Abstract
Retroperitoneal liposarcoma (RLPS) is one of the most common subtypes of retroperitoneal soft tissue sarcomas and lacks effective treatment. This study aimed to provide a thorough profile of immune characteristics of RLPS. This study included 56 RLPS patients. Multisite tumor tissues were collected from 16 patients. Immunohistochemistry was carried out to identify CD4+, CD8+, FoxP3+, CD20+, or programmed cell death‐1 (PD‐1)+ tumor infiltrating lymphocytes (TILs) and Programmed cell death ligand‐1 (PD‐L1) expression in tumor tissues. Ultradeep sequencing of T‐cell receptor (TCR) β‐chain gene was carried out in 42 tumor samples as well as peripheral blood samples collected from 6 patients. In RLPS, TILs were distributed in 3 patterns and T cells were more prevalent than B cells. Generally, the proportion of TILs decreased and PD‐L1 expression increased with tumor progression. Patients with higher PD‐1/PD‐L1 expression tended to have poorer prognosis, whereas patients with tertiary lymphoid structure tended to have a favorable disease‐free survival. Although T‐cell clones in tumors were quite different from those in peripheral blood, TCR sequencing showed low TCR repertoire reads as well as polyclonal status within tumors, which indicated limited T cell response in the tumors. Both TILs distribution and TCR repertoires suggested spatial immune heterogeneity in RLPS. Our research described the immune landscape of RLPS, and suggested RLPS might be a kind of tumor with low T cell infiltration as well as great immune heterogeneity. Therefore, strategies that can facilitate lymphocytic infiltration and immune reactivity need to be developed in the future to improve the efficacy of immunotherapy.
Keywords: heterogeneity, immune landscape, retroperitoneal liposarcoma, T‐cell receptor, Tumor‐infiltrating lymphocytes (TILs)
1. INTRODUCTION
Retroperitoneal soft tissue sarcoma (STS) is a type of heterogeneous malignancy with an incidence of 0.5‐1/100 000; liposarcoma is the most common subtype and accounts for 45% of retroperitoneal STSs.1, 2 Patients with retroperitoneal liposarcoma (RLPS) often manifest a painless enlarging mass, and thus tumors tend to be bulky and difficult to remove completely when diagnosed, which might contribute to a poorer prognosis in RLPS patients than in patients with extremity liposarcoma.3 Surgery remains the only curative method for RLPS patients, and the effect of radiotherapy and chemotherapy is limited.2 Therefore, research on the treatment of RLPS is essential.
Immune therapy has been extensively studied as a promising treatment. Immune checkpoint pathways play a role in assisting the evasion of tumor cells from immune surveillance.4, 5, 6 Therefore, immune checkpoint inhibitors can activate cytotoxic T lymphocytes, leading to destruction of cancer cells.5 This immune checkpoint inhibitor therapy can improve the overall survival and maintain a durable therapeutic response, and it has been proved a huge success in several solid tumors.7, 8, 9, 10 However, most cancer patients failed to respond to this treatment and some of them even suffered from hyperprogressive disease.11 Consequently, full understanding of the adaptive immune response in tumors could help to screen the potential beneficial population and improve the efficacy of immune therapy.
Due to the multiple aspects involved in the adaptive immune response of tumor patients.12 Blank et al proposed the term “cancer immunogram”—which is similar to “immune landscape”—to describe interaction between cancer cells and the host immune system. Specifically, this term includes tumor immunity, host immune status, immune cell infiltration, and immune checkpoint expression.13 Recent studies found that higher programmed cell death‐1 (PD‐1) ligand‐1 (PD‐L1) expression was correlated with worse prognosis in several malignancies.14, 15 Tumor mutational burden was identified as an independent predictor of immunotherapy.16 Host immune status was also identified as relevant to the immunotherapeutic response.16, 17 These results suggested the influence of the “cancer immunogram” on efficacy of immune therapy. Thus, evaluation of the immune landscape in RLPS could predict the response of immune therapy in patients with RLPS.18
There are several techniques to evaluate the immune landscape. Here, we used sequencing of T‐cell receptor (TCR) β chain complementary determining region 3 (CDR3) to evaluate the quantity and diversity of T‐cell clones. In tumor immunity, T cells recognize and combine with tumor antigens‐MHC complex through TCR. For most T cells, the TCR consists of an α and β chain, within which rearrangement of V, D, and J genes generates various specific CDR3 sequences to recognize specific antigens. Therefore, TCRs represent various subclones of antigen‐specific T cells.19 Previous studies of ovarian and pancreatic cancers assessed multisite samples of a single tumor using TCR β CDR3 sequencing and the results suggested that the composition of TCR repertoires was homogenous.20, 21 However, in hepatocellular carcinoma, lung adenocarcinoma, and esophageal squamous cell carcinoma (ESCC), TCR repertoires were heterogeneous.22, 23, 24 These results suggested that different types of tumors could have quite unique TCR characteristics.
In STS patients, current clinical trials have indicated that the response to PD‐1/PD‐L1 blockade therapy varied in different subtypes and only a few liposarcoma patients had objective responses to pembrolizumab.25, 26 Due to the small sample size and uncertain effectiveness, further application of immune checkpoint inhibitors in RLPS is limited to date.27 Several studies have been undertaken to evaluate the immune landscape in patients with STS. Programmed cell death‐1 was overexpressed in osteosarcoma, chondrosarcoma, all variants of liposarcoma, and rhabdomyosarcoma, whereas PD‐L1 expression was relatively rare.28 Another 2 studies on well‐/dedifferentiated liposarcoma (WDLPS/DDLPS) described composition and distribution of the tumor‐infiltrating lymphocytes (TILs) in inflammatory WDLPS and suggested that PD‐1 was expressed in 65% patients, which provided limited clues for the immune landscape in RLPS due to the relatively small sample size.29, 30 Pollack et al31 characterized the TCR repertoires of infiltrating lymphocytes in STSs, and the results suggested that undifferentiated pleomorphic sarcoma was supposed to be a potential subtype that might respond effectively to immunotherapy. However, despite a number of studies that were carried out in STS patients, investigations that specifically aimed at RLPS with relatively larger sample size have not been undertaken. In addition, given the bulky mass and tumor evolution, the heterogeneity of immune profiles remains unexplored. This study aimed to provide a detailed immune landscape as well as immune heterogeneity within RLPS and to provide reference for future immunotherapy.
2. MATERIALS AND METHODS
2.1. Patients and samples
A total of 56 RLPS patients who underwent surgical resection in Peking University Cancer Hospital from 2011 to 2017 were included into this retrospective study (Figure S1). Patients who received chemotherapy or radiotherapy were excluded. For 16 of these patients, we collected a total of 98 tumor specimens from different sites of a single tumor. The average age of these patients was 54.8 ± 11.2 years old. The tumors were classified as WDLPS, DDLPS, myxoid/round cell liposarcoma (MLPS) and pleomorphic liposarcoma (PLPS) according to the WHO classification and graded according to French Fédération Nationale des centres de Lutte Contre le Cancer (FNCLCC) grading system. Detailed information is listed in Tables S1 and S2. An inflammatory gastric cancer specimen and human placental tissue were used as positive control of immunohistochemistry (IHC) staining.
For 6 of 16 patients from whom multisite specimens were collected, a total of 42 fresh frozen tumor samples were collected for subsequent TCR β CDR3 region ultradeep sequencing (Table S2 and Figure S2). Preoperative peripheral venous blood was also collected as control.
All patients signed informed consent. The research was approved by the ethics committee of Peking University Cancer Hospital (2019KT19) and was carried out in accordance with the principles of the Declaration of Helsinki.
2.2. Immunohistochemistry staining of CD4, CD8, FoxP3, CD20, PD‐1, and PD‐L1 in RLPS
Formalin‐fixed and paraffin‐embedded specimens were cut into 4‐μm‐thick slices for IHC, which was undertaken as previously described.21 The concentration of Abs and antigen retrieval methods are listed in Table S3.
2.3. Evaluation of immunohistochemistry staining
Expression of PD‐L1 and PD‐1 in tumor cells was evaluated using immunoreactivity score (IRS) by 2 pathologists who were blinded to the clinical data of the patients. The IRS was calculated by multiplying the percentage of positive cells and staining intensity. The percentage of positive cells was scored as: 0, negative; 1, less than 25%; 2, 25% to 75%; and 3, more than 75%. Staining intensity was scored as: 0, negative (−); 1, weak (+); 2, moderate (++); and 3, strong (+++).The IRS was recorded as 0‐1, 2‐4, 5‐8, and 9‐12, respectively. We classified the patients as having “negative” or “positive” expression, and IRS was classified as 0 or more than 0 for further analyses.
Lymphocytes with CD4, CD8, FoxP3, CD20, and PD‐1 staining were quantified by an Aperio ImageScope (version 12.3.3) using Aperio Nuclear Algorithm (Leica) (Figure S3). Two independent pathologists were invited to undertake the tuning process, and subsequent quantification processes were carried out under their unanimous consensus. An area‐fixed square tool annotated 1 228 000.00 μm2 was used to select quantification sites. For each specimen, various sites covering at least 50% area of the specimen were chosen for quantification according to tissue dimensions, and the average results of all sites were taken as the final count of lymphocytes. For each patient, the average quantity of multiple specimens was calculated.
2.4. DNA extraction
Total genomic DNA of liposarcoma tissues and whole blood cells was extracted using an EasyPure Genomic DNA Kit (EE101; Transgen Biotech) and a Whole Blood Genomic DNA Kit (DP1801; Bioteke) following the manufacturer's instructions. At least 2 μg total genomic DNA was sent for subsequent ultradeep TCR β CDR3 sequencing analysis (MyGenostics Gene Technology).
2.5. Ultradeep sequencing of TCR β CDR3 region
Both terminals of DNA fragments were filled in and artificially linked with adaptors using Oligonucleotide sequences for TruSeq DNA Sample Prep Kits (MG0033/25, MyGenostics). The sequence of the 5′ adaptor was 5′‐AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT‐3′ and the 3′ adaptor was 3′‐GATCGGAAGAGCACACGTCTGAACTCCAGTCACATCTCGTATGCCGTCTTCTGCTTG‐5′. Next, gene segments were enriched through multiplex PCR using adaptor‐specific primers and sequenced using an Illumina NextSeq500 sequencer. Sequences with poor‐quality control and those including stop codons or frameshift mutations were excluded from the raw reads data that were used for further analyses. Finally, productive sequences of TCR β CDR3 regions were identified by BLAST alignment between the sequencing reads and the International ImMunoGeneTics (IMGT) database, and a standard algorithm was used to identify the specific V, D, and J segments.32
2.6. T‐cell receptor β CDR3 sequence analysis
Sequences with identical CDR3 amino sequence and V‐J gene were considered as the same type of clone. Clonality analysis, which can reflect both the number and the distribution of T cell clones, was calculated as described previously.21, 23, 24 Only the top 100 clones were included into subsequent analyses to reduce the possibility of heterogeneity estimation caused by enormous diversity of the host TCR repertoire.20, 21, 22, 23, 24 To compare TCR repertoire overlap between 2 specimens, Jaccard similarity coefficient as used and then transformed into a distance value using: . A distance‐based neighbor joining algorithm in MEGA (version 7.0) was used to draw a phylogenic tree in accordance with previous studies.20, 21, 24
2.7. Statistical analysis
The Shapiro‐Wilk normality test was used to verify whether data were normally distributed. The Mann‐Whitney U test and Kruskal‐Wallis H test were used to evaluate the difference between 2 or more groups. Friedman consistency examination was used to assess the heterogeneity of TILs among multisite specimens collected from a single patient. To analyze correlations between immune factors and clinicopathological features, Spearman's rank correlation analysis and the χ2 test were used. Kaplan‐Meier survival analysis was used to test disease‐free survival (DFS) and overall survival (OS) of liposarcoma patients, and the Cox proportional hazard regression model was applied to evaluate independent prognostic factors. P < .05 was considered statistically significant. Statistical analyses were undertaken using SPSS 22.0.
2.8. Accession number
T cell 8 receptor CDR3 raw sequencing data were submitted to the Sequence Read Archive database (BioProject accession number is PRJNA516984).
3. RESULTS
3.1. Quantity and distribution pattern of TILs in RLPS
In the 56 RLPS patients, T cells were more prevalent than CD20+ B cells (P < .001). CD8+ T cells were the most prevalent lymphocytes, followed by CD4+ T cells and CD20+ B cells, whereas FoxP3+ regulatory T cells (Tregs) were rare (Figure 1). The median number of CD8+ T cells was 61.7 (1.9‐1534.4), CD4+ T cells was 38.2 (1.6‐784.4), FoxP3+ Tregs was 7.5 (0.6‐245.5), and CD20+ B cells was 18.4 (1.3‐977.6). The median percentages of CD8+ T cells, CD4+ T cells, and FoxP3+ Treg cells were 1.91 (0.17‐30.15)%, 1.53 (0.09‐14.21)%, and 0.86 (0.02‐4.08)%, respectively. The median proportion of CD20+ B cells was 0.67 (0.03‐15.05)%. (Table S1). To reduce the bias caused by various sizes of tumor cells, we undertook subsequent analyses using the proportion of TILs. The typical staining for each subgroup of cells is shown in Figure 2.
Figure 1.
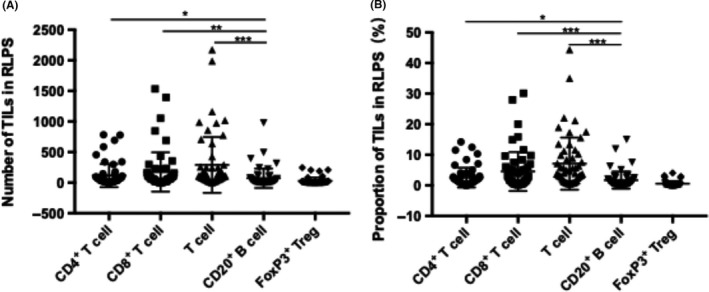
Quantity of various subgroups of tumor‐infiltrating lymphocytes (TILs) in tumors of 56 retroperitoneal liposarcoma (RLPS) patients. (A) Absolute numbers and (B) proportions of various subgroups of TILs showed T cells were more prevalent than B cells (P < .001). CD8+ TILs were the most prevalent subtype, and both their number (P = .001) and proportion (P < .001) were significantly higher than CD20+ B cells. CD4+ TILs and CD20+ TILs were moderate, whereas FoxP3+ regulatory T cells (Tregs) were rare. *P < .05; **P < .01; ***P < .001
Figure 2.

Typical immunohistochemistry staining of tumor‐infiltrating lymphocytes (TILs) within retroperitoneal liposarcoma. Specific TILs were stained brown, and the typical cells are (A) CD4+ TILs, (B) CD8+ TILs, (C) CD20+ TILs, and (D) FoxP3+ TILs (arrow). Magnification, ×400
In RLPS tumors, lymphocytes were distributed in both tumor parenchyma and fibrous septum. Three lymphocyte distribution patterns were detected: scattered distribution, clustered distribution, and tertiary lymphoid structure (TLS) (Figure S4). Tertiary lymphoid structures were found in 9 of the 56 patients. Higher proportions of CD4+ TILs (P = .199), CD8+ TILs (P < .001), FoxP3+ Tregs (P = .014), and CD20+ B cells (P < .001) were detected in patients with TLS (Figure S5 and Table S4). In general, the distribution of CD4+ T cells and CD8+ T cells showed all of the 3 patterns. CD20+ B cells tended to aggregate in TLSs. FoxP3+ Tregs were rare and had a scattered distribution within TLS areas.
3.2. Expression of PD‐1 and PD‐L1 in RLPS
Programmed cell death‐1 was expressed in both TILs and tumor cells, and it was typically located in cytoplasm and on the cell membrane (Figure 3A). The PD‐1 staining was weaker in tumor cells than in TILs. Thus, for quantification of PD‐1+ TILs, only TILs with 3+ and 2+ staining intensity were included. The median proportion of PD‐1+ TILs was 0.26% (range, 0.03%‐5.12%). The frequency of PD‐1+ cells was low, as only 23.2% patients (13/56) showed 1% or higher PD‐1+ TILs. In the remaining 76.8% patients (43/56), PD‐1+ TILs accounted for less than 1%. For tumor cells, PD‐1 was expressed in 25% patients (14/56). Programmed cell death‐1 ligand‐1 was expressed in 13 of 56 RLPS patients (23.2%), and was mainly located in cytoplasm and on the membrane of tumor cells (Figure 3B).
Figure 3.

Typical staining of programmed cell death‐1 (PD‐1) and its ligand (PD‐L1) in retroperitoneal liposarcoma. A, Typical staining of PD‐1, which was expressed in both tumor cells and tumor‐infiltrating lymphocytes (TILs). The staining intensity was stronger in TILs than in tumor cells. B, Typical cytoplasmic and membrane staining of PD‐L1 in tumor cells. A clear boundary of PD‐L1 expression can be seen under this microscopic image. Magnification, ×200 (left) and ×400 (right)
Expression of PD‐1 in tumor cells was significantly correlated with PD‐L1 expression (P = .001), as only 11.90% (5/42) of PD‐1− patients expressed PD‐L1, whereas 57.14% of PD‐1 positive patients (8/14) expressed PD‐L1 (Figure 4). Mann‐Whitney analyses showed that PD‐L1 expression was associated with CD4+ T cells (P = .003) and FoxP3+ Tregs (P = .008; Figure 4 and Table S4). No other significant correlation was found between these immune parameters (Table S4).
Figure 4.
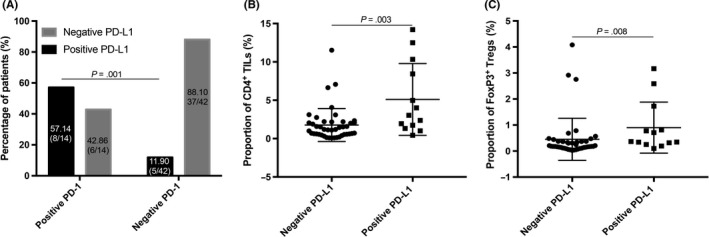
Correlations between pairwise characteristics of the retroperitoneal liposarcoma immune landscape. A, Programmed cell death‐1 (PD‐1) ligand‐1 (PD‐L1) was markedly higher in patients with positive PD‐1 expression in tumor cells than those with negative PD‐1 expression (P = .001). B,C, Proportions of (B) CD4+ tumor‐infiltrating lymphocytes (TILs) (P = .003) and (C) FoxP3+ regulatory T cells (Tregs) were significantly higher in tumors with PD‐L1 expression than those without PD‐L1 expression (P = .008)
3.3. Correlations between immune characteristics and clinicopathological features or prognosis in RLPS patients
Higher frequency of CD4+ T cells was detected in patients with newly diagnosed tumors than in those with relapsed tumors (P = .024). Higher proportions of CD20+ B cells were detected in patients with a single tumor (P = .04) or without tumor necrosis (P = .002). Proportions of CD4+ T cells (r = −0.272, P = .043), CD8+ T cells (r = −.314, P = .018), and CD20+ B cells (r = −.307, P = .023) were negatively correlated with tumor size. No significant correlation was found between FoxP3+ Tregs or TLS presence and clinical features. Moreover, PD‐1+ TILs were the lowest in grade 2 tumors (P = .014). Expression of PD‐L1 was higher in patients with multiple tumors (P = .035) or necrotic tumors (P = .028), and it was also associated with FNCLCC grade (P = .006). Similar with PD‐L1, PD‐1 expression in tumor cells was also higher in patients with multiple tumors (P = .014) (Figure 5, Tables S4 and S5).
Figure 5.
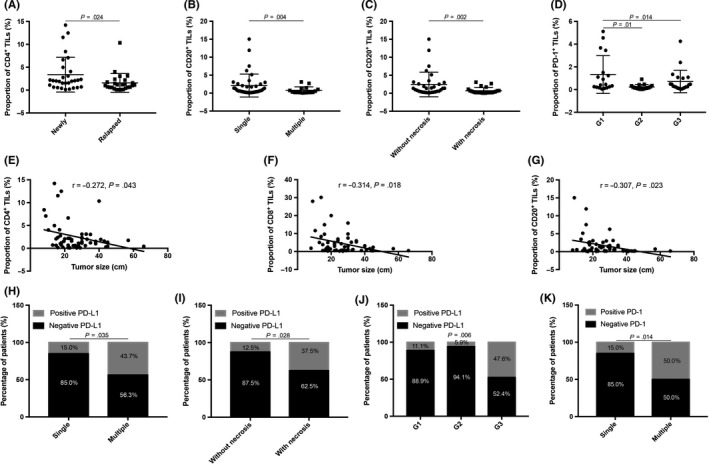
Significant correlations between immune characteristics and clinicopathological features of patients with retroperitoneal liposarcoma. A, CD4+ tumor‐infiltrating lymphocytes (TILs) decreased in relapsed tumors (P = .024). B,C, Higher proportions of CD20+ TILs were detected in patients with (B) single tumor (P = .004) or (C) without tumor necrosis (P = .002). D, Patients with grade 2 tumors had the lowest proportion of programmed cell death‐1 (PD‐1)+ TILs (P = .014). (E) CD4+ (P = .043), (F) CD8+ (P = .018), and (G) CD20+ TILs were negatively correlated with tumor size (P = .023). H‐J, Expression of PD‐1 ligand‐1 (PD‐L1) markedly decreased in patients with (H) multiple tumors (P = .035), (I) necrotic tumors (P = .026), or (J) G3 tumors (P = .006). K, Similar to PD‐L1, PD‐1 expression in tumor cells decreased in patients with multiple tumors compared to those with single tumors (P = .014)
In the survival analysis, no immune characteristics were found to be significantly associated with DFS or OS of RLPS patients. However, patients with higher FoxP3+ TILs, PD‐1+ TILs, and PD‐L1 tended to have shorter DFS and OS. Patients with TLS presence tended to have favorable DFS and OS (Table S6). Univariate and multivariate analyses indicated that in this group of RLPS patients only tumor size was an independent predictor of DFS (hazard ratio, 1.067; 95% confidence interval, 1.021‐1.116; P = .004) and OS (hazard ratio, 1.073; 95% confidence interval, 1.015‐1.135; P = .013).
3.4. Quantity and expansion of T‐cell clones in RLPS
Furthermore, ultradeep sequencing was used to analyze the quantity and expansion of T‐cell clones in 42 tumor specimens of 6 patients. During the process, 2 specimens (case1 T2‐3 and case3 T6) were not qualified for further analyses. Results of IHC showed that the average absolute number of T cells (the sum of CD4+ T cells and CD8+ T cells) was dramatically lower in these 2 specimens (30.27 and 50.60, respectively) than in other specimens (127.76 ± 96.90).
As shown in Figure 6 and Table S7, the total unique TCR β reads varied from site to site with a median value of 7231 (range, 1986‐47 521). In peripheral blood samples and tumor specimens, the average productive TCR β sequence reads were similar (tumor vs blood, 192 675.13 ± 209 294.58 vs 601 783.00 ± 611 009.48; P = .163), whereas the unique TCR reads were significantly less in tumor specimens than in blood samples (tumor vs blood, 12 276.00 ± 12 237.27 vs 23 499.83 ± 133 50.44; P = .044). Clonality analyses showed that TILs in tumor tissues were not more oligoclonal than peripheral blood lymphocytes (tumor vs blood, 0.22 ± 0.13 vs 0.20 ± 0.09, P = .796). The clone expansion of the top 100 TCR clones was not significantly different between tumor and blood samples, as the average frequency was 46.65 ± 20.58% in tumor and 33.92 ± 19.64% in blood samples (P = .163).
Figure 6.
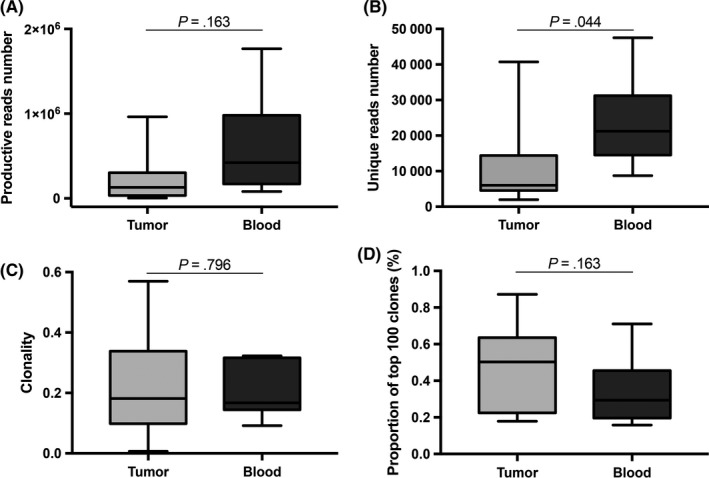
Quantity and diversity of T cells in retroperitoneal liposarcoma (RLPS) tumors. A, Number of productive reads was higher in peripheral blood than in RLPS tumor, but the difference was not statistically significant. B, Number of unique reads was markedly higher in blood samples than in RLPS tissues. C, Clonality was similar in RLPS tissues and blood samples. D, Top 100 T‐cell clones in RLPS accounted for approximately 0.22%, a little higher than in blood
3.5. T‐cell receptor repertoire of RLPS differed from peripheral blood lymphocytes
To illustrate the similarities and discrepancies between TCR repertoires in liposarcoma tissues and peripheral blood samples in 6 RLPS patients, the overlaps of pairwise specimens were assessed by calculating the Jaccard similarity coefficient, which represents the proportion of intersection reads number in the union set. Neighbor‐joining phylogenic trees were then delineated. As shown in Figure 7, the intratumoral TCR repertoire overlaps were significantly higher than that between tumor specimens and blood samples (tumor and blood vs tumor and tumor, 0.24 ± 0.16 vs 0.38 ± 0.23; P < .001). The distances between tumor specimens and blood samples were great, except that the TCR repertoire of T2 specimen in case 2 showed great similarity with that in blood samples.
Figure 7.
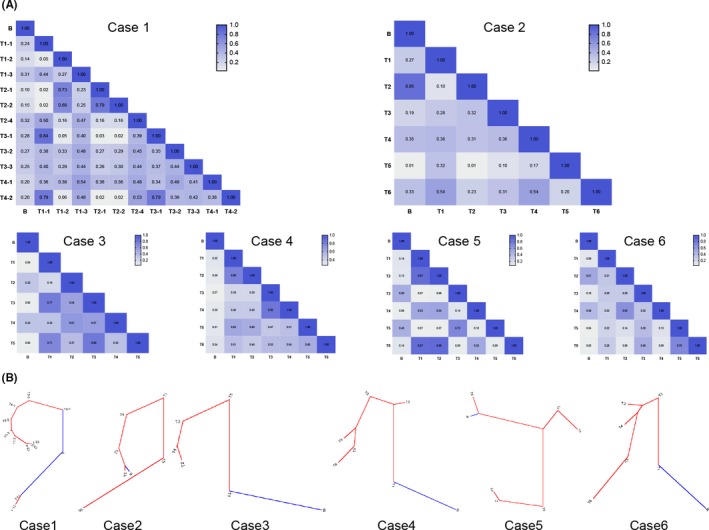
Composition difference of T‐cell receptor (TCR) β repertoires between different specimens from 6 patients with retroperitoneal liposarcoma (RLPS). A, Pairwise TCR β repertoire overlaps of 6 RLPS patients. Jaccard similarity coefficient was calculated to compare similarity between pairwise TCR β repertoires of tumor samples and peripheral blood samples, and the coefficient was closer to 1 when the 2 specimens were more similar. B, Neighbor‐joining tree of each RLPS patient was generated by transforming the Jaccard similarity coefficient into pairwise distance. Blue, peripheral blood; red, tumor tissue. The distance between 2 nodes indicates the difference between 2 specimens
3.6. T‐cell receptor repertoire of RLPS was lower than other tumors
We also compared the TCR repertoire of RLPS with that in studies of pancreatic cancer and ESCC.21, 24 As shown in Figure S6, the average productive reads of RLPS were significantly lower than that of pancreatic cancer (192 675 ± 209 295 vs 2 093 023 ± 685 896; P < .001) and ESCC (192 675 ± 209 295 vs 1 307 119 ± 719 871; P < .001), and the average unique reads of RLPS were 12 276 ± 12 237, which was also significantly lower than that in pancreatic cancer (15 646 ± 5984; P = .0016) and ESCC (112 833 ± 53 982; P < .001).
3.7. Correlation between TCR repertoire and T cell presence
The proportion of CD4+, CD8+, and total T cells were correlated with productive TCR β reads (CD4+: r = .459, P = .003; CD8+: r = .386, P = .014; total T cells: r = .485, P = .002). For unique TCR β reads, only CD4+ T cells (r = .402, P = .01) and total T cells (r = .385, P = .014) were correlated to unique TCR reads, whereas CD8+ T cells were irrelevant to unique TCR β reads (r = .263, P = .101) (Figure S7).
3.8. Spatial heterogeneity of TILs and TCR repertoires within a single tumor
To describe the spatial heterogeneity within liposarcoma tissues, both TILs distribution and TCR repertoires were assessed. Proportions of CD4+, CD8+, FoxP3+, CD20+, and PD‐1+ TILs among multiple tumor sites were compared in each case using Friedman consistency examination (S8). The TILs distribution was heterogeneous in 8 of 16 patients (Figure Table S2). The TILs heterogeneity was not significantly associated with clinicopathological features or prognosis of RLPS patients, but patients with heterogeneous TILs tended to have favorable DFS (Figure S9).
T‐cell receptor repertoires at multiple sites of each tumor were also compared in 6 patients. The median Jaccard similarity coefficient was 0.38 (0.01‐0.88) in all tumor specimens (Figure 7), within which only 35 of 125 pairwise Jaccard coefficients were 0.5 or higher. Sequences that could be detected in all sites were considered as ubiquitous whereas others were heterogeneous. As shown in Figure 8, in the 100 most abundant T‐cell clones across all specimens, ubiquitous sequences were relatively rare in multiple specimens of each patient. Moreover, the most abundant T‐cell clones in peripheral blood were not identical with those in tumor specimens.
Figure 8.
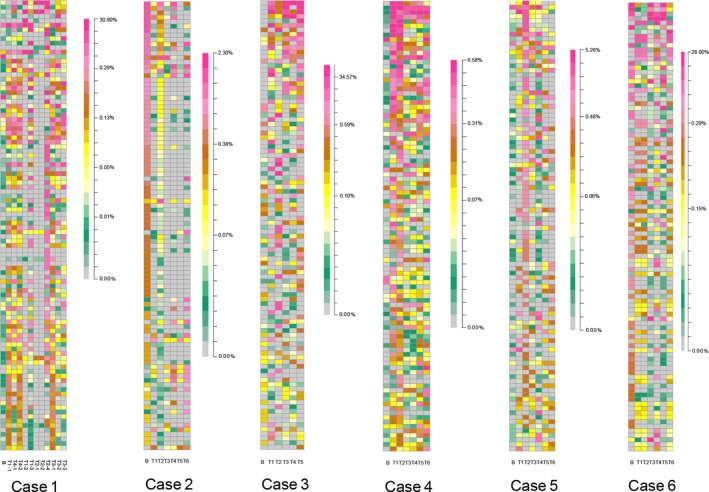
Heat maps of the top 100 T‐cell receptor (TCR) clones across all specimens of each patient with retroperitoneal liposarcoma (RLPS). The colors of each grid represent different frequencies of the top 100 TCR clones across all samples obtained from both RLPS tumor and peripheral blood. The heat map reveals heterogeneous TCR β repertoire in RLPS patients
4. DISCUSSION
In this study, we retrospectively studied subpopulations and distribution of TILs in 56 RLPS patients. In RLPS, CD8+ T cells were the most prevalent, followed by CD4+ T cells and CD20+ B cells, with FoxP3+ Tregs the rarest. T cells were more prevalent than CD20+ B cells, which was not consistent with the findings of Tseng et al.29 Controversial conclusions were also mentioned in their article. We supposed that individual difference in immune status was the main cause of this situation. Tumor grade and subtypes can also contribute to TILs infiltration. To further explore the influence of RLPS subtypes, we focused on a group of patients with TLS. Tertiary lymphoid structures were found in 16.1% patients (9/56) and correlated with higher CD8+ T cells, FoxP3+ Tregs, and CD20+ B cells, suggesting that RLPS with TLS presence might be an immune active subtype and could benefit from immunotherapy. Interestingly, we observed all slices and found another 9 patients with TILs aggregation but without follicle‐like structures, indicating that the formation of TLS is a dynamic process. Thus, understanding the mechanism of TLS formation could help to develop new immunotherapeutic strategies with better efficacy. In the 9 patients with TLS, 4 were WDLPS, 2 were DDLPS, and 3 were MLPS; none of the 5 PLPS patients showed TLS presence. Moreover, the proportion of TILs was highest in DDLPS, lower in WDLPS and MLPS, and the lowest in PLPS, which is the subtype with the highest tumor mutational burden.2 These findings suggested that there might be different causes that drive lymphocyte infiltration in different RLPS subtypes, but given the small sample size, further studies are needed.
In RLPS, the median proportion of PD‐1+ TILs was 0.26% (0.03%‐5.12%), and the PD‐1+ TILs accounted for less than 1% in 43/56 patients. Programmed cell death‐1 was also expressed on RLPS tumor cells in 14/56 patients and PD‐L1 was expressed in 13/56 patients. Unlike a previous study in 81 STS patients, which found 59% PD‐L1 and 90% PD‐1 expression in STS patients, the PD‐1 and PD‐L1 expression in our study were much lower.31 Given that 80% of patients in that study were G2 or G3, we compared the expression level in tumors with different FNCLCC grade and found that G2 tumors had the lowest PD‐1+ TILs and G3 tumors had significantly higher PD‐L1 expression. In addition, TILs proportions were generally lower in patients with relapsed, larger, or multiple tumors or in tumors with necrosis. The results indicated that immune effector cells were weakened and immune suppressive factors were enhanced as the tumor gradually advanced. Although the proportions of TILs or immune suppressive factors were not significant prognostic factors, we observed a tendency that patients with higher FoxP3+ Tregs or PD‐1/PD‐L1 expression had a poorer prognosis, and patients with TLS presence had a favorable DFS. These results were similar to previous studies in gastric cancer and intrahepatic cholangiocarcinoma,14, 15 suggesting that PD‐1/PD‐L1 blockade or other treatments to improve immune activity might have therapeutic potential in advanced RLPS patients.
Despite the infiltration of TILs and potential subtypes that could benefit from immunotherapy, TCR CDR3 sequencing suggested low T cell infiltration in RLPS patients compared with previous studies. Both the productive TCR β reads and the unique TCR β reads of RLPS specimens were lower than those in corresponding peripheral blood samples and other tumors.20, 21, 22, 24 Low productive reads were associated with less T cell infiltration, which can be confirmed by IHC results. Low unique reads suggested fewer T cell clonal subpopulations. In particular, CD8+ T cells were correlated with productive reads but not associated with unique reads, indicating limited proliferation of effector T cells. Further clonality analyses showed that intratumoral T cells were polyclonal in liposarcoma patients, which was similar to those in ESCC patients.24 For the host immune system, oligoclonal status often indicates the expansion of pathogen‐specific clonal subpopulations. Therefore, these results suggested limited T cell infiltration and expansion in RLPS, which might be the obstacles that need to be overcome in immunotherapy of RLPS patients.
Liposarcoma is characterized by its giant tumor size as well as heterogeneous composition and clinical behavior in a single tumor.33 In our study, we analyzed immune landscape heterogeneity in RLPS. Heterogeneous TILs distribution was found in 50.0% patients and tended to correlate with favorable DFS. In addition, although the intratumoral TCR repertoires were less heterogeneous than the differences between tumor and blood, the median pairwise intratumoral TCR repertoires overlap was 0.38, and only 35 of 125 pairwise overlaps were 0.5 or higher. These results suggested a larger heterogeneity in RLPS than in other tumors.20, 21, 22, 23, 24 Analyses of TCR repertoires showed that ubiquitous sequences were rare across different sites of a single tumor, which also indicated a great spatial heterogeneity within RLPS. Previous studies showed heterogeneous gene expression profile in RLPS, which may generate various tumor‐specific antigens and contribute to the immune heterogeneity.34, 35
Despite the challenge posed by low TILs and immunogenicity as well as large intratumoral heterogeneity, immunotherapy still has its own advantages in treating such complex tumors, as it can simultaneously activate multiple T cell subgroups. Our results suggested a higher clone expansion in RLPS than in blood samples. The most abundant T cell clones in tumors were not identical with those in peripheral blood, suggesting the existence of antitumor immune response. Thus, it is crucial to explore how to increase lymphocyte infiltration in the future, and comparison across a large population could help to identify liposarcoma‐specific antigens for further immunotherapy. Larger population studies from multiple centers and longer follow‐up are needed to improve our research.
In conclusion, for the first time, we evaluated the composition and distribution of TILs as well as PD‐1 and PD‐L1 expression in 56 RLPS patients. Our results suggested that low lymphocytic infiltration and PD‐1/PD‐L1 expression might limit the application of immunotherapy in liposarcoma patients. However, the complex heterogeneity of liposarcoma makes immunotherapy an ideal therapeutic strategy. Thus, strategies such as the combination of immunotherapy and other treatments that could increase infiltration of lymphocytes in tumors could improve the efficacy of liposarcoma treatment.
DISCLOSURE
All authors declare that there is no conflict of interest in this study.
Supporting information
ACKNOWLEDGMENTS
This study was supported by the Beijing Municipal Administration of Hospital's Ascent Plan (Grant No. DFL20181104); National Natural Science Foundation (Grant No. 31770836); Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (Grant No. XMLX201708); Beijing Municipal Natural Science Foundation (Grant No. 7153161); and Beijing Municipal Administration of Hospitals’ Youth Programme (Grant No. QML20181104).
Yan L, Wang Z, Cui C, et al. Comprehensive immune characterization and T‐cell receptor repertoire heterogeneity of retroperitoneal liposarcoma. Cancer Sci. 2019;110:3038–3048. 10.1111/cas.14161
Funding information
Beijing Municipal Natural Science Foundation, Grant/Award Number: 7153161; Beijing Municipal Administration of Hospital's Ascent Plan, Grant/Award Number: DFL20181104; Beijing Municipal Administration of Hospitals’ Youth Programme, Grant/Award Number: QML20181104; Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support, Grant/Award Number: XMLX201708; National Natural Science Foundation, Grant/Award Number: 31770836
Contributor Information
Xiuyun Tian, Email: xiuyunt@126.com.
Chunyi Hao, Email: haochunyi@bjmu.edu.cn.
REFERENCES
- 1. Dumitra S, Gronchi A. The diagnosis and management of retroperitoneal sarcoma. Oncology (Williston Park). 2018;32:464‐469. [PubMed] [Google Scholar]
- 2. Lee ATJ, Thway K, Huang PH, Jones RL. Clinical and molecular spectrum of liposarcoma. J Clin Oncol. 2018;36:151‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gootee J, Aurit S, Curtin C, Silberstein P. Primary anatomical site, adjuvant therapy, and other prognostic variables for dedifferentiated liposarcoma. J Cancer Res Clin Oncol. 2019;145:181‐192. [DOI] [PubMed] [Google Scholar]
- 4. Frydenlund N, Mahalingam M. PD‐L1 and immune escape: insights from melanoma and other lineage‐unrelated malignancies. Hum Pathol. 2017;66:13‐33. [DOI] [PubMed] [Google Scholar]
- 5. Muenst S, Soysal SD, Tzankov A, Hoeller S. The PD‐1/PD‐L1 pathway: biological background and clinical relevance of an emerging treatment target in immunotherapy. Expert Opin Ther Targets. 2015;19:201‐211. [DOI] [PubMed] [Google Scholar]
- 6. Zou W, Wolchok JD, Chen L. PD‐L1 (B7‐H1) and PD‐1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016; 8: 328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti‐PD‐1) in melanoma. N Engl J Med. 2013;369:134‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373:123‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gettinger SN, Horn L, Gandhi L, et al. Overall survival and long‐term safety of nivolumab (Anti‐Programmed Death 1 Antibody, BMS‐936558, ONO‐4538) in patients with previously treated advanced non‐small‐cell lung cancer. J Clin Oncol. 2015;33:2004‐2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fuentes‐Antras J, Provencio M, Diaz‐Rubio E. Hyperprogression as a distinct outcome after immunotherapy. Cancer Treat Rev. 2018;70:16‐21. [DOI] [PubMed] [Google Scholar]
- 12. Chen DS, Mellman I. Elements of cancer immunity and the cancer‐immune set point. Nature. 2017;541:321‐330. [DOI] [PubMed] [Google Scholar]
- 13. Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016; 352: 658‐660. [DOI] [PubMed] [Google Scholar]
- 14. Li Z, Lai Y, Sun L, et al. PD‐L1 expression is associated with massive lymphocyte infiltration and histology in gastric cancer. Hum Pathol. 2016;55:182‐189. [DOI] [PubMed] [Google Scholar]
- 15. Sabbatino F, Villani V, Yearley JH, et al. PD‐L1 and HLA class I antigen expression and clinical course of the disease in intrahepatic cholangiocarcinoma. Clin Cancer Res. 2016;22:470‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598‐2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor‐based immunotherapy. Lancet Oncol. 2016;17:e542‐e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galluzzi L, Chan TA, Kroemer G, Wolchok JD, Lopez‐Soto A. The hallmarks of successful anticancer immunotherapy. Sci Transl Med. 2018;10:eaat7807. [DOI] [PubMed] [Google Scholar]
- 19. Mahe E, Pugh T, Kamel‐Reid S. T cell clonality assessment: past, present and future. J Clin Pathol. 2018;71:195‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emerson RO, Sherwood AM, Rieder MJ, et al. High‐throughput sequencing of T‐cell receptors reveals a homogeneous repertoire of tumour‐infiltrating lymphocytes in ovarian cancer. J Pathol. 2013;231:433‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cui C, Tian X, Wu J, et al. T cell receptor beta‐chain repertoire analysis of tumor‐infiltrating lymphocytes in pancreatic cancer. Cancer Sci. 2019;110:61‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi L, Zhang Y, Feng L, et al. Multi‐omics study revealing the complexity and spatial heterogeneity of tumor‐infiltrating lymphocytes in primary liver carcinoma. Oncotarget. 2017;8:34844‐34857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang C, Ding H, Huang H, et al. TCR repertoire intratumor heterogeneity of CD4(+) and CD8(+) T cells in centers and margins of localized lung adenocarcinomas. Int J Cancer. 2019;144:818‐827. [DOI] [PubMed] [Google Scholar]
- 24. Chen Z, Zhang C, Pan Y, et al. T cell receptor beta‐chain repertoire analysis reveals intratumour heterogeneity of tumour‐infiltrating lymphocytes in oesophageal squamous cell carcinoma. J Pathol. 2016;239:450‐458. [DOI] [PubMed] [Google Scholar]
- 25. Groisberg R, Hong DS, Behrang A, et al. Characteristics and outcomes of patients with advanced sarcoma enrolled in early phase immunotherapy trials. J Immunother Cancer. 2017;5:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tawbi HA, Burgess M, Bolejack V, et al. Pembrolizumab in advanced soft‐tissue sarcoma and bone sarcoma (SARC028): a multicentre, two‐cohort, single‐arm, open‐label, phase 2 trial. Lancet Oncol. 2017;18:1493‐1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wisdom AJ, Mowery YM, Riedel RF, Kirsch DG. Rationale and emerging strategies for immune checkpoint blockade in soft tissue sarcoma. Cancer. 2018;124:3819‐3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torabi A, Amaya CN, Wians FH Jr, Bryan BA. PD‐1 and PD‐L1 expression in bone and soft tissue sarcomas. Pathology. 2017;49:506‐513. [DOI] [PubMed] [Google Scholar]
- 29. Tseng WW, Demicco EG, Lazar AJ, Lev DC, Pollock RE. Lymphocyte composition and distribution in inflammatory, well‐differentiated retroperitoneal liposarcoma: clues to a potential adaptive immune response and therapeutic implications. Am J Surg Pathol. 2012;36:941‐944. [DOI] [PubMed] [Google Scholar]
- 30. Tseng WW, Malu S, Zhang M, et al. Analysis of the intratumoral adaptive immune response in well differentiated and dedifferentiated retroperitoneal liposarcoma. Sarcoma. 2015;2015:547460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pollack SM, He Q, Yearley JH, et al. T‐cell infiltration and clonality correlate with programmed cell death protein 1 and programmed death‐ligand 1 expression in patients with soft tissue sarcomas. Cancer. 2017;123:3291‐3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V‐J and V‐D‐J JUNCTIONs. Bioinformatics. 2004;20(Suppl 1):i379‐i385. [DOI] [PubMed] [Google Scholar]
- 33. Wang GY, Lucas DR. Dedifferentiated Liposarcoma With Myofibroblastic Differentiation. Arch Pathol Lab Med. 2018;142:1159‐1163. [DOI] [PubMed] [Google Scholar]
- 34. Skubitz KM, Pambuccian S, Manivel JC, Skubitz AP. Identification of heterogeneity among soft tissue sarcomas by gene expression profiles from different tumors. J Transl Med. 2008;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanojia D, Nagata Y, Garg M, et al. Genomic landscape of liposarcoma. Oncotarget. 2015;6:42429‐42444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


