISPRi and TSNARE1 CRISPRa, confirming the trends we observed in our manual analysis (Supplementary Fig. 3G). Despite changes in SYP+ puncta number, we did not observe significant changes in population-wide neuronal activity by MEA (Supplementary Fig. 3H). Although we had expected to prioritize electrophysiological studies through MEA analysis, we proceeded instead with intracellular recording, guided by our genomic analyses, which linked increased SNAP91 expression (the SZ-relevant direction 12) to novel post-synaptic effects (Fig. 3B).
In order to specify consistent pre-synaptic inputs, electrophysiology experiments were conducted on sparse reciprocally labeled isogenic neurons grown on unlabeled control (dCas9-effector+scrambled gRNA) or SNAP91 CRISPRa/i (dCas9-effector+ SNAP91 gRNA) NGN2-excitatory neuronal lawns (Fig. 3D). Comparisons of control (mCherry-positive) and SNAP91 CRISPRa/i (GFP-positive) neurons on a control lawn tested effects on the postsynaptic neuron, whereas contrasting rare same-colored isogenic neurons across different lawns (either control or CRISPRa/i) queried impacts on the presynaptic neuron (Supplementary Fig. 3E). Strikingly, we observed reciprocal changes in spontaneous excitatory postsynaptic currents (sEPSCs) when perturbing SNAP91 in neurons cultured on the control lawn, indicating effects in the postsynaptic neuron: frequency (1.9-fold up, P = 1 × 10−5) and amplitude (1.1-fold up, P = 6.2 × 10−3) both increased following SNAP91 CRISPRa, whereas frequency decreased following SNAP91 CRISPRi (3.3-fold down, P = 1 × 10−5) (Fig. 3E, F). Perturbations of SNAP91 in presynaptic neurons in either direction reduced sEPSC frequency (SNAP91 CRISPRa: 3.2-fold down, P = 1 × 10−4; SNAP91 CRISPRi: 2-fold down, P = 1 × 10−4) (Fig. 3E, F). Interestingly, perturbation of SNAP91 did not produce genome-wide significant differentially expressed genes (Supplementary Fig. 3A), suggesting the observed synaptic phenotypes to be a consequence of multiple subtle changes in gene expression (Fig. 3A) or might reflect disruptions at the protein level. Altogether, in addition to the effects in presynaptic neurons, our findings hint at a second heretofore undescribed role for SNAP91 in postsynaptic receptor stabilization and/or retrograde signaling.
After summarizing all synaptic phenotypes in a heat map (Fig. 3F), rather than observing a correlation between samples with SZ-relevant (upregulated) perturbations in SNAP91 and TSNARE1, we unexpectedly found a striking congruence between SNAP91 CRISPRi and TSNARE1 CRISPRa (Fig. 3G). This was apparent in the gene set enrichment patterns (Fig. 3B) and confirmed in the correlation of the SNAP91 CRISPRi and TSNARE1 CRISPRa t statistics (Fig. 3A, Supplementary Fig. 3I), suggesting that although not necessarily relevant to the study of SZ, unexpected convergent effects between SNAP91 and TSNARE1 may occur.
Overall, these findings highlight the complexity of the possible consequences of SZ-associated eQTLs, whereby subtle changes in target gene expression in either direction can converge on synaptic level phenotypes.
Combinatorial perturbation of SZ-eQTL genes improves correlation with differential expression in neuropsychiatric disorders
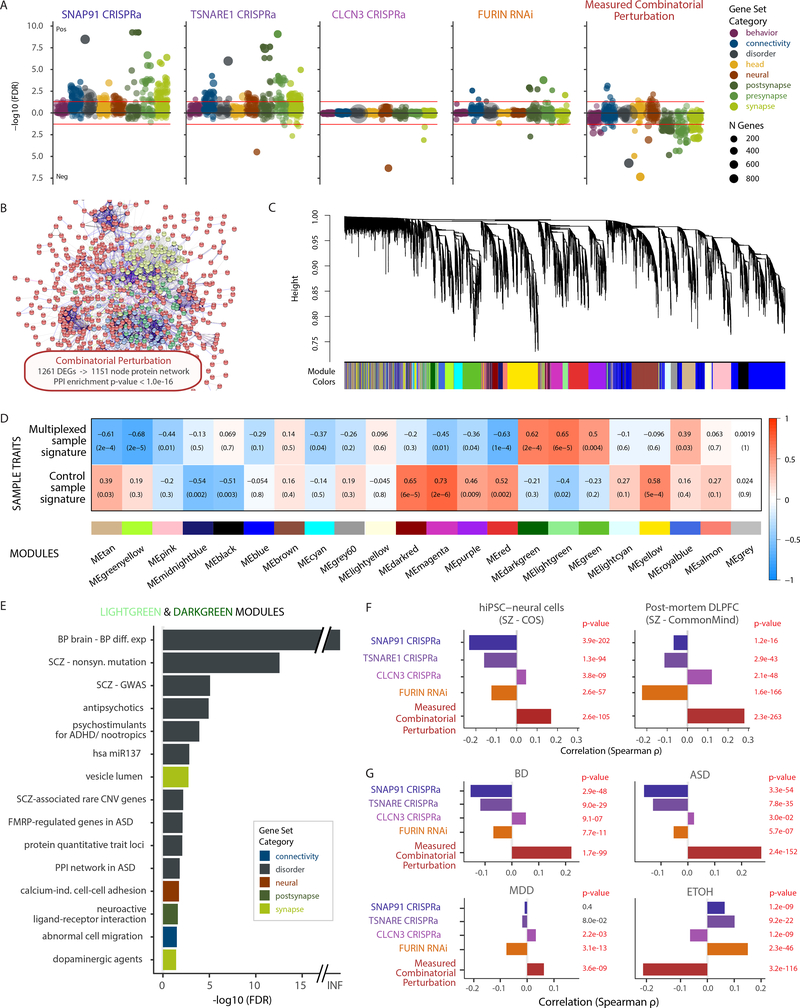
We combinatorially perturbed four SZ-eQTL genes in a better approximation of the polygenic nature of SZ (Fig. 4; Supplementary Fig. 4), in the direction predicted for disease risk, using CRISPRa to upregulate SNAP91, TSNARE1 and CLCN4 and RNAi to repress FURIN (Supplementary Fig. 4A–C, Supplementary Dataset 1, Supplementary Table 2). Combinatorial perturbation led to differential expression of 1,261 genes (DEGs, FDR < 5%, 665 up, 596 down) and impacted co-regulated downstream genes and proteins. Competitive gene set enrichment analysis, consistent with our phenotypic analyses, found strong enrichment of synaptic gene sets following individual SNAP91 and TSNARE1 CRISPRa, while CLCN3 CRISPRa and FURIN RNAi showed more moderate outcomes, and the combinatorial perturbation in fact yielded negative correlation with synaptic gene sets (Fig. 4A). The 1,261 combinatorial perturbation DEGs formed a protein network of 1,151 nodes with highly significant protein-protein interaction (PPI) enrichment in the STRING database (http://string-db.org) (P < 1 × 10−16) (Fig. 4B) and weighted gene co-expression network analysis (WGCNA) revealed 21 co-expression modules (Fig. 4C). Two WGCNA modules were highly significantly associated with the combinatorially perturbed sample signature (ME-lightgreen and ME-darkgreen) (Fig. 4D), showing significant enrichment (FDR<5%) in gene sets commonly associated with psychiatric disorders, such as miR137, common and rare SZ-variant genes, differential expression in bipolar disorder (BD) and proteinprotein interaction networks and FMRP-regulated genes in autism spectrum disorder (ASD), but also, interestingly, SZ-relevant drug classes (i.e. antipsychotic, nootropic and dopaminergic) (Fig. 4E).
Figure 4. Transcriptomic analysis of combinatorial perturbation of SNAP91, TSNARE1, CLCN3 and FURIN in the direction predicted for their SZ-eQTLs.
A. Competitive gene set enrichment analysis using limma camera (see methods), based on 698 neural gene sets, stratified by eight neural categories. B. Protein network (predicted using STRING database (http://string-db.org)) with 1,151 nodes from 1,261 differentially expressed genes (DEGs, FDR < 5%) resulting from the combinatorial perturbation. Enrichment P value P = 1 × 10−16 by random permutation. C. Hierarchical clustering of genes through topological overlap-based dissimilarity (top) and assigned module colors (bottom). D. Sample trait-module association heatmap. Rows correspond to sample groups (multiplexed CRISPR and controls), columns to module eigengenes. Cell labels denote weighted Pearson correlation (supported through color legend) and Student asymptotic P value of corresponding module and group (n = 4 independent multiplexed CRISPR samples, and n = 12 independent control samples). E. Over-representation analysis (ORA), using a hypergeometric test, of 698 curated gene sets and ranked genes in the two modules, ME-lightgreen (521 genes) and ME-darkgreen (258 genes), with highest correlation to combinatorial perturbation samples. Only gene sets with FDR < 5% shown. F. Analysis of concordance between the current study and two SZ RNA-seq studies (hiPSC-derived NPCs and FB-neurons (COS cohort, top) and post-mortem DLPFC (CMC cohort, bottom), based on Spearman correlation between genome-wide t statistics of 11,245 genes. P values computed through one-sided hypothesis test for the Spearman correlation coefficients being greater than zero. G. Concordance between this study with post-mortem RNA-seq datasets of five neuropsychiatric disorders 56 determined through Spearman correlation between the t statistics of 11,245 genes. P values were computed through a one-sided hypothesis test for the Spearman correlation coefficients being greater than zero. BD Bipolar Disorder, MDD Major Depressive Disorder, ASD Autism Spectrum disorder, ETOH Alcohol dependence.
To test for convergence of our SZ-eQTL perturbations with differential expression in an hiPSC neural dataset generated from control and childhood onset SZ (COS) cases 10 as well as a post-mortem SZ analysis (CMC) 12, which originally identified our common variant target genes, we calculated the Spearman correlation of their t statistics (Fig. 4F). All but one individual gene perturbation (CLCN3) actually correlated negatively with both data sets. However, remarkably, the combinatorial perturbation displayed highly significant positive correlation with the differential expression in SZ from both the postmortem CMC and hiPSC-based COS cohort (Fig. 4F). Very similar results were observed when comparing the t statistics with those from multiple post-mortem brain comparisons of BD, ASD and major depressive disorder (MDD) (NIMH HBCC and UCLA datasets), but not alcohol dependence (Fig. 4G). We also compared our differential expression t statistics to genetically regulated gene expression associations (GREX) of SZ (prediXcan 8). Although we did not see a significant correlation between overall sets of summary statistics (P > 0.05), genes that were nominally significant in our combinatorial analysis were more likely to also be nominally significant in the DLPFC-GREX analysis (binomial test; P < 7 × 10−92) (Supplementary Table 3).
Our observations suggest considerable downstream effects specific to a combinatorial perturbation of SZ-eQTL genes that go beyond what would be expected from an additive effect of individually perturbed genes. These synergistic effects emphasize the importance of considering the polygenic nature of SZ and other neuropsychiatric disorders, where a combination of variants contributes to disease.
Synergistic effects beyond the additive impact of individual SZ-eQTL genes are enriched in synaptic as well as common and rare SZ variant genes
In an effort to explore the nature of this synergistic effect, we modeled the additive effect of differential expression in singly perturbed SZ-eQTL genes computationally (Fig. 5A), which led to similar numbers of expected DEGs as found in the combinatorial approach (641 up, 604 down vs. 665 up, 596 down). Fig. 5B illustrates this for three representative genes, which exemplify the different synergistic effects that can be found when comparing additive model and combinatorial perturbation. Interestingly, the correlation between nominally significant synergistic DE gene t-statistics and DLPFC-GREX Z-scores was significant (P = 0.0097, Supplementary Table 3).
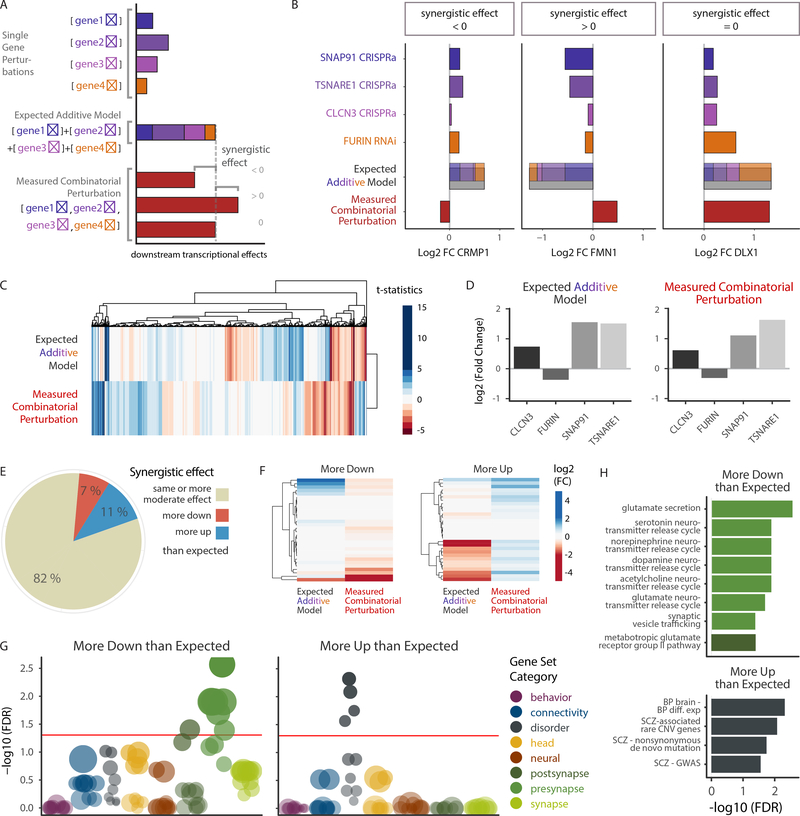
Figure 5. Synergistic effects of SZ-eQTL genes converge on synaptic function and common and rare variant-signatures.
A. Schematic of differential expression analysis. Individual gene modifications, the implementation of the expected additive model based on the latter and the measured combinatorial perturbation allowing for the detection of synergistic effects through comparison with the additive model. B. log2 fold changes of three representative genes (CRMP1, FMN1, DLX1) in individual SZ-eQTL gene perturbations, their computed additive model and their combinatorial perturbation, illustrating different possible synergistic effects (negative, positive and none, respectively). C. Hierarchical clustering of the t statistics for the additive model and the combinatorial perturbation. Color gradient represents t statistic values. D. Differential expression log2 (fold changes) of SNAP91, TSNARE1, CLCN3 and FURIN in the additive model and the combinatorial perturbation. E. Pie chart showing percentages of genes that exhibit similar or more moderate differential expression (beige) following combinatorial perturbation in comparison with the expected additive model, as well as genes that are more downregulated (red) and more upregulated (blue). F. Hierarchical clustering of the differential expression log2 (fold changes) of “more down” and “more up” genes, in the additive model vs. the combinatorial perturbation. FMN1, as seen in (B), is part of the “more up” category. G-H. Over-representation analysis (ORA) using a hypergeometric test, of 698 curated gene sets and those “more down” and “more up” genes with significant synergistic differential expression (FDR < 10%, n(more down) = 36 genes, n(more up) = 132 genes), ranked by adjusted significance.
We identified those genes that showed larger changes following combinatorial perturbation than predicted by our additive model (Fig. 5C–F; Supplementary Fig. 4D, E). For both the additive model and the combinatorial perturbation, we performed hierarchical clustering of the t statistics for each contrast (Fig. 5C). The differences between the predicted and observed cumulative effects (including inverse differential expression of some genes) was not explained by unequal gene perturbation magnitudes of SNAP91 (FC = 2.9 vs. 2.2), TSNARE1 (FC = 3.1 vs. 2.9), CLCN3 (FC = 1.5 vs. 1.7), or FURIN (FC = 0.8 vs. 0.86) between the individual and combinatorial perturbations (Fig. 5D, for CMC info see Supplementary Table 4).
To examine synergistic effects in more detail, we grouped genes based on differential expression between the additive model and the combinatorial perturbation (Fig. 5E). enes were classified as “more” differentially expressed in the combinatorial perturbation than predicted if their logarithmic fold change differed by at least 0.3 (a conservative estimation of the maximum standard deviation in all samples) (Fig. 5F). Most genes (82%) were altered approximately as predicted or less (Supplementary Fig. 4D, E), while 7% (1430 genes) were more downregulated and 11% (2107 genes) more upregulated than expected (Fig. 5E, F). As might be expected, the latter were mainly overrepresented in WGCNA modules that showed correlation with the transcriptional profile in the combinatorial perturbation samples, and vice versa (Supplementary Fig. 4F, Fig. 4D). Furthermore, testing for overrepresentation in our curated neural gene sets showed that genes more downregulated than expected from the additive model were significantly enriched for pre- and postsynaptic gene sets (Fig. 5G), particularly those involved in the secretion of glutamate and other neurotransmitters, synaptic vesicle trafficking and a postsynaptic glutamate receptor pathway (Fig. 5H). Genes more upregulated than expected in the additive model correlated with disorder signatures (Fig. 5G), particularly genes harboring rare CNVs or nonsynonymous de novo mutations associated with SZ, as well as SZ GWAS genes (Fig. 5H). This latter result is striking, as it links common and rare variants more broadly associated to psychiatric disease.
Here we show that combinatorial perturbation of four SZ-eQTL genes, in an approximation of the presumed transcriptional alterations they cause in SZ, results in a synergistic effect that culminates in the downstream alteration of both rare and common risk variant genes.
DISCUSSION
We demonstrated human-specific functional validation of a putative causal SNP (FURIN rs4702) by CRISPR editing and multi-SNP candidate genes (SNAP91, TSNARE1, CLCN3) via single and combinatorial CRISPRa/i manipulations. CRISPR editing of a single non-coding SNP altered neuronal expression of the cis-gene target and impacted a variety of neuronal phenotypes, while CRISPRa/i effected small changes in target genes that resulted in convergent downstream transcriptomic differences capturing effects observed in the post-mortem brain. Our isogenic hiPSC-based strategies manipulated common variant loci and genes in human neurons and suggest that synergy between risk variants may impact SZ risk.
CRISPR editing at FURIN rs4702 led to significant transcriptomic and cellular effects when altering even a single non-coding SNP, but also identified a surprisingly large cell-type-specific effect not detected by post-mortem studies. Located in the 3’UTR of the FURIN gene and within the binding site of miR-338, the regulatory activity of FURIN rs4702 is dependent upon miR-338 expression levels, which in itself is sufficient to mediate the thalamocortical disruptions observed in SZ-associated 22q11.2 deletion syndrome 35. CRISPRa/i studies further revealed the impact of SZ-eQTL genes on neuronal branching, synaptic puncta density, and synaptic activity. Although genetic, genomic and proteomic studies previously implicated both pre- 6 and post-synaptic 36–38 processes in SZ; here, our electrophysiological analyses demonstrated that even a single SZ-eQTL gene (SNAP91) can impact both pre- and postsynaptic neuronal function.
Our study integrated existing PGC GWAS 1, fine-mapping and c/s-eQTL 12 analyses to prioritize the SNPs and genes tested here. Advances in analytic strategies, such as new multi-SNP cis-eQTL analyses (i.e. Colocalization 12, PrediXcan 8) coupled with their application to larger PGC2-SZ GWAS genetic and genomic datasets continue to expand and refine the list of SZ-associated common variants (Supplementary Table 5) and genes (Supplementary Table 6) suitable for functional validation. The release of the unpublished PGC3-SZ GWAS (65,205 cases and 87,919 controls 39) and larger postmortem RNA-seq datasets will further resolve the list of putative causal variants and genes linked to SZ. Moreover, as more types of QTL studies become available, this will inform new avenues for functional validation; for example, a SNAP91 splice QTL is in high linkage disequilibrium with a SZ risk index SNP, suggesting that differential splicing may influence expression differences in SZ 40.
This work underscores the technical difficulties in adapting CRISPR-based systems as scalable platforms to test SZ-eQTL genes. First, not all genes (notably FURIN) proved equally amenable to CRISPRa/i, particularly in mature neurons, reinforcing that all gRNAs must be independently validated in each neural cell type and each donor 21. Second, although MEA is a widely used, convenient and scalable method to record population-wide neuronal activity 41, here it did not detect phenotypes identified by electrophysiology (similar phenomenon reported 42–44). We therefore recommend screening synaptic characteristics across multiple assays whenever possible. Although labor-intensive, patch clamp electrophysiology remains the gold-standard technique for studying synaptic function. Long-term, recent advances in automated patch clamping technology 45 may yet improve the scalability of this approach for screening large numbers of SZ-eQTL genes, alone and in combination. Towards this, advanced engineering of gRNA structure 46 and gRNA expression systems 47 will improve the efficiency of multiplexed gene regulation.
We previously hypothesized that, relative to post-mortem analyses, isogenic experiments would show comparable effect sizes but decreased standard deviation as a reflection of reduced biological and technical variation 48; instead, we report that the cis-eQTL effect sizes observed through isogenic comparisons of FURIN rs4702 were substantially larger than observed in post-mortem brain analyses 12. This suggests that the cell-type-specific effects may have been diluted in post-mortem cis-eQTL studies of brain homogenate. As CRISPR-based SNP edits are repeated across larger numbers of donors (particularly those with extreme polygenic risk scores (PRS)), we predict that observed cis-eQTLs should remain relatively consistent between individuals, meaning that all individuals will have similar (expected) cis-effect size from the CRISPR allelic conversion. There is currently no specific reason to reject this null model and suspect widespread epistasis, although empirical studies may prove otherwise. Nonetheless, we speculate that downstream transcriptomic and cellular (i.e. synaptic) effects may vary as a result of interactions with pre-existing donor-specific risk alleles, a hypothesis that may prove true more broadly across complex genetic disorders. Moreover, manipulating SZ-eQTL genes across donors with high and low PRS might make it possible to better distinguish the additive, epistatic and omnigenic models of inheritance.
Convergence between the various risk variants linked to psychiatric diseases, including SZ, ASD and BD has long been hypothesized 36,49–54 but remains yet unproven. We observed a striking convergence and synergy downstream even of our small sampling of four SZ-eQTL genes. First, CRISPRa/i-based gene expression analyses revealed a surprising degree of overlap between the downstream differential expression patterns and neuronal phenotypes of two SZ-eQTL genes, SNAP91 and TSNARE1. Second, combinatorial perturbation of four SZ-eQTL genes in the SZ-relevant direction (up: SNAP91, TSNARE1, CLCN3; down: FURIN) revealed negative synergy converging on synaptic function and positive synergy linking the rare and common variant genes implicated in psychiatric disease risk. These observations agree with a highly complex, polygenic etiology of SZ and other psychiatric disorders 55,56 and are interesting to consider in the context of additive 57,58, epistatic 59, and omnigenic 60,61 models of inheritance. For example, it is tempting to speculate as to whether the synergy we observed is consistent with individual risk being additive on a liability scale, or, if our evidence instead suggests that SNAP91, with reciprocal effects on synaptic function and seemingly lacking downstream transcriptional targets, might represent a core gene for SZ as conceptualized in the omnigenic model. High-throughput CRISPR-based perturbation methods 62,63 will help to infer gene regulatory networks, resolve coregulation of core disease genes and/or identify the existence of peripheral master genes in psychiatric disease. Overall, our observations, coupled with findings that high polygenic risk increases disease liability in carriers of rare mutations 64,65, suggest cumulative effects between rare and common risk variants. Our hope is that hiPSC-based models will illuminate the synergistic impact of common variants on cellular and molecular phenotypes, leading us towards precision psychiatry 56 and more personalized medicine 66.
METHODS
(see Supplementary Notes for more detailed experimental methods)
Prioritization of candidate genes
Candidate causal gene selection was informed by a previous SZ fine mapping analysis 12 utilizing GWAS and eQTL data. Briefly, summary statistics derived from the 2014 Psychiatric Genomics Consortium (PGC) SZ GWAS 1,2 were compared to dorsolateral prefrontal cortex (DLPFC) eQTL statistics from the CommonMind Consortium (CMC) in order to identify co-localized GWAS and eQTL signatures. This co-localization analysis, implemented in Sherlock 67, identified 33 genes for which there was significant overlap (Sherlock adjusted P < 5 × 10−2) in eQTL and GWAS signatures, and therefore evidence of disease risk being driven by expression regulation. Of these 33 genes, five (CLCN3, CNTN4, FURIN, SNAP91, TSNARE1) were within loci for which no other genes showed co-localization; we prioritized these five genes for functional follow-up. Furthermore, sgRNA target selection for FURIN was informed by results from a separate fine mapping analysis of the 2014 PGC SCZ GWAS loci, carried out with FINEMAP 68. FINEMAP was run using the GWAS SNP-level z-scores and LD matrices generated from samples within the PGC SZ cohort as input, and posterior probabilities of causality were estimated for all SNPs within GWAS loci (as defined by r2 > 0.1 region). The lead eSNP for FURIN, rs4702, was the variant with greatest posterior probability (0.94) across all loci, and was chosen for CRISPR editing.
hiPSC CRISPR targeting and validation
Validated control hiPSCs for CRISPR editing and CRISPRa/i were selected from a previously reported case/control hiPSC cohort of childhood onset SZ (COS) 10. The following controls 10 were used for CRISPR editing (hiPSCs: NSB3182–1 (female, for deletion), NSB3182–3 (female, for rs4702 G knock-in)) and CRISPRa/i (hiPSC NPCs NSB553-S1–1 (male), NSB2607–1-4 (male), NSB690–2-1 (male)). hiPSCs were cultured in StemFlex media (Gibco, #A3349401).
i. gRNA and HDR template design and cloning
Using the Benchling (www.benchling.com) web tool, four gRNAs were designed and selected based on their overlapping location/ distance of their PAM to the target SNP as well as predicted specificity and efficiency. gRNA candidates were synthesized as single-strand oligonucleotides with BbsI overhangs, phosphorylated/annealed (37 °C for 30 min, 95 °C for 5 min, ramp down to 25 °C at 5 °C per minute), diluted 1:200 and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene #62988) performing 1-step BbsI (NEB, #R3539) restriction and ligation (37 °C for 5 min, 23 °C for 5 min, 10 cycles). The single-stranded HDR template was designed to bind the sense strand (opposite the gRNA), span 36 nucleotides upstream and 91 nucleotides downstream the target site and contained a C in place of a T at the SNP site as well as an A instead of a C at the PAM site (Supplementary Table 7).
ii. hiPSC nucleofection
hiPSCs were transfected using the Lonza P4 Primary Cell 4D-Nucleofector Kit (V4XP-4024) according to the manufacturer’s instructions. Briefly, cells were dissociated following accutase incubation at 37 °C and 1.5 × 106 cells were centrifuged at 800 × g for 5 min. Pellet was resuspended in 100 μl nucleofector solution containing 4 μg PX459-gRNA plasmid. The suspension was transferred to a nucleofection cuvette, transfected in the Lonza 4D nucleofector and seeded onto a Matrigel-covered 12-well plate in StemFlex containing 10 pm Rock Inhibitor, Y27632 (Axxora #ALX-270–333-M025). After incubation for 36 h, 1 μg/ml puromycin was added for an additional 36 h.
iii. gRNA validation
The cells were then harvested, pelleted and lysed for 1 h at 55 °C in Sarkosyl lysis buffer (10 mM Tris pH 7.5 (Sigma #T5941), 10 mM EDTA pH 8.0 (Life Technologies #15575–020), 0.5% Sarkosyl (Sigma #L9150), 1 mg/ml Proteinase K (Invitrogen #25530049; added just before use)). 1 μl of lysate was used for amplification of a 600-bp region around the target site (for primers see Supplementary Table 7). The amplicon was Sanger-sequenced and the resulting trace file analyzed using TIDE (www.tide.deskgen.com). The gRNA showing the highest nuclease efficiency was chosen for all further experiments (Supplementary Table 7).
iv. Pooled ddPCR screening
96-well DNA isolation:_Following nucleofection of 4 μg PX459-rs4702 plasmid/1.8 μg single-stranded HDR template and puromycin selection as above, cells were harvested using EDTA and seeded onto a Matrigel-coated 96-well plate. Cells at 70–80% confluence were split onto 2 new Matrigel-coated 96-well plates. At 60% confluence DNA was isolated from one plate by 30 μl/well Sarkosyl lysis buffer and incubating at 55 °C overnight. The following day, condensation was collected by centrifugation at 3,000 × g for 2 min and 100 μl/well 75 mM NaCl/100% Ethanol was added to the lysate and incubated at −20 °C for 1 h. The plate was then inverted over the sink, blotted on a paper towel and rinsed with 100 μl 70% Ethanol three times. After the final wash, the plate was blotted until no liquid was left soaking into the paper towel and air dried for 5 min. 30 μl/well H2O were added and incubated at 65 °C for 1 h. DNA was resuspended by pipetting up and down 20 times.
Taqman ddPCR: 3 μl/well of each row were pooled. ddPCR mix (1× ddPCR Supermix for Probes (Bio-Rad #1863024), 1× TaqMan SNP Genotyping Assay (rs4702 A/G, Thermo Fisher Scientific #4351379, H2O to 25 μl) was added to 100 ng DNA of each pooled sample as well as H2O, rs4702-AA and rs4702-GG controls. ddPCR was performed and analyzed with the QX100 Droplet Digital PCR System (Bio-Rad #186–3001) according to the manufacturer’s instructions. For the pooled row sample showing the highest G allele concentration, ddPCR was now performed for each well as above. The resulting well with the highest G allele concentration was noted and the same well was found on the duplicated 96-well plate still in culture. Cells in that well were then seeded onto a new 96-well plate using EDTA to allow for non-homogenous distribution. This 96-well plate in return was duplicated, DNA was isolated and pooled Taqman ddPCR performed as described above. Enrichments of 40–60% can be expected by round 2. If the enrichment is too low, a third round can be performed, though enrichments here often reach close to 100% and therefore lack a heterogeneity between clones.
v. Restriction fragment polymorphism screening and Sanger sequencing
At 60% confluence DNA was isolated from one plate as described above. 2 μl of each well was used for amplification of a 600-bp region around the target site in a 20 μl reaction as above. Restriction enzyme digest was performed by adding digest mix (3 μl 10× NEB 3.1, 6.7 μl H2O and 0.3 μl SfaNI restriction enzyme (NEB #R0172) each) directly to the PCR products and incubating for 1.5 h at 37 °C and 20 min at 65 °C. SfaNI displays an additional cutting site when the amplicon has rs4702 genotype G, and can therefore be used to pre-screen candidates. DNA samples that showed a G allele restriction band pattern after agarose gel electrophoresis were Sanger-sequenced and aligned to the amplicon sequence. Clones with a clean G allele edit were marked on the duplicated plate still in culture. At 70% confluence these clones were expanded to 6-well plates and stocks were frozen.
vi. Neural cell generation (more details in Supplementary Note)
NGN2-excitatory 20, ASCL1/DLX2-GABAergic neurons 24, NF/S-astrocytes 25, hiPSC-NPCs 26 (although newly generated isogenic AA and GG hiPSC-NPCs were purified by MACS to enrich for CD271−/CD133+ cells (Miltenyi Biotech #130–097-127 and #130–091-895), and hCS 27 and hSS 28 organoids were generated as previously described.
CRISPRa/i manipulation of endogenous gene expression (more details in Supplementary Note)
hiPSC-NPCs were generated as previously described 69,26. CRISPRa/i gRNA design and cloning, antibiotic-selection of dCas9-VPR and dCas9-KRAB hiPSC-NPCs, gRNA lentiviral transduction and NGN2-induction20 were conducted as previously described 21.
Molecular and synaptic phenotyping
i. Real-time quantitative PCR
Real-time qPCR was performed as previously described 21. Cells were harvested with Trizol and total RNA extraction was carried out following the manufacturer’s instructions. Quantitative transcript analysis was performed using a QuantStudio 7 Flex Real-Time PCR System with the Power SYBR Green RNA-to-Ct Real-Time qPCR Kit (all Thermo Fisher Scientific). Total RNA template (25 ng per reaction) was added to the PCR mix, including primers (Supplementary Table 7). qPCR conditions were as follows; 48 °C for 15 min, 95 °C for 10 min followed by 45 cycles (95 °C for 15 s, 60 °C for 60 s). All qPCR data were collected from at least 3 independent biological replicates of one experiment. Data analyses were performed using GraphPad PRISM 6 software.
ii. Immunostaining and microscopy
NGN2-excitatory neurons (23- or 24-days-old) (on coverslips) were washed with ice-cold PBS and fixed with 4% PFA/sucrose PBS solution at pH 7.4 for 20 mins, room temperature. Then, fixative solution was replaced with permeabilizing solution (ice-cold 0.1% Triton-X PBS supplemented with 5% donkey serum), followed by incubation on the ice for 20 mins and another 20 mins at room temperature. After washed with PBS 3 times, NGN2-excitatory neurons were incubated with blocking solution (0.1% Tween-20, 5% donkey serum in PBS) for 1 hour, room temperature. The blocking solution was aspirated and replaced with the same solution with primary antibodies (GFP-Rb, Abcam, ab6556, 1:500; MAP2-Ms, Thermo Fisher Scientific, MA5–12826, 1:500; SYNAPTOPHYSIN1-Rb, Synaptic Systems, 101 002, 1:500; SYNAPSIN1/2-Rb, Synaptic Systems, 106 006, 1:500; CTIP2-Ck, Abcam, ab18465, 1:500; SATB2-Ms, Abcam, satba4b10, 1:5; GABA-Rb, Sigma A2052, 1:500; GAD67-Ms, Millipore, MAB5406, 1:1,000) and incubated 2 days at 4 °C. Neurons were then incubated with secondary antibodies (Alexa 488 anti-Rabbit, Dk, Jackson immunoResearch, 711–545-152, 1:500; Alexa 568 anti-Chicken, Gt, Thermo FIsher Scientific, A-11041, 1:500; Alexa 647 anti-mouse, Dk, Jackson immunoResearch, 715–605-150, 1:500), prepared in blocking solution, for two hours at room temperature, followed by PBS washing 3 times.
20 μl of AquaPolymount mounting solution (Polysciences Inc., #18606–20) was added per coverslip and air-dried for two days at ambient temperature. Neurons on the coverslips were imaged with a Zeiss LSM 780 microscope.
For presynaptic ICC imaging, images were acquired (five images each from three biological replicates per condition and cell line in three different experiment sets) using a confocal microscope (LSM 780, Zeiss) with 10× ocular lens and 63× objective lens. After uniform thresholding of all SYP, SYN1/2 and MAP2 images, SYP, SYN1 and SYN1/2 puncta number or their size were measured. Total SYP and SYN1/2 puncta number per image were divided by that image’s respective MAP2AB-positive area in order to calculate SYP and SYN1/2 puncta counts normalized to MAP2AB levels. Average puncta size of SYP and SYN1/2 per image was used for statistical analysis. Data from 51–54 images from 3 independent experiments were analyzed using GraphPad PRISM 6 software.
For neurite tracing, GFP-labeled NGN2-excitatory neurons (D5), cultured as described above, were seeded at very low density (50–100 cells/coverslip) on a lawn of same age unlabeled NGN2-excitatory neurons (5.0–6.0 × 105 cells/coverslip). Co-culture was carried out until day 14 to minimize GFP-positive neurites from overgrowth and tangling, thus easing imaging and analysis. Immunostaining was performed against GFP. For neurite tracing imaging, GFP NGN2-excitatory neurons were captured at a magnification of 20× using a confocal microscope (LSM 780, Zeiss). Nine images containing one neuron each were captured per coverslip per condition (2 technical replicates of two isogenic cell lines in two independent experiments). Images were stitched using NIH open-source image processing software Image J (Version 2.0.0-rc-69/1.52n), at 5% overlap and transformed as maximum projection. Using the ImageJ plug in “simple neurite tracer”, each neuron’s cell soma was identified and neurites traced from primary neurites outwards. Tracings were exported for each individual neuron and analyzed using R. Total neurite length was defined as the sum of all primary and non-primary traces within one neuron. Neurite tracings from neurons belonging to the same coverslip were averaged for boxplots but overlaid individually in scatter plots.
iii. Neurosphere assay
Control rs4702 AA and edited rs4702 GG NPC lines were dissociated using accutase and cultured in low adherence plates for 48 h to generate neurospheres. Neurospheres were manually picked and replated into individual wells of a Matrigel-coated 96-well plate. Images of neurospheres were taken 1 hour post plating using a brightfield microscope with a 4× objective (N.A. 0.1). 48 hours after plating neurospheres were fixed in 4% FA, stained using DAPI and imaged using an epifluorescence microscope with a 4× objective (N.A. 0.1). Images of each neurosphere were traced at each time point and the average radial migration was calculated using ImageJ software by subtracting the average radius of each neurosphere 1 h post-plating from the radius 48 h post plating. 28 neurospheres taken from 3 separate passages were analyzed for each experimental condition. Results were analyzed with an unpaired Student’s t test.
iv. Serine-threonine kinase activity profiling
Profiling of serine-threonine kinase activity was performed using the PamStation12 microarray (PamGene International) and STK (4-well) PamChips containing 144 consensus phosphopeptide sequences (3 of which are internal controls) per well, immobilized on porous ceramic membranes. Each PamChip well was blocked with 2% bovine serum albumin (BSA) before 2 μg of protein in the manufacturer’s kinase buffer (PamGene), 157 μM adenosine triphosphate (ATP), and FITC-labeled anti-phospho serine-threonine antibodies (PamGene) were added in each well. The homogenized samples containing the active kinases and assay mix were pumped through the wells to facilitate interaction between kinases in the sample and specific peptide substrates immobilized on the chip. The degree of phosphorylation per well was measured in real time using Evolve (PamGene) kinetic image capture software. The software captures FITC-labeled anti-phospho antibodies binding to each phosphorylated peptide substrate every 6 sec for 60 min. Peptide spot intensity was captured across multiple exposure times (10, 20, 50, 100, 200 ms) during post-wash, and the linear regression slope was calculated and used as the signal (i.e. peptide phosphorylation intensity) in comparative analyses. The signal ratio between pairs of samples was used to calculate fold change (FC) for each peptide. Peptides with a FC of at least 30% (i.e. FC > 1.30 or FC < 0.70) were considered changed in degree of phosphorylation. Peptides that were undetectable or non-linear in the post-wash phase (i.e. the coefficient of determination R2 of the corresponding linear regression less than 0.90) were excluded from subsequent analyses.
Significant upstream kinases were determined through permutation analysis using 2,000 random sets of reporter peptide substrates. A kinase signaling network was created using the STRING database 70 (Supplementary Fig. 2H).
v. Multiple Electrode array (MEA)
In order to evaluate electrical activity of NGN2-excitatory neurons by MEA, density-matched isogenic NGN2-neuronal populations, co-cultured with pHAs, were prepared as described above. Specifically, at day 3, pHAs were split as 17K cells/well in a Matrigel-coated 48W MEA plate (Axion Biosystems, M768-tMEA-48W) and maintained as above. At day 7, NGN2-excitatory neurons were detached, spun down and seeded on the pHA culture. Outer space of each well in the plate was filled with autoclaved/deionized water to minimize the evaporation of marginal wells during long-term culture. Half volume of neuronal medium (supplemented with 2% FBS) was replaced with fresh medium including 2 μM Ara-C from day 9 until the end of MEA recording. Electrical activity of neurons was recorded at 37 °C once every week from day 15 (~Day 15, 23, 29, 35 and 41). On recording day, plate was loaded into the Axion Maestro MEA reader (Axion Biosystems). Recording was performed via AxiS 2.4. Batch mode/statistic compiler tool was run following the final recording. Quantitative analysis of the recording was exported as Microsoft excel sheet. Data from 6–12 biological replicates were analyzed using GraphPad PRISM 6 software or R.
vi. Whole-cell patch clamp electrophysiology
NGN2-excitatory neurons (23–24-days-old), grown on pHA, with SNAP91 CRISPRa/i were prepared as described above. Especially, at day 6, very low titer of EF1a-dTomato or EF1a-eGFP lentiviruses were added to a well either of dCas9-effector NGN2-excitatory neurons with scramble gRNA or SNAP91 gRNA. At day 7, unlabeled dCas9-effector NGN2-excitatory neurons with scramble gRNA or SNAP91 gRNA (grown separately from the labeled NGN2-excitatory neurons above) were split on pHA culture as 4.5–6.0 × 105 cells/coverslip, serving as neuronal lawns. Additionally, NGN2-excitatory neurons labeled with dTomato or eGFP were seeded into each neuronal lawn as 4.5–6.0 × 103 cells/coverslip. Feeding the cultures was done same as described above. Patch clamp recordings were performed from these neurons at day 29–31 using borosilicate glass electrodes (3–5 MQ). Whole-cell voltage-clamp recordings were obtained with the internal solution containing (in mM): 130 mM K-gluconate, 6 mM KCl, 4 mM NaCl, 10 mM Na-HEPES, 0.2 mM K-EGTA; 0.3 mM GTP, 2 mM Mg-ATP, 10 mM D-glucose. The pH and osmolarity of the internal solution were close to physiological conditions (pH 7.3, 290–300 mOsmol). Excitatory postsynaptic currents (EPSCs) were recorded in voltage-clamp mode to monitor the spontaneous EPSC frequency and amplitude. Spontaneous EPSCs were recorded in BrainPhys (Stem Cell Technologies) at −60 mV. Data were low-pass filtered at10 kHz and acquired at 10 kHz using Multiclamp 700B (Axon Instruments) and pClamp 10 (Molecular Devices). Series and membrane resistance was continuously monitored, and recordings were discarded when these measurements changed by > 20%. Recordings in which series resistance exceeded 25 MQ were rejected. Detection and analysis of EPSCs were performed using MiniAnalysis (Synaptosoft). For current-clamp recordings, series resistance was monitored and canceled using a bridge circuit, and pipette capacitance was compensated. Voltage signals were low-pass filtered at 10 kHz. The baseline membrane potential was maintained near −70 mV with a current injection (5 pA). We recorded membrane potential responses to hyperpolarizing and depolarizing current pulses (500 ms in duration) and then, we examined action potential and subthreshold membrane properties by using Signal 4 (Cambridge Electronic Design).
vii. Data analysis
Data from all phenotypic assays above were first organized in a Microsoft Excel spreadsheet and analyzed using GraphPad PRISM 6 software or R. For qPCR data analysis and synaptic imaging analysis, values are expressed as mean ± SEM. Statistical significance was tested using either one-sided Student’s t test or one-way ANOVA with Tukey’s post-hoc test for comparison of all sample means. For presynaptic imaging, MEA and whole-cell patch clamp electrophysiology, statistical significance was tested using multiple comparisons testing for two-way ANOVA (Sidak’s post-hoc test for comparison of all sample means).
RNA-seq analysis (more details in Supplementary Note)
RNA sequencing libraries were prepared using the Kapa Total RNA library prep kit. Paired-end sequencing reads (125 bp) were generated on an Illumina HiSeq2500 platform (Coverage/Reads: 40M), aligned to hg19 using STAR aligner and uniquely mapping reads counted with featureCounts. The main drivers of gene expression variance were determined using the variancePartition package 71. The limma voom function was used to compute weights for heteroscedasticity adjustment by estimating the mean-variance trend for log2 counts. Linear models were fit to the expression values of each gene using the lmFit function. Supplementary Figure 4B summarizes the contrasts used for the combinatorial perturbation experiment. Empirical Bayes moderation was applied using the eBayes function to obtain more precise estimates of gene-wise variability. P values were adjusted for multiple hypotheses testing using false discovery rate (FDR) estimation, and differentially expressed genes were determined as those with an estimated FDR < 5%, unless stated otherwise.
Gene set enrichment analysis (GSEA) was performed on a curated neural subset (Supplementary Dataset 2) of the MAGMA 72 collection using the limma package 73 camera function 74, which tests if genes are ranked highly in comparison to other genes in terms of differential expression, while accounting for inter-gene correlation. Overrepresentation analysis (ORA) was performed when subsets of DEGs were of interest, such as the synergistic genes. The genes of interests were ranked by –log10 (P value) and enrichment was performed against a background of all expressed genes using the WebGestaltR package.
The expected additive effect was modeled through addition of the individual coefficient comparisons: (TSNARE1 - ctrl) + (SNAP91 - ctrl) + (CLCN3 - ctrl) + (FURIN - ctrl). The synergistic effect was modeled by subtraction of the additive effect from the combinatorial perturbation comparison: (Multiplexed genes - multiplexed controls) - (TSNARE1 - Ctrl) + (SNAP91 - ctrl) + (CLCN3 - ctrl) + (FURIN - ctrl). We categorized all genes by the direction of their change in both models and their log2 (fold change) in the synergistic model (Supplementary Fig. 4E, Supplementary Dataset 3). log2 (FC) standard deviations were calculated for all samples and never exceeded 0.3. Genes were grouped into ‘positive synergy’ if their FC was larger than 0.3 and ‘negative synergy’ if smaller than −0.3. If the corresponding additive model log2 (FC) showed the same or no direction, the gene was classified as “more” differentially expressed in the combinatorial perturbation than predicted.
Integration with external datasets and hierarchical clustering to confirm neural cell identity was performed as described previously 10. Counts data was log2 (RPKM) transformed followed by PCA analysis and plotting in R. RNA-seq datasets were obtained from GTEx (www.gtexportal.org), CMC (www.synapse.org/CMC), BrainSpan (www.brainspan.org/), and GEO (www.ncbi.nlm.nih.gov/geo/) (Supplementary Fig. 3C, D).
DATA AVAILABILITY
All source donor hiPSCs have already been deposited at the Rutgers University Cell and DNA Repository (study 160; http://www.nimhstemcells.org/): CRISPR-edited hiPSCs are in the process of being submitted in advance of publication. RNA-seq data are available at www.synapse.org/#!Synapse:syn20502314; additionally, we make available the following resource by which neural cell-type-specific expression of any gene can be easily cross-referenced in our hiPSC datasets as well as case/control post-mortem and hiPSC-NPC & hiPSC-neuron RNA-seq datasets (schroden.shinyapps.io/BrennandLab-ExpressionApp-limited). Owing to constraints reflecting the original consents, which are restricted to the study of neuropsychiatric disease only, the raw RNA-seq data will be made available by the authors upon reasonable request and IRB approval.
Code Availability
Code is available at github.com/nadschro/SZvariant-synergy.
Oversight
All hiPSC research conducted under the oversight of the Institutional Review Board (IRB) and Embryonic Stem Cell Research Overview (ESCRO) committees at ISSMS.
Supplementary Material
ACKNOWLEDGEMENTS
This manuscript is dedicated to Pamela Sklar; a scientist, mentor and friend who advanced the field of psychiatric genetics. This work was partially supported by National Institute of Health (NIH) grants R56 MH101454 (K.J.B., E.S. and H.M.), R01 MH106056 (K.J.B.) and R01 MH109897 (P.S. and K.J.B.), the New York Stem Cell Foundation (K.J.B.) and Project ALS #2017-03 (J.G. and H.P.). We thank the Neuroscience and Stem Cell Cores at Icahn School of Medicine at Mount Sinai. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Paul O’Reilly provided thoughtful feedback on the manuscript.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 01/04/2019.
Footnotes
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare no conflicts of interest.
REFERENCES
- 1.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardinas AF et al. Common schizophrenia alleles are enriched in mutationin-tolerant genes and in regions under strong background selection. Nat Genet (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaffe AE et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall LS et al. A Transcriptome Wide Association Study implicates specific pre- and post-synaptic abnormalities in Schizophrenia. 384560 (2018). [DOI] [PMC free article] [PubMed]
- 7.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huckins LM et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51,659–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbyn A et al. Landscape of Conditional eQTL in Dorsolateral Prefrontal Cortex and Co-localization with Schizophrenia GWAS. Am J Hum Genet 102, 1169–1184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman GE et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun 8, 2225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet 50, 54–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest MP et al. Open Chromatin Profiling in hiPSC-Derived Neurons Prioritizes Functional Noncoding Psychiatric Risk Variants and Highlights Neurodevelopmental Loci. Cell Stem Cell (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium GT et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho SM et al. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods 101, 113–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho SM et al. Evaluating Synthetic Activation and Repression of Neuropsychiatric-Related Genes in hiPSC-Derived NPCs, Neurons, and Astrocytes. Stem Cell Reports (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paquet D et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–9 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Yang N et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canals I et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods 15, 693–696 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Bowles KR, T CWJ, Qian L, Jadow BM & Goate AM Reduced variability of neural progenitor cells and improved purity of neuronal cultures using magnetic activated cell sorting. PLoS One 14, e0213374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou Y et al. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338–3p reduces BDNF production. Schizophr Res 199, 176–180 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Mowla SJ et al. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem 276, 12660–6 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Campenot RB Local control of neurite development by nerve growth factor. Proc Natl Acad Sci U S A 74, 4516–9 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuire JL et al. Altered serine/threonine kinase activity in schizophrenia. Brain Res 1568, 42–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petralia RS et al. Reduction of AP180 and CALM produces defects in synaptic vesicle size and density. Neuromolecular Med 15, 49–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith JJ, Sumiyama K & Amemiya CT A living fossil in the genome of a living fossil: Harbinger transposons in the coelacanth genome. Mol Biol Evol 29, 985–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun S et al. Thalamic miR-338–3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion. Nat Med 23, 39–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromer M et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirov G et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 17, 142–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Focking M et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry (2014). [DOI] [PubMed] [Google Scholar]
- 39.Ripke S, Group SW & O’Donovan M Current Status of Schizophrenia Gwas. European Neuropsychopharmacology 27, S415 (2017). [Google Scholar]
- 40.Takata A, Matsumoto N & Kato T Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat Commun 8, 14519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardy C et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A 112, E2725–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon HB et al. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci 15, 1667–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dityatev A, Dityateva G & Schachner M Synaptic strength as a function of post-versus presynaptic expression of the neural cell adhesion molecule NCAM. Neuron 26, 207–17 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Burrone J, O’Byrne M & Murthy VN Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature 420, 414–8 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Suk HJ et al. Closed-Loop Real-Time Imaging Enables Fully Automated Cell-Targeted Patch-Clamp Neural Recording In Vivo. Neuron 96, 244–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866 e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Port F & Bullock SL Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods 13, 852–4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman GE, Schrode N, Flaherty E & Brennand KJ New considerations for hiPSC-based models of neuropsychiatric disorders. Mol Psychiatry (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talkowski ME et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell SM et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Network, Pathway Analysis Subgroup of the Psychiatric Genomics, C., International Inflammatory Bowel Disease Genetics, C. & International Inflammatory Bowel Disease Genetics Consortium, I. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18, 199–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ballouz S & Gillis J Strength of functional signature correlates with effect size in autism. Genome Med 9, 64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia P, Chen X, Fanous AH & Zhao Z Convergent roles of de novo mutations and common variants in schizophrenia in tissue-specific and spatiotemporal co-expression network. Transl Psychiatry 8, 105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brainstorm C et al. Analysis of shared heritability in common disorders of the brain. Science 360(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gandal MJ et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wray NR, Wijmenga C, Sullivan PF, Yang J & Visscher PM Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 173, 1573–1580 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Wainschtein P et al. Recovery of trait heritability from whole genome sequence data. bioRxiv, 588020 (2019). [Google Scholar]
- 59.Zuk O, Hechter E, Sunyaev SR & Lander ES The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A 109, 1193–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, Li YI & Pritchard JK Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 177, 1022–1034 e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyle EA, Li YI & Pritchard JK An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubin AJ et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 176, 361–376 e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mimitou EP et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiner DJ et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49, 978–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tansey KE et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry 21, 1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMahon FJ & Insel TR Pharmacogenomics and personalized medicine in neuropsychiatry. Neuron 74, 773–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 67.He X et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet 92, 667–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brennand KJ et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffman GE & Schadt EE variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu D & Smyth GK Camera: a competitive gene set test accounting for intergene correlation. Nucleic Acids Res 40, e133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source donor hiPSCs have already been deposited at the Rutgers University Cell and DNA Repository (study 160; http://www.nimhstemcells.org/): CRISPR-edited hiPSCs are in the process of being submitted in advance of publication. RNA-seq data are available at www.synapse.org/#!Synapse:syn20502314; additionally, we make available the following resource by which neural cell-type-specific expression of any gene can be easily cross-referenced in our hiPSC datasets as well as case/control post-mortem and hiPSC-NPC & hiPSC-neuron RNA-seq datasets (schroden.shinyapps.io/BrennandLab-ExpressionApp-limited). Owing to constraints reflecting the original consents, which are restricted to the study of neuropsychiatric disease only, the raw RNA-seq data will be made available by the authors upon reasonable request and IRB approval.
Code Availability
Code is available at github.com/nadschro/SZvariant-synergy.
Oversight
All hiPSC research conducted under the oversight of the Institutional Review Board (IRB) and Embryonic Stem Cell Research Overview (ESCRO) committees at ISSMS.