Abstract
Polycomb Repressive Complex 2 (PRC2) maintains repression of cell type-specific genes but also associates with genes ectopically in cancer. While it is currently unknown how PRC2 is removed from genes, such knowledge would be useful for the targeted reversal of deleterious PRC2 recruitment events. Here, we show that G-tract RNA specifically removes PRC2 from genes in human and mouse cells. PRC2 preferentially binds G-tracts within nascent pre-mRNAs, especially within predicted G-quadruplex structures. G-quadruplex RNA evicts the PRC2 catalytic core from the substrate nucleosome. PRC2 transfers from chromatin to RNA upon gene activation and chromatin-associated G-tract RNA removes PRC2, leading to H3K27me3 depletion from genes. Targeting G-tract RNA to the tumor suppressor gene CDKN2A in malignant rhabdoid tumor cells reactivates the gene and induces senescence. These data support a model in which pre-mRNA evicts PRC2 during gene activation and provides the means to selectively remove PRC2 from specific genes.
Chromatin structure is responsive to changes in transcriptional state but the mechanisms for this are unclear. Nascent pre-mRNA has primarily been considered to be a passive intermediary but a potential regulatory role for nascent pre-mRNA may explain some of the changes in chromatin structure that occur upon gene expression 1.
The chromatin regulator PRC2 prevents inappropriate activation of genes specific for other cell types and other stages of cell differentiation 2,3. The PRC2 subunit EZH2 methylates histone H3 lysine 27 (H3K27me3) and, together with PRC1, induces formation of a repressive chromatin conformation. PRC2 is essential for cell differentiation, both during embryogenesis and throughout life. Dysregulation of PRC2 function occurs in a range of cancers and can drive cancer cell proliferation, invasion and metastasis 4. EZH2 methyltransferase inhibitors block proliferation of a number of cancer cell types, including malignant rhabdoid tumors (MRT), germinal centre B-cell diffuse large B-cell lymphoma, and diffuse intrinsic pontine glioma (DIPG) 5–10, and are currently being evaluated in clinical trials.
The binding of PRC2 to genes is dynamic. During cell differentiation, PRC2 is lost from genes that become activated and gained at genes that become repressed 11–14. Changes in PRC2 occupancy and H3K27me3 are also observed during cell transformation and in cancer 8,10,15–21. The oncogenic effects of PRC2 have been linked to ectopic repression of particular genes, for example CDKN2A (encoding p16INK4A) in MRT and DIPG 5,6,10,22. However, rather than targeting these key genes specifically, EZH2 inhibition leads to the reactivation of polycomb target genes across the genome 5,9, which may alter tumor cell identity and promote tumor progression 23.
PRC2 is recruited to chromatin through CpG islands (CGIs). Insertion of CGIs into the genome is sufficient to induce PRC2 recruitment 24–26. The recruitment of PRC2 to CGIs is consistent with the binding of the accessory factors PHF1 (PCL1) and MTF2 (PCL2) to non-methylated CpG DNA 27,28 and the binding of JARID2 to H2AK119ub, deposited by PRC1 29,30. Although recognition of CGIs offers an explanation for the spatial pattern in which PRC2 is associated with the genome, this mechanism does not account for changes in PRC2 occupancy that occur during cell differentiation or during cell transformation. Knowledge of the mechanisms responsible for these dynamic patterns of PRC2 chromatin binding is necessary to understand how cell differentiation programs are regulated and may allow the development of methods to inhibit polycomb activity at specific genes.
In addition to interacting with chromatin, PRC2 also binds RNA but the impact of this on PRC2 function remains unclear. Although first identified to bind specific non-coding RNAs (ncRNAs), UV-crosslinking-based methods have revealed that PRC2 directly interacts with the majority of nascent pre-mRNAs and nascent ncRNAs in embryonic stem cells (ESC) 31,32. When binding short RNA oligonucleotides in vitro, recombinant forms of PRC2 display a preference for repeated G-tracts, especially when folded into G-quadruplex (G4) structures 33–36, but the relevance of this for PRC2 RNA function in cells is unknown. First postulated to promote the recruitment of PRC2 to chromatin, it has recently been found that RNA blocks the interaction of PRC2 with nucleosomes 31,36 and inhibits its methyltransferase activity 33,37–39. Potentially consistent with this, global inhibition of RNA polymerase II 40 or global RNA degradation 31 triggers PRC2 recruitment to chromatin at active genes in cells. Similarly, insertion of premature poly(A) signals 33 or promoter or enhancer inactivation 24,41 increases PRC2 binding and H3K27me3 in cis. However, whether these results reflect loss of antagonistic RNA, loss of RNA polymerase II, or depletion of antagonizing chromatin modifications such as H3K4me3, is unknown.
We hypothesized that nascent RNA plays a role in the temporal regulation of PRC2 occupancy at its target genes. Specifically, we considered that nascent, chromatin-associated RNA may remove PRC2 from chromatin. To address this, we sought to identify the RNA sequences preferentially bound by PRC2 in cells and determine the impact of these RNA elements on PRC2 occupancy at genes. Our results support a model in which chromatin-associated G-tract RNA evicts PRC2 from chromatin during gene activation and provides the means to remove PRC2 from specifically targeted genes. These data also support the broader consideration of nascent pre-mRNA as a regulatory molecule that modulates chromatin state at active genes.
Results
PRC2 binds G-tracts within nascent RNAs in cells
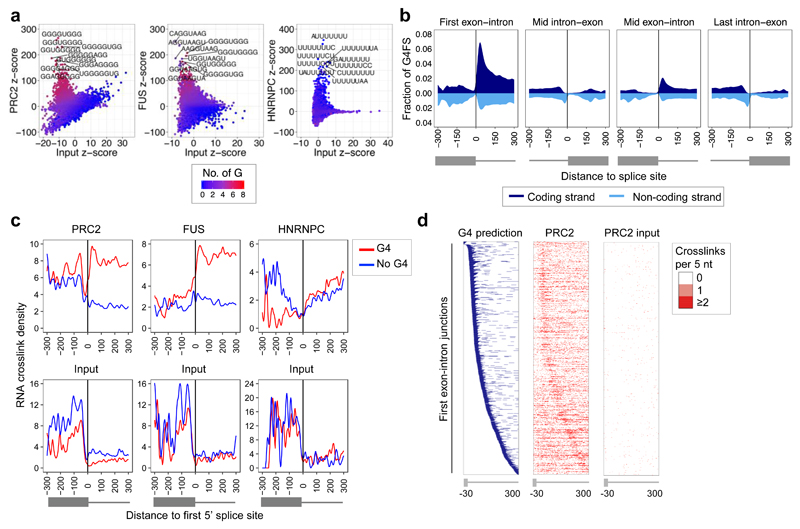
Using iCLIP, we have previously found that PRC2 directly interacts with the majority of nascent pre-mRNAs and ncRNAs in mouse ESC 31. We sought to determine whether PRC2 favored any particular sequence within nascent transcripts. To ensure identified sequences were specific for PRC2, we also mapped background protein crosslink sites on input RNA. Comparing PRC2 and input RNA crosslink sites, we identified a strong enrichment of G-tracts at PRC2 RNA binding sites (Fig. 1a and Supplementary Fig. 1a). Enrichment of these sequences was also observed at crosslink sites for the RNA binding protein FUS, as has been observed previously in mouse brain 42, but was not apparent for HNRNPC, which binds poly(U) sequences 43.
Fig. 1. PRC2 binds G-tracts with the potential to form G4 structures in nascent RNA.
(a) Enrichment of 8-mer sequences at PRC2, FUS and HNRNPC RNA crosslink sites identified by iCLIP (vs input controls). Gs per 8-mer are indicated by color. The ten 8-mers with the highest z-score are labeled.
(b) Average G4 prediction score (G4-forming sequences (G4FS)) for the coding (dark blue, above x-axis) and non-coding (cyan, below x-axis) strands around mouse gene splice sites.
(c) RNA crosslink density for PRC2, FUS, HNRNPC, and their input controls at the set of first 5’ splice sites that are predicted (red, n=942) or not predicted (blue, n=760) to be able to form G4 structures (PRC2 P<2.2x10-16, FUS P<2.2x10-16, Wilcoxon rank-sum test).
(d) Left: Heat map (blue) showing the position of sequences predicted to be able to form G4 structures -30 to +300 nt around the first 5’ splice site of nascent RNAs expressed in mouse ESC. Right: Heat maps (red) showing the position of PRC2 and input RNA crosslink sites at the same 5’ splice sites. The number of crosslink sites per 5 nt window is indicated by color.
In vitro, the binding of PRC2 to G-tract sequences has been reported to increase when these RNAs are folded into a G4 structure 35 and we confirmed this to be the case (Supplementary Fig. 1b-d). We therefore explored whether PRC2 maintained this binding preference in cells. Calculating the propensity for G4 formation across all genes using G4Hunter 44 revealed a peak in predicted G4 formation 50 nt into the first intron (Fig. 1b). This is consistent with previous reports of G-tract enrichment at the 5’ end of introns 45,46. Although PRC2 bound these sequences near the first 5’ splice site, we did not observe any effects of PRC2 on splicing (Supplementary Fig. 2a).
To explore whether the potential for G4 formation increased PRC2 RNA binding, we measured PRC2 crosslink site density at first 5’ splice sites predicted to be able to form G4 structures versus those that were not. First 5’ splice sites predicted to form G4 structures exhibited significantly higher PRC2 RNA binding (Fig. 1c and Supplementary Table 1). This increased binding was localized at the site of predicted G4 formation (Fig. 1d and Supplementary Fig. 2b) and was observed even when normalizing for G content (Supplementary Fig. 2c). The presence of a predicted G4 structure was also associated with increased FUS RNA binding at the beginning of the first intron, consistent with previous reports of FUS binding to G4 RNA in vitro 47, but no change in HNRNPC binding (Fig. 1c). We found that PRC2 bound across the range of predicted G4 structures but displayed a slight but significant preference for those formed from smaller numbers of G-tracts (Supplementary Fig. 2d). We conclude that PRC2 preferentially binds G-tracts within nascent RNAs in cells, especially when predicted to form G4 structures.
Embedded G4 structures inhibit the interaction of PRC2 with nucleosomes
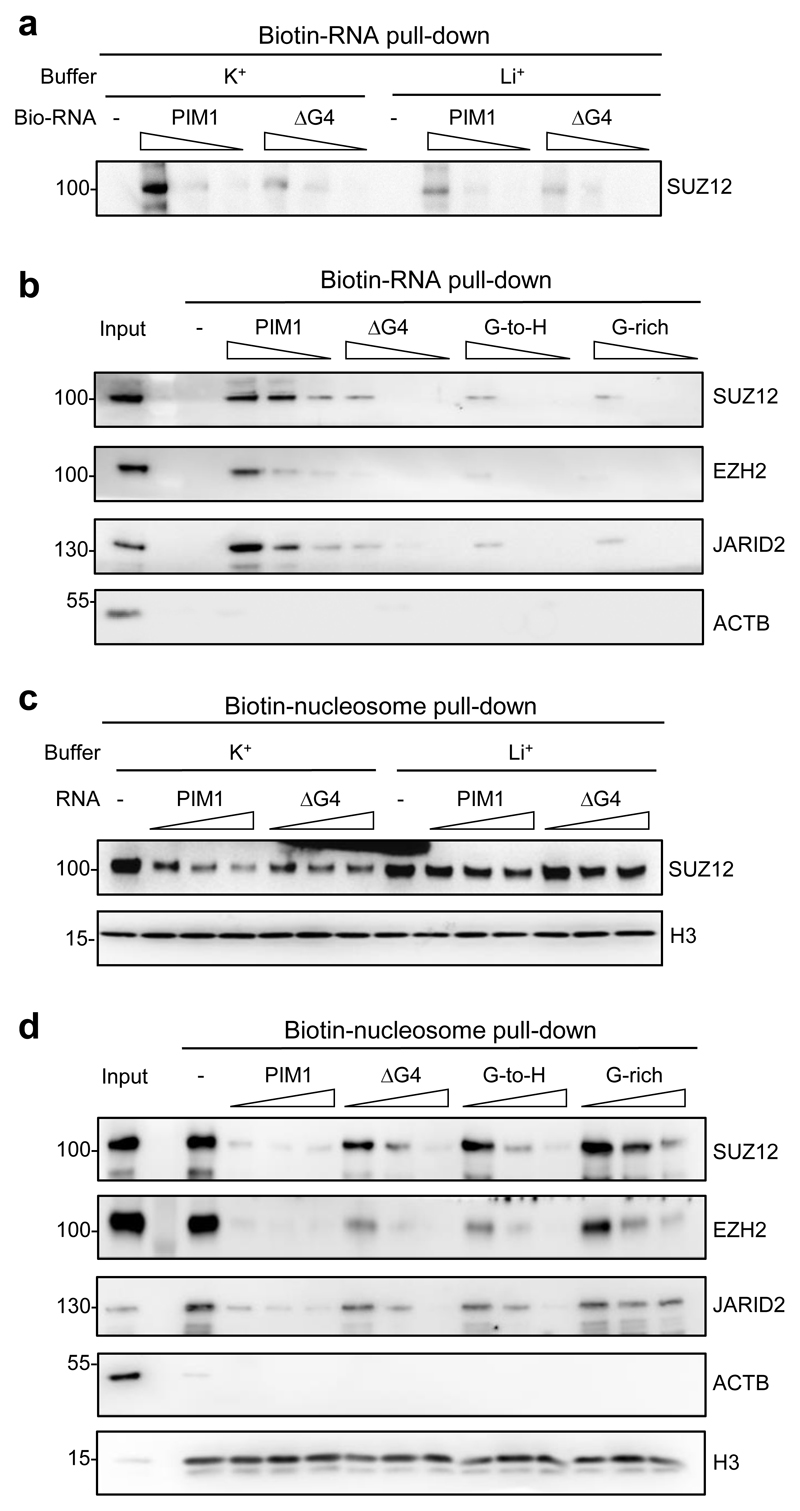
PRC2 has been found to exhibit high affinity for short G4-forming RNAs in vitro but whether it can recognize endogenous G4-forming sequences embedded within longer physiological RNAs is not clear. Thus, to verify that PRC2 recognizes G4 structures within the context of longer RNA molecules, we synthesized a previously described 150 nt sequence from the gene PIM1 that contains a central 23 nt G4-forming sequence, and a control RNA lacking this region (ΔG4) 48 (Supplementary Figs. 3a and b). Examination of iCLIP data showed that PRC2 bound to this region of PIM1 RNA in mouse ESC (Supplementary Fig. 3c). Incubation of PRC2 with these RNAs demonstrated that the embedded G4-forming sequence increased RNA binding by recombinant PRC2 (SUZ12–EZH2–EED–RBBP4 or RBBP7) (Fig. 2a, Supplementary Fig. 3d). Binding was stronger in buffer containing KCl, which allows G4 formation, compared to buffer containing LiCl, which does not. Endogenous PRC2 in ESC nuclear extract also bound more strongly to PIM1 RNA than to ΔG4 RNA or to control RNAs in which the G nucleotides within the G4-forming sequence were mutated (Fig. 2b). Then, testing whether PIM1 RNA blocked the binding of PRC2 to nucleosomes, we found that G4-formation increased the ability of PIM1 RNA to block both recombinant and endogenous PRC2 binding to nucleosomes (Fig. 2c, d and Supplementary Fig. 3e and f). We conclude that PRC2 recognizes G4 structures embedded within longer transcripts and this inhibits its interaction with nucleosomes.
Fig. 2. G4 structures within longer RNAs block PRC2 binding to nucleosomes.
(a) Immunoblot for SUZ12 after pull-down of recombinant PRC2 (EZH2–SUZ12–EED –RBBP4 or RBBP7) with pre-folded biotinylated PIM1 RNA or control PIM1 RNA lacking G4-forming sequence (ΔG4) in KCl or LiCl-containing buffer. Streptavidin beads were incubated with 500, 50 or 5 ng/ul of RNA, washed, and then incubated with PRC2. Representative of three independent experiments (others shown in Supplementary Fig. 3d).
(b) Immunoblot for SUZ12, EZH2, JARID2 and ACTB after pull-down of PRC2 from ESC nuclear extract with 10-fold dilutions of biotinylated wild-type PIM1 RNA, ΔG4 RNA, G-to-H RNA (G4-forming G nucleotides mutated to non-G) and G-rich RNA (G-to-H RNA with an equal number of non-G to G mutations outside of the G4-forming region). Representative of two independent experiments.
(c) Immunoblot for SUZ12 and H3 after pull-down of recombinant PRC2 with biotinylated nucleosomes (reconstituted with 185 bp DNA) in the presence of PIM1 or ΔG4 RNA (2, 20 or 200 ng/μl). Representative of three independent experiments (others shown in Supplementary Fig. 3e).
(d) Immunoblot for SUZ12, EZH2, JARID2, ACTB and H3 after pull-down of PRC2 from ESC nuclear extract with biotinylated nucleosomes in the presence of biotinylated wild-type PIM1, ΔG4, G-to-H or G-rich PIM1 RNA (2, 20 or 200 ng/μl). Representative of two independent experiments. Uncropped blot images are shown in Supplementary Data Set 1.
G4 RNA blocks interaction of the PRC2 catalytic core with the substrate core nucleosome particle
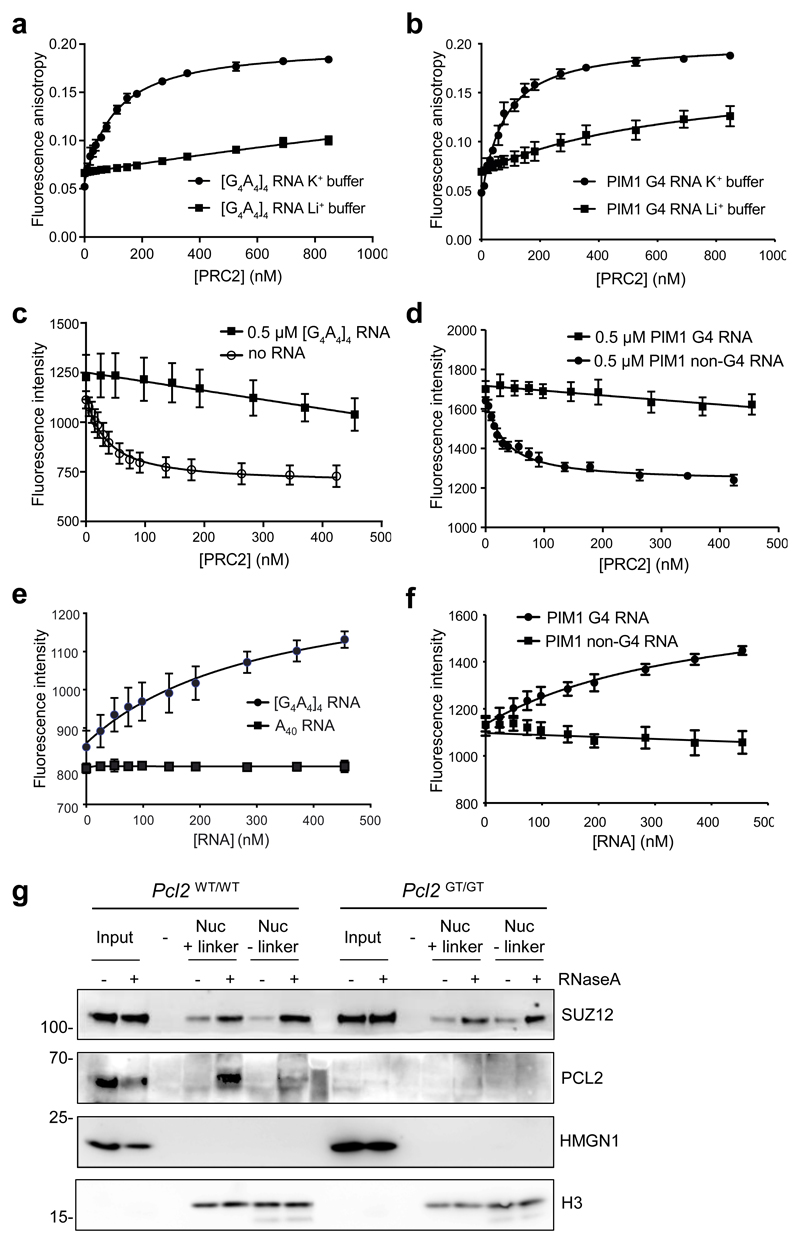
We next sought to explore the basis for the antagonism between RNA and nucleosomes for PRC2 binding. We reasoned that because the PRC2 catalytic core has been reported to be competent for G4 RNA binding 34, G4 RNA may block the interaction of the PRC2 core with nucleosomes. We purified a recombinant catalytic core complex comprising EZH2, EED and the SUZ12 VEFS domain 49, and, using fluorescence anisotropy, found that it bound to an archetypal G4-forming RNA ([G4A4]4) and to the endogenous G4-forming sequence within PIM1 RNA in KCl but with no significant binding in LiCl buffer (Fig. 3a and b).
Fig. 3. G4 RNA inhibits interaction of the PRC2 catalytic core with the substrate core nucleosome particle.
(a) Fluorescence anisotropy measuring binding of the PRC2 catalytic core (EZH2–EED–SUZ12 VEFS domain) directly to fluorescein labelled [G4A4]4 RNA in either 100 mM K+ or Li + buffer (mean and S.E., n=3 independent experiments).
(b) As (a) except for the 24 nt G4-forming sequence within PIM1 RNA.
(c) Fluorescence intensity measuring binding of the PRC2 catalytic core directly to MDCC-labeled H3K27M obligate substrate core nucleosome particles (reconstituted with 147 bp DNA) in the presence of 500 nM [G4A4]4 RNA or no RNA (mean and S.E., n=3 independent experiments).
(d) As (c), except with 500 nM PIM1 G4 RNA or a control non-G4-forming 24 nt portion of PIM1 RNA.
(e) Titration of [G4A4]4 and control A40 RNAs into a pre-formed complex of core PRC2 and MDCC-labeled substrate core nucleosome particle. The increase in fluorescence intensity with [G4A4]4 RNA is interpreted as release of PRC2 from the nucleosome (mean and S.E., n=3 independent experiments).
(f) As (e), except with G4 and non-G4 forming PIM1 RNAs.
(g) Immunoblot for SUZ12, PCL2, HMGN1 and H3 after co-immunoprecipitation of PRC2 from Pcl2GT/GT or Pcl2WT/WT ESC with nucleosomes containing biotin-tagged histone H2A (reconstituted with either 185 bp or 147 bp DNA) from mock or RNaseA-treated nuclear extract. Representative of 2 independent experiments. Uncropped blot images are shown in Supplementary Data Set 1.
In order to measure PRC2 binding to the nucleosome we engineered a nucleosome with a fluorescently-tagged histone H3 and 147 bp DNA. After confirming that robust binding to [G4A4]4 and PIM1 G4 RNA was also observed in the low-salt nucleosome binding buffer required for the fluorescent assay (16.7 ± 1.2 nM and 22.5 ± 1.8 nM, respectively; Supplemental Fig. 4a and b), we then measured the effect of the RNA on the binding of the catalytic core to nucleosomes. In the absence of G4 RNA, the PRC2 catalytic core interacted with nucleosomes with high affinity (25.9 ± 10.7 nM) but in the presence of 500 nM [G4A4]4 RNA, PRC2 binding to the nucleosome was effectively blocked (Supplementary Fig. 4c).
The PRC2 catalytic core interacts with the substrate H3 tail through EZH2 and the K27-methylated H3 tail of the allosteric nucleosome through EED. To specifically test the effect of G4 RNA on the binding of the PRC2 catalytic core to its substrate, we used nucleosomes containing a fluorescently-labeled H3K27M-modified tail, which engages EZH2 but not EED 49,50. In the absence of RNA, the PRC2 catalytic core interacted with this obligate substrate core nucleosome particle with high affinity (29.9 ± 3.9 nM). In the presence of [G4A4]4 or PIM1 G4 RNA the interaction was blocked, whereas a non-G4-forming portion of PIM1 RNA had no effect (Fig. 3c, d and Supplementary Fig. 4d). Given this antagonistic effect of G4 RNA on the interaction of the PRC2 core with its substrate, we considered that the RNA may also be able to displace PRC2 from the nucleosome. Strikingly, we found that both [G4A4]4 and PIM1 G4 RNA, but neither control non-G4 PIM1 RNA nor poly(A) RNA, was also able to remove PRC2 from a pre-formed core-PRC2:substrate nucleosome complex (Fig. 3e and f).
To validate these findings, we measured the effect of RNA degradation on the binding of endogenous PRC2 in nuclear extract to wild-type mononucleosomes either lacking linker DNA (reconstituted with 147 bp DNA) or containing linker DNA (reconstituted with 183 bp DNA). We found that RNA depletion increased PRC2 binding to nucleosomes independently of linker DNA and independently of the DNA-binding accessory factors PCL2, AEBP2 and JARID2 (Fig. 3g and Supplementary Fig. 4e and f). Together, these data show that G4 RNA evicts PRC2 from the substrate core nucleosome particle via interactions with the PRC2 catalytic core independently of accessory factors.
Chromatin-associated G-tract RNA removes PRC2 from genes
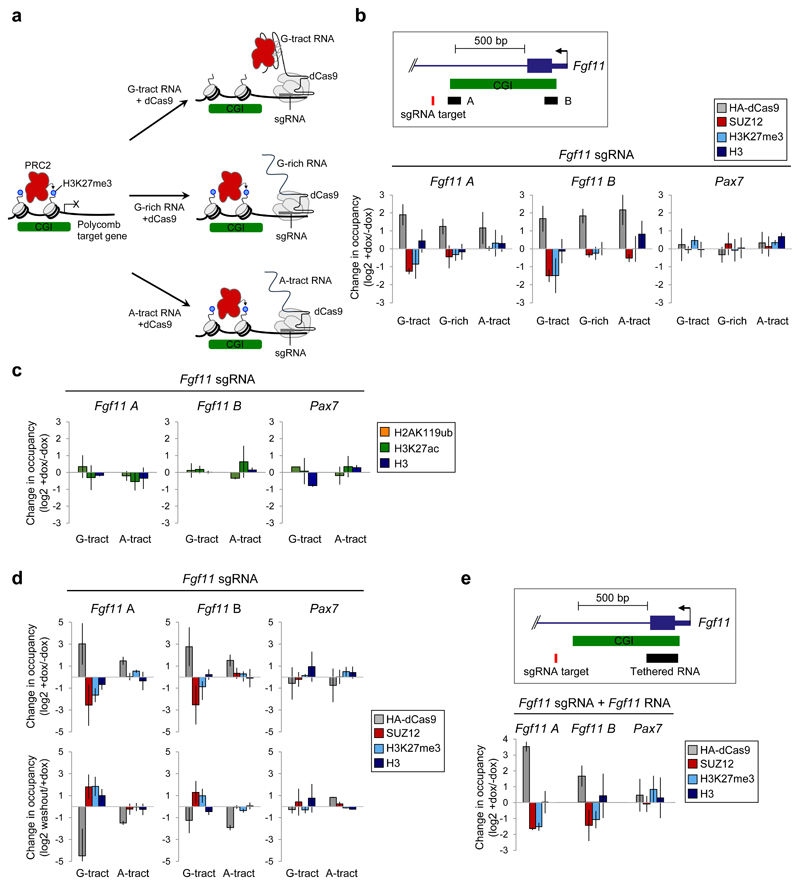
The binding of PRC2 to G-tracts within nascent pre-mRNA in cells and the ability of G4 RNA to evict PRC2 from nucleosomes suggested that G-tracts within nascent RNAs remove PRC2 from chromatin at genes. We considered that if this hypothesis was correct then mimicking chromatin-associated, nascent RNA by tethering G-tract RNA to the 5’-end of genes with dCas9 should remove PRC2 from chromatin (Fig. 4a). To test this, we generated a doxycycline (dox)-inducible HA-dCas9 NIH-3T3 cell line and co-expressed short guide RNAs (sgRNAs) to recruit dCas9 to the first intron of the PRC2 target gene Fgf11 (Fig. 4b and Supplementary Fig. 5a and b). We appended to the 3’-end of the sgRNA 51–53 either a 220 nt sequence composed of repeated G-tracts, an equal length sequence with the same overall G-content (50%) but lacking sequential runs of Gs, or a control RNA in which the G-tracts were replaced with A-tracts, which PRC2 binds only weakly in vitro 33,35 and which are depleted from PRC2 binding sites in cells (Fig. 1a and Supplementary Fig. 1a). We then performed ChIP for HA-dCas9, SUZ12, H3K27me3, total H3 and non-specific IgG control, before and after induction of dCas9 (Fig. 4b and Supplementary Fig. 5c). As predicted, dCas9 induction led to specific recruitment of the dCas9-G-tract-RNA, dCas9-G-rich-RNA, and the dCas9-A-tract-RNA ribonucleoproteins to Fgf11. dCas9-tethered G-tract RNA significantly reduced PRC2 binding and H3K27me3 at Fgf11 but not at other genes. No change was observed in total histone H3 occupancy. In contrast, dCas9-tethered G-rich or A-tract RNAs had no effect on PRC2 chromatin binding or H3K27me3. The loss of PRC2 occupancy was not caused indirectly by induction of Fgf11 transcription (Supplementary Fig. 5d). We conclude that chromatin-associated G-tract RNA is sufficient to remove PRC2 and deplete H3K27me3 from genes.
Fig. 4. Chromatin-associated G-tract RNA removes PRC2 from specific genes in cells.
(a) Hypothesis: G-tract RNA, tethered to chromatin with dCas9, should compete with CGI chromatin for PRC2, reducing H3K27me3. The same length RNA that is equally G-rich but lacking G-tracts or RNA in which the G-tracts are replaced with A-tracts, both of which bind PRC2 only weakly, should both have no effect.
(b) Top: Position of the Fgf11 sgRNA and primer pairs A and B. Bottom: Change in HA-dCas9, SUZ12, H3K27me3 and total H3 occupancy at Fgf11 and Pax7 measured by ChIP-qPCR after dox-mediated induction of HA-dCas9 expression in cells containing the Fgf11 sgRNA, to which G-tract, G-rich or A-tract RNA is appended (mean and S.D., n=3 independent dox inductions. P-values: Fgf11-A G-tract RNA SUZ12=0.0018, H3K27me3=0.14. Fgf11-B G-tract RNA SUZ12=0.0052, H3K27me3=0.03. Fgf11-B G-rich RNA SUZ12=0.03, Welch’s one-tailed t-test).
(c) Change in H2AK119ub, H3K27ac and total H3 at Fgf11 and Pax7 before and after incubation with dox (mean, S.D., n=3 independent dox inductions, no significant changes, Welch’s one-tailed t-test).
(d) Change in HA-dCas9, SUZ12, H3K27me3 and total H3 occupancy at Fgf11 and Pax7 before and after dox treatment (day 6) and after subsequent dox washout (day 12) (mean and S.D., n=3 independent dox inductions. P-values: Dox induction Fgf11-A G-tract RNA: SUZ12=0.02, H3K27me3=0.0066. Fgf11-B G-tract RNA SUZ12=0.046, H3K27me3=0.49. Dox washout Fgf11-A G-tract RNA: SUZ12=0.041, H3K27me3=0.0094. Fgf11-B G-tract RNA SUZ12=0.052, H3K27me3=0.091, Welch’s one-tailed t-test).
(e) Top: Fgf11 RNA sequence spanning the first exon-intron junction was appended to Fgf11 sgRNA. Bottom: As (b), except using Fgf11 sgRNA to which the Fgf11 RNA sequence has been appended (mean and S.D., n=3 independent dox inductions. P-values: Fgf11 A SUZ12=2.9x10-4, H3K27me3=0.0019. Fgf11 B SUZ12=0.045, H3K27me3=0.026, Welch’s one-tailed t-test).
We sought to determine whether the effect of chromatin-associated G-tract RNA was specific to PRC2. No changes were observed in the levels of H3K27ac or H2AK119ub (Fig. 4c and Supplementary Fig. 5e), demonstrating that loss of PRC2 was not simply due to occlusion of chromatin modifying enzymes by G-tract RNA. We also examined whether proximity to the PRC2 binding site on chromatin was important for the effect of chromatin-associated G-tract RNA. Tethering G-tract RNA to a non-PRC2-bound site at the 3’ end of Fgf11, 2.25 kb from the PRC2-bound CGI at the 5’ end of the gene, had no effect on PRC2 or H3K27me3 occupancy at the CGI, suggesting proximity of the RNA to the site of PRC2 binding on chromatin is important for PRC2 removal (Supplementary Fig. 5f).
We next asked whether the continued presence of a G-tract RNA was required to prevent PRC2 recruitment to its target genes. We removed dox from the cells, causing loss of dCas9 expression (Supplementary Fig. 5a), and repeated the measurements of PRC2 occupancy and H3K27me3. We found that dox removal led to a partial restoration of PRC2 chromatin binding and full restoration of H3K27me3 (Fig. 4d and Supplementary Fig. 5g). We conclude that while it is present, chromatin-associated G-tract RNA actively prevents PRC2 binding to CGI chromatin and that loss of the RNA subsequently allows PRC2 recruitment and H3K27me3 at the gene.
We sought to determine whether endogenous G-tract RNA sequence spanning the first exon-intron junction could also remove PRC2 from genes. As we had found for the artificial G-tract RNA, tethering RNA sequence from the 5’ end of Fgf11 to the Fgf11 gene resulted in depletion of PRC2 and loss of H3K27me3 (Fig. 4e). Thus, endogenous G-tracts located around the first 5’ splice site of nascent RNA can also remove PRC2 from chromatin.
PRC2 transfers from chromatin to RNA upon gene activation
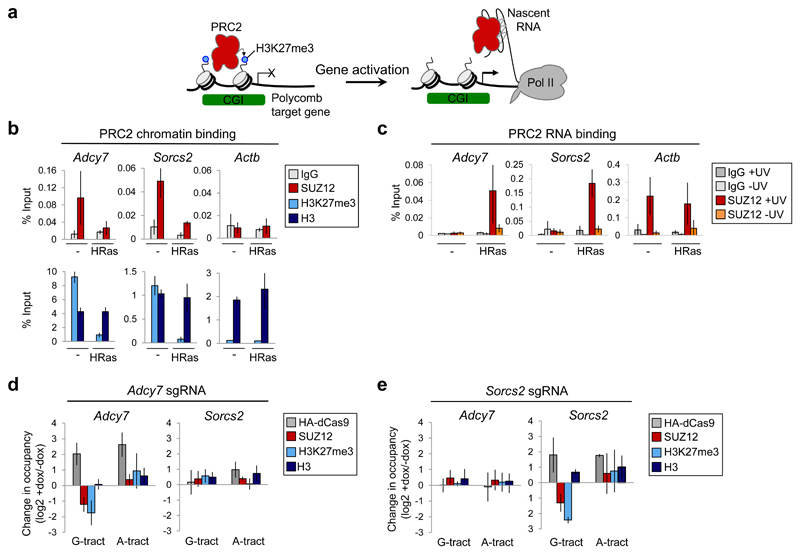
The transfer of PRC2 from chromatin to chromatin-associated G-tract RNA suggested that PRC2 also transfers from chromatin to nascent pre-mRNA upon gene activation (Fig. 5a). Cell transformation induced by oncogenic HRasV12 is accompanied by dynamic changes in PRC2 chromatin occupancy 15,16,18,19,41. Notably, expression of HRasV12 leads to activation of Adcy7 and Sorcs2 and the subsequent loss of PRC2 18. Loss of PRC2 from Sorcs2 is dependent on the Sorcs2 TSS 41, consistent with a role for the nascent pre-mRNA. Consistent with this hypothesis, we found that activation of Adcy7 and Sorcs2 downstream of HRasV12 was accompanied by a change in PRC2 from binding chromatin to binding the pre-mRNA (Fig. 5b,c and Supplementary Fig. 6a-c). We next tested whether chromatin-associated G-tract RNA recapitulated the effect of gene activation on PRC2 binding at these genes. Tethering G-tract RNA, but not A-tract RNA, to Adcy7 reduced PRC2 binding and H3K27me3 at this gene but had no effect on PRC2 occupancy at Sorcs2 (Fig. 5d and Supplementary Fig. 6d). Reciprocally, tethering G-tract RNA to Sorcs2 reduced PRC2 occupancy and H3K27me3 at this gene but had no effect on Adcy7 (Fig. 5e and Supplementary Fig. 6e). Thus, PRC2 transfers from chromatin to nascent pre-mRNA upon gene activation and this can be recapitulated by tethering G-tract RNA to genes.
Fig. 5. PRC2 transfers from chromatin to nascent pre-mRNA during gene activation.
(a) Hypothesis: Upon activation of polycomb target genes, PRC2 switches from binding chromatin to binding nascent RNA.
(b) SUZ12, H3K27me3 and total H3 chromatin occupancy (with IgG control) at Adcy7, Sorcs2 and Actb before and after HRasV12 expression (mean and S.D., n=3 independent ChIPs. Adcy7 SUZ12 P=0.0051, H3K27me3 P=2.1x10-4. Sorcs2 SUZ12 P=0.0041, H3K27me3 P=4.9x10-4, Welch’s one-tailed t-test).
(c) SUZ12 binding to Adcy7, Sorcs2 and Actb nascent pre-mRNA before and after HRasV12 expression, measured by RIP-qPCR with and without UV-crosslinking of cells (mean and S.D., n=3 independent RIPs. Adcy7 SUZ12 +/-Ras P=0.011, Sorcs2 SUZ12 +/-Ras P=0.0019, Welch’s one-tailed t-test).
(d) Change in HA-dCas9, SUZ12, H3K27me3 and total H3 occupancy at Adcy7 and Sorcs2 after dox-mediated induction of HA-dCas9 expression in cells containing sgRNA specific for Adcy7 (mean and S.D., n=3 independent dox inductions. Adcy7 G-tract RNA: SUZ12 P=0.014, H3K27me3 P=0.024, Welch’s one-tailed t-test).
(e) As (d), except in cells containing sgRNA specific for Sorcs2. Sorcs2 G-tract RNA: SUZ12 P=0.017, H3K27me3 P=2.9x10-4, Welch’s one-tailed t-test).
G-tract RNA reverses ectopic recruitment of PRC2 triggered by oncogenic HRas
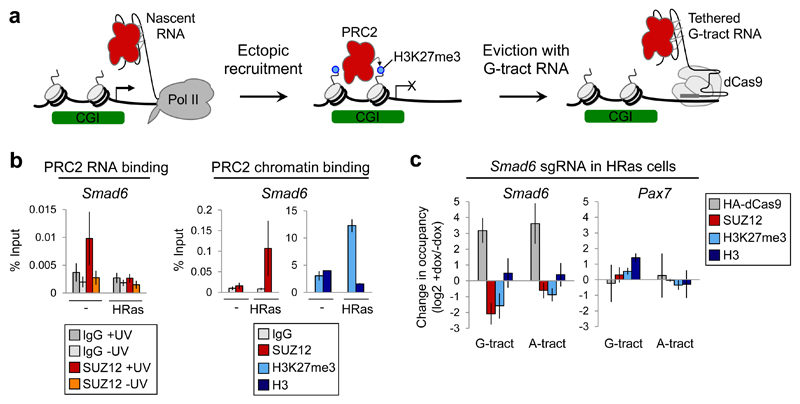
Cell transformation mediated by oncogenes such as HRasV12 causes changes in PRC2 association with chromatin, including ectopic recruitment to specific genes. PRC2 activity can be inhibited in cancer cells with small molecules but this reactivates PRC2 target genes non-specifically. We postulated that G-tract RNA tethering would instead allow the specific reversal of deleterious PRC2 recruitment events (Fig. 6a). HRasV12-mediated recruitment of PRC2 to Smad6 is necessary for Ras-induced senescence 19 and is dependent on transcriptional repression 41. Consistent with this dependence on transcription repression reflecting loss of the competing nascent pre-mRNA, we found that expression of HRasV12 resulted in a switch in PRC2 binding from nascent pre-mRNA to chromatin at Smad6 (Fig. 6b). We next asked whether chromatin-associated G-tract RNA could reverse this recruitment of PRC2 to chromatin at Smad6 in HRasV12-expressing cells. We found that tethering G-tract RNA to Smad6 countered HRasV12-mediated PRC2 recruitment and reduced H3K27me3 at the gene (Fig. 6c and Supplementary Fig. 6f). As we had found for the other genes tested, G-tract RNA tethering and the resultant PRC2 loss was not sufficient to activate Smad6 transcription (Supplementary Fig. 6g). We conclude that G-tract RNA tethering allows the reversal of oncogene-mediated PRC2 recruitment events.
Fig. 6. G-tract RNA reverses PRC2 recruitment triggered by oncogenic HRasV12.
(a) Hypothesis: Tethered G-tract RNA can reverse the ectopic recruitment of PRC2 to specific genes that occurs during cell transformation.
(b) Left: SUZ12 binding to Smad6 nascent pre-mRNA before and after HRasV12 expression, measured by RIP-qPCR with and without UV-crosslinking of cells (mean and S.D., n=3 independent RIPs. Smad6 SUZ12 +/-Ras P=0.043, Welch’s one-tailed t-test). Right: SUZ12, H3K27me3 and total H3 chromatin occupancy (with IgG control) at Smad6 before and after HRasV12 expression (mean and S.D., n=3 independent ChIPs. SUZ12 P=0.036, H3K27me3 P=0.021, Welch’s one-tailed t-test).
(c) Change in HA-dCas9, SUZ12, H3K27me3 and total H3 occupancy at Smad6 and Pax7 after dox-mediated induction of HA-dCas9 in HRasV12-expressing cells containing sgRNA specific for Smad6 (mean and S.D., n=3 independent dox inductions. G-tract RNA: SUZ12 P=0.0082, H3K27me3 P=0.02, Welch’s one-tailed t-test).
G-tract RNA tethering activates the tumor suppressor gene CDKN2A in MRT cells
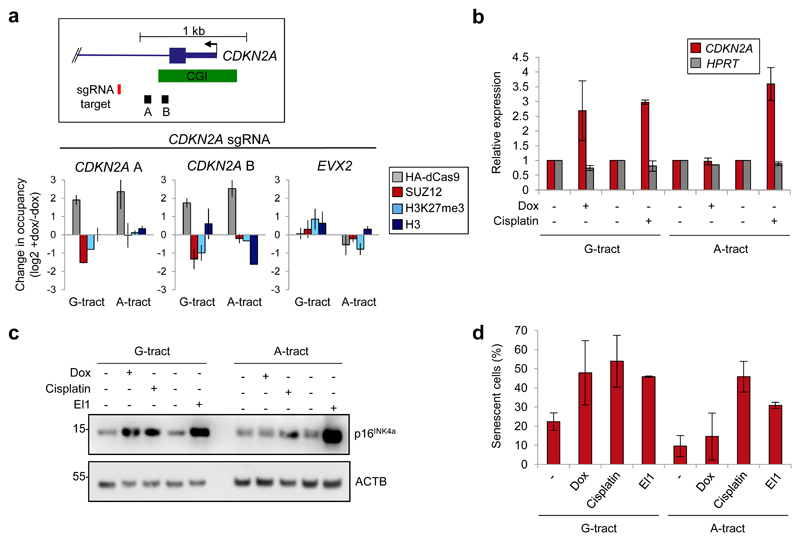
PRC2 silences tumor suppressor genes in a number of cancer types, including CDKN2A (p16INK4a) in MRT and DIPG 5,6,10,22. We therefore sought to determine the effect of tethering G-tract RNA to this gene in MRT cells. We found that recruitment of G-tract, but not A-tract, RNA caused loss of PRC2 and H3K27me3 (Fig. 7a and Supplementary Fig. 7a and b). Strikingly, this was sufficient to activate CDKN2A and increase p16INK4a protein levels to a similar extent to the chemotherapeutic agent cisplatin and the EZH2 inhibitor EI1 (Figs. 7b and c). Furthermore, CDNK2A upregulation was mirrored by an increase in the proportion of senescent cells (Fig. 7d and Supplementary Fig. 7c). We conclude that G-tract RNA tethering can be used to reverse polycomb-mediated silencing of specific tumor suppressor genes in cancer cells.
Fig. 7. G-tract RNA tethering activates CDNK2A and induces cell senescence.
(a) Top: Position of the CDNK2A sgRNA and primer pairs A and B. Bottom: Change in HA-dCas9, SUZ12, H3K27me3 and total H3 occupancy at CDKN2A and EVX2 after dox-mediated induction of HA-dCas9 in G-401 cells containing sgRNA specific for CDKN2A, to which is appended G-tract or A-tract RNA (mean and S.D., n=2 independent dox inductions).
(b) Change in CDKN2A mRNA abundance in cells described in (a) with and without treatment with dox or cisplatin (3.3 μM, 24 hrs) (mean and S.D., n=3 independent experiments. G-tract +/-dox P=0.022, Welch’s one-tailed t-test).
(c) Immunoblot for p16INK4a and ACTB in cells described in (a) with and without treatment with dox, cisplatin (3.3 μM, 24 hrs) or the EZH2 inhibitor EI1 (10 μM, 6 days). Representative of three independent experiments. Uncropped blot images are shown in Supplementary Data Set 1.
(d) Proportion of senescent cells (β-galactosidase staining) in cultures treated as in (c) (mean and S.D., n=3 independent dox inductions or n=2 cisplatin and EI1 treatments. G-tract +/-Dox P=0.032, Student's one-tailed t-test).
Discussion
Current models of how PRC2 interacts with chromatin provide an explanation for the spatial distribution of PRC2 within the genome, but they do not account for the changes in PRC2 gene occupancy that occur during cell differentiation or in cancer. We have discovered that chromatin-associated G-tract RNA removes PRC2 from its target genes. We found that PRC2 directly binds G-tracts within nascent RNAs, especially those at the first 5’ splice site predicted to form G4 structures. G4 RNA binds to the PRC2 catalytic core and antagonizes its interaction with the substrate core nucleosome particle. Consistent with this, PRC2 is transferred from chromatin to nascent pre-mRNA during gene activation and chromatin-associated G-tract RNA is sufficient to remove PRC2 from chromatin and deplete H3K27me3. These results support a model in which G-tracts within nascent RNA remove PRC2 from chromatin during activation of polycomb-repressed genes. We also demonstrate that this mechanism can be exploited to allow the targeted removal of PRC2 from tumor suppressor genes in cancer cells.
The mechanisms responsible for the removal of PRC2 from chromatin have been unclear. A number of recent studies have demonstrated that PRC2 recruitment is responsive to the activation state of the gene. Inhibition of RNA polymerase II or insertion of premature poly(A) signals triggers PRC2 recruitment to CGIs at active genes 33,40. Similarly, the removal of PRC2 from genes during HRasV12-mediated cell transformation is dependent on their transcriptional activation 41. Furthermore, PRC2 is recruited to CGIs inserted into the genome, but not if they contain binding sites for transcriptional activators present in the cell 26 or if they are positioned between an active promoter-enhancer pair 24. These results show that PRC2 is only able to stably associate with chromatin in the absence of transcriptional activity. Based on experiments showing that global RNA degradation triggers PRC2 recruitment to transcribed genes and that RNA inhibits PRC2 nucleosome interaction and methyltransferase activity 31,33,36–39, we and others have suggested that one of the features of active genes that inhibits PRC2 function is the nascent pre-mRNA itself. Our results support this model, demonstrating that chromatin-associated RNA can prevent PRC2 recruitment to active genes and, in addition, that G-tract RNA removes stably-associated PRC2 from genes.
Our data clarifies the nature of PRC2 RNA binding specificity and provides an explanation as to its function. PRC2 RNA binding activity was first identified through its association with specific ncRNAs. Systematic measurement of direct RNA binding in cells later revealed that PRC2 binds the majority of nascent pre-RNAs and ncRNAs in a promiscuous manner 31, a conclusion also drawn from lower-stringency native RNA IP experiments 54. Although PRC2 was initially observed to bind a broad range of RNAs in vitro 54, later studies using more homogenous short oligonucleotides revealed specificity for repetitive G-tract sequences, especially when folded into G4 structures 34–36. Potentially consistent with this, it was also reported that G-tract sequences were enriched in RNAs that co-precipitated with EZH2, but not with SUZ12, from formaldehyde-crosslinked HeLa cells 35. However, questions remained regarding the discordant results between EZH2 and SUZ12, whether the detected interactions were direct or indirect, the inability of the method to distinguish RNA-strandedness, whether G-tract sequences were the most enriched sequences at PRC2 RNA binding sites, and the locations of these sequences within RNAs. Our measurements of PRC2 RNA crosslinking in cells at single-nucleotide resolution reveals that G-tract sequences with the potential to form G4 structures are the preferred RNA binding sites for PRC2 in cells and that these are predominantly localised just downstream of the first 5’ splice site. The concentration of these sequences at the 5’ end of nascent RNAs, in close proximity to the site of PRC2 binding on chromatin, may aid the removal of PRC2 from genes. However, although PRC2 displays a preference for G-tract RNAs, it can bind other RNA sequences in vitro and in cells and thus other nascent RNA elements may also be able to remove PRC2 from chromatin, albeit less efficiently. Additional studies will be required to determine whether G-tract sequences located near the 5’ end of RNAs are required for the removal of PRC2 from chromatin. Such experiments will need to avoid disrupting the functions of G-tract sequences in splicing 55,56 and the function of the corresponding DNA sequences as transcription factor and PRC2 and PRC1 binding sites within CGI promoters.
PRC2, augmented by the accessory subunits PHF1, MTF2 or PHF19 (in PRC2.1) or JARID2 and AEBP2 (in PRC2.2), forms multivalent interactions with the nucleosome core, modified histone tails and DNA 2. Recent structural analysis showed the details of the interaction between the catalytic EZH2 SET domain and the substrate nucleosome and between EED and the K27-methylated nucleosome 57, whilst the non-catalytic lobe of PRC2 has recently been shown to cooperate with AEBP2 and JARID2 to form a further nucleosome interaction surface 58. Consistent with previous results 34,39, we found that the minimal catalytically active PRC2 core (EZH2, EED and the SUZ12 VEFS domain) binds RNA and does so preferentially in conditions favoring G4 formation. We therefore focused on potential antagonism between G4 RNA and the substrate core nucleosome particle for binding to the PRC2 catalytic core. Using a well-defined system consisting of the PRC2 catalytic core and an obligate substrate nucleosome reconstituted with 147 bp DNA, we found that G4 RNA blocks the binding of the PRC2 catalytic core to the substrate core nucleosome particle. Significantly, titration of G4 RNA disrupted a preformed complex of the PRC2 catalytic core and the substrate nucleosome, which is consistent with our finding that chromatin-associated G-tract RNA evicts PRC2 from chromatin in cells. The competitive effect of RNA on PRC2 nucleosome binding in nuclear extracts was also unaffected by the absence of PRC2 accessory factors. These experiments demonstrate that G4 RNA blocks a fundamental aspect of PRC2 function that is common to both PRC2.1 and PRC2.2. Other PRC2 RNA binding surfaces have been identified in JARID2 36,37,59 and AEBP2 35 in PRC2.2 and RNA blocks binding of PRC2.2 to histone-free DNA 36 and to non-histone substrates 39. Thus, further studies will be required to determine whether additional PRC2-chromatin interactions are also antagonized by G4 RNA in cells.
Our results show that chromatin-associated G-tract RNA can remove PRC2 from chromatin and deplete repressive chromatin modification. Other studies have also demonstrated a role for nascent pre-mRNA in countering the function of negatively-acting chromatin regulators. For example, nascent pre-mRNA interacts with DNMT1 and RNA blocks DNMT1 activity 60. On the other hand, nascent pre-mRNA promotes the interaction of positively-acting regulators with chromatin, including the transcription factor YY1 52 and the histone methyltransferases Set1 and Set2 61. Unspliced, chromatin-associated RNA has also been found to promote HNRNPU oligomerisation and chromatin decompaction 62. Together with the data shown here, these studies argue that nascent pre-mRNA is not merely a passive intermediary but plays a direct role in altering chromatin state to promote its own production 1.
By showing that tethered G-tract RNA removes PRC2 from chromatin, we have discovered a means to selectively remove PRC2 from specifically targeted genes. PRC2 removal can have no effect on gene expression (as is the case for Adcy7, Sorcs2 and Smad6) or can induce gene activation (as is the case for CDKN2A), demonstrating that the requirement for PRC2 in the maintenance of gene silencing is context-dependent. Unlike small molecule inhibitors of PRC2 activity that block PRC2 function genome-wide, G-tract RNA tethering allows the selective reversal of deleterious PRC2 recruitment events, which, in the case of CDKN2A, allows gene activation and the induction of senescence. This ability to reverse PRC2 recruitment at specific genes may also allow the identification of individual gene silencing events critical for oncogenesis. Other methods, such as CRISPR activation 63, enable targeted transcriptional activation of specific genes. However, the ectopic recruitment of activators results in non-physiological levels of expression that are no longer responsive to endogenous regulatory cues. In contrast, G-tract RNA tethering selectively removes the repressive regulatory layer. Tethering of G-tract RNA may thus facilitate the physiological re-activation of specific polycomb target genes that are inappropriately silenced in disease.
Methods
Cell culture
E14, Ezhfl/fl, Aebp2WT/WT and Aebp2GT/GT (kind gifts from Neil Brockdorff) and Jarid2GT/GT (gift from Amanda Fisher) mouse ESCs were maintained on 0.1% gelatin in KO-DMEM, 10% FCS, 5% knockout serum replacement, non-essential amino acids, L-glutamine, 2-mercaptoethanol, penicillin-streptomycin and 1000 U/ml leukemia inhibitory factor (03-0011-100, Stemgent). Pcl2GT/GT and Pcl2WT/WT (gifts from Adrian Bracken) were maintained in GMEM with the same supplements, except with no serum replacement and replacing L-glutamine with GlutaMAX. G-401 cells were acquired from Sigma with certification from the European Collection of Authenticated Cell Cultures (ECACC) and maintained in McCoy 5A media supplemented with 10% FBS, 2 mM L-glutamine and penicillin/streptomycin. NIH-3T3 cells (gift from Bart Vanhaesebroeck) were cultured in DMEM, 10% FBS and penicillin-streptomycin. All cell lines were tested negative for mycoplasma. A NIH-3T3 cell line expressing H-RasV12 was generated by transfecting pWZL hygro H-Ras V12 (gift from Scott Lowe, Addgene plasmid # 1874 62) with Fugene HD (Promega) and selection in hygromycin (2 μg/ml). For RNA tethering, cells were transfected with pHAGE TRE dCas9 (Addgene plasmid # 50915, a gift from Rene Maehr and Scot Wolfe) and selected with 2 μg/ml G418. The dCas9 cell lines was then transfected as before with pLKO.1-puro U6 sgRNA constructs and selected with puromycin at 1 μg/ml. dCas9 expression was induced using doxycycline (2 μg/ml) for 6 days (with media changed every 2 days). For washout experiments, fresh media was added on day 6, changed every 2 days, until day 12. When indicated, cells were treated with cisplatin (Sigma) at 3.3 μM for 24 hours or with EI1 (Generon) 10 μM for 6 days
RNA tethering
Tethered sequences were placed at the 3’-end of the sgRNA sequence (taken from pLKO.1-puro U6 sgRNA), separated by a spacer. The sequences were synthesized as gBlocks (Integrated DNA Technologies (IDT)) that also comprised BfuAI-stuffer (taken from pLKO.1-puro U6 sgRNA) and a Pol III T6 terminator (sequences in Supplementary Table 2). The gBlocks were digested with AgeI and EcoRI (New England Biolabs) and ligated into pLKO.1-puro U6 sgRNA BfuAI stuffer (Addgene plasmid #50920, a gift from Rene Maehr and Scot Wolfe 64).
The single guide RNA (sgRNA) targeting sequence for Sorcs2 was previously described 41. Other sgRNAs were designed using CHOPCHOPv2 65 (sequences in Supplementary Table 2), synthesized as oligonucleotides, annealed, and inserted into the vector using the BfuAI site.
A G-rich sequence with the same G-content as the tethered G-tract sequence, appended to the Fgf11 sgRNA and a 5’ spacer sequence, was ordered as a gBlock (sequence in Supplementary Table 2), digested with NdeI and EcoRI and cloned into pLKO.1-puro U6 sgRNA BfuAI stuffer.
A G-tract RNA sequence spanning the Fgf11 exon-intron junction (chr11:69,801,412-69,801,633 in mm10), appended to the Fgf11 sgRNA and a 5’ spacer sequence, was ordered as a gBlock (sequence in Supplementary Table 2), digested with NdeI and EcoRI and cloned into pLKO.1-puro U6 sgRNA BfuAI stuffer.
Input iCLIP
iCLIP data for PRC2 (antibody to SUZ12), FUS and HNRNPC were taken from 31. For input samples, we adapted the iCLIP protocol 66 to allow measurement of background RNA crosslinking. 2.5 x 107 Ezh2fl/fl cells per sample were irradiated with 254 nm UV-C light in a Stratalinker 2400 (Stratagene). We used 0.2 J/cm2 for SUZ12 and 0.15 J/cm2 for FUS and HNRNPC inputs to match energies used previously for the respective RNPs. Cells were lysed in 50 μl of lysis buffer and treated with RNaseI and Turbo DNase (Thermo Fisher Scientific) following the standard iCLIP protocol 66. Lysates were mixed with NuPAGE loading buffer plus reducing agent and resolved on a 4-12% NuPAGE Bis-Tris gel (Invitrogen) in MOPS buffer. RNPs were transferred to a nitrocellulose membrane (Hybond, GE Healthcare) and washed twice with 1x PBS, following the standard iCLIP protocol, but without the immunoprecipitation step. Using the autoradiograph previously used for PRC2, FUS and HNRNPC iCLIPs as a mask, we extracted from the membrane RNPs at 110-130 kDa for PRC2 input samples, 70-120 kDa for FUS input samples and 55-110 kDa for HNRNPC input samples. We then treated the membrane as normal to extract the RNA. The RNA pellet was de-phosphorylated using PNK (New England Biolabs), purified and ligated to the L3 linker according to the iCLIP protocol. Library construction was performed as described 31. Libraries were quantified using the KAPA Universal Library Quantification kit. Single-end 50 bp reads were generated on a HiSeq 2500.
UV RNA immunoprecipitation (UV-RIP)
2x108 cells per UV-RIP were irradiated with 0.2 J/cm2 of 254 nm UV-C light in a Stratalinker 2400. RIP was performed as described 67 with antibodies to SUZ12 (Cell Signaling #3737), HA-dCas9 (3F10, Roche 11867423001), or non-specific IgG control (Abcam ab46540). Beads were washed 6x with cold NT2 buffer with 1M urea, pelleted and then incubated in 200 μl PK buffer (100 mM Tris-HCl pH 7.4, 50 mM NaCl, 10 mM EDTA) with 10 μl proteinase K (Roche 03115828001) for 20 mins at 1,100 rpm and 37°C. An equal volume of PK buffer containing 7M urea was added and a second incubation performed. Supernatant was collected and RNA purified by phenol/cholorform extraction.
RNA quantification
RNA was purified using TRIsure (Bioline), treated with Turbo DNase (Thermo Fisher Scientific) for 30 mins at 37°C and reverse transcribed using SuperScript III (Life Technologies) and random hexamer primers. Specific RNAs were quantified by qPCR (Applied Biosystems) using QuantiTect SYBR Green PCR Kit (Qiagen) with the primers shown in Supplementary Table 2.
Chromatin IP (ChIP)
Cells were trypsinised from the plate, washed with PBS and cross-linked with 1% formaldehyde for 20 mins. ChIP was performed as described 31, except that cells were sonicated for 5 cycles for NIH-3T3 cells or 4 cycles for G-401 cells (30 s “on”, 30 s “off”) using a Diagenode Picoruptor. ChIP was performed using antibodies to SUZ12 (Cell Signaling 3737), HA-dCas9 (3F10, Roche 11867423001), H3K27me3 (Abcam ab6002 or ab192985), H3K27ac (ab4729), H2AK119ub (CST 8240s), total H3 (Abcam #ab1791) or non-specific IgG control (Abcam ab46540). Enrichment of specific gene sequences was measured relative to input DNA by qPCR (Applied Biosystems) using QuantiTect SYBR Green PCR Kit (Qiagen) with the primers shown in Supplementary Table 2.
Histone methyltransferase assays
30 nM PRC2 (EZH2–SUZ12–EED–RBBP4 or RBBP7; Active Motif 31387) was incubated in the presence of 0.8 μM nucleosomes in 20 mM Hepes pH 7.9, 150 mM NaCl/KCl/LiCl, 20% glycerol 0.05% IGEPAL CA-630, 0.25 mM EDTA 1mM DTT, 320 μM SAM and Complete protease inhibitor for 30 mins at 25oC.
RNAs for binding experiments
G4-forming PIM1 and control ΔG4 sequences were taken from 48. Two additional control RNAs, one for which the Gs within the G4-forming sequence were mutated to non-Gs (G-to-H) and a second for which the Gs within the G4-forming sequence were mutated to non-Gs and an equal number of non-G nucleotides outside of the G4-forming sequence were mutated to Gs (G-rich), were synthesized as gBlocks (IDT, sequences in Supplementary Table 2) and cloned into pcDNA3.1. Linearized vectors were transcribed using the MAXIscript T7 Transcription Kit (Thermo Fisher Scientific) and RNA treated with Turbo DNase (Thermo Fisher Scientific). Biotin-14-CTP (19519016 Life Technologies) was added in a 0.4:1 ratio relative to CTP. RNA integrity was verified by polyacrylamide gel electrophoresis. G4 structure formation was confirmed using a reverse transcriptase stalling assay 68. [rG4rA4]5, 5’-biotinylated-[rG4rA4]5, [rGrA]20 and 5’-biotinylated-[rGrA]20 40-mer RNA oligonucleotides were obtained from IDT. Native gel electrophoresis to measure formation of secondary structure was performed as described 35. RNA was folded either as described 35 or in pull-down buffer to confirm maintenance of RNA structure during PRC2 pull-down assays. Radiolabeled RNA was visualised using a Typhoon phosphorimager (GE) and ImageQuantTL (GE).
RNA pull-downs
Biotinylated RNAs were incubated in pull-down buffer containing 10 mM HEPES pH 7.9, 150 mM KCl or LiCl, 0.25 mM EDTA (pH 8.0), 1 mM DTT, 5% Glycerol, 0.05% IGEPAL CA-630, 33 ng/μl BSA, RNaseOUT (Invitrogen) and Complete protease inhibitor and G4 formation promoted by heating to 95°C before cooling on ice and incubation at 37°C for 30 mins. 500, 50 or 5 ng/μl folded biotinylated-RNA was bound to MyOne Streptavidin T1 Dynabeads (Thermo Fisher Scientific) for 1 hr at 4°C, washed, and then incubated with 1.5 ng/μl of recombinant PRC2 (Active Motif 31387) for 3 hrs at 4oC. Beads were washed 3x with binding buffer and then resuspended in NuPAGE loading buffer. In vitro transcribed biotinylated PIM1 or PIM1-ΔG4 RNA were folded in pull-down buffer containing NaCl and G4 formation was promoted as above. RNA was bound to MyOne Streptavidin T1 Dynabeads, added to 0.1 μg/μl ESC nuclear extract, prepared as described 69, and the pull-down allowed to proceed as above.
Nucleosome pull-downs
Recombinant human histones were expressed in E. coli and purified as described 69. Nucleosomes were assembled by salt deposition dialysis using a biotinylated 601 sequence-containing 185 bp DNA fragment, as described 69. 50 nM nucleosomes were incubated with 1.5 ng/μl recombinant PRC2 (Active Motif 31387), 10 μl MyOne Streptavidin T1 Dynabeads (Thermo scientific) and 200, 20 or 2 ng/μl of pre-folded RNA, in pull-down buffer (10 mM HEPES pH 7.9, 150 mM LiCl or KCl, 0.25 mM EDTA (pH 8.0), 1 mM DTT, 5% Glycerol, 0.05%, IGEPAL CA-630, 320 μM SAM, 33 ng/μl BSA, and Complete protease inhibitor) for 3 hrs at 4oC. Beads were washed 3x at 4oC in Li+ or K+ pull-down buffer supplemented with 1M urea. For nucleosome pull-downs using nuclear extract, 50 nM nucleosomes were incubated with 0.2 μg/μl mESC nuclear extract, in nucleosome pull-down buffer containing NaCl instead of KCl or LiCl.
When measuring the effect of linker DNA, in order to ensure pull-down of PRC2 binding to intact nucleosomes and not to any potential free DNA, we used nucleosomes containing biotinylated H2A (Abcam ab200286) assembled using 147 bp or 185 bp non-biotinylated 601 sequence-containing DNA. Beads were washed twice in pull-down buffer with 1 M NaCl and then twice in pull-down buffer with 150 mM NaCl. Samples were then resuspended in 1x LDS buffer (Thermo Fisher), heated, spun-down before resolution by SDS-PAGE.
Immunoblotting
Immunoblotting was performed for SUZ12 (Santa Cruz sc-46264), EZH2 (CST 3147), JARID2 (CST 13594), AEBP2 (CST 14129), PCL2 (Proteintech 16208-1-AP), p16INK4a (Santa Cruz sc-56330), ACTB (CST 4967), HMGN1 (Bethyl Laboratories A302-363A), HRAS G12V (D2H12, CST 14412), H3K27me1 (Abcam 61015) and H3 (Abcam ab1791). Proteins were visualised using Amersham ECL Western Blotting Detection Reagent (GE) and detected using an ImageQuantLAS 4000 imager and ImageQuantTL (GE). Contrast and brightness was altered in a linear fashion equally across the whole image. The main figures present cropped images; uncropped images are presented in Supplementary Data Set 1.
Recombinant protein production
PRC2 core complex (EZH2–EED–SUZ12 VEFS domain) was purified as described 49. Yeast histone octamer containing wild-type H3 or H3 with the K27M mutation was expressed in E. coli and purified using a two-step method as described 70. To label the octamer, the mutation K18C was introduced to histone H3 and the fluorophore (7-Diethylamino-3-((((2-Maleimidyl)ethyl)amino)carbonyl)coumarin (MDCC)) attached mixing 40 μM octamer with 200 μM MDCC under non-reducing conditions for 30 mins in the dark, after which labeled nucleosomes were purified using a PD10 column (GE Healthcare). The completeness of the labelling reaction was verified by mass spectrometry. Nucleosomes were reconstituted with 147 bp DNA containing the Widom 601 sequence using standard procedure 71.
Fluorescence binding experiments
Direct binding between RNA and PRC2 was analysed by fluorescence anisotropy using a fluorescein-labeled (G4A4)4 RNA (AAAAAAGGGGAAAAGGGGAAAAGGGGAAAAGGGGAAAAAA) or a 28 nt G4-forming portion of PIM1 RNA (ATCCCGGGGGUGGGGGGUGGGGGUGGGU). RNA was heated to 95°C in 100 mM KCl or LiCl, cooled on ice and incubated at 37°C. All binding experiments were performed at 20°C, and fluorescence measured on a Jasco FP-8500 spectrofluorometer with excitation at 495 nm and emission at 525 nm. PRC2 was titrated into 20 nM labeled RNA in assay buffer (50 mM Tris-HCl pH 7.5, 0.01 % Brij-35, 400 μM SAM) with either 100 mM KCl or 100 mM LiCl. Fluorescence anisotropy data were analysed using GraphPad Prism (GraphPad Software, USA) and DynaFit (BioKin Ltd).
The binding of PRC2 to nucleosomes were performed using fluorescence intensity titrations utilising MDCC-labeled nucleosome (excitation 430 nm, emission 476 nm). PRC2 was titrated into 10 nM labeled nucleosomes in 25 mM Tris-HCl pH 7.5, 40 mM KCl, 0.01 % Brij-35, 10 μM BSA, and 400 μM SAM. Binding was indicated by a decrease in fluorescence intensity. For the competition experiment, PRC2 was titrated into labeled nucleosomes in the presence of 500 nM unlabelled (G4A4)4 RNA, PIM1 G4 RNA (CGGGGGUGGGGGGUGGGGGUGGGU) or a control non-G4-forming portion of PIM1 RNA (GAGUUCUGCUGAAUGCCGCGAAGAU) using the buffer conditions detailed above. For the PRC2 eviction experiment, the PRC2-nucleosome complex was pre-formed by mixing 50 nM PRC2 and 10 nM MDCC-labeled nucleosomes and then a titration performed with either (G4A4)4, A40, PIM1 G4 or PIM1 non-G4-forming control RNA. Binding affinities were determined in DynaFit (BioKin Ltd) by applying a simple 1:1 binding model.
Measuring cell senescence
The proportion of senescent cells was measured using the Senescence Assay Kit (Abcam ab228562). Cells were treated as described above, stained for 2 hours and washed according to the manufacturer’s instructions. Cells were harvested by trypsinization and signal was measured using the FL-1 channel on a Fortessa X20 flow cytometer and quantified with FlowJo (BD Biosciences).
G4 structure prediction
G4 scores were calculated across the mm9 genome using G4Hunter 44 using a 25 nt sliding window. Sequences with a G4 score above a threshold of 1.2 were selected and overlapped with splice sites defined by Ensembl 59. Profile plots represent the fraction of G4 forming sequences at each position, divided by the total number of junctions covering the position. The G4 score was smoothened over a 30 nt sliding window using the smth.gaussian function from the smoother package in R with smoother.gaussianwindow.alpha=2.3 and plotted with the ggplot2 package in R.
iCLIP data analysis
iCLIP data were processed using iCount (https://github.com/tomazc/iCount) as described 31. The unique molecular identifiers (UMIs) were registered and experimental barcodes removed before mapping the sequences to mm9 using Bowtie version 0.12.7 (command line: -v 2 -m 1 -a --best --strata) in iCount. Reads indicative of PCR duplicates (reads mapping to the same position with the same UMI) and reads aligning to multiple positions were removed. Crosslinks overlapping a RepeatMasker feature or ncRNAs under 200 nt in length or annotated as a snoRNA were also removed 31. High-confidence crosslink sites were identified using the peaks function in iCount (FDR<0.05), using the RegionsAsOne setting and with a 50 nt flank 43. These were then used as input into the iCount k-mers function. The frequencies of all possible 8-mers were calculated for a -30 to +30 nt region around each crosslink site. Enrichment of each k-mer was calculated in iCount as the actual frequency (ftrue) relative to the average frequency in a set of 100 random permutations (frandom_avg) and expressed as a z-score Z=(ftrue–frandom_avg)/σrandom 42. The enrichment of G-rich sequences was also observed if the 20 nt region around the crosslink site was masked (and thus was not an artifact of crosslinking).
Crosslink sites were assigned to the nearest splice site junction by iCount (Ensembl59 annotation). First exon-intron junctions were defined as those both annotated by Ensembl59 and from the de novo transcript assembly obtained from mouse ESC total RNA-seq data (GSM1632634, GSM16326345, GSM16326346) 31 using Cufflinks and Cuffmerge 72. First exon-intron junctions with a predicted G4-forming RNA structure (G4Hunter 1.75 threshold) within -30 to +300 nt around the first 5´ splice site were identified (942 junctions). Junctions were classified as non-G4 forming if no G4 structures were predicted by G4Hunter above a threshold of 1 and there were no G4 regular expression matches ((G[2,20] N[1,7])4-20) -300 to +300 nt around the splice site (760 junctions). The number of crosslink sites at each position were normalized by the total number of exons or introns at that position and by the total number of crosslink sites in the dataset multiplied by 10^9. The data points were smoothened over a 30 nt sliding window as above. To normalise the crosslink density for G content, the G frequency at each position was calculated for both groups, and the crosslink density for the non-G4 group divided by the non-G4/G4 G-frequency ratio. The number of crosslink sites per 5 nt window was displayed using the heatmap.2 function from the gplots package in R.
Characteristics of G4-forming sequences
The number of G-tracts in each sequence, the number of Gs within each G-tract, the number of nucleotides in the loops, the base composition within the loops, and the position of the crosslinked Gs within G-tracts, were calculated using custom scripts and plotted in R. The expression level of the genes (RPKM) in each group was obtained from total RNA-seq data 31 and log10 transformed.
Alternative splicing
RNA-seq data from 40 (WT ESC: GSM1399452, GSM1399453, GSM1399454 and SUZ12-/- ESC: GSM1399458, GSM1399459, GSM1399460) were filtered to remove adapters and low-quality bases as before. Reads were then trimmed to a uniform length of 40 nt and aligned to mm9 using TopHat2 73 with default parameters. Insert size mean and SD were calculated using Picard. Splicing events were defined using MISO 74 (http://genes.mit.edu/burgelab/miso/). Alternative splicing events were filtered using the following thresholds: num-inc 1 --num-exc 1 --num-sum-inc-exc 10 --delta-psi 0.20 --bayes-factor 2. For comparison, the number of alternative splicing events that occur during differentiation of ESC to neural precursor cells was calculated using MISO with the same thresholds using data from 75 ESC: GSM1180294 & GSM1180295; NPC day 3: GSM1184609 & GSM1184610).
Statistical analysis
The significance of the increase in the crosslink site density across the set of G4-forming first exon-intron junctions (n=942) versus the set of non-G4-forming first exon-intron junctions (n=760) was estimated using a Wilcoxon rank-sum test. The significance of the decrease in the number of G-tracts per crosslinked, predicted G4 versus non-crosslinked, predicted G4 was estimated using a Wilcoxon rank-sum test. Measurements of PRC2 RNA or nucleosome binding were performed in triplicate and data plotted in GraphPad Prism (GraphPad Software, USA) with error bars representing the standard error of the mean. The significance of changes in HA-dCas9, SUZ12, H3K27me3, H3K27ac and H2AK119ub occupancy after addition of dox relative to untreated cells was estimated using an unpaired one-tailed Welch’s t-test (n=3 independent dox treatments and ChIP experiments). The significance of changes in HA-dCas9, SUZ12 and H3K27me3 occupancy after washout of dox relative to dox-treated cells was estimated using an unpaired one-tailed Welch’s t-test (n=3 independent treatments and ChIP experiments). The significance of changes in SUZ12 and H3K27me3 occupancy of genes in Ras-expressing NIH-3T3 cells versus parental cells was estimated using an unpaired one-tailed Welch’s t-test (n=3 independent ChIPs). The significance of changes in SUZ12 binding to RNA in Ras-expressing NIH-3T3 cells versus parental cells was estimated using an unpaired one-tailed Welch’s t-test (n=3 independent UV-RIPs). The significance of the difference in gene expression between Ras-expressing NIH-3T3 cells versus parental cells and for G-401 cells treated with dox or with cisplatin was estimated using a one-tailed paired Student’s t-test (n=3 independent RNA purifications). The significance of the difference in the proportion of senescent cells with and without treatment with dox, cisplatin or EI1 was estimated using a one-tailed paired Student’s t-test (n=3 independent treatments). A confidence interval of 95% was used to assess significance. A normal distribution was assumed for all populations subjected to t-tests. Supplementary Data Set 2 contains t-statistics, confidence intervals, effect sizes, degrees of freedom and p-values for all t-tests.
Supplementary Material
Acknowledgements
We thank the UCL Cancer Institute Genomics Core Facility and Bill Lyons Informatics Centre, both supported by the Cancer Research UK– UCL Centre (award C416/A18088). We would like to thank A. Bracken (Trinity College Dublin), N. Brockdorff (University of Oxford), A. Fisher (London Institute for Medical Sciences) and B. Vanhaesebroeck (UCL) for cell lines. Thanks also to I. Ruiz de los Mozos and J. Ule for assistance with iCount and feedback on the manuscript and to M. Vila de Mucha for assistance with flow cytometry. The research was funded by grants from the European Research Council (ERC, 311704), Worldwide Cancer Research (13-0256) and Bloodwise (18008) to RGJ, CoNaCyT (411064) to MT, ERC (309952) and the Helmholtz Society to TB, and Cancer Research UK (FC001078), Medical Research Council (FC001078) and Wellcome Trust (FC001078) grants to the Francis Crick Institute (funding NJ, SK, SJG and JRW).
Footnotes
Reporting summary statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Input iCLIP sequencing data have been deposited in the Gene Expression Omnibus (GEO) with accession code GSE120696. Previously published iCLIP sequencing data and RNA-seq data are available in GEO under accession code GSE66829. The positions of predicted G-quadruplex RNA structures and the positions of PRC2 crosslink sites around first 5' splice sites are provided in Supplementary Table 1. Supplementary Data Set 2 contains t-statistics, confidence intervals, effect sizes and degrees of freedom for all significance tests. Raw quantitative PCR data and all other data are available upon request. Requests for data and materials should be addressed to RGJ (r.jenner@ucl.ac.uk).
Author Contributions
MB co-designed and performed all experiments, unless where noted below. MT performed the nucleosome IPs with different linker DNA lengths. NJ, assisted by SK, measured competition between G4 RNA and the substrate core nucleosome particle for the PRC2 catalytic core in experiments co-designed by JRW. GK performed bioinformatics analysis, assisted by JA and RGJ. KBW helped with qRT-PCR experiments. BMF and AT produced nucleosomes. JH, TB, SJG, JRW and RGJ supervised the research. RGJ co-designed experiments and wrote the paper with help from all authors.
Competing Interests
The authors declare no competing interests.
References
- 1.Skalska L, Beltran-Nebot M, Ule J, Jenner RG. Regulatory feedback from nascent RNA to chromatin and transcription. Nat Rev Mol Cell Biol. 2017;18:331–337. doi: 10.1038/nrm.2017.12. [DOI] [PubMed] [Google Scholar]
- 2.Holoch D, Margueron R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 5.Qi W, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–5. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caganova M, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123:5009–22. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–92. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 10.Mohammad F, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 11.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 12.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–82. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg T, et al. A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood. 2014;123:3914–24. doi: 10.1182/blood-2012-12-473439. [DOI] [PubMed] [Google Scholar]
- 18.Hosogane M, Funayama R, Nishida Y, Nagashima T, Nakayama K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 2013;9:e1003698. doi: 10.1371/journal.pgen.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneda A, et al. Activation of Bmp2-Smad1 signal and its regulation by coordinated alteration of H3K27 trimethylation in Ras-induced senescence. PLoS Genet. 2011;7:e1002359. doi: 10.1371/journal.pgen.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo Y et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 21.Souroullas GP, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med. 2016;22:632–40. doi: 10.1038/nm.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–64. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries NA, et al. Prolonged Ezh2 Depletion in Glioblastoma Causes a Robust Switch in Cell Fate Resulting in Tumor Progression. Cell Rep. 2015;10:383–397. doi: 10.1016/j.celrep.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 24.Jermann P, Hoerner L, Burger L, Schubeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci U S A. 2014;111:E3415–21. doi: 10.1073/pnas.1400672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch MD, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. Embo J. 2011;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perino M, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet. 2018;50:1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- 29.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7 doi: 10.1038/ncomms13661. 13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 31.Beltran M, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–8. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long Y, et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife. 2017;6:e31558. doi: 10.7554/eLife.31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell. 2017;65:1056–1067 e5. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–85. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herzog VA, et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 2014;46:973–981. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q et al. RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat Struct Mol Biol. 2019;26:237–247. doi: 10.1038/s41594-019-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–60. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Hosogane M, Funayama R, Shirota M, Nakayama K. Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell Rep. 2016;16:696–706. doi: 10.1016/j.celrep.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 42.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedrat A, Lacroix L, Mergny JL. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–59. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–33. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engelbrecht J, Knudsen S, Brunak S. G+C-rich tract in 5' end of human introns. J Mol Biol. 1992;227:108–13. doi: 10.1016/0022-2836(92)90685-d. [DOI] [PubMed] [Google Scholar]
- 47.Takahama K, et al. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol. 2013;20:341–50. doi: 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Kwok CK, Marsico G, Sahakyan AB, Chambers VS, Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat Methods. 2016;13:841–4. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 49.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7 doi: 10.1038/ncomms11316. 11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown ZZ, et al. Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J Am Chem Soc. 2014;136:13498–501. doi: 10.1021/ja5060934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–70. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–81. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–50. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–71. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–7. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol. 2018;25:154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S, Jiao L, Shubbar M, Yang X, Liu X. Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding. Mol Cell. 2018;69:840–852 e5. doi: 10.1016/j.molcel.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Ruscio A, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–6. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sayou C, et al. RNA Binding by Histone Methyltransferases Set1 and Set2. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00165-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nozawa RS, et al. SAF-A Regulates Interphase Chromosome Structure through Oligomerization with Chromatin-Associated RNAs. Cell. 2017;169:1214–1227 e18. doi: 10.1016/j.cell.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–23. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–6. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huppertz I, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–87. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bouwman RD, et al. Human immunodeficiency virus Tat associates with a specific set of cellular RNAs. Retrovirology. 2014;11:53. doi: 10.1186/1742-4690-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwok CK, Balasubramanian S. Targeted Detection of G-Quadruplexes in Cellular RNAs. Angew Chem Int Ed Engl. 2015;54:6751–4. doi: 10.1002/anie.201500891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saravanan M, et al. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proc Natl Acad Sci U S A. 2012;109:20883–8. doi: 10.1073/pnas.1214735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 72.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–15. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56:286–97. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.