Abstract
In the last few years, immunotherapy has transformed the way we treat solid tumors, including melanoma, lung, head neck, breast, renal, and bladder cancers. Durable responses and long-term survival benefit has been experienced by many cancer patients, with favorable toxicity profiles of immunotherapeutic agents relative to chemotherapy. Cures have become possible in some patients with metastatic disease. Additional approvals of immunotherapy drugs and in combination with other agents are anticipated in the near future. Multiple additional immunotherapy drugs are in earlier stages of clinical development, and their testing in additional tumor types is under way. Despite considerable early success and relatively fewer side effects, the majority of cancer patients do not respond to checkpoint inhibitors. Additionally, while the drugs are generally well tolerated, there is still the potential for significant, unpredictable and even fatal toxicity with these agents. Improved biomarkers may help to better select patients who are more likely to respond to these drugs. Two key biologically important predictive tissue biomarkers, specifically, PD-L1 and mismatch repair deficiency, have been FDA-approved in conjunction with the checkpoint inhibitor, pembrolizumab. Tumor mutation burden, another promising biomarker, is emerging in several tumor types, and may also soon receive approval. Finally, several other tissue and liquid biomarkers are emerging that could help guide single-agent immunotherapy and in combination with other agents. Of these, one promising investigational biomarker is alteration or deficiency in DNA damage response (DDR) pathways, with altered DDR observed in a broad spectrum of tumors. Here, we provide a critical overview of current, emerging, and investigational biomarkers in the context of response to immunotherapy in solid tumors.
Keywords: Biomarkers, Cytotoxic T-lymphocyte antigen 4 (CTLA-4), DNA damage response (DDR), Immunotherapy, Mismatch repair deficiency (MMR), Programmed death 1 (PD-1), Tumor mutation burden (TMB)
Introduction
Cancer immunotherapy has demonstrated clear benefit in patients with a wide range of malignancies, and has revolutionized the way we approach and treat cancer. This field of immune oncology has flourished in the past few years with the rapid development and FDA-approval of several novel immunotherapy drugs. Currently available immunotherapy drugs are monoclonal antibodies targeting programmed cell death protein (PD-1), programmed cell death protein ligand 1 (PD-L1) and cytotoxic T-lymphocyte antigen 4 (CTLA-4) [1, 2]. CTLA-4 is an inhibitor of T-cell function that interacts with its ligands, such as CD80 and CD86. PD-1 is another key T-cell immune regulator belonging to the co-stimulatory receptor family CD28:7 [3]. PDL-1 ligand is found on tumor cells, while PD-1 receptors are selectively expressed on CD4+ and CD8+ T-cells, monocytes, natural killer (NK) T-cells, B cells, and dendritic cells [1, 3]. These ligand–receptor interactions can be readily blocked by CTLA-4 or PD-1/PD-L1 antibodies, respectively, that augment T-cell activation and proliferation and elicit antitumor responses [2, 4]. Conceptually, both pathways serve as negative regulators (“checkpoints”) of T-cell activation and/or function. Blockade of this checkpoint therefore reverses the negative regulation and results in immune activation.
A biomarker is defined as a measurable substance in tissue, blood, or other body fluids, and is an indicator of a clinical condition. All biomarkers should undergo thorough validation, and any biomarker intended to be used clinically should have been prospectively demonstrated to be of value in a randomized trial, or, at a minimum, prospectively defined and analyzed in real time as part of a trial. Biomarkers can be prognostic or predictive. Prognostic biomarkers correlate with the natural progression or aggressiveness of a cancer and can be quite useful for informing about the risk of recurrence or survival for particular tumor types. Predictive biomarkers can predict a response to a given treatment. Cancer biomarkers can serve as excellent surrogates for monitoring cancer response to treatments. Therefore, these are most useful when assessed before the initiation of treatment. This review will mainly emphasize predictive biomarkers across various solid tumor types, most relevant to decision-making regarding immunotherapy selection. In addition, we will briefly discuss an emerging prognostic value of tumor-infiltrating lymphocytes (TILs) assessment based on several published colon, breast cancer and melanoma studies.
At this time (March 2019), the FDA has approved six checkpoint inhibitors (CKIs): one monoclonal antibody targeting CTLA-4 pathway, ipilimumab, and six antibodies targeting PD-L/PD-L1, including atezolizumab, avelumab, durvalumab, nivolumab, cemiplimab and pembrolizumab, for the treatment of patients with multiple cancer types [1]. These approvals have dramatically changed the landscape of cancer treatment [1, 2]. However, the majority of cancer patients, including the majority with malignancies considered “sensitive’ to these agents, e.g., melanoma, non-small cell lung cancer and others, either do not respond or become resistant to these agents. Additionally, toxicity, primarily in the form of autoimmunity, can be significant and even fatal. Financial issues must also be considered, as the cost of these agents is substantial (approximately $10,000/dose in the United States). Therefore, restricting use of these agents to those most likely to benefit is of considerable interest to patients, physicians and many health care systems. Biomarkers have the potential to play a critical role in clinical practice, as they can identify the patient population appropriate for receiving a particular treatment [3, 5]. Here, we will overview clinically relevant FDA-approved immunotherapy biomarkers, such as PD-L1, and mismatch repair (MMR) deficiency, as well as emerging biomarkers, such as tumor mutation burden (TMB), TILs, and several other experimental biomarkers.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Standard Approved Biomarkers: Pd-L1 and MMR
PD-L1 as a Prognostic and Predictive Biomarker, Expression Cutoffs, Immunohistochemistry (IHC) Testing
Prognostic cancer biomarkers determine a patient’s outcomes, regardless of therapy, and are often determined by observational clinical studies. Predictive biomarkers determine the patient’s potential to respond to a specific therapy and are usually established in the context of a clinical trial [6]. Testing for clinical efficacy of a drug can be divided into two categories: companion diagnostics and complementary diagnostics. Companion diagnostics are used to identify patients who will most likely benefit from therapy, to determine which patients will most likely suffer from side effects of therapy, and are also helpful in predicting response rates. Such companion diagnostics tend to be tied to a specific drug and its licensing [7]. In contrast, complementary diagnostics may be used for patient treatment selection, but are not required to evaluate safe and effective use of the corresponding therapy in practice [8]. Due to the significance of PD-L1 pathway in the development of cancer and for drug development, the value of PD-L1 expression as a potential predictive and prognostic biomarker has been extensively studied. Some FDA-approved PD-1/PD-L1 antibodies now have companion diagnostic IHC antibody tests to detect PD-L1 expression. The efforts to incorporate these tests into clinic practice are ongoing.
Evaluation of PD-L1 expression as a biomarker dates from the initial trials with PD-1/L1-targeted therapies. In 2012, Topalian led a large pivotal phase 1 nivolumab study that enrolled patients with various advanced solid tumors. This study demonstrated single-agent PD-L1 immunotherapy activity in melanoma, renal cell carcinoma (RCC), and non-small cell lung cancer (NSCLC), while no activity was observed in metastatic castration-resistant prostate cancer and metastatic colorectal cancer (mCRC). From the patients tested, 60% of the tumors expressed > 5% PD-L1 expression (by using the IHC-5H1 antibody). The patients with > 5% PD-L1 expression had a 36% overall response rate compared to the patients with negative PD-L1 expression who had a 0% overall response rate [9].
Different PD-L1 expression cutoffs and scoring systems have been used for PD-L1-/PD-1-directed FDA-approved drugs in different trials, and are summarized in Table 1. The standard cutoff and scoring system includes tumor proportion scoring (TPS), which counts the total number of PD-L1 positive tumor cells divided by the total number of positive and negative PD-L1 tumor cells. TPS is currently the scoring system used for pembrolizumab to quantify PDL1 expression in NSCLC [10, 11]. Another potential scoring system for pembrolizumab is the combined positive score (CPS) which divides the number of PD-L1-positive cells (tumor cells, lymphocytes, macrophages) by the number of tumor cells. The CPS system is used in PD-L1 testing in metastatic gastric and gastroesophageal junctional carcinoma, in bladder cancer, and in cervical cancer to identify which patient will be more likely to benefit from pembrolizumab therapy [12, 13].
Table 1.
Key PD-L1 testing platforms and cutoffs
| Companion diagnostic antibody | Checkpoint inhibitor | Biomarker platform | Staining cutoff | Main epitope location |
|---|---|---|---|---|
| 22C3 MAb | Pembrolizumab | Dako Link48 | > 1% tumor cell | Extracellular |
| 28-8 RAb | Nivolumab | Dako Link48 | > 50% tumor cells | Extracellular |
| SP142 RAb | Atezolizumab | Ventana Benchmark |
TC3/IC3 ≥ 10% TC2/IC2 ≥ 5% TC1/IC1 ≥ 1% |
Intracellular |
| SP263 RAb | Durvalumab | Ventana Benchmark | > 25% | Extracellular |
| 73-10 RAb | Avelumab | Dako | ≥ 1% ≥ 50% ≥ 80% | Intracellular |
MAb mouse antibody, RAb rabbit antibody, TC tumor cells, IC immune cells
Clinical trials using atezolizumab reported PD-L1 expression in tumor cells (TC) and tumor-infiltrating immune cells (IC), with tumors being characterized into four different groups: TC3/IC3 (at least 10% expressing PD-L1), TC2/IC2 (at least 5% expressing PD-L1), TC1/IC1 (at least 1% expressing PD-L1), and TC0/IC0 (less than 1% expressing PD-L1) [14]. Trials using durvalumab have segregated patients into two groups based on tumor expression cutoffs: > 25% of cells expressing PD-L1 versus < 25% of cells expressing PD-L1 [14, 15]. Across clinical trials performed in advanced lung cancer patients with both the anti-PD-L1 antibodies (atezolizumab, durvalumab) and with anti-PD-1 antibodies (nivolumab, pembrolizumab), higher PD-L1 levels corresponded with improved clinical outcomes.
In summary, a wide range of commercially available PD-L1 IHC tests for routine clinical testing are currently available. These tests utilize different antibodies, different manufacturers and different cutoff scores to detect or quantify tumor PD-L1 expression. This has resulted in considerable confusion and uncertainty regarding the interpretation of various tests and their utility. Consequently, several groups have performed prospective comparisons of the different assays. As one example, the Blueprint PD-L1 IHC assay comparison study sponsored by the International Association for the Study of Lung Cancer (IASLC) evaluated four different antibody clones, namely, 22C3, 28-8, SP142, and SP263 [5]. In this study, 39 NSCLC tumors were stained with the four FDA-approved IHC assay antibody clones as listed above. Interestingly, 22C3, 28-8, and SP263 were found to be comparable to each other in the staining of tumor tissue. In contrast, SP142 antibody used in diagnostic testing for atezolizumab was found to underscore tumor tissue and overscore immune cell PD-L1 expression. It has been proposed that SP142 clone is more sensitive in detecting intracellular epitopes compared to extracellular epitopes [5]. There have been several other studies comparing the various PD-L1 assays with similar results, [16, 17]. The lack of a standardized approach to PD-L1 expression testing makes comparisons between studies extremely difficult.
PD-L1 FDA-Approved Applications in Solid Tumors
As noted above, the FDA has approved several agents concurrently with specific tests for PD-L1 expression. These approvals have varied in terms of the antibodies employed, the cutoffs and whether expression is determined on the tumor cells alone or in combination with infiltrating immune cells. The FDA defines two different types of diagnostic tests that can be used with a therapeutic agent or device [18]. A companion diagnostic is a test which provides information that is essential for the safe and effective use of a corresponding drug or biological product. A complementary diagnostic is a test that aids in the benefit–risk decision-making about the use of the therapeutic product, where the difference in benefit–risk is clinically meaningful. The information on the companion diagnostic is included in the therapeutic product labeling. Currently, the FDA does not have a formal definition for complementary diagnostics [18].
Non-small Cell Lung Cancer
The initial trials evaluating anti-PD-1/L1 agents were carried out in the setting of second-line therapy, i.e., in patients whose disease had progressed after initial platinum-based chemotherapy (Table 2). CheckMate 057 was a phase 3 study that compared nivolumab with docetaxel in patients who had failed standard platinum-based doublet therapy. This study demonstrated an increased overall survival for nivolumab compared to docetaxel [19]. PD-L1 expression, as measured by the 28-8 antibody, correlated with response to nivolumab. The patients who received nivolumab and had higher PD-L1 tumor expression exhibited an improved response rate (19% vs. 9%) compared to patients with no PD-L1 tumor expression [19]. In contrast, another phase 3 study, CheckMate 17 in squamous NSCLC, comparing docetaxel to nivolumab, again demonstrated benefit for immunotherapy over chemotherapy, but failed to show a significant correlation between PD-L1 and clinical outcomes. PD-L1 tumor expression was neither predictive nor prognostic. Among all the patients, nivolumab was superior in overall survival, response rate, and progression-free survival to docetaxel, regardless of PD-L1 levels [20].
Table 2.
Recent phase 2/3 NSCLC, urothelial cancer, melanoma studies utilizing PD-L1 as a biomarker
| Study, year | Treatment Arms | Total pts | OS, months or % OS (or PFS) | PD-L1 predicts outcome? |
|---|---|---|---|---|
| CKI alone 2nd line for NSCLC stage IV | ||||
| CheckMate 017, 2015 | Nivolumab vs. docetaxel | 272 | 9.2 vs. 6.0 | No |
| CheckMate 057, 2015 | Nivolumab vs. docetaxel | 582 | 12.2 vs. 9.4 | Yes |
| Keynote 010, 2016 | Pembrolizumab vs. docetaxel | 1033 | 10.4 vs. 8.5 | Yes |
| OAK, 2017 | Atezolizumab vs. docetaxel | 850 | 13.8 vs. 9.6 | Yes |
| CKI alone 1st line for NSCLC stage IV | ||||
| Keynote 024, 2016 | Pembrolizumab vs. PltD | 305 | 30.2 vs. 14.2 | Yes |
| CheckMate 026, 2017 | Nivolumab vs. PltD | 541 | 13.2 vs. 14.4 | No |
| CKI with chemotherapy and/or bevacizumab for stage IV NSCLC in 1st line | ||||
| Keynote 189, 2018 | Pembrolizumab + PltD vs. PltD | 616 | 69% vs. 49% at 12 months | Yes |
| Keynote 407, 2018 | Pembrolizumab + PltD vs. PltD | 559 | 15.9 vs. 11.3 | No |
| IMpower 131, 2018 | Atezolizumab + PltD vs. PltD | 1021 | 12-months PFS 25% vs. 12% | No |
| IMpower 150, 2018 | Atezolizumab + bevacizumab + PltD vs. bevacizumab + PltD | 692 | Median OS, 19 vs. 15 months | No |
| CKI after initial chemoradiation for NSCLC stage III | ||||
| PACIFIC, 2017 | chemoXRT, followed by durvalumab × 1 year vs. observation | 709 | 66% vs. 55% at 24 months | Maybe |
| CKI after chemotherapy for metastatic urothelial carcinoma | ||||
| Phase 2 study, 2016 | Atezolizumab after Plt | 315 | 11.4 months in IC2/3; 8.8 months in IC1/2/3; 7.9 in all patients | Yes |
| CKI alone for previously treated advanced melanoma | ||||
| Keynote-001, 2016 | Pembrolizumab | 655 | Hazard ratio 0.76 in PD-L1 + melanoma | Yes |
NSCLC non-small cell lung carcinoma, CKI checkpoint inhibitor, Pts patients, PltD platinum doublet chemotherapy
Recently, several interesting single-gene biomarkers have emerged, which are also of potential clinical use and may aid in treatment decisions with CKIs. It was observed that NSCLCs that harbor EGFR mutations or ALK rearrangements have lower PD-L1 expression and lower CD8+ T-cell tumor infiltration, which translates into decreased efficacy, and low response rates with single-agent checkpoint inhibitors [21]. Interestingly, most EGFR-mutant NSCLC patients with high PD-L1 do not respond to single-agent pembrolizumab, which implies that CKIs alone are not effective in these patients, raising questions about the biologic significance of the PD-L1 checkpoint in this patient subset, and implying that the PD-L1 IHC score is not a reliable biomarker in this population [22].
The Keynote 024 trial compared pembrolizimab to standard-of-care platinum-based chemotherapy as initial therapy in previously untreated advanced NSCLC and PD-L1 expression ≥ 50%. Patients receiving pembrolizumab had significantly longer progression-free and overall survival over patients receiving platinum-based chemotherapy [23]. Following results of the Keynote trial 024, PD-L1 was FDA-approved as the companion diagnostic for pembrolizumab use. The companion test uses the PD-L1 IHC 22C3 assay and was FDA-approved for NSCLC clinical use in stage IV NSCLC [18].
In recent first-line studies of pembrolizumab in combination with platinum doublets in advanced or metastatic NSCLC, patients with all PD-L1 expression cutoffs benefited from the addition of CKI to chemotherapy. Therefore, PD-L1 expression has limited clinical utility for patients receiving CKI and chemotherapy. Finally, PACIFIC, a randomized trial of 1-year of adjuvant durvalumab after concurrent chemoradiation for locally advanced (stage III) NSCLC, demonstrated a significant overall survival benefit with this CKI compared to observation. A post hoc analysis of PD-L1 status (informative samples were available on 74% of patients) demonstrated significant efficacy for all levels of PD-L1 expression. The exception was for PD-L1 < 1%, where there was numerical superiority, ut not statistical significance [hazard ratio (HR) 0.73, 0.48–1.11]. The major NSCLC trials in regards to the PD-L1 biomarker are summarized in Table 2.
Urothelial Cancer
The PD-L1 cutoff employing the Sp142 antibody for atezolizumab in urothelial cancers was 1% using an immune score of IC1 or greater [13]. The reported ORR is 27% and 13% in PD-L1-positive and PD-L1-negative tumors, respectively [24]. In comparison, the durvalumab cutoff for use in this malignancy is 25% for PD-L1 staining with the SP263 antibody. The ORR was 27% for PD-L1-positive tumors and 5% for PD-L1-negative tumors [25, 26]. In 2018, the FDA mandated that PD-L1 status to be determined in patients with bladder cancer for frontline drug use for cisplatin-ineligible patients receiving pembrolizumab or atezolizumab. Specifically, frontline FDA approval of these drugs is now limited to the ≥ 5% PD-L1 IC patient population with atezolizumab and the ≥ 10% CPS patient population [27].
Renal Cell Carcinoma
In 2015, the FDA approved nivolumab for treatment of RCC patients progressing on prior TKI. In the pivotal CheckMate 025 phase III study, nivolumab reduced the risk of death by 27% versus everolimus, representing a 5.4-month improvement in median overall survival (OS). However, this large phase 3 study led by Motzer et al. showed no significant differences in survival between advanced RCC patients treated with nivolumab in those who had high PD-L1 ≥ 1%vs. low expression < 1% tumor expression [28].
Melanoma
A PD-L1 (Dako 22C3) positivity cutoff of ≥ 1% was used in two recent KEYNOTE-001 and KEYNOTE-006 clinical trials in advanced melanoma [29, 30]. Daud et al. have found that the overall response rate (ORR) is 39% with pembrolizumab for PD-L1-positive tumors and the average ORR-is 10% for patients with PD-L1-negative melanoma tumors [29]. A recent phase 3 KEYNOTE-006 study analysis by PD-L1 status showed 24-month OS of 58% in PD-L1-positive tumors and 43% in PD-L1-negative tumors, superior to the respective 24-month OS with ipilimumab (45% in PD-L1-positive and 32% in PD-L1-negative tumors) [30]. Nonetheless, the FDA has not yet recommended using PD-L1 in melanoma for treatment decisions.
Triple-Negative Breast Cancer (TNBC)
In TNBC patients, both pembrolizumab and atezolizumab have recently been investigated to assess their efficacy, tolerability and side effects in several clinical trials [31]. Preliminary data from a phase 1 study on metastatic TNBC treated with atezolizumab (JAVELIN solid tumor study) showed that patients who had higher ORR also had tumors expressing high levels of PD-L1 [32]. In 2018, the Impassion130 phase 3 clinical trial investigated atezolizumab + nab-paclitaxel versus placebo + nab-paclitaxel as first-line treatment in untreated metastatic or locally advanced triple-negative breast cancer (TNBC) [33]. All patients in this study (n = 902) had prospective immunohistochemical testing of PD-L1 expression on tumor specimens (SP142 PD-L1 assay). The authors defined PD-L1-positive tumors as those with ≥ 1% of tumor area having PD-L1-expressing tumor-infiltrating immune cells. Patients were treated until disease progression or until they experienced an unacceptable level of toxicity.
Within the intention-to-treat group, atezolizumab-nab-paclitaxel treatment provided a longer median progression-free survival (PFS) [median, 7.2 months vs. 5.5 months; stratified hazard ratio for progression or death, 0.80; 95% confidence interval (CI) 0.69–0.92; P = 0.002] compared to placebo-nab-paclitaxel. The median PFS of patients treated with atezolizumab-nab-paclitaxel in the PD-L1-positive subgroup was 7.5 months versus 5.0 months (stratified hazard ratio for progression or death, 0.62; 95% CI 0.49–0.78; P < 0.001). The median OS of atezolizumab-nab-paclitaxel treated patients was 21.3 months (P = 0.08) in the PD-L1-negative subgroup and 25.0 months in the PD-L1-positive subgroup. These results demonstrated a prognostic benefit of a PD-L1-positive TNBC. The response rate to atezolizumab-nab-paclitaxel was 56.0% in the intention-to-treat group to and 58.9% in the PD-L1-positive subgroup. PD-L1-positive status produced improved outcomes in patients treated with anti-PD-L1 therapy.
In March 2019, the FDA approved atezolizumab + nab-paclitaxel for the treatment of PD-L1-positive metastatic, locally advanced, or unresectable TNBC, citing data from the Impassion130 trial [34]. FDA has also approved the Ventana PD-L1 (SP142) assay as a companion diagnostic device for selecting TNBC patients for atezolizumab.
Other Solid Tumors
The FDA approved the CPS scoring system (the number of PD-L1 staining cells including tumor cells, lymphocytes, macrophages, divided by the total number of viable tumor cells, multiplied by 100) for recurrent locally advanced or metastatic gastric and gastroesophageal junction adenocarcinoma, and cervical cancer as a companion diagnostic test [13]. The current standard cutoff for head neck squamous cell cancer (HNSCC), cervical, as well gastric cancer PD-L1 positivity is 1%. Based on recent clinical studies, the FDA has approved pembrolizumab for the treatment of patients with recurrent locally advanced or metastatic gastric or gastroesophageal junction adenocarcinoma with disease progression after two or more prior lines of therapy including fluoropyrimidine- and platinum-containing chemotherapy and if appropriate, HER2/neu-targeted therapy, for patients with tumors expressing PD-L1 [Combined Positive Score (CPS) ≥ 1]. Similarly, the FDA has approved pembrolizumab for the treatment of patients with recurrent or metastatic cervical cancer with disease progression on or after chemotherapy whose tumors express PD-L1 (CPS ≥ 1) [35, 36]. In addition, pembrolizumab as a single agent has demonstrated some activity. As an example, ORR for pembrolizumab-treated patients with advanced HNSCC PD-L1 positive and negative cancers are 22% and 4%, respectively [37, 38].
Complementary diagnostics have now also been approved by the FDA for nivolumab in melanoma and NSCLC, for atezolizumab in NSCLC and bladder cancer, and for durvalumab in urothelial cancer [17, 39].
PD-L1 IHC Tissue Testing: Limitations, Expression Heterogeneity
Limitations in using PD-L1 expression as a single predictive and prognostic biomarker for immunotherapy efficacy may include heterogenous expression of PD-L1 in tumors, intratumoral heterogeneity, differences in expression between primary and metastatic tumors and significant variability in PD-L1 antibodies and in scoring systems used [5]. In one study, PD-L1 expression of 73-paired cases of primary lung cancers and matching brain metastases was studied. The results showed a significant 14% discordance in PD-L1 expression between primary lung cancer tumor and paired brain metastasis; most discordant cases had tissues that were obtained more than 6 months apart [40]. Additional research using paired biopsies or synchronous samples are needed to clarify prevalence of intra-patient PD-L1 staining discrepancies.
Soluble PD-L1
Approximately 30% of NSCLC patients do not have adequate tissue at diagnosis for standard clinical tests [41]. Thus, there is an immediate need to develop peripheral blood-based biomarkers for standard biomarker testing. Recent studies have shown that both PD-1 and PD-L1 have soluble forms (sPD-1 and s-PD-L1 or sB7-H1) in the serum, and that their increasing levels that can be measured by enzyme-linked immunosorbent assays (ELISA) may correlate with responses to immunotherapy and survival [42–44]. Experimental approaches have speculated that sPD-1 may play an adjuvant role in enhancing T-cell responses [45, 46]. Frigola et al. reported that higher sB7-H1 or sPD-L1 were found in clear cell renal cell carcinoma patients with larger tumors, advanced stage and grade, and necrosis [47]. They also identified that exposure of CD4+ and CD8+ lymphocytes to sPD-L1 (tumor cell or immune cell) led to cell death, suggesting a mechanism to regulate immune homeostasis [48].
Current studies suggest that low levels of sPD-L1 may correlate with longer survival. These have been observed in patients with advanced NSCLC, multiple myeloma, renal cell carcinoma [49]. Zhou et al. reported that, in melanoma patients treated with ipilimumab (n = 23) or pembrolizumab (n = 35), high baseline levels of sPD-L1 correlated with progressive disease. They also reported that, after 5 months of treatment, an increase in sPD-L1 associated with partial responses in either arm [50] (Table 3).
Table 3.
Current investigational liquid biomarkers of ICB response
| Marker | Drug | Malignancy | End-point results | References |
|---|---|---|---|---|
| LDH |
Ipilimumab Pembrolizumab Nivolumab |
Melanoma |
Elevated baseline LDH = lower ORR P = 0.0292 Elevated baseline LDH = decreased response rate of 22.3, 95% CI (17.1–28.1) compared to 42.0, 95% CI (36.6–47.5) |
Diem et al. [153] Ribas et al. [8] |
| Neutrophil-lymphocyte ratio (NLR) | Nivolumab | NSCLC |
Baseline NLR > 3 shorter PFS predictive marker at 2 and 4 weeks P = 0.484 2 weeks P = 0.00528 4 weeks P = 0.00515 |
Nakaya et al. [154] |
| Ipilimumab | Melanoma |
Baseline NLR > 5 worse PFS and OS PFS P = 0.0006 OS P < 0.0001 |
Ferrucci et al. [155] | |
| Absolute eosinophil count | Pembrolizumab | Melanoma |
High count–low response rate P < 0.001 |
Weide et al. [126] |
| Ipilimumab | Melanoma |
High count–low response rate P < 0.0001 |
Ferrucci et al. [127] | |
| Monocyte count and myeloid derived suppressor cells (MDSCs) | Ipilimumab | Melanoma |
Low baseline levels show a favorable response P < 0.001 |
Martens et al. [147] |
| T-cell markers and sPD-L1 | Ipilimumab | Melanoma |
High CD4(+)CD25(+)FoxP3(+)-Treg better survival P < 0.001 |
Martens et al. [147] |
| Ipilimumab | Melanoma |
Increased baseline T-cell receptor diversity associated with improved response, no survival difference P = 0.01 |
Postow et al. [156] | |
| Nivolumab | NSCLC |
Increased SOX-2 reactive T-cells in periphery better response P = 0.04 |
Dhodapkar et al. [157] | |
|
PD-1 and PD-L1 Antibodies |
NSCLC |
Increased PD-1, Ki-67 + CD8 T-cells 4 weeks into treatment correlated with clinical benefit. P < 0.0001 |
Kamphorst et al. [158] | |
|
PD-1 and PD-L1 Antibodies |
NSCLC |
Baseline elevated PD-L1 as a poor prognostic marker P = 0.002 |
Boffa et al. [159] | |
|
PD-1 and PD-L1 Antibodies |
OSCC |
Elevated PD-L1 mRNA expression in peripheral blood could contribute to increased metastatic behavior (higher grade cancer, node positive status) P < 0.05 |
Weber et al. [160] | |
|
Ipilimumab Pembrolizumab |
Melanoma |
High pretreatment levels of sPD-L1 were associated with increased likelihood of progressive disease P = 0.0015 |
Zhou et al. [50] | |
| B cell-antibody markers | Ipilimumab | Melanoma |
NYESO antibody seropositive have better ORR P = 0.02 |
Yuan et al. [161] |
| Ipilimumab | Melanoma |
Soluble CTLA4 antibody associated with improved response P = 0.02 |
Leung et al. [162] | |
| Soluble CD25 | Ipilimumab | Melanoma |
Elevated baseline CD25 associated with shorter OS P = 0.056 |
Hannani et al. [144] |
| bTMB | Atezolizumab | NSCLC |
bTMB correlated with TMB, bTMB correlated with PFS, bTMB did not associate with high PD-L1 expression bTMB P = 0.035 PD-L1 P = 0.160 |
Gandara et al. [41] |
NSCLC non-small cell lung carcinoma, LDH lactate dehydrogenase, OSCC oral squamous cell cancer, ORR objective response rate, OS overall survival, PFS progression-free survival, bTMB blood–tumor mutational burden, sPD-L1 soluble PD-L1
Mismatch Repair Deficiency (MMR) as a Companion Diagnostic with Pembrolizumab Regardless of Tumor Type
MMR is an essential DNA repair mechanism which edits DNA mismatches, which can develop due to misincorporation of bases during DNA replication and recombination repair, or be acquired while repairing damage, such as cisplatin adducts, O6-methylguanine, 8-oxoguanine, UV photo products, etc. [51, 52]. There is evidence to suggest that, when DNA damage cannot be repaired, MMR machinery can also directly signal for cell cycle arrest and apoptosis [53]. MMR defect (dMMR) leads to an increased rate of mismatch errors, and results in microsatellite instability (MSI), which is used to clinically detect dMMR [54]. MSI is detected in about 15% of all colorectal cancers: 3% are associated with Lynch syndrome and about 12% are caused by sporadic, acquired hypermethylation of the promoter of the MLH1 gene, which occurs in tumors with the CpG island methylator phenotype [55]. MMR deficiency is also common in endometrial cancer, and other tumor types [56]. In a recent study, 42 of 149 tumor specimens exhibited loss of MMR protein by IHC. The tumors that exhibited loss of MMR protein were characterized by an increased presence of tumor-infiltrating CD8+ T-cells and high PD-L1/PD-1 expression [57].
Given that mutations have the potential to result in abnormal proteins which may be antigenic, it is not surprising that MMR deficient tumors exhibit high tumor mutation burden (TMB), neoantigen load, and T-cell infiltration, [58, 59] and respond well to immune checkpoint blockade. A landmark study in 2015 tested the activity of pembrolizumab in 41 patients with metastatic carcinomas who had either MMR-deficient or -proficient tumors. In this trial, 40% of colorectal cancer (CRC) patients with MMR-deficient tumors treated with pembrolizumab had measurable responses, compared to 0% CRC patients with MMR-proficient tumors. Similarly, CRC patients with MMR-deficient tumors had immune-related progression-free survival of 78%, compared to 11% of CRC patients with MMR-proficient tumors, at the 20-month time point. This study also established that the immune-related objective response rate and progression-free survival were similar in patients who had MMR-deficient CRC or another MMR-deficient tumor type [60]. A second study tested the activity of pembrolizumab in 12 different MMR-deficient tumor types, and showed that disease control was reached in 77% of the patients (66 of 86 patients), with 21% patients experiencing a complete response. This trial also showed objective radiographic responses in 53% of the patients (46 of 86 patients) [61]. The 12 MMR-deficient tumor types tested in this study included CRC, endometrial cancer, gastroesophageal cancer, neuroendocrine tumors, osteosarcoma, pancreatic cancer, prostate cancer, small intestine cancer, thyroid cancer, tumor of ampulla of vater, cholangiocarcinoma, and carcinoma of unknown primary.
The results from the above studies led to accelerated tissue agnostic FDA approval of pembrolizumab for patients with MMR-deficiency or MSI-high tumors in 2017 [62]. This approval is for adult and pediatric unresectable or metastatic solid tumors that have progressed after previous treatments or for CRC patients who have progressed after treatment with oxaliplatin, irinotecan and fluoropyrimidine. For the first time in its history, the FDA approved the use of a cancer treatment based solely on the genetic profile and not on the tumor type [62]. Currently, a polymerase chain reaction (PCR)-based test for MSI or an IHC-based test for MMR deficiency can identify appropriate patients for MMR-positive patient pembrolizumab treatment [12]. In clinical trials, appropriate patients have been identified using both the clinical tests, despite differences in histology. The PCR-based test tests the length of repetitive DNA which are known as microsatellites in the normal and tumor tissue (five markers of MSI are employed: BAT25, BAT26, D2S123, D5S346, and D17S250) [63, 64]. Based on this, tumors are characterized as MSI-high, MSI-low or MSI-stable (MSS). IHC testing is performed by staining for all four MMR proteins (MLH1, MSH2, MSH6, and PMS2), and reported as intact or positive staining, protein absence or negative staining, and uninterpretable [64]. Studies suggest that IHC-based and MSI-based tests are both equally sensitive, with recommendations to use IHC-testing as a first line of screening because it is inexpensive and readily available [65]. In terms of dMMR testing, both PCR and IHC have ~ 90% sensitivity; combining both tests can increase sensitivity further [56].
While the discovery of the association between MMR defects and response to immune checkpoint blockade is truly groundbreaking, only a few patients, primarily a subset of colon and endometrial cancer patients, are currently receiving the benefit of immunotherapy. More work needs to be carried out to completely understand the differences between germline and somatic MMR defects, and to identify more MMR and MMR-like tumors. This is important, because it has been observed that patients with Lynch Syndrome (germline MMR-defect) experience lower response rates when compared to patients with sporadic MMR defects. It has been speculated that a constitutional defect in MMR may regulate the immune system to be more tolerant to the tumor and thereby translate into a reduced response to immune checkpoint blockade in clinic [66]. Finally, while MMR-deficient tumors are responsive to immune checkpoint blockade, majority of colon cancers do not carry this defect. In case of CRCs, ~ 85% tumors are MMR-proficient [67]. Thus, it is essential to identify other tumor DDR defects that produce a similar phenotype as an MMR defect in order to identify suitable candidates for clinical trials with immunotherapy checkpoint inhibitors. Finally, broader application of IHC and/or PCR MMR testing across multiple tumor types and not only colon cancer is warranted based on MMR-specific FDA approval [56]. A recent study in CRC showed that Fusobacterium nucleatum, a bacterium found in the gut, regulates tumor immune response based on the tumor MSI status [68]. This recent work suggests that the relationship between the gut microbiome and immune response may be dependent on tumor characteristics (such as MSI status) (see “Microbiome”).
Tumor Mutation Burden (TMB) as an Emerging Tissue Biomarker and Tumor Neoantigen Load
Acquisition of both driver and passenger mutations in the DNA is an inherent property of tumor cells. These genetic mutations are facilitated by cancer proliferation and cancer cell selection pressures. The mutations acquired by the tumor cells quite often alter protein function and expression, resulting in the formation of neoantigens that are expressed on the surface of the tumor cells. T-cells recognize these neoantigens, eliciting an anti-tumor response. Therefore, the neoantigen load mediated by a high mutation burden plays a key role in antitumor immune responses. Highly mutated tumors are more likely to generate tumor-specific mutant epitopes, which may function as neoantigens recognized as non-self by the immune system. Therefore, increased activation of immune cells by treatment with immunotherapy may lead to improved immune-mediated tumor cell clearance and clinical responses. A significant association between neoantigen production and immune-mediated clinical response has indeed been observed in several studies [69]. However, accurate measurement of this neoantigen production is expensive and time-consuming. Most research has estimated overall neoantigen load based only on somatic nonsynonymous coding mutations, called whole-exome sequencing (WES).
In comparison with overall neoantigen load, tumor mutational burden (TMB) is a good measure for neoantigen load assessment as it is easier to measure and use in clinic. Although not all mutations can give rise to tumor immunogenic peptides, their number influences the amount of neoantigens potentially produced. Thus, TMB may serve as either a predictor of clinical benefit or a prognostic factor, with the potential to play a critical role in predicting CKI efficacy [70, 71]. TMB is an estimate of somatic mutations which can be assessed from WES data or from sequencing a select panel of genes, typically several hundred genes. TMB testing usually implies testing for a panel of cancer-related genes (usually, at least 200 genes) assessed in the entire tumor using next-generation sequencing (NGS). Clinical NGS testing platforms (Foundation Medicine, Caris Life Sciences, and others) can be utilized to estimate TMB; see Table 4 for TMB cutoffs and methodologies. The number of acquired somatic mutations that are present in the tumors can be compared to the DNA from adjacent healthy, normal tissue or the DNA from normal whole blood, although it is not mandatory.
Table 4.
TMB cutoffs and methodology
| Method of TMB detection | TMB cutoff | Malignancy | Therapy | Result | References |
|---|---|---|---|---|---|
| Whole-exome sequencing of DNA (tumors and matched normal blood) | Cut-off: number of nonsynonymous mutations high > 100 and low < 100 per tumor | Melanoma | Anti-CTLA-4 |
Increased mutational burden correlated with benefit from therapy. OS for long-term benefit 4.4 year, for minimal or no benefit 0.9 year P = 0.01 |
Snyder et al. [11] |
| Whole-exome sequencing of DNA (tumors and matched normal blood) | Cut-offs were high (> 200) and low (< 200) nonsynonymous mutation burden | NSCLC | Anti-PD-1 |
Nonsynonymous mutation burden significantly associated with clinical benefit from anti-PD-1 therapy ORR and PFS were higher in patients with high nonsynonymous burden [ORR 63% vs. 0%; median PFS 14.5 vs. 3.7 months) P = 0.03 |
Rizvi et al. [70] |
|
Whole-exome sequencing Tested nonsynonymous mutations in genes on the cancer gene panel (CGP): foundation medicine panel (FM-CGP) and institutional panel (HSL-CGP) |
Nonsynonymous mutations: high (≥ 7 for FM-CGP and ≥ 13 for HSL-CGP) and a low (< 7 for FM-CGP and < 13 for HSL-CGP) |
Melanoma NSCLC Melanoma |
Anti-PD-1 Anti-CTLA-4 |
CGP mutational load significantly associated with durable clinical benefit, PFS Median PFS 14.5 vs. 3.4 months P = 0.008 No clinical benefit with CGP-mutational load P = 0.24 |
Campesato et al. [163] |
| FoundationOne assay—hybrid capture-based next-generation sequencing (base substitutions, indels, gene rearrangements, copy number changes). TMB detected from FoundationOne assay was extrapolated to whole-exome data | Cut-off: low: < 3.3 mutations/mb intermediate: 0.3–23.1 mutations/mb high: > 23.1 mutations/mb | Melanoma | PD-1 blockade |
High mutation load was also associated with superior OS and PFS using Cox proportional hazards model, adjusted for age, gender, stage, and prior ipilimumab (high vs. low HR 0.14, for PFS; HR 0.09, for OS) P < 0.001 |
Johnson et al. [164] |
| FoundationOne assay | Low (1–5 mutations/mb), intermediate (6–19 mutations/mb), and high (≥ 20 mutations/mb) |
Melanoma NSCLC and other tumor types |
PD-1 or PD-L1 monotherapy Combination of anti-CTLA4 & anti-PD-1 therapy Anti-CTLA4 and IL2 |
The RR for patients with high (≥ 20 mutations/mb) vs. low to intermediate TMB was 22/38 (58%) vs. 23/113 (20%) (P = 0.001); median PFS, 12.8 vs. 3.3 months (P < 0.0001); median OS, not reached vs. 16.3 months (P = 0.0036). | Goodman et al. [76] |
| FoundationOne assay | TMB high: ≥ 10 mutations/mb | NSCLC (CheckMate 227 trial) |
Anti-PD-1 Anti-CTLA-4 |
Significantly longer PFS in patients with ≥ 10 mutations/mb TMB treated with anti-PD-1 and anti-CTLA-4 therapy. The 1-year PFS rate was 42.6% with anti-PD-1 and anti-CTLA-4 therapy vs. 13.2% with chemotherapy; median PFS was 7.2 months vs. 5.5 months. ORR was 45.3% with anti-PD-1 and anti-CTLA-4 therapy and 26.9% with chemotherapy P < 0.01 |
Hellmann et al. [78] |
| FoundationOne assay | Median TMB ≥ 9 mutations/mb, High TMB ≥ 13.5 mutations/mb | NSCLC | Anti-PDL1 |
Five-year RFS and OS of DEL, PM, and WT were 67.3/85.9%, 76.4/88.6%, 59.2/71.5%, respectively, and both survivals of each mutant were significantly better than those of WT P = 0.027 |
Kowanetz et al. [172] |
| TMB: total number of somatic missense mutations. Used whole-exome sequence data, and compared to FoundationOne assay profile. | Low: 0 to < 143 mutations; medium: 143–247 mutations; high: ≥ 248 mutations | SCLC |
Anti-PD-1 Anti-CTLA-4 |
Within both the nivolumab monotherapy and nivolumab plus ipilimumab treatment groups, ORR were higher in those patients with high tumor mutational burden (21.3% and 46.2%, respectively) than in patients with low (4.8% and 22.2%, respectively) or medium (6.8% and 16.0%, respectively) TMB P = not reported |
Hellmann et al. [165] |
| bTMB: hybridization-capture panel as the tumor FoundationOne TMB test | bTMB cut-points (≥ 10, ≥ 16 and ≥ 20) | NSCLC | Anti-PDL1 | Improved OS and PFS for all bTMB cut-points (P = 0.035). Cut-point ≥ 16 had stronger PFS (0.036). bTMB correlated with TMB, bTMB correlated with PFS (P = 0.013), bTMB did not associate with high PD-L1 expression | Gandara et al. [41] |
bTMB blood-tumor mutational burden, NSCLC non-small cell lung carcinoma, TMB tumor mutational burden, ORR objective response rate, OS overall survival, PFS progression-free survival, RR response rate, SCLC small cell lung carcinoma, RFS recurrence-free survival, DEL exon 19 deletions (EGRF), PM and exon 21 L858R (EGRF), WT wild type
It is not surprising that tumors harboring high TMB, such as melanoma, NSCLC, urothelial tumors and tumors with defects in DNA repair mechanisms have shown a significantly positive outcome with CKIs [11, 24, 60, 70]. These tumors tend to be associated with carcinogen exposure.
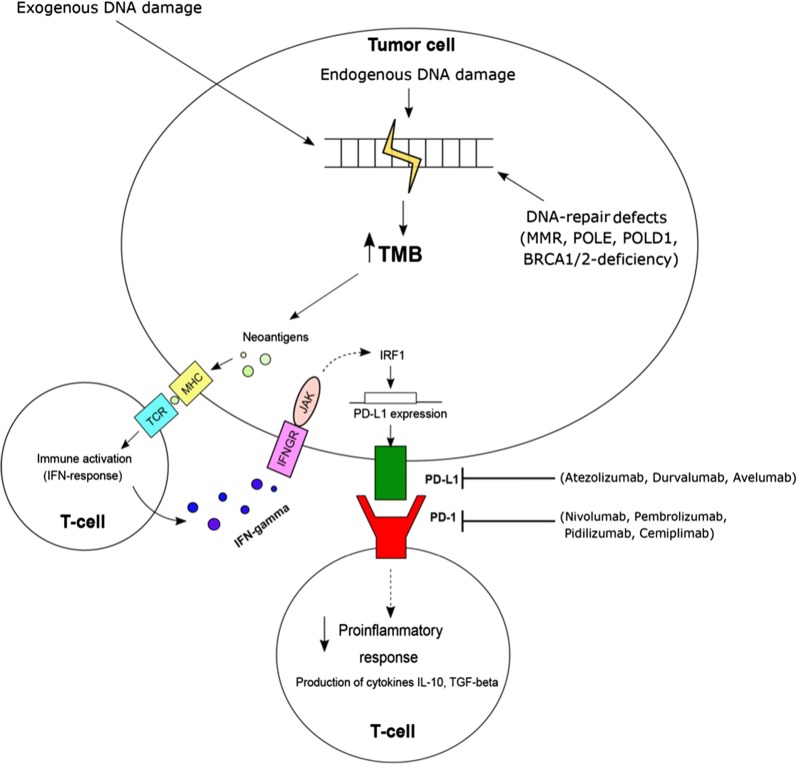
As shown in Fig. 1, high TMB increases the foreignness or neoantigen load of different tumor types. A high TMB will increase the probability of neoantigen generation; neoepitopes generated from mutated genes, when bound to MHC molecules can be seen as foreign by T-cells leading to effective antitumor immune response.
Fig. 1.
Cancer cell biomarkers and checkpoint inhibitor response. (1) Mutations in tumor cells, mostly related to smoking, generate neo-antigens, (2) neo-antigens are expressed on the cancer cell surface, (3) antigen presenting cells (APCs) recognize neo-antigens, and present them to CD8+ T-cells, inducing cytotoxic T-cell responses. (4) Cytotoxic CD8+ T-cell activation occurs, resulting in robust neo-antigen-dependent tumor cell death. Checkpoint inhibitors are effective against tumors with high PD-L1, MMR-positive tumors, or TMB-high tumors that reach a threshold for robust CD8+ cytotoxic T-cell activation. MHC major histocompatibility complex, TCR T-cell receptor. B7.1/CD80 and B7.2/CD86 are proteins expressed on APC that bind to CTLA-4 on cytotoxic CD8+ T-cells
TMB Estimation, Clinical Data and Neoantigen Load
Van Rooji et al. showed that tumor mutations may impact neoantigen and predict responses to immunotherapy in patients with melanoma. They performed whole-exome sequence analysis of the tumor and found increased mutational load (somatic mutations, nonsynonymous and synonymous mutations, frameshifts and deletions). They also established the presence of neoantigen-specific T-cell reactivity towards mutant ATR (S > L peptide) and found that 5-week after treatment with ipilimumab, there was a fivefold increase in ATR S > L peptide-specific-T-cell response [72]. In the setting of metastatic melanoma, Hugo et al. showed that a high tumor mutation load correlated with an improved OS, but not tumor response to anti-PD1 therapy. Tumors that responded had more nonsynonymous single-nucleotide variants versus tumors that did not respond; however, their data did not achieve statistical significance [73].
Rizvi et al. employing WES, has shown that a high non-synonymous mutation burden and neoantigen burden is associated with durable clinical benefit (partial or stable response lasting > 6 months) in patients with NSCLC who were treated with pembrolizumab [70]. High TMB (≥ 178 non-synonymous mutations) and neoantigen burden were both associated with significantly longer progression-free survival. In this retrospective analysis, and drug efficacy correlated with a smoking-associated mutation signature, higher neoantigen burden, and DNA repair pathway mutations; each factor was associated with high TMB. Van Allen et al. and Snyder et al. noted that nonsynonymous tumor mutational load was significantly associated with benefit from ipilimumab in the setting of metastatic melanoma [11, 74]. Both studies performed WES studies to analyze mutational loads.
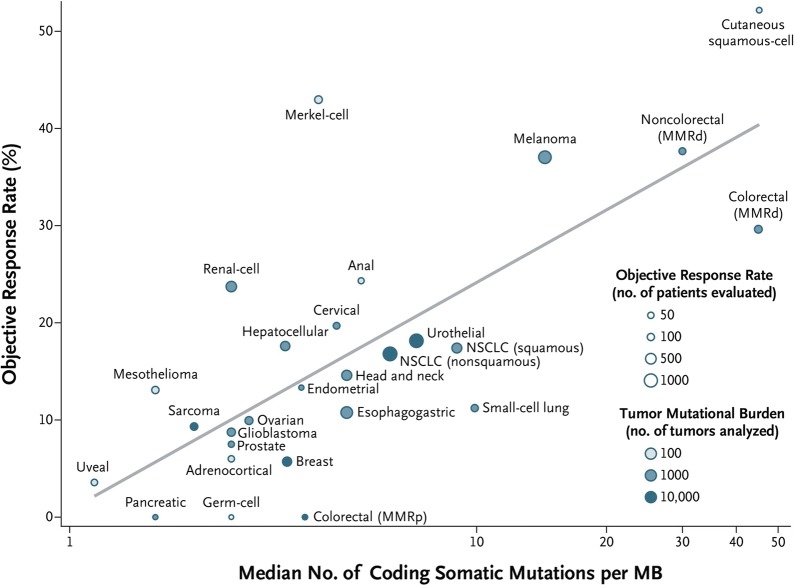
More recently, Yarchoan et al. analyzed the data from several PD-1 and PD-L1 clinical studies and found a significant correlation between TMB and the objective response rate (P < 0.001) to immunotherapy across 27 solid tumor types (Fig. 2) [75]. Here, the tumor mutation burden data was provided by Foundation Medicine profiling and a TMB profile was established for 27 solid tumor types. Some of the tumor types had a better response than would have been predicted by TMB, while others did worse. Goodman et al. included multiple cancer types in their study and observed that TMB can independently predict response to immune therapy (see Table 4 and [76]). In this study, the cancer types were divided as melanoma, NSCLC and other tumor types. Their analysis revealed that patients with an increased TMB exhibited clinical benefit for the multiple tumor types studied.
Fig. 2.
Correlation between tumor mutational burden and objective response rate with anti-PD-1 or anti-PD-L1 therapy in 27 tumor types. [Reprinted with permission from https://www.nejm.org/doi/10.1056/NEJMc1713444]. Shown are the median numbers of coding somatic mutations per megabase (MB) of DNA in 27 tumor types or subtypes among patients who received inhibitors of programmed death 1 (PD-1) protein or its ligand (PD-L1), as described in published studies for which data regarding the objective response rate are available. The number of patients who were evaluated for the objective response rate is shown for each tumor type (size of the circle), along with the number of tumor samples that were analyzed to calculate the tumor mutational burden (degree of shading of the circle). Data on the x axis are shown on a logarithmic scale. MMRd denotes mismatch repair-deficient, MMRp mismatch repair-proficient, and NSCLC non-small cell lung cancer. A significant correlation between the tumor mutational burden and the objective response rate (P < 0.001) to the IO was demonstrated by the above study [75]
Recent CheckMate 026 and CheckMate 227 NSCLC studies have shown improved outcomes in TMB-high NSCLC subgroups treated with nivolumab or nivolumab and ipilimumab combination, regardless of PD-L1 status. In CheckMate 026, the response rate was higher in the nivolumab group than in the chemotherapy group (47% vs. 28%), and median PFS was longer (median, 9.7 months vs. 5.8 months), while in the CheckMate 227, the 1-year progression-free survival rate was 42.6% with nivolumab plus ipilimumab versus 13.2% with chemotherapy [77, 78]. This is encouraging and, therefore, TMB might be considered as a NSCLC immunotherapy biomarker in the near future.
Interestingly, as predicted by Schumacher and Schreiber, melanoma and lung cancer have the highest TMB, hence exhibit the highest neoantigen load, and thus are extremely responsive to checkpoint inhibitor immunotherapies [79]. It has been shown that mutations or defects in DNA replication and repair genes, such as the MMR genes and DNA polymerase epsilon (POLE) and DNA polymerase delta 1 (POLD1), have been implicated in the generation of a high tumor mutation burden and therefore are associated with better response to immunotherapy [80, 81]. Mutations in POLD1 and POLE have been identified in patients diagnosed with NSCLC, and these patients responded well to pembrolizumab [60]. McGranahan and colleagues have also demonstrated that a high burden of clonal tumor neoantigens correlated with improved NSCLC and melanoma patient survival, an increased presence of tumor-infiltrating lymphocytes, and durable response to immunotherapy [82].
TMB Limitations, Clinical Application Using Targeted Gene Panels
Standardization of TMB as a biomarker is challenging because it is caused by so many diverse variables or insults on the tumor genome. This is discussed at length in a recent review [83]. Currently, WES is considered the gold standard for estimation of TMB [83]. Adoption of WES to routine clinical practice is difficult due to technical requirements, such as DNA amount, coverage and analysis time [83]. Recent studies suggest that TMB analyzed per megabase of genome is more relevant than the number of genes analyzed for somatic mutations [84]. There is concordance between results of targeted panels and whole-exome data when a megabase of genome is analyzed, as in the MSK-IMPACT trial [85].
Currently, TMB estimation assays are offered through different clinical vendors and need to be assessed for consistency. TMB cut-offs are currently clustered in the range of ~ 10–12 mutations per megabase, which needs to be standardized and a consistent cutoff needs to be established. As discussed above and shown in Table 4, TMB assessment through routine clinical testing, using next-generation sequencing panels, can provide robust data that correlates TMB with response to immune therapy. A recent study evaluated three commercially available NGS-targeted gene panels (that covered the genomic area of 0.39 megabase, 0.53 megabase and 1.7 megabase) with matching WES data from non-small cell lung cancer patients. The authors observed a strong correlation when they compared the whole-exome data to the data from the panel that covered > 1 megabase of the genome, strongly suggesting that the TMB results from commercial NGS panels are comparable to those obtained from WES [86].
TMB: Future Directions and Blood-Based TMB Testing Development
TMB as a companion diagnostic might receive an FDA approval in the near future in select tumor types [75, 76]. In the future, clinical trials that test TMB in multiple tumor types are needed to establish it as a biomarker of response to immune therapy. TMB as a biomarker may provide useful insight into why certain cancers respond to CKIs and can be used independently of PD-L1. TMB, alone or combined with specific tumor mutation signatures, may provide insight into why some cancers respond to CKIs and others do not. A recent study identified that the mutational signature associated with concurrent loss of POLE or POLD1 and MMR is distinct from that of POL or MMR loss alone [82]. Thus, a better understanding of mutational signatures and patterns might ultimately impact the clinical standardization of TMB as a biomarker.
Availability of tumor tissue to assess TMB, PD-L1 by IHC, tumor mutations and other biomarkers is usually limited. Researchers from Foundation Medicine (Cambridge, MA, USA) recently published a retrospective analysis of > 1000 plasma samples from previously treated NSCLC patients. In this study, they developed, tested and validated a novel blood-based TMB (bTMB) assay, which uses cell-free DNA, and reported that bTMB correlated with TMB, and progression-free survival in NSCLC patients treated with atezolizumab [41]. They also determined that a cutoff point of bTMB ≥ 16 correlated with meaningful clinical outcomes. Finally, bTMB was observed as an independent biomarker of progression-free survival and did not correlate with PD-L1 expression. Currently, to obtain a valid bTMB score requires a minimum amount of circulating DNA in the blood, with mutations at an allele frequency of ≥ 1%. Further, this assay only incorporates single nucleotide variants and not indels. Further development and validation of this bTMB assay will change the landscape of available tissue types to assess biomarkers of response to immunotherapy [41].
Investigational Tissue Biomarkers
Single Gene Mutations and Amplification as Baseline Predictors of Response to CKIs
Recently, several interesting single-gene biomarkers have emerged, which are also of potential clinical use and may aid in treatment decisions with CKIs (Table 5, see gene alterations). It was observed that NSCLCs that harbor EGFR mutations or ALK rearrangements have lower PD-L1 expression and lower CD8+ T-cell tumor infiltration, which translates into decreased efficacy and low response rates with single-agent checkpoint inhibitors [21]. Interestingly, most EGFR-mutant NSCLC patients with high PD-L1 do not respond to single-agent pembrolizumab, which implies that CKIs alone are not effective in these patients, and that the PD-L1 IHC score is a less reliable biomarker in this population [22].These recent clinical results with PD-1/PD-L1 inhibitors contradict earlier pre-clinical studies that suggested that EGFR and ALK regulate PD-L1 expression and that blocking PD-1/PD-L1 axis could be of clinical use [87, 88]. This clinical experience implies that the significance of PD-1/PD-L1 expression in driving immune escape and of EGFR- and ALK-driven NSCLC is unclear.
Table 5.
Current investigational tissue biomarkers of ICB response
| Marker | Drug | Malignancy | End-point results | References |
|---|---|---|---|---|
| Gene expression | ||||
| IFN- γ, IDO1, CXCL9 | Atezolizumab | Melanoma, NSCLC, RCC |
Pre-treatment tumors—elevated expression of IFN-γ and IFN-γ-inducible genes (e.g., IDO1 and CXCL9) for melanoma only P = 0.024 |
Herbst et al. [145] |
| CCL4, CCL5, CXCL9, CXCL10, CXCL11 | Ipilimumab | Melanoma |
High cytolytic activity, best response—correlated with high expression of such chemokines P = 2.3 × 10−52 |
Ji et al. [166] |
| PD-L1 |
Nivolumab Pembrolizumab Atezolizumab |
Melanoma, NSCLC, GU cancer |
PD-L1 expression is associated with response for these cancer types P < 0.0001 |
Carbognin et al. [167] |
| CD8, CD4, CD3, PD-1, FOXP3, LAG3 |
Nivolumab Ipilimumab |
Melanoma |
Higher level of expression of immune-related biomarkers in responders P < 0.05 |
Chen et al. [168] |
| PD-L2, CTLA-4, Granzyme A, B, Perforin-1 | Ipilimumab | Melanoma | PD-L2 (P = 0.041), CTLA-4 (P = 0.033), granzyme A, B, perforin 1 (P = 0.042)—higher expressed in responders | Van Allen et al. [74] |
| CD40, CD27, HVEM |
Nivolumab Pembrolizumab |
Melanoma |
High expression of HVEM, CD27, CD40 is associated with a better response to ICB P = 0.004 |
Auslander et al. [114] |
| Gene alterations (tumor) | ||||
| EGFR, MDM2, MDM4 | CTLA-4, PD-1/PD-L1 | Lung, bladder, breast tumors | Patients with EGFR aberrations or MDM amplifications were hyper-progressors. EGFR (P = 0.002), MDM2 (P = 0.001), MDM4 (P = 0.03) | Kato et al. [97] |
| ALK, EGFR | PD-1/PD-L1 | NSCLC | EGFR mutations or ALK rearrangements associated with low response rate. P = 0.053 | Gainor et al. [21] |
| KRAS/TP53 | Pembrolizumab | NSCLC | KRAS/TP53 mutations associated with increased expression of PD-L1, highest proportion of PD-L1+ and CD8+ T-cells, increased TMB, better clinical outcome. P < 0.001 | Dong et al. [89] |
| STK11/LKB1 in KRAS tumors | PD-1 or CTLA-4 | LUAC | Low PD-L1 expression, resistance to therapy. P < 0.001 | Skoulidis et al. [90] |
| PBRM1 | PD-1 or CTLA-4 | ccRCC | Clinical benefit for patients with PBRM1 loss of function mutations. P = 0.012 | Miao et al. [91] |
| IFN-gamma pathway genes | Ipilimumab | Melanoma | Non-responders have genomic defects in IFN-gamma genes. P = 0.015 | Gao et al. [92] |
| ATM, POLE, BRCA2, ERCC2, FANCA, MSH6 |
Nivolumab Atezolizumab |
Advanced urothelial cancers | Presence of any DDR alteration was associated with a higher response rate. P < 0.001 | Teo et al. [120] |
| JAK1, JAK2, B2M | Pembrolizumab | Melanoma | JAK1 or JAK2 and beta-2-microglobulin (B2M) truncating mutations associated with acquired resistance to PD-1 blockade | Zaretsky et al. [95] |
| MSH2, MSH6, PMS2, MLH1 | Pembrolizumab | 12 solid tumor types | Objective radiographic responses observed in 53% (95% CI 42–64%) of patients, and complete response in 21% of patients | Le et al. [61] |
| Tumor-infiltrating lymphocytes (TILs) |
Pembrolizumab Ipilimumab |
Melanoma |
High level of CD8+ TILs, expressions in the tumor and at the invasive tumor margin in responders P = 0.005 P = 0.0002 |
Hamid et al. [169] Tumeh et al. [170] |
| Ipilimumab | Melanoma | Association between clinical activity and increased TILs | Hamid et al. [169] | |
| Nivolumab |
Melanoma NSCLC RCC |
P = 0.005 Presence of TILs not sufficient to induce PD-L1 and not an independent factor associated with clinical response |
Taube et al. [171] | |
ccRCC clear cell renal cell carcinoma, LUAC lung adenocarcinoma, NSCLC non-small cell lung carcinoma, SCLC small cell lung carcinoma, RCC renal cell carcinoma, GU genitourinary cancer
In NSCLC, KRAS/TP53 mutant NSCLC subset is associated with increased expression of PD-L1 on tumor cells, increased TMB and a higher degree of tumor infiltration by CD8+ T-cells, and thereby with improved clinical outcomes. In contrast, STK11/LKB1 aberrations in KRAS-mutated lung tumors are associated with low PD-L1 expression and resistance to checkpoint inhibition [89, 90]. Recently, multiple genomic correlates have been linked to better clinical benefit with single-agent checkpoint inhibitors in a large study with melanoma and renal cancer patients [74, 91]. Thus, PBRM1 loss-of-function mutations correlated with clinical benefit from checkpoint inhibition in patients with renal cancer [91].
Additionally, genomic defects, namely, copy number alterations and mutations in IFN-γ pathway genes, are associated with primary resistance to checkpoints in melanoma [92]. In this study, 12 non-responders were tested, and, on average, non-responders were found to have ~ 15.33 mutations in IFN-gamma pathway genes versus 1 mutation in responders. Also, non-responders were found to have a higher rate of copy number alterations (namely, in IFNGR1 and 2, IRF-1, JAK-2) and amplifications (namely, in SOCS1, PIAS4) than the responders in IFN-gamma-related genes. Patients with copy number alternations in IFN-gamma-related genes exhibited a significantly shorter OS (40 months) versus patients with unaltered tumors (48.2 months; P = 0.0018). Larger prospective studies are required to validate the findings as well as standardized analysis that can allow for actual clinical use. These studies highlight the importance of INF-gamma signaling in immune checkpoint blockade [93]. While little is known about CKI resistance mechanisms, some early data are currently emerging. Mutations in JAK1, JAK2, and B2M have been reported in melanoma patients who become resistant to single-agent checkpoint inhibitors [94, 95]. Upregulation of TIM-3 inhibitory checkpoint has been associated with resistance to checkpoint inhibitors in a few NSCLC patients, and TIM-3 and PD-1 combined inhibition might be a novel treatment strategy [96].
Hyperprogression is an entity that has been recently described in which disease grows rapidly after treatment with a CKI. Kato et al. have studied tumor mutations in this setting, which were defined as patients with time-to-treatment failure (TTF) < 2 months, 50% increase in tumor burden compared with pre-treatment imaging, and > twofold increase in progression pace, using next generation sequencing [97]. They found that patients with lung, bladder, and breast tumors, who had EGFR, MDM2, and MDM4 gene amplifications exhibited tumor hyper-progression. Future research is needed to validate these findings.
Tumor Infiltrating Lymphocytes (TILs): Emerging Role as Prognostic and Predictive Biomarkers and T-Cell-Related Tissue-Based Tests
Several studies have shown that the presence of TILs may be predictive of immunotherapeutic response and prognosis in several types of cancer, including melanoma, breast cancer, and CRC. TILs are thought to represent the tumor–immune system interaction, hence the widespread interest in their potential as a biomarker for immune checkpoint inhibitor response. The presence of TILs is associated with MSI and high neoantigen load. Thus, evaluating the presence of TILs may help to identify patients who have a higher probability of benefiting from immune therapy [98]. Hamada et al. [98] employed TIME (tumor immunity in the micro-environment) classification, based on CD274 (PD-L1) expression and TILs presence, to predict response to immunotherapy. The TIME model was developed during studies of melanoma, but their study investigated its utility in CRC. Using molecular pathological epidemiology data of 812 CRC cases from the Nurses’ Health Study and the Health Professionals Follow-up Study, the authors evaluated the association between tumor characteristics and outcomes. The following characteristics were observed in TIL-positive patients: they were MSI-high, had CpG island methylator phenotype (CIMP), BRAF mutation, high neoantigen load, low levels of long interspersed nucleotide element-1 (LINE-1) hypomethylation, and nuclear CTNNB1 expression. These findings in TIL-positive patients were significant compared to TIL-negative patients (P < 0.001). Finally, the study did not find any significant association between PD-L1 expression or TIL status and overall survival (P > 0.67–1.27).
Research by Galon et al. has greatly improved our understanding of the role of TILs in adjuvant settings, predicting survival in patients with colon cancer [99, 100]. Recently, his group has developed an immunoscore that is based on density, location, phenotype and functionality of T-cells in colon cancer, which is independent of tumor size, depth and nodal status, as a prognostic tool which may have significant prognostic value [101]. Efforts are ongoing to standardize the immunoscore with immunotherapy responses, and facilitate its introduction into routine clinical practice [102, 103]. It has been proposed to add an immune-based assay to the current tumor, lymph nodes and metastasis (TNM) cancer classification system to enhance clinicians’ ability to develop effective treatment plans and provide a more accurate prognosis [103]. While many studies have utilized the presence of all subsets of lymphocytes within a tumor as a biomarker, Pagès et al. digitally quantified the density of CD3+ and CD8+ T-cells within the tumor and the invasive margin, referred to as the immunoscore. An international consortium was created to validate the consensus immunoscore in clinical practice for patients with stage I-III CRC. Following analysis of 2681 patients, the immunoscore was broken into categories based on immune-cell density: low (0–25%), intermediate (25–70%), and high (70–100%). They found that OS at 5 years in patients with a high immunoscore was 82%, compared to 77% in intermediate and 62% in low immunoscore (unadjusted HR for high vs. low immunoscore 0.53, 95% CI 0.38–0.75; P = 0.0004). Patients with a high immunoscore also had the lowest risk of recurrence at 5 years (8%), compared to patients with intermediate (18%) and low immunoscore (32%) (HR for high vs. low immunoscore 0.20, 95% CI 0.10–0.38; P < 0·0001). Data presented by the authors showed that immunoscore is predictive of prognosis in stages I–III CRC.
TILs have also been explored as a biomarker for prognosis and response rate in breast cancer, with TIL-positive status associated with better response rate to immunotherapy and prognosis in certain breast cancer subtypes [32]. Currently, TNBC and HER2-positive breast cancer subtypes have been most associated with TILs. These breast cancer subtypes typically harbor high genetic instability, which leads to high TMB and increased tumor immune response. As one recent example, among patients treated with neoadjuvant chemotherapy, 10% increase in TILs was associated with increased disease-free survival in TNBC [HR 0.93 (95% CI 0.87–0.98), P = 0.011] and in HER2-positive breast cancer [HR 0.94 (95% CI 0.89–0.99), P = 0.017], but not in HER2-negative breast cancer [HR 1.02 (95% CI 0.96–1.09), P = 0.46] [104]. Positive TILs were also associated with longer OS in TNBC [HR 0.92 (95% CI 0.86–0.99), P = 0.032]. Additional studies in other tumor types revealed that TILs may have prognostic role in other tumor types, such as triple-negative and HER2+ breast cancer and in melanoma [105–107]. High stromal TIL levels were associated with improved ORR in patients with metastatic TNBC receiving pembrolizumab (PD-1 inhibitor) in a phase II trial (KEYNOTE-086) (P = 0.496); therefore, TILs are emerging as potentially important biomarkers of prediction of response to immunotherapeutic agents in TNBC [108].
The presence of TILs in tumors is continuing to be investigated as a prognostic tool, and as a predictive tool for immunotherapeutic response in several cancers. Validating observed associations between TIL positivity and patient outcomes, and standardizing assays are necessary to implement immune-based tumor characterization in the clinic.
In addition to TILs estimation in tumors, both T-cell receptor sequencing and analysis of neoantigens has been recently emerging. In a melanoma vaccine study, the functional status of tumor peptide-specific CD8+ T-cells correlated with clinical response [109]. Furthermore, results of T-cell receptor (TCR) sequencing have also been studied as a potential marker. Cha et al. showed that CTLA-4 inhibitors lead to TCR diversification and evolution, thereby leading to improved clinical responses in patients with increased TCR heterogeneity and reduced clonal loss [110]. Patient-specific neoantigens derived from tumor sequencing and autologous T-lymphocyte recognition of these neoantigens have been studied in patients treated with ipilimumab in melanoma and pembrolizumab in NSCLC [11, 70]. In patients responding to immunotherapies, previously predicted HLA peptides were used to screen T-lymphocytes ex vivo and individual peptide sequences responsible for T-cell activation were found. The population of T-cells that were identified was a small fraction of the total T-lymphocyte population, but intriguingly, their numbers increased substantially with the duration of treatment. These studies therefore demonstrated that the neoantigen recognition by peripheral T-lymphocytes is the major mechanism of immune response against tumors. The widespread application of these techniques still needs to be standardized and validated.
Gene Expression Signatures
Gene expression signatures associated with immunotherapy responses are an area of active investigations. Both the β-catenin signaling pathway signature and the 10-gene interferon-gamma signature have been shown to be predictive of resistance and responses to checkpoint inhibitors in melanoma [111, 112]. Additionally, elevated APOBEC gene expression levels may correlate with improved overall survival in melanoma patients [113]. A recent study performed RNA profiling of melanoma, identified a novel immuno-predictive score [IMPRES], which outperformed previously published immune transcriptomic signatures, and was very significantly associated with a better response to immunotherapy [114]. These and other gene expression signatures need to be validated in independent studies, and in other tumor types.
Tumor DDR as a Biomarker
Tumor DDR defects have been known as important in determining the response to chemotherapeutic and radiation therapy (Table 5). With the FDA approval of immune checkpoint blockade for MMR-deficient tumors, other DDR defects under investigation are BRCA and DNA polymerase alterations (POLE and POLD1) in the tumor. Strickland et al. showed that high-grade serous ovarian cancers (n = 316, ovarian TCGA dataset) that have genetic alterations in BRCA 1 and 2 genes have increased neoantigen loads, compared to those with alterations in other genes involved in homologous recombination (HR). This study also showed that HR-deficient tumors when compared to HR-proficient tumors, have an increased expression of PD-1, PD-L1, CD3+ and CD8+ tumor-infiltrating lymphocytes and that these are independently associated with better overall survival [115].
Mechanistically, it was shown that PD-L1 expression is upregulated in response to double-strand break damage in cancer cell lines (U2OS, osteosarcoma; H1299, NSCLC; A549, NSCLC; DU145, prostate cancer). They further showed that PD-L1 upregulation requires the activity of the ATM/ATR and their downstream kinases and the STAT 1/STAT3/IRF1 signaling pathway [116]. It has also been shown that ATM and ATR activation on DNA damage leads to the increased levels of inflammatory cytokines, such as TNF-α and IL-6 [117, 118]. Also, activation of DNA damage response pathways can directly activate STING and IRF3, leading to host immune responses [119]. Depletion of BRCA2, Ku 70/80, PALB2 and XRCC5 in cancer cell lines (U2OS, H1299, A549, DU-145) enhanced PD-L1 expression on induction of DNA damage. Analysis of TCGA data showed that solid tumors (CRC, stomach, breast, uterine) carrying mutations in DNA repair proteins such as BRCA2, PALB2 and Ku 70/80 have higher expression of PD-L1 [116].
In a recent study in the setting of metastatic urothelial carcinoma (n = 15 patients) ,alterations in DNA damage response and repair genes were associated with an increased response to immune checkpoint blockade (atezolizumab or nivolumab). The authors performed a retrospective analysis of three trials and determined that a better response rate and overall survival were observed in patients who had tumors carrying deleterious or likely deleterious DDR mutations, when compared to patient tumors with DDR alterations of unknown significance. Patient tumors carrying DDR alterations had an overall better survival when compared to patient tumors without DDR alterations. Interestingly, the genes that were altered commonly were ATM (n = 7), POLE (n = 3), BRCA2, ERCC1, FANCA and MSH6 (n = 2 for each) [120].
In another endometrial cancer study, it was observed that POLE-mutated tumors had the highest mutational load and proportionally the highest neoantigen load, when compared to hypermutated MMR-deficient tumors and MSS tumors [121]. Immunohistochemical analysis of lung squamous cell carcinoma tissue from 41 patients showed that expression of PD-L1 correlated with nuclear staining of DNA damage response marker, phosphorylated H2AX (γ-H2AX). This suggests that increased DNA damage, which may be unrepaired damage or persistent DNA damage, may upregulate expression of PD-L1 on the tumor cells and thereby sensitize such tumors to immune checkpoint blockade [122].
A recent review summarized the close relationship between DDR and immune response [66]. This paper delineated the likely association between tumor DDR status and response to immunotherapies. Taken together, a combination of DNA damaging therapeutics or DNA repair inhibitors with immunotherapy could be a novel strategy that might benefit cancer patients [118]. Several DNA repair inhibitors, targeted at PARP (NCT02660034, NCT03308942, NCT02849496) or ATR (AZD6738, NCT02264678), are currently being investigated in clinic in combination with immune checkpoint inhibitors.
Microbiome
Microbiome can modulate immunity and affect anticancer immune responses [123]. Matson et al. and Gopalakrishnan et al. studied melanoma patients receiving PD-1 blockade and found a greater abundance of so-called good or favorable bacteria in the guts of responding patients, while nonresponders had an imbalance in gut flora composition [124, 125]. The gut microbiome has been implicated in regulating immune responses in the tumor micro-environment [68]. One species, Fusobacterium nucleatum, may play a role in CRC carcinogenesis and mediate immune response to tumors. Hamada et al. showed that F. nucleatum-mediated immune response is dependent on tumor MSI status [68]. Their interesting large colon cancer (CRC) patient study used a molecular pathological epidemiology database of 1041 CRC cases from two prospective studies (Nurses’ Health Study and Health Professionals Follow-up Study). They studied tumor molecular characteristics, such as MSI status and the presence or absence of F. nucleatum (as determined by qPCR). The authors found that MSI-high status was associated with the presence of F. nucleatum in CRC. However, F. nucleatum was negatively associated with TIL positivity in MSI-high CRC tumors [multivariable odds ratio (OR) 0.45; 95% confidence interval (CI) 0.22–0.92], but positively associated with TIL-positive tumors in MSS CRC tumors (multivariable OR 1.91; 95% CI 1.12–3.25). Likewise, the presence of F. nucleatum was negatively associated with intratumoral periglandular reaction (i.e., lymphocytic reaction in tumor stroma within the tumor) in MSI-high CRC (multivariable OR 0.43; 95% CI 0.21–0.87), but positively associated with intratumoral periglandular reaction in MSS cancer (multivariable OR 1.97, 95% CI 1.00–3.86). Overall, the results from this study suggest that F. nucleatum may suppress adaptive antitumor immune response in MSI-high CRC and promote inflammation in MSS cancer. However, this study did not investigate the potential utility of positive intratumoral F. nucleatum to predict response to immunotherapy.
The interplay between the microbiome and tumor molecular characteristics in the context of antitumor immune response requires further investigation. A relationship between the microbiome and cancer is an ongoing area of research. Several studies are exploring how the microbiome is a related to carcinogenesis, cancer treatment response, and prognosis, but much work remains to probe the role of microbiota as a cancer immunology-related prognostic and predictive biomarker.
Investigational Liquid Biomarkers
Routine Peripheral Blood Biomarkers
It has become increasingly evident that the host immune response is critical in determining the magnitude of benefit from immunotherapy. Thus, multiple studies have examined routine blood-based parameters with response to immunotherapy. These blood-based parameters include absolute neutrophil counts (ANC), absolute or relative lymphocyte counts, total leucocyte counts, neutrophil-to-lymphocyte ratio (NLR), absolute or relative eosinophil counts, relative basophils, absolute monocyte counts (AMC), serum lactate dehydrogenase (LDH), and C-reactive protein (Table 3). These markers are frequently obtained in the course of routine clinical care and do not incur any additional expense.
Elevated leukocyte dehydrogenase (LDH) levels at baseline can indicate high tumor burden, and have been shown to correlate with low response rate to immunotherapy such as pembrolizumab. The overall response rate (ORR) to pembrolizumab in melanoma was noted to be 26% for patients with high LDH levels as compared to 40% in a general study patient population. Similarly, response rates were 40% when LDH was in the normal range, 34% when LDH was up to 2 times the upper normal limit, and 8% when LDH is elevated over 2 times the upper normal limit in a first-line setting [8]. Pretreatment high eosinophil count and low neutrophil count have also been correlated with improved response to ipilimumab and pembrolziumab in melanoma patients [126, 127]. LDH and vascular endothelial growth factor (VEGF) levels have been associated with poor outcomes from chemotherapy. LDH in particular is clearly associated with inferior outcomes and is part of the testicular cancer (NSGCT) staging system and was once proposed to be added to SCLC by the SWOG [128, 129]. Soluble serum-based markers such as C-reactive protein (CRP), LDH, VEGF, soluble CD25 and others may predict immunotherapy treatment efficacy. Krajsova et al. found that advanced, refractory melanoma patients treated with ipilimumab, baseline serum LDH and CRP levels correlated with improved overall survival [130]. In another study with 95 metastatic melanoma patients treated with ipilimumab, decreasing levels of both LDH and CRP were correlated with disease control and survival [131]. Several studies have summarized that a normal baseline serum LDH associates with improved overall survival, as recently reviewed in [132].
A recent retrospective study of advanced melanoma patients (n > 209), treated with a combination of nivolumab and ipilimumab, found that baseline levels of several routine peripheral blood parameters (high relative basophils, high relative eosinophils, low absolute monocytes, low LDH, and low NLR ratio) correlated with overall survival and overall response [133].
A large study with melanoma patients (n = 616), treated with pembrolizumab, also found that increased relative lymphocyte count (RLC) at baseline was associated with improved overall survival [126]. Kuzman et al. reported that metastatic renal cell carcinoma patients (n = 81), treated with high-dose interleukin-2 (IL-2) therapy, with an objective response had a median NLR at 2.3 versus 3.4 in those without. In these patients, NLR associated significantly with overall survival [134]. A study of resectable stage IIIB/C melanoma (n = 35), treated with ipilimumab and followed at baseline, 6 weeks, and at 3, 6 and 9 months found that lower baseline levels of T-regs and myeloid-derived suppressor cells (MDSC) levels are associated with relapse-free survival [135]. If some of these basic peripheral blood markers are validated in randomized clinical trials, it may become relatively easier and more cost-effective to integrate them as biomarkers to inform responses to immunotherapy.
DDR Status of Peripheral Blood Cells
It has become increasingly clear that DDR pathways interact closely with the immune response [136]. Interestingly, a previous study has shown an inverse relationship between DNA repair status in peripheral lymphocytes and patient overall survival has been observed in NSCLC patients (n = 591) that were treated with platinum-based chemotherapy [137]. In our work, we have shown that patients with early-onset familial CRC, who carry germline alterations in DNA double-strand break repair pathways, exhibit increased levels of γ-H2AX in their primary T-cells on induction of DNA damage when compared to cancer-free healthy controls [138]. Similarly, T-cells from familial, early-onset prostate cancer patients also exhibited elevated levels of γ-H2AX and germline alterations in DNA repair pathways [139]. These studies suggest that a constitutional DNA repair defect or an individual’s innate ability to repair DNA damage might also regulate immune responses to the tumor and other tissues. Further studies are warranted to understand if DNA repair capacity can serve as possible biomarker of response to agents such as immune checkpoint blockade.
Soluble Serum-Based Biomarkers and Cytokines
High pre-treatment serum levels of VEGF have been associated with decreased overall survival in advanced melanoma patients who were treated with ipilimumab as well as in lung and several other tumors for patients treated with chemotherapy [140, 141]. Yuan et al. found that pre-treatment serum VEGF levels correlated with clinical benefit from ipilimumab in patients with metastatic melanoma. Here, a VEGF assay (from Meso Scale Discovery) was used to measure levels in the sera and a cutoff of < 43 pg/ml was associated with clinical benefit in 159 out of 176 cases [140]. In an earlier study, serum levels of VEGF were also associated with durable benefit from high-dose IL-2 therapy in patients with metastatic melanoma and renal cell carcinoma [142]. Zhao et al. studied serum cytokines and VEGF levels in 50 NSCLC cases, before and after immunotherapy. Here, high serum levels of VEGF were significantly correlated with increasing levels of IL-4, suggesting that VEGF might shift the balance towards Th2 cytokines in the serum of patients following immunotherapy [143].
In another study with melanoma patients (n = 262) treated with ipilimumab, baseline level of sCD25 was an independent predictor of overall survival, with high levels correlating with resistance to therapy [144]. Increased serum cytokine levels of interferon gamma (IFN-γ), IL-18, ITAC (IFN-γ inducible chemokine), and decreased levels of IL-6 have been associated with treatment with anti-PD-L1 antibody (MPDL3280A) [145, 146].
Emerging ELISA, Flow Cytometry, Single-Cell Cytometry Assays
Improvements in techniques such as flow cytometry are providing a better window into immune signatures of cancer patients. Multicolor flow cytometry-based approaches are now employed to determine the population and subpopulation of Tregs, T-cells, NK cells, monocytes, MDSCs and other immune cell subtypes from peripheral blood collections and can give us important insights into tumor biology and prognosis. So far, variability in study designs and assessed markers has made it difficult to combine the observations from these studies [132]. A study with advanced melanoma patients (n = 209), treated with ipilimumab, found that low LDH, AMC, MDSCs and high AEC, Treg frequency and RLC were associated with improved overall survival [147]. As another example, a recent study showed that upregulation of PD-1 on T-cells, CD14+ monocytes, and NK cells correlates with renal cell carcinoma staging [148]. The cytotoxic effector CD8+ T- and NK-cells in these patients showed significantly increased expression of perforin and granzyme B, which correlated with PD-1 expression. Strikingly, the elevated levels of all biomarkers reverted to baseline levels within weeks of primary tumor resection, indicating that circulating PD-1+ cells are supported by the tumor, possibly through expression of cytokines or PD-1 ligands in the micro-environment. Current limitations of flow cytometry-based tests application include the costs of these tests and lack of standardization or cutoffs.
B Lymphocytes, Tumor Antigens and Neoantigens
Auto-antibodies against tumor-derived antigens have been reported in multiple different malignancies such as pancreatic, ovarian and prostate cancer [149, 150] (Table 3). These auto-antibodies usually represent intracellular antigens and, thus, their role in antibody-dependent cell-mediated cytotoxicity is controversial at best. They may, however, act as surrogate markers for T-cell activation. For example, melanoma patients with NY-ESO 1-specific antibodies have shown greater clinical benefit to treatment with ipilimumab [151]. Also, in a lung cancer study, it was shown that increased B-cell and dendritic cell aggregates at the tumor site lead to an increased progression-free and overall survival in these patients post-resection [151]. B-cells thus may play a role in identification of tumor antigens and improve T-cell-specific immune surveillance. Finally, attempts are also being made to identify specific immunogenic peptides in these circulating auto-antibodies with the goal of possibly developing targeted immune therapies [150].
Conclusions
In summary, clinically useful FDA-approved and robust biomarkers, PD-L1 expression and MMR are not sufficient to predict response to CKIs for all patients with solid tumors, and nor can they estimate response or duration of response in any patient. Given the large number of novel predictive biomarkers that are in development, several vigorous criteria should be used, including association of a biomarker with clinical efficacy, especially with overall survival across several independent clinical trials, minimal complexity, high reproducibility, and minimal cost. Standard biomarker testing procedures, including assay harmonization and standardization should also be used [152]. Currently, TMB and TILs are emerging as PD-L1-independent biomarkers that are expected to receive FDA approvals and enter routine clinical practice. While some novel biomarkers show promise, and are currently being actively investigated, further careful studies are needed to avoid redundancy between biomarkers. DDR deficiency or alteration is also a promising biomarker and future studies are warranted. There is still clearly much to learn from retrospective and prospective studies evaluating the value of both approved and developmental immunotherapy biomarkers.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article. Yanis Boumber was supported by the NIH R01 CA218802, NIH R21 CA223394 grants, and V Foundation translation award program T2018-013. Sanjeevani Arora was supported by the DOD W81XWH-18-1-0148. Martin Edelman was supported by the DOD W81XWH-18-1-0196 grant. Randy W. Lesh was supported by an Alpha Omega Alpha Carolyn L. Kuckein Student Research Fellowship. All Fox Chase Cancer affiliated authors are in part supported by the NCI Core Grant, P30 CA006927, to Fox Chase Cancer Center.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Yanis Boumber has served on advisory boards of Astra Zeneca, AbbVie, and Caris Life Sciences. Hossein Borghaei has received research support for clinical trials from Millennium, Merck/Celgene, BMS/Lilly, has served on advisory board or as a consultant for BMS, Lilly, Genentech, Celgene, Pfizer, Merck, EMD-Serono, Boehringer Ingelheim, Astra Zeneca, Novartis, Genmab, Regeneron, BioNTech, Cantargia AB, Amgen, Abbvie, Axiom, PharmaMar, and on data and safety monitoring board for the University of Pennsylvania, CAR T Program. Martin J. Edelman has served on scientific advisory board for Biomarker strategies, as an advisor for Windmil Therapeutics, Syndax, on advisory boards for Armo, Bergen; data safety monitoring boards for Astra-Zeneca, Takeda, Boehringer Ingelheim and has received research funding from Apexigen, BMS, Nektar, Precision Oncology, and Merck. Sanjeevani Arora, Rodion Velichinskii, Randy W. Lesh, Usman Ali, Michal Kubiak, and Pranshu Bansal have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Abbreviations
- ALC
Absolute lymphocyte count
- CRP
C-reactive protein
- ELISA
Enzyme-linked immunosorbent assay
- HD
High-dose
- ICOS
Inducible T-cell costimulator
- IHC
Immunohistochemistry
- IL
Interleukin
- LDH
Lactate dehydrogenase
- MDSC
Myeloid-derived suppressor cell
- MMR
Mismatch repair deficiency
- NY-ESO-1
NY-esophageal cancer 1
- ORR
Overall response rate
- OS
Overall survival
- PFS
Progression-free survival
- RCC
Renal cell carcinoma
- TIL
Tumor-infiltrating lymphocyte
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.9199421.
Contributor Information
Sanjeevani Arora, Email: Sanjeevani.Arora@fccc.edu.
Yanis Boumber, Email: Yanis.Boumber@fccc.edu.
References
- 1.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal P, Osman D, Gan GN, Simon GR, Boumber Y. Recent advances in immunotherapy in metastatic NSCLC. Front Oncol. 2016;6:239. doi: 10.3389/fonc.2016.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24:26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, Richardson W, Towne P, Hanks D, Vennapusa B, Mistry A, Kalamegham R, Averbuch S, Novotny J, Rubin E, Emancipator K, McCaffery I, Williams JA, Walker J, Longshore J, Tsao MS, Kerr KM. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. 2017;12:208–222. doi: 10.1016/j.jtho.2016.11.2228. [DOI] [PubMed] [Google Scholar]
- 6.Aguiar PN, Jr, Santoro IL, Tadokoro H, de Lima Lopes G, Filardi BA, Oliveira P, Mountzios G, de Mello RA. The role of PD-L1 expression as a predictive biomarker in advanced non-small-cell lung cancer: a network meta-analysis. Immunotherapy. 2016;8:479–488. doi: 10.2217/imt-2015-0002. [DOI] [PubMed] [Google Scholar]
- 7.Lim C, Tsao MS, Le LW, Shepherd FA, Feld R, Burkes RL, Liu G, Kamel-Reid S, Hwang D, Tanguay J, da Cunha Santos G, Leighl NB. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26:1415–1421. doi: 10.1093/annonc/mdv208. [DOI] [PubMed] [Google Scholar]
- 8.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu WJ, Weber JS, Gangadhar TC, Hersey P, Dronca R, Joseph RW, Zarour H, Chmielowski B, Lawrence DP, Algazi A, Rizvi NA, Hoffner B, Mateus C, Gergich K, Lindia JA, Giannotti M, Li XN, Ebbinghaus S, Kang SP, Robert C. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315:1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 9.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butterfield LH, Ribas A, Dissette VB, Amarnani SN, Vu HT, Oseguera D, Wang HJ, Elashoff RM, McBride WH, Mukherji B, Cochran AJ, Glaspy JA, Economou JS. Determinant spreading associated with clinical response in dendritic cell-based immunotherapy for malignant melanoma. Clin Cancer Res. 2003;9:998–1008. [PubMed] [Google Scholar]
- 11.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Information on PD-L1 Expression and Testing for Oncologists. https://www.keytruda.com/hcp/gastric-adenocarcinoma/pd-l1-expression-testing/. Accessed 9 Aug 2019.
- 13.Udall M, Rizzo M, Kenny J, Doherty J, Dahm S, Robbins P, Faulkner E. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol. 2018;13:12. doi: 10.1186/s13000-018-0689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigg C, Rizvi NA. PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction? J Immunother Cancer. 2016;4:48. doi: 10.1186/s40425-016-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diggs LP, Hsueh EC. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark Res. 2017;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsao MS, Kerr KM, Kockx M, Beasley MB, Borczuk AC, Botling J, Bubendorf L, Chirieac L, Chen G, Chou TY, Chung JH, Dacic S, Lantuejoul S, Mino-Kenudson M, Moreira AL, Nicholson AG, Noguchi M, Pelosi G, Poleri C, Russell PA, Sauter J, Thunnissen E, Wistuba I, Yu H, Wynes MW, Pintilie M, Yatabe Y, Hirsch FR. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. J Thorac Oncol. 2018;13:1302–1311. doi: 10.1016/j.jtho.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, Homer R, West WW, Wu H, Roden AC, Fujimoto J, Yu H, Anders R, Kowalewski A, Rivard C, Rehman J, Batenchuk C, Burns V, Hirsch FR, Wistuba II. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol. 2017;3:1051–1058. doi: 10.1001/jamaoncol.2017.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheerens H, Malong A, Bassett K, Boyd Z, Gupta V, Harris J, Mesick C, Simnett S, Stevens H, Gilbert H, Risser P, Kalamegham R, Jordan J, Engel J, Chen S, Essioux L, Williams JA. Current status of companion and complementary diagnostics: strategic considerations for development and launch. Clin Transl Sci. 2017;10:84–92. doi: 10.1111/cts.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhaufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crino L, Blumenschein GR, Jr, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, Huynh TG, Zhao L, Fulton L, Schultz KR, Howe E, Farago AF, Sullivan RJ, Stone JR, Digumarthy S, Moran T, Hata AN, Yagi Y, Yeap BY, Engelman JA, Mino-Kenudson M. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res. 2016;22:4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T, Coluzzi P, Ledezma B, Mendenhall M, Hunt J, Wolf B, Jones B, Madrigal J, Horton J, Spiegel M, Carroll J, Gukasyan J, Williams T, Sauer L, Wells C, Hardy A, Linares P, Lim C, Ma L, Adame C, Garon EB. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor naive patients with advanced NSCLC. J Thorac Oncol. 2018;13:1138–1145. doi: 10.1016/j.jtho.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR. Investigators K-: pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massard C, Gordon MS, Sharma S, Rafii S, Wainberg ZA, Luke J, Curiel TJ, Colon-Otero G, Hamid O, Sanborn RE, O’Donnell PH, Drakaki A, Tan W, Kurland JF, Rebelatto MC, Jin X, Blake-Haskins JA, Gupta A, Segal NH. Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J Clin Oncol. 2016;34:3119–3125. doi: 10.1200/JCO.2016.67.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brower V. Anti-PD-L1 inhibitor durvalumab in bladder cancer. Lancet Oncol. 2016;17:e275. doi: 10.1016/S1470-2045(16)30242-X. [DOI] [PubMed] [Google Scholar]
- 27.Suzman DL, Agrawal S, Ning YM, Maher VE, Fernandes LL, Karuri S, Tang S, Sridhara R, Schroeder J, Goldberg KB, Ibrahim A, McKee AE, Pazdur R, Beaver JA. FDA approval summary: atezolizumab or pembrolizumab for the treatment of patients with advanced urothelial carcinoma ineligible for cisplatin-containing chemotherapy. Oncologist. 2018;24(4):563–569. doi: 10.1634/theoncologist.2018-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, Choueiri TK, Gurney H, Donskov F, Bono P, Wagstaff J, Gauler TC, Ueda T, Tomita Y, Schutz FA, Kollmannsberger C, Larkin J, Ravaud A, Simon JS, Xu LA, Waxman IM, Sharma P, CheckMate I. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daud AI, Wolchok JD, Robert C, Hwu WJ, Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, Joseph R, Gangadhar TC, Dronca R, Patnaik A, Zarour H, Roach C, Toland G, Lunceford JK, Li XN, Emancipator K, Dolled-Filhart M, Kang SP, Ebbinghaus S, Hamid O. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34:4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A, Investigators K Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 31.Katz H, Alsharedi M. Immunotherapy in triple-negative breast cancer. Med Oncol. 2017;35:13. doi: 10.1007/s12032-017-1071-6. [DOI] [PubMed] [Google Scholar]
- 32.Basile D, Pelizzari G, Vitale MG, Lisanti C, Cinausero M, Iacono D, Puglisi F. Atezolizumab for the treatment of breast cancer. Expert Opin Biol Ther. 2018;18:595–603. doi: 10.1080/14712598.2018.1469619. [DOI] [PubMed] [Google Scholar]
- 33.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA, Investigators IMT Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 34.FDA approves atezolizumab for PD-L1 positive unresectable locally advanced or metastatic triple-negative breast cancer. https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-atezolizumab-pd-l1-positive-unresectable-locally-advanced-or-metastatic-triple-negative. Accessed 9 Aug 2019. [DOI] [PubMed]
- 35.Fashoyin-Aje L, Donoghue M, Chen H, He K, Veeraraghavan J, Goldberg KB, Keegan P, McKee AE, Pazdur R. FDA approval summary: pembrolizumab for recurrent locally advanced or metastatic gastric or gastroesophageal junction adenocarcinoma expressing PD-L1. Oncologist. 2018;21(1):103–109. doi: 10.1634/theoncologist.2018-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.FDA approves pembrolizumab for advanced cervical cancer with disease progression during or after chemotherapy. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-pembrolizumab-advanced-cervical-cancer-disease-progression-during-or-after-chemotherapy. Accessed 9 Aug 2019.
- 37.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, Cheng JD, Chow LQ. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 38.Chow LQM, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, Berger R, Eder JP, Burtness B, Lee SH, Keam B, Kang H, Muro K, Weiss J, Geva R, Lin CC, Chung HC, Meister A, Dolled-Filhart M, Pathiraja K, Cheng JD, Seiwert TY. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol. 2016;34:3838–3845. doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durvalumab (Imfinzi). https://www.fda.gov/drugs/resources-information-approved-drugs/durvalumab-imfinzi. Accessed 9 Aug 2019.
- 40.Mansfield AS, Aubry MC, Moser JC, Harrington SM, Dronca RS, Park SS, Dong H. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Ann Oncol. 2016;27:1953–1958. doi: 10.1093/annonc/mdw289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G, Malboeuf C, Lieber DS, Lipson D, Silterra J, Amler L, Riehl T, Cummings CA, Hegde PS, Sandler A, Ballinger M, Fabrizio D, Mok T, Shames DS. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 42.Shi MH, Xing YF, Zhang ZL, Huang JA, Chen YJ. Effect of soluble PD-L1 released by lung cancer cells in regulating the function of T lymphocytes. Zhonghua Zhong Liu Za Zhi. 2013;35:85–88. doi: 10.3760/cma.j.issn.0253-3766.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Xing YF, Zhang ZL, Shi MH, Ma Y, Chen YJ. The level of soluble programmed death-1 in peripheral blood of patients with lung cancer and its clinical implications. Zhonghua Jie He He Hu Xi Za Zhi. 2012;35:102–106. [PubMed] [Google Scholar]
- 44.Nagato T, Ohkuri T, Ohara K, Hirata Y, Kishibe K, Komabayashi Y, Ueda S, Takahara M, Kumai T, Ishibashi K, Kosaka A, Aoki N, Oikawa K, Uno Y, Akiyama N, Sado M, Takei H, Celis E, Harabuchi Y, Kobayashi H. Programmed death-ligand 1 and its soluble form are highly expressed in nasal natural killer/T-cell lymphoma: a potential rationale for immunotherapy. Cancer Immunol Immunother. 2017;66:877–890. doi: 10.1007/s00262-017-1987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng H, Zhang GM, Xiao H, Yuan Y, Li D, Zhang H, Qiu H, He YF, Feng ZH. HSP70 vaccine in combination with gene therapy with plasmid DNA encoding sPD-1 overcomes immune resistance and suppresses the progression of pulmonary metastatic melanoma. Int J Cancer. 2006;118:2657–2664. doi: 10.1002/ijc.21795. [DOI] [PubMed] [Google Scholar]
- 46.Song MY, Park SH, Nam HJ, Choi DH, Sung YC. Enhancement of vaccine-induced primary and memory CD8(+) T-cell responses by soluble PD-1. J Immunother. 2011;34:297–306. doi: 10.1097/CJI.0b013e318210ed0e. [DOI] [PubMed] [Google Scholar]
- 47.Frigola X, Inman BA, Lohse CM, Krco CJ, Cheville JC, Thompson RH, Leibovich B, Blute ML, Dong H, Kwon ED. Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clin Cancer Res. 2011;17:1915–1923. doi: 10.1158/1078-0432.CCR-10-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frigola X, Inman BA, Krco CJ, Liu X, Harrington SM, Bulur PA, Dietz AB, Dong H, Kwon ED. Soluble B7-H1: differences in production between dendritic cells and T cells. Immunol Lett. 2012;142:78–82. doi: 10.1016/j.imlet.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X, Lang J. Soluble PD-1 and PD-L1: predictive and prognostic significance in cancer. Oncotarget. 2017;8:97671–97682. doi: 10.18632/oncotarget.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Mahoney KM, Giobbie-Hurder A, Zhao F, Lee S, Liao X, Rodig S, Li J, Wu X, Butterfield LH, Piesche M, Manos MP, Eastman LM, Dranoff G, Freeman GJ, Hodi FS. Soluble PD-L1 as a biomarker in malignant melanoma treated with checkpoint blockade. Cancer Immunol Res. 2017;5:480–492. doi: 10.1158/2326-6066.CIR-16-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 52.Arora S, Huwe PJ, Sikder R, Shah M, Browne AJ, Lesh R, Nicolas E, Deshpande S, Hall MJ, Dunbrack RL, Jr, Golemis EA. Functional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methods. Cancer Biol Ther. 2017;18:519–533. doi: 10.1080/15384047.2017.1326439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Negureanu L, Salsbury FR., Jr The molecular origin of the MMR-dependent apoptosis pathway from dynamics analysis of MutSalpha-DNA complexes. J Biomol Struct Dyn. 2012;30:347–361. doi: 10.1080/07391102.2012.680034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch HT, Jascur T, Lanspa S, Boland CR. Making sense of missense in Lynch syndrome: the clinical perspective. Cancer Prev Res (Phila) 2010;3:1371–1374. doi: 10.1158/1940-6207.CAPR-10-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(2073–87):e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratner D, Lennerz JK. Implementing keytruda/pembrolizumab testing in clinical practice. Oncologist. 2018;23:647–649. doi: 10.1634/theoncologist.2017-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita H, Nakayama K, Ishikawa M, Nakamura K, Ishibashi T, Sanuki K, Ono R, Sasamori H, Minamoto T, Iida K, Sultana R, Ishikawa N, Kyo S. Microsatellite instability is a biomarker for immune checkpoint inhibitors in endometrial cancer. Oncotarget. 2018;9:5652–5664. doi: 10.18632/oncotarget.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, Zhang M, Papadopoulos N, Kinzler KW, Vogelstein B, Sears CL, Anders RA, Pardoll DM, Housseau F. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 60.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA., Jr PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA., Jr Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type-FDA approval of pembrolizumab for mismatch repair-deficient solid cancers. JAMA Oncol. 2018;4:157–158. doi: 10.1001/jamaoncol.2017.4182. [DOI] [PubMed] [Google Scholar]
- 63.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 64.Hampel H, de la Chapelle A. How do we approach the goal of identifying everybody with Lynch syndrome? Fam Cancer. 2013;12:313–317. doi: 10.1007/s10689-013-9611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675–693. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 68.Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, Kosumi K, Masugi Y, Twombly TS, Cao Y, Song M, Liu L, da Silva A, Shi Y, Gu M, Li W, Koh H, Nosho K, Inamura K, Keum N, Wu K, Meyerhardt JA, Kostic AD, Huttenhower C, Garrett WS, Meyerson M, Giovannucci EL, Chan AT, Fuchs CS, Nishihara R, Giannakis M, Ogino S. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol Res. 2018;6:1327–1336. doi: 10.1158/2326-6066.CIR-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fancello L, Gandini S, Pelicci PG, Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer. 2019;7:183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, Nieuwland M, Stratton MR, Kerkhoven RM, Kesmir C, Haanen JB, Kvistborg P, Schumacher TN. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR, Jr, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA, CheckMate I. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, Borghaei H, Ramalingam SS, Brahmer J, Reck M, O’Byrne KJ, Geese WJ, Green G, Chang H, Szustakowski J, Bhagavatheeswaran P, Healey D, Fu Y, Nathan F, Paz-Ares L. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 80.Nicolas E, Golemis EA, Arora S. POLD1: central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene. 2016;590:128–141. doi: 10.1016/j.gene.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, Zahurancik WJ, Suo Z, Lipman T, Wimmer K, Kratz CP, Bowers DC, Laetsch TW, Dunn GP, Johanns TM, Grimmer MR, Smirnov IV, Larouche V, Samuel D, Bronsema A, Osborn M, Stearns D, Raman P, Cole KA, Storm PB, Yalon M, Opocher E, Mason G, Thomas GA, Sabel M, George B, Ziegler DS, Lindhorst S, Issai VM, Constantini S, Toledano H, Elhasid R, Farah R, Dvir R, Dirks P, Huang A, Galati MA, Chung J, Ramaswamy V, Irwin MS, Aronson M, Durno C, Taylor MD, Rechavi G, Maris JM, Bouffet E, Hawkins C, Costello JF, Meyn MS, Pursell ZF, Malkin D, Tabori U, Shlien A. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171(1042–56):e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fenizia F, Pasquale R, Roma C, Bergantino F, Iannaccone A, Normanno N. Measuring tumor mutation burden in non-small cell lung cancer: tissue versus liquid biopsy. Transl Lung Cancer Res. 2018;7:668–677. doi: 10.21037/tlcr.2018.09.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N, Hollmann T, Schalper KA, Gainor JF, Shen R, Ni A, Arbour KC, Merghoub T, Wolchok J, Snyder A, Chaft JE, Kris MG, Rudin CM, Socci ND, Berger MF, Taylor BS, Zehir A, Solit DB, Arcila ME, Ladanyi M, Riely GJ, Schultz N, Hellmann MD. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Endris V, Buchhalter I, Allgauer M, Rempel E, Lier A, Volckmar AL, Kirchner M, von Winterfeld M, Leichsenring J, Neumann O, Penzel R, Weichert W, Glimm H, Frohling S, Winter H, Herth F, Thomas M, Schirmacher P, Budczies J, Stenzinger A. Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer. 2019;144(9):2303–2312. doi: 10.1002/ijc.32002. [DOI] [PubMed] [Google Scholar]
- 87.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, Zhang Y, He X, Zhou T, Qin T, Huang Y, Yi X, Zhang L. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 88.Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, Harada T, Matsumoto K, Takayama K, Takamori S, Kage M, Hoshino T, Nakanishi Y, Okamoto I. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res. 2015;21:4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 89.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q, Yang JJ, Yang XN, Lin JX, Yan HH, Zhai HR, Yan LX, Liao RQ, Wu SP, Wu YL. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 90.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, Schrock AB, Hartmaier RJ, Trabucco SE, Gay L, Ali SM, Elvin JA, Singal G, Ross JS, Fabrizio D, Szabo PM, Chang H, Sasson A, Srinivasan S, Kirov S, Szustakowski J, Vitazka P, Edwards R, Bufill JA, Sharma N, Ou SI, Peled N, Spigel DR, Rizvi H, Aguilar EJ, Carter BW, Erasmus J, Halpenny DF, Plodkowski AJ, Long NM, Nishino M, Denning WL, Galan-Cobo A, Hamdi H, Hirz T, Tong P, Wang J, Rodriguez-Canales J, Villalobos PA, Parra ER, Kalhor N, Sholl LM, Sauter JL, Jungbluth AA, Mino-Kenudson M, Azimi R, Elamin YY, Zhang J, Leonardi GC, Jiang F, Wong KK, Lee JJ, Papadimitrakopoulou VA, Wistuba II, Miller VA, Frampton GM, Wolchok JD, Shaw AT, Janne PA, Stephens PJ, Rudin CM, Geese WJ, Albacker LA, Heymach JV. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D, Horak C, Wind-Rotolo M, Tracy A, Giannakis M, Hodi FS, Drake CG, Ball MW, Allaf ME, Snyder A, Hellmann MD, Ho T, Motzer RJ, Signoretti S, Kaelin WG, Jr, Choueiri TK, Van Allen EM. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, Chen PL, Hwu P, Allison JP, Futreal A, Wargo JA, Sharma P. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(397–404):e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ni L, Lu J. Interferon gamma in cancer immunotherapy. Cancer Med. 2018;7:4509–4516. doi: 10.1002/cam4.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, Le DT, Pardoll DM, Diaz LA, Jr, Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, Ribas A. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, Jones RE, Kulkarni MM, Kuraguchi M, Palakurthi S, Fecci PE, Johnson BE, Janne PA, Engelman JA, Gangadharan SP, Costa DB, Freeman GJ, Bueno R, Hodi FS, Dranoff G, Wong K-K, Hammerman PS. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23:4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamada T, Soong TR, Masugi Y, Kosumi K, Nowak JA, da Silva A, Mu XJ, Twombly TS, Koh H, Yang J, Song M, Liu L, Gu M, Shi Y, Nosho K, Morikawa T, Inamura K, Shukla SA, Wu CJ, Garraway LA, Zhang X, Wu K, Meyerhardt JA, Chan AT, Glickman JN, Rodig SJ, Freeman GJ, Fuchs CS, Nishihara R, Giannakis M, Ogino S. TIME (Tumor Immunity in the MicroEnvironment) classification based on tumor CD274 (PD-L1) expression status and tumor-infiltrating lymphocytes in colorectal carcinomas. Oncoimmunology. 2018;7:e1442999. doi: 10.1080/2162402X.2018.1442999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 100.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Galon J. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 101.Galon J, Pages F, Marincola FM, Thurin M, Trinchieri G, Fox BA, Gajewski TF, Ascierto PA. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Batista L, Brandone N, Garcia S, Prestat E, Malifarge E, Legrand A, Laugé C, Boumber Y, Dulaimi E, Tanière P, Hofman P, Ilié M, Adam M, Galon J, Gerbon C, Fieschi J. Abstract 5529: PD-L1 and CD8 IHC dual assessment in NSCLC using the Halioseek® assay: a multicentric study [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2018, vol. 78(Suppl 13). Chicago, IL. Philadelphia (PA): AACR; Cancer Res; 2018;Abstract nr 5529.
- 103.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, Nagtegaal ID, Vink-Borger E, Hartmann A, Geppert C, Kolwelter J, Merkel S, Grutzmann R, Van den Eynde M, Jouret-Mourin A, Kartheuser A, Leonard D, Remue C, Wang JY, Bavi P, Roehrl MHA, Ohashi PS, Nguyen LT, Han S, MacGregor HL, Hafezi-Bakhtiari S, Wouters BG, Masucci GV, Andersson EK, Zavadova E, Vocka M, Spacek J, Petruzelka L, Konopasek B, Dundr P, Skalova H, Nemejcova K, Botti G, Tatangelo F, Delrio P, Ciliberto G, Maio M, Laghi L, Grizzi F, Fredriksen T, Buttard B, Angelova M, Vasaturo A, Maby P, Church SE, Angell HK, Lafontaine L, Bruni D, El Sissy C, Haicheur N, Kirilovsky A, Berger A, Lagorce C, Meyers JP, Paustian C, Feng Z, Ballesteros-Merino C, Dijkstra J, van de Water C, van Lent VS, Knijn N, Musina AM, Scripcariu DV, Popivanova B, Xu M, Fujita T, Hazama S, Suzuki N, Nagano H, Okuno K, Torigoe T, Sato N, Furuhata T, Takemasa I, Itoh K, Patel PS, Vora HH, Shah B, Patel JB, Rajvik KN, Pandya SJ, Shukla SN, Wang Y, Zhang G, Kawakami Y, Marincola FM, Ascierto PA, Sargent DJ, Fox BA, Galon J. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 104.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, Budczies J, Huober J, Klauschen F, Furlanetto J, Schmitt WD, Blohmer JU, Karn T, Pfitzner BM, Kummel S, Engels K, Schneeweiss A, Hartmann A, Noske A, Fasching PA, Jackisch C, van Mackelenbergh M, Sinn P, Schem C, Hanusch C, Untch M, Loibl S. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 105.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, Wolff AC, Wood WC, Davidson NE, Sledge GW, Sparano JA, Badve SS. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salgado R, Denkert C, Campbell C, Savas P, Nuciforo P, Aura C, de Azambuja E, Eidtmann H, Ellis CE, Baselga J, Piccart-Gebhart MJ, Michiels S, Bradbury I, Sotiriou C, Loi S. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the NeoALTTO trial. JAMA Oncol. 2015;1:448–454. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, Lonardi S, Zavadil J, Osman I, Bhardwaj N. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loi S, Adams S, Schmid P, Cortés J, Cescon DW, Winer EP, Toppmeyer DL, Rugo HS, De Laurentiis M, Nanda R, Iwata H, Awada A, Tan A, Wang A, Aktan G, Karantza V, Salgado R. LBA13Relationship between tumor infiltrating lymphocyte (TIL) levels and response to pembrolizumab (pembro) in metastatic triple-negative breast cancer (mTNBC): results from KEYNOTE-086. Ann Oncol. 2017;28(Suppl 5):mdx440.005. [Google Scholar]
- 109.Schaefer C, Butterfield LH, Lee S, Kim GG, Visus C, Albers A, Kirkwood JM, Whiteside TL. Function but not phenotype of melanoma peptide-specific CD8(+) T cells correlate with survival in a multiepitope peptide vaccine trial (ECOG 1696) Int J Cancer. 2012;131:874–884. doi: 10.1002/ijc.26481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, Fong L. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med. 2014;6:23ra870. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 112.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mullane SA, Werner L, Rosenberg J, Signoretti S, Callea M, Choueiri TK, Freeman GJ, Bellmunt J. Correlation of apobec mrna expression with overall survival and PD-L1 expression in urothelial carcinoma. Sci Rep. 2016;6:27702. doi: 10.1038/srep27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, Tian T, Wei Z, Madan S, Sullivan RJ, Boland G, Flaherty K, Herlyn M, Ruppin E. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strickland KC, Howitt BE, Shukla SA, Rodig S, Ritterhouse LL, Liu JF, Garber JE, Chowdhury D, Wu CJ, D’Andrea AD, Matulonis UA, Konstantinopoulos PA. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016;7:13587–13598. doi: 10.18632/oncotarget.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, Nuryadi E, Sekine R, Oike T, Kakoti S, Yoshimoto Y, Held KD, Suzuki Y, Kono K, Miyagawa K, Nakano T, Shibata A. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8:1751. doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Karakasilioti I, Kamileri I, Chatzinikolaou G, Kosteas T, Vergadi E, Robinson AR, Tsamardinos I, Rozgaja TA, Siakouli S, Tsatsanis C, Niedernhofer LJ, Garinis GA. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 2013;18:403–415. doi: 10.1016/j.cmet.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br J Cancer. 2018;118:312–324. doi: 10.1038/bjc.2017.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, Cipolla CK, Bluth MJ, Chaim J, Al-Ahmadie H, Snyder A, Carlo MI, Solit DB, Berger MF, Funt S, Wolchok JD, Iyer G, Bajorin DF, Callahan MK, Rosenberg JE. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S, Stover E, Strickland KC, D’Andrea AD, Wu CJ, Matulonis UA, Konstantinopoulos PA. Association of polymerase e-Mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319–1323. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 122.Osoegawa A, Hiraishi H, Hashimoto T, Takumi Y, Abe M, Takeuchi H, Miyawaki M, Okamoto T, Sugio K. The positive relationship between gammaH2AX and PD-L1 expression in lung squamous cell carcinoma. In Vivo. 2018;32:171–177. doi: 10.21873/invivo.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and anticancer immunosurveillance. Cell. 2016;165:276–287. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 124.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Sanchez EMR, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weide B, Martens A, Hassel JC, Berking C, Postow MA, Bisschop K, Simeone E, Mangana J, Schilling B, Di Giacomo AM, Brenner N, Kahler K, Heinzerling L, Gutzmer R, Bender A, Gebhardt C, Romano E, Meier F, Martus P, Maio M, Blank C, Schadendorf D, Dummer R, Ascierto PA, Hospers G, Garbe C, Wolchok JD. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin Cancer Res. 2016;22:5487–5496. doi: 10.1158/1078-0432.CCR-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ferrucci PF, Ascierto PA, Pigozzo J, Del Vecchio M, Maio M, Antonini Cappellini GC, Guidoboni M, Queirolo P, Savoia P, Mandala M, Simeone E, Valpione S, Altomonte M, Spagnolo F, Cocorocchio E, Gandini S, Giannarelli D, Martinoli C. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann Oncol. 2016;27:732–738. doi: 10.1093/annonc/mdw016. [DOI] [PubMed] [Google Scholar]
- 128.Milose JC, Filson CP, Weizer AZ, Hafez KS, Montgomery JS. Role of biochemical markers in testicular cancer: diagnosis, staging, and surveillance. Open Access J Urol. 2011;4:1–8. doi: 10.2147/OAJU.S15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Albain KS, Crowley JJ, LeBlanc M, Livingston RB. Determinants of improved outcome in small-cell lung cancer: an analysis of the 2580-patient Southwest Oncology Group data base. J Clin Oncol. 1990;8:1563–1574. doi: 10.1200/JCO.1990.8.9.1563. [DOI] [PubMed] [Google Scholar]
- 130.Krajsova I, Arenberger P, Lakomy R, Kubala E, Brezinova I, Poprach A, Stastny M, Muzik J, Melichar B. Long-term survival with ipilimumab: experience from a national expanded access program for patients with melanoma. Anticancer Res. 2015;35:6303–6310. [PubMed] [Google Scholar]
- 131.Simeone E, Gentilcore G, Giannarelli D, Grimaldi AM, Caraco C, Curvietto M, Esposito A, Paone M, Palla M, Cavalcanti E, Sandomenico F, Petrillo A, Botti G, Fulciniti F, Palmieri G, Queirolo P, Marchetti P, Ferraresi V, Rinaldi G, Pistillo MP, Ciliberto G, Mozzillo N, Ascierto PA. Immunological and biological changes during ipilimumab treatment and their potential correlation with clinical response and survival in patients with advanced melanoma. Cancer Immunol Immunother. 2014;63:675–683. doi: 10.1007/s00262-014-1545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hopkins AM, Rowland A, Kichenadasse G, Wiese MD, Gurney H, McKinnon RA, Karapetis CS, Sorich MJ. Predicting response and toxicity to immune checkpoint inhibitors using routinely available blood and clinical markers. Br J Cancer. 2017;117:913–920. doi: 10.1038/bjc.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rosner S, Kwong E, Shoushtari AN, Friedman CF, Betof AS, Brady MS, Coit DG, Callahan MK, Wolchok JD, Chapman PB, Panageas KS, Postow MA. Peripheral blood clinical laboratory variables associated with outcomes following combination nivolumab and ipilimumab immunotherapy in melanoma. Cancer Med. 2018;7:690–697. doi: 10.1002/cam4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuzman JA, Stenehjem DD, Merriman J, Agarwal AM, Patel SB, Hahn AW, Alex A, Albertson D, Gill DM, Agarwal N. Neutrophil-lymphocyte ratio as a predictive biomarker for response to high dose interleukin-2 in patients with renal cell carcinoma. BMC Urol. 2017;17:1. doi: 10.1186/s12894-016-0192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Retseck J, Nasr A, Lin Y, Lin H, Mendiratta P, Butterfield LH, Tarhini AA. Long term impact of CTLA4 blockade immunotherapy on regulatory and effector immune responses in patients with melanoma. J Transl Med. 2018;16:184. doi: 10.1186/s12967-018-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakad R, Schumacher B. DNA damage response and immune defense: links and mechanisms. Front Genet. 2016;7:147. doi: 10.3389/fgene.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang LE, Yin M, Dong Q, Stewart DJ, Merriman KW, Amos CI, Spitz MR, Wei Q. DNA repair capacity in peripheral lymphocytes predicts survival of patients with non-small-cell lung cancer treated with first-line platinum-based chemotherapy. J Clin Oncol. 2011;29:4121–4128. doi: 10.1200/JCO.2010.34.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arora S, Yan H, Cho I, Fan HY, Luo B, Gai X, Bodian DL, Vockley JG, Zhou Y, Handorf EA, Egleston BL, Andrake MD, Nicolas E, Serebriiskii IG, Yen TJ, Hall MJ, Golemis EA, Enders GH. Genetic variants that predispose to DNA double-strand breaks in lymphocytes from a subset of patients with familial colorectal carcinomas. Gastroenterology. 2015;149(1872–83):e9. doi: 10.1053/j.gastro.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nicolas E, Arora S, Zhou Y, Serebriiskii IG, Andrake MD, Handorf ED, Bodian DL, Vockley JG, Dunbrack RL, Ross EA, Egleston BL, Hall MJ, Golemis EA, Giri VN, Daly MB. Systematic evaluation of underlying defects in DNA repair as an approach to case-only assessment of familial prostate cancer. Oncotarget. 2015;6:39614–39633. doi: 10.18632/oncotarget.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yuan J, Zhou J, Dong Z, Tandon S, Kuk D, Panageas KS, Wong P, Wu X, Naidoo J, Page DB, Wolchok JD, Hodi FS. Pretreatment serum VEGF is associated with clinical response and overall survival in advanced melanoma patients treated with ipilimumab. Cancer Immunol Res. 2014;2:127–132. doi: 10.1158/2326-6066.CIR-13-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zang J, Hu Y, Xu X, Ni J, Yan D, Liu S, He J, Xue J, Wu J, Feng J. Elevated serum levels of vascular endothelial growth factor predict a poor prognosis of platinum-based chemotherapy in non-small cell lung cancer. Oncotargets Ther. 2017;10:409–415. doi: 10.2147/OTT.S124124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sabatino M, Kim-Schulze S, Panelli MC, Stroncek D, Wang E, Taback B, Kim DW, Deraffele G, Z Pos, Marincola FM, Kaufman HL. Serum vascular endothelial growth factor and fibronectin predict clinical response to high-dose interleukin-2 therapy. J Clin Oncol. 2009;27:2645–2652. doi: 10.1200/JCO.2008.19.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao P, Bu X, Wei X, Sun W, Xie X, Li C, Guo Q, Zhu D, Wei X, Gao D. Dendritic cell immunotherapy combined with cytokine-induced killer cells promotes skewing toward Th2 cytokine profile in patients with metastatic non-small cell lung cancer. Int Immunopharmacol. 2015;25:450–456. doi: 10.1016/j.intimp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 144.Hannani D, Vetizou M, Enot D, Rusakiewicz S, Chaput N, Klatzmann D, Desbois M, Jacquelot N, Vimond N, Chouaib S, Mateus C, Allison JP, Ribas A, Wolchok JD, Yuan J, Wong P, Postow M, Mackiewicz A, Mackiewicz J, Schadendorff D, Jaeger D, Zornig I, Hassel J, Korman AJ, Bahjat K, Maio M, Calabro L, Teng MW, Smyth MJ, Eggermont A, Robert C, Kroemer G, Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade: obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 2015;25:208–224. doi: 10.1038/cr.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 147.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M, Ascierto PA, Di Giacomo AM, Maio M, Schilling B, Sucker A, Schadendorf D, Hassel JC, Eigentler TK, Martus P, Wolchok JD, Blank C, Pawelec G, Garbe C, Weide B. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res. 2016;22:2908–2918. doi: 10.1158/1078-0432.CCR-15-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.MacFarlane AW, Jillab M, Plimack ER, Hudes GR, Uzzo RG, Litwin S, Dulaimi E, Al-Saleem T, Campbell KS. PD-1 expression on peripheral blood cells increases with stage in renal cell carcinoma patients and is rapidly reduced after surgical tumor resection. Cancer Immunol Res. 2014;2:320–331. doi: 10.1158/2326-6066.CIR-13-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gnjatic S, Ritter E, Buchler MW, Giese NA, Brors B, Frei C, Murray A, Halama N, Zornig I, Chen YT, Andrews C, Ritter G, Old LJ, Odunsi K, Jager D. Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:5088–5093. doi: 10.1073/pnas.0914213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mintz PJ, Kim J, Do KA, Wang X, Zinner RG, Cristofanilli M, Arap MA, Hong WK, Troncoso P, Logothetis CJ, Pasqualini R, Arap W. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 151.Yuan J, Adamow M, Ginsberg BA, Rasalan TS, Ritter E, Gallardo HF, Xu Y, Pogoriler E, Terzulli SL, Kuk D, Panageas KS, Ritter G, Sznol M, Halaban R, Jungbluth AA, Allison JP, Old LJ, Wolchok JD, Gnjatic S. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc Natl Acad Sci USA. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van der Burg SH, Kalos M, Gouttefangeas C, Janetzki S, Ottensmeier C, Welters MJ, Romero P, Britten CM, Hoos A. Harmonization of immune biomarker assays for clinical studies. Sci Transl Med. 2011;3:108ps44. doi: 10.1126/scitranslmed.3002785. [DOI] [PubMed] [Google Scholar]
- 153.Diem S, Kasenda B, Spain L, Martin-Liberal J, Marconcini R, Gore M, Larkin J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br J Cancer. 2016;114:256–261. doi: 10.1038/bjc.2015.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nakaya A, Kurata T, Yoshioka H, Takeyasu Y, Niki M, Kibata K, Satsutani N, Ogata M, Miyara T, Nomura S. Neutrophil-to-lymphocyte ratio as an early marker of outcomes in patients with advanced non-small-cell lung cancer treated with nivolumab. Int J Clin Oncol. 2018;23:634–640. doi: 10.1007/s10147-018-1250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ferrucci PF, Gandini S, Battaglia A, Alfieri S, Di Giacomo AM, Giannarelli D, Cappellini GC, De Galitiis F, Marchetti P, Amato G, Lazzeri A, Pala L, Cocorocchio E, Martinoli C. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. Br J Cancer. 2015;112:1904–1910. doi: 10.1038/bjc.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Postow MA, Manuel M, Wong P, Yuan J, Dong Z, Liu C, Perez S, Tanneau I, Noel M, Courtier A, Pasqual N, Wolchok JD. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J Immunother Cancer. 2015;3:23. doi: 10.1186/s40425-015-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dhodapkar KM, Gettinger SN, Das R, Zebroski H, Dhodapkar MV. SOX2-specific adaptive immunity and response to immunotherapy in non-small cell lung cancer. Oncoimmunology. 2013;2:e25205. doi: 10.4161/onci.25205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, Behera M, Wu H, McCausland M, Chen Z, Zhang C, Khuri FR, Owonikoko TK, Ahmed R, Ramalingam SS. Proliferation of PD-1 + CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA. 2017;114:4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boffa DJ, Graf RP, Salazar MC, Hoag J, Lu D, Krupa R, Louw J, Dugan L, Wang Y, Landers M, Suraneni M, Greene SB, Magana M, Makani S, Bazhenova L, Dittamore RV, Nieva J. Cellular expression of PD-L1 in the peripheral blood of lung cancer patients is associated with worse survival. Cancer Epidemiol Biomark Prev. 2017;26:1139–1145. doi: 10.1158/1055-9965.EPI-17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weber M, Wehrhan F, Baran C, Agaimy A, Buttner-Herold M, Preidl R, Neukam FW, Ries J. PD-L1 expression in tumor tissue and peripheral blood of patients with oral squamous cell carcinoma. Oncotarget. 2017;8:112584–112597. doi: 10.18632/oncotarget.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan J, Adamow M, Ginsberg BA, Rasalan TS, Ritter E, Gallardo HF. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc Natl Acad Sci USA. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Leung AM, Lee AF, Ozao-Choy J, Ramos RI, Hamid O, O’Day SJ, Shin-Sim M, Morton DL, Faries MB, Sieling PA, Lee DJ. Clinical Benefit from ipilimumab therapy in melanoma patients may be associated with serum CTLA4 Levels. Front Oncol. 2014;4:110. doi: 10.3389/fonc.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Campesato LF, Barroso-Sousa R, Jimenez L, Correa BR, Sabbaga J, Hoff PM, Reis LF, Galante PA, Camargo AA. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget. 2015;6:34221–34227. doi: 10.18632/oncotarget.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Johnson DB, Frampton GM, Rioth MJ, Yusko E, Xu Y, Guo X, Ennis RC, Fabrizio D, Chalmers ZR, Greenbowe J, Ali SM, Balasubramanian S, Sun JX, He Y, Frederick DT, Puzanov I, Balko JM, Cates JM, Ross JS, Sanders C, Robins H, Shyr Y, Miller VA, Stephens PJ, Sullivan RJ, Sosman JA, Lovly CM. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4:959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, Rizvi NA, Hirsch FR, Selvaggi G, Szustakowski JD, Sasson A, Golhar R, Vitazka P, Chang H, Geese WJ, Antonia SJ. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33(853–61):e4. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, Shahabi V. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Carbognin L, Pilotto S, Milella M, Vaccaro V, Brunelli M, Calio A, Cuppone F, Sperduti I, Giannarelli D, Chilosi M, Bronte V, Scarpa A, Bria E, Tortora G. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS ONE. 2015;10:e0130142. doi: 10.1371/journal.pone.0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, De Macedo MP, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba II, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gomez H, Bastholt L, Chasalow SD, Berman D. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. doi: 10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kowanetz M, Zou W, Shames DS, Cummings C, et al. Tumor mutation burden (TMB) is associated with improved efficacy of Atezolizumab in 1L and 2L+ NSCLC patients. J Thorac Oncol. 2017;12:S321–S342. [Google Scholar]