Abstract
Understanding how neurons form, extend, and navigate their finger-like axonal and dendritic processes is crucial for developing therapeutics for the diseased and damaged brain. Although less well appreciated, many other types of cells also send out similar finger-like projections. Indeed, unlike neuronal specific phenomena such as synapse formation or synaptic transmission, an important issue for thought is that this critical long-standing question of how a cellular process like an axon or dendrite forms and extends is not primarily a neuroscience problem but a cell biological problem. In that case, the use of simple cellular processes – such as the bristle cell process of Drosophila – can aid in the fight to answer these critical questions. Specifically, determining how a model cellular process is generated can provide a framework for manipulations of all types of membranous process-containing cells, including different types of neurons.
Introduction
Our brains control such diverse abilities as movement, sensation, intelligence, speech, emotion, and memory only because our neurons communicate with one another and the rest of our body through highly organized networks. These neuronal networks or connections are assembled when neurons send out their stalk-like axon and dendrite appendages (Fig 1A). How do these appendages form? Extend? Change shape? What controls their length and directionality? How might we get them to regrow if they are damaged or diseased? These questions have captivated scientists for over a century [1–5] and are of the utmost importance to human health, yet they still remain largely unanswered. Compounding this problem is that researchers in these areas have also become interested in other related (but completely different) phenomena such as: What molecules are involved in setting-up particular neuronal connections and circuits? What are the neuronal circuits underlying specific behaviors? What differentiates one neuronal class from another? What range of morphological/biological events do specific axon guidance cues specify? While these are also questions of the utmost importance, they are not designed to uncover the mechanisms of neuronal process shape, extension, and navigation.
Figure 1. Neurons and other membranous process-containing cells.
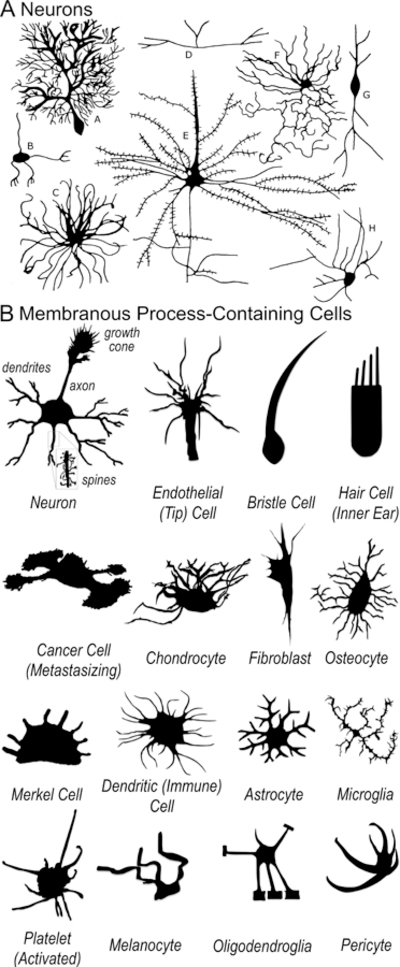
(A) Neurons are the best-known of the membranous process-containing cells – but are also the most complex. Adapted from [88]. (B) Many different types of cells – including those within the nervous, cardiovascular, immune, and musculoskeletal systems – send out membranous extensions/processes. Many of these other process-containing cells are much simpler than neurons. The mechanisms of this process extension and the means to control it, so as to stimulate re-extension of neuronal processes or inhibit the extensions of metastasizing cancer cells, for example, are poorly understood. Adapted from [89–103].
The Difficulties of Deciphering the Mechanisms of Axon Formation, Extension, and Navigation
Over the years, a large number of diverse labs have made critical inroads into the mechanisms of neuronal process extension and pathfinding (e.g., reviewed in [1–21]). For example, ground-breaking work into the identification of extracellular guidance cues and their cell-surface receptors has provided a molecular basis for axon growth and guidance (reviewed in [3–5,7,12,18]). Likewise, elegant studies have begun to identify intracellular molecules required for transducing these extracellular signals into changes in the growth and orientation of neuronal extensions (reviewed in [9,11,13,16,20]). Adding to these critical molecular and biochemical studies is a long history of research that has sought to define the cell biology of neuronal extension and guidance, including the cytoskeletal and cell/substrate adhesive dynamics that accompany axon/dendrite elongation (e.g., reviewed in [6,8,14,15,17,19]). Collectively, this work has provided exciting insights into the mechanisms of axon extension and guidance, with important biomedical implications.
Despite these important advances, however, much remains to be learned about the mechanisms of neuronal growth and guidance [21]. Additionally, a sampling of recent results (e.g., [22–30]), including a revised view of the axon guidance cue Netrin [25,27,28], indicates that some of our long-held views on these mechanisms may be incomplete – further highlighting the importance of continuing to examine/reexamine these complex mechanisms and the fundamental functions of specific guidance cues (including by employing higher resolution approaches to study them). Moreover, similar to other cells, neuronal process extension and pathfinding require the dynamic assembly and disassembly of the structural elements – actin and tubulin cytoskeletal proteins – that control cellular shape (Fig 2A–C; [31,32]). Yet, while extracellular guidance signals have been identified that activate receptors on the surface of axons/dendrites (Fig 2D; [4,5]), how these cytoskeletal elements are precisely controlled by guidance cues/receptors remains enigmatic. Thus, it remains incompletely understood how neurons form a membranous process and how these membranous processes extend, orient in space, shape themselves, reach a certain diameter, modulate their extension velocity, form branches, know when to stop extending, and transition to a stable structure.
Figure 2. Control of F-actin and microtubule dynamics drive neuronal form and function.
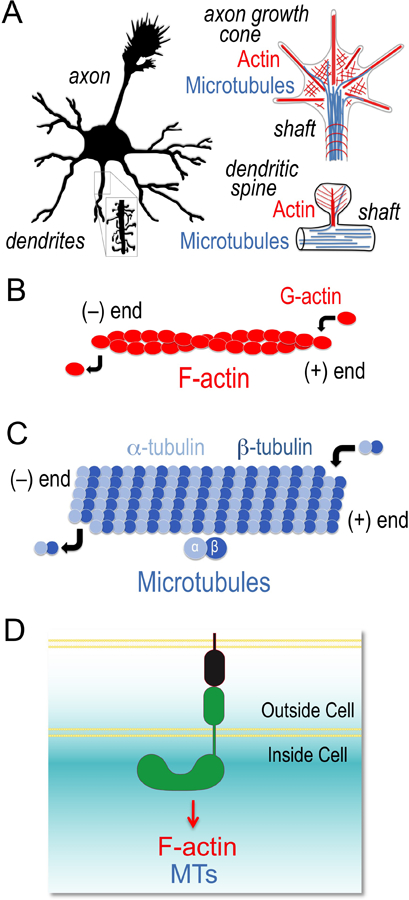
(A–C) Neuronal extension, shape, connectivity, and function (A) is driven by regulating the ability of single actin proteins (G-actin) to form filaments (F-actin) (B) and single alpha (α) and beta (β) tubulin proteins to form microtubules (C). (D) Signals from the cell surface, such as from different guidance cues and their receptors control F-actin and microtubule (MT) dynamics through poorly understood means.
What has complicated our ability to clearly understand these events? While in many instances, employing neurons as a model can be seen as a “rosetta stone” for cell biology and is highly advantageous for deciphering fundamental biological mechanisms (e.g., the study of neuronal synaptic transmission provides great insight into membrane/vesicle fusion and is applicable interdisciplinarily to all cells/tissue systems [33]), the morphological complexity of neurons makes them more challenging when deciphering the mechanisms of axon formation, extension, and navigation. For example, neurons have the unusual ability to place parts of themselves (i.e., axons and dendrites, or collectively referred to as neurites) at locations far from their cell body. Consequently, far less is known of the organization and dynamics of F-actin and microtubules in these extending processes than in the cell body of a typical migrating cell. Furthermore, the movement (elongation) of axons and dendrites is much different than the movement (migration) of cells. Whereas migrating cells need to drag along their soma and nucleus as they move from place to place, neurons do not. Instead, neurons send out long cylindrical extensions that stabilize and continue to elongate. Additionally, axons and dendrites often undergo branching and form complex arborizations that are much different than what is present in a typical cell (e.g., Fig 1A). Moreover, the in vitro and in vivo settings often utilized for studying these growth and guidance events are multicellular and complex, and therefore results are not always easy to interpret (e.g., as highlighted above for [22–30]). Likewise, although cell culture experiments are a crucial tool that have provided invaluable insights into the cytoskeletal dynamics of extending neurites and the mechanisms regulating them, they also have their limitations (e.g., see [8,14]), including that neurons are post-mitotic, for which there are a paucity of cell lines, and less work has been done to define the molecular and biochemical mechanisms of axonal cytoskeletal responses in vivo. Indeed, given the historical difficulties in labeling/following the extension of axons in vivo [34], significantly more is known about cell migration and the biology of the cytoskeleton and organelles in the soma than about their dynamic changes in neurites. Additionally, although progress is being made (e.g., [22,25]) and new technologies such as Super-resolution microscopy have been developed, the technical difficulties associated with seeing and defining what is happening inside of tiny axons and their extending growth cone tips in vivo (including the dynamics of their actin and microtubule cytoskeletons, the coordinated movement of proteins within them, the spatiotemporal activation of specific signaling cascades, etc.) have slowed our understanding of how neurons are specifically controlled to enable neurite formation, growth, and guidance.
Not just Neurons: other types of cells also extend membranous processes
So, are there additional strategies that might aid in the goal of further elucidating these mechanisms? Although less well appreciated, many other types of cells – from endothelial to immune to glial to bone to cancer cells – also send out similar finger-like processes that are crucial for their specialized functions (Fig 1B). Like neurons, these cells also depend on actin and microtubules to extend their processes. Indeed, it has long been observed that many different types of cells contain similarly shaped processes/extensions (Fig 1B; e.g., [35–38]) – and that motile cells have a common cell biological feature: the polarized formation of cytoplasmic protrusions that determine the direction of movement (e.g., Fig 3A–D; [38–44]). Likewise, not only do cellular extensions, including axons, form as the result of these protrusive events (e.g., Fig 3C–D), but the tips of elongating processes, including axons/growth cones, continually send out new protrusions (filopodia), which allow these cellular extensions to change directions (e.g., Fig 3E). Moreover, at least some of these non-neuronal cell types, such as endothelial cells (and their involvement in angiogenesis/vasculogenesis), use many of the same cues and signaling systems to alter their cellular morphology in ways similar to neurons [45–47]. Yet, as in the case of neurons and their processes, there is not a complete understanding of the mechanisms generating and shaping them. Indeed, despite the fact that some of these cells are much simpler in form than neurons or endothelial cells (see Fig 1B), for what appears to be a combination of reasons – e.g., a limited appreciation of the importance of studying these questions in a simple system, the absence of a unified research focus aimed at using a simple system to answer these questions, or the “right” simple system not being employed, etc. – there are no actionable answers to these basic cell biological questions of how membranous processes form, extend, elongate, orient, and stop growing in any system.
Figure 3. Development and morphology of membranous process extension and navigation.
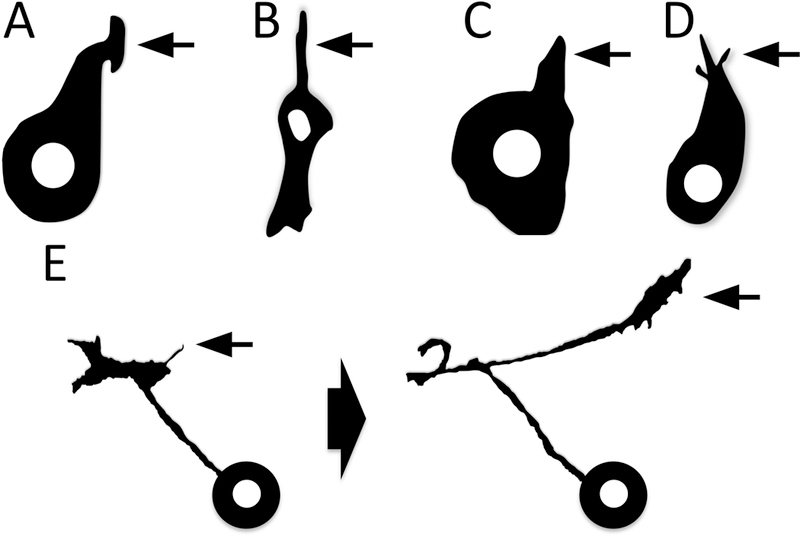
(A–D) The extension of cellular processes (arrows) – from neutrophil (A) and mesenchymal (B) cells in vitro to bristle (C) and neuronal (D) cells in vivo. Adapted from [74,104–106]. (E) Note also the means by which axons extend “new” processes/change the direction of their process extension – via a new protrusion/filopodium (E, left arrow), which then forms the new growing cellular process/axon (E, right arrow). Adapted from [87].
A Complementary Strategy: other types of cellular process-containing cells as models/prototypes
Therefore, unlike neuronal specific phenomena such as synapse formation or synaptic transmission, cellular extensions are not unique to neurons. Thus, critical long-standing questions that the field is interested in answering – How do cellular processes such as axons form? Extend? Change shape? What controls their length and directionality? How might we get them to regrow if damaged? – may be thought of broadly as cell biological problems that can also be investigated in simpler non-neuronal membranous process-containing cells. Although these events in neurons are undoubtedly more complex and bring additional elements into the mix (e.g., the coordination of cytoskeletal dynamics with substrate/cell adhesion and the guidance of axons over long distances with multiple choice points, rather than the simple orientation/reorientation of membranous extensions), this reductionist approach of using simple cellular process-containing cells as a prototype to help address important long-standing questions takes a page from the long history of the elegant work using the simple eukaryote yeast as a prototype (“reference model”) to answer important biological questions [48], so that one can then decipher more complex events than those occurring in yeast. Thus, a simple process-containing cell – such as the simple bristle cell of the fruit fly Drosophila (Figs 1B, 4A–B), as one example – is a similar powerful model that can be brought to bear and add to the research being done on these biomedically-critical questions.
Figure 4. The bristle cell as a simple membranous process-containing cell.
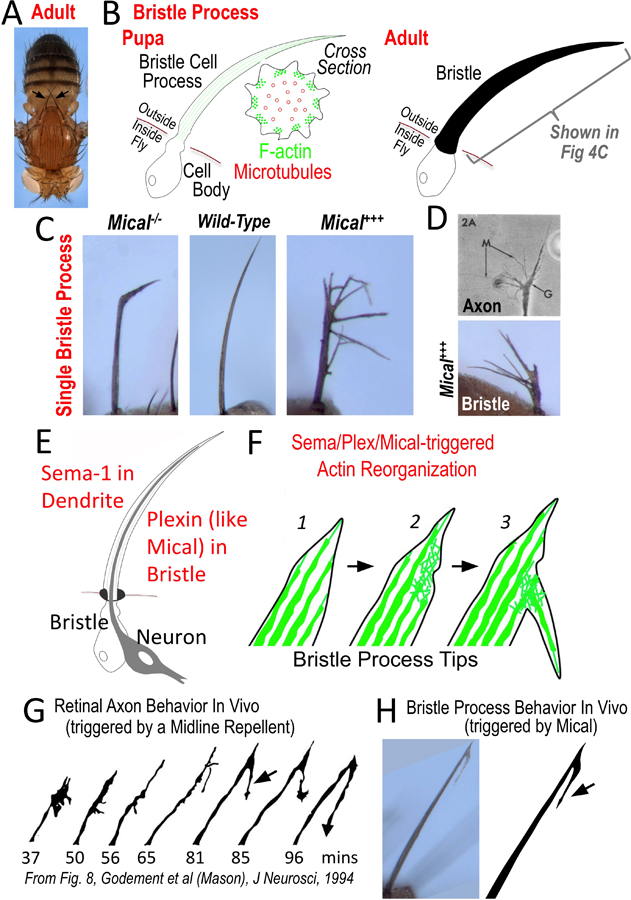
(A–B) The bristle cell with its long process provides a model cellular process. (A) Note the long single cell bristle processes on the body of the adult fly (e.g., each arrow points to a single bristle cell with its process). Images reproduced with permission from [74]. (B, left) The stereotypical arrangement of F-actin and microtubules pushes out the bristle process during pupal development. (B, right) The bristle cell also secretes a chitin cuticle that wraps around its cellular process – preserving a record in the adult of its developmental history and allowing a rapid initial characterization (in different contexts and genetic backgrounds) without requiring tissue processing. (C–D) Changing Mical levels alters bristle process extension and morphology – including (D) inducing it to resemble an axon growth cone. Images reproduced with permission from [74,76,107]. (E–F) Sema (on dendrite) interacts with Plexin (on bristle), to activate Mical within the bristle to induce cellular remodeling and branching (F; [74]). (F) This remodeling and branching occurs through the disassembly of F-actin (green) within bristle processes (2), which then allows currently unknown factors to form this actin-rich branch (3). (G–H) The Sema/Plexin/Mical-induced bristle branching (H, arrow) is remarkably similar to an axon’s response in vivo to a repellent (G, arrow). In particular, the axon’s (growth cone’s) response is to decrease in size (G, compare 37mins to 50 and 56mins) and then form a new “back” branch (G, arrow) [87] – which is the same response seen in the bristle upon Mical activation (H; [74]). Image reproduced with permission from [87].
A Simple Model: employing the bristle cell to decipher the molecular and cellular biology that builds a membranous process
Due to its simplicity, accessibility, large-size, and stereotypical actin and microtubule organization [49–52], the bristle process (Figs 1B, 4A–B) provides an attractive complementary tool for deciphering the mechanisms that generate and shape a cellular process. Specifically: 1) The bristle cell is a simple isolated single cell with only one extending long process (Fig 4A–B; [49,52]), thereby making it morphologically less complex than a neuron or other membranous process-containing cells (e.g., compare the bristle cell in Fig 1B to the other membranous process-containing cells). 2) The bristle cell’s anatomy and development have been well-defined and are indistinguishable from cell-to-cell/animal-to-animal (Fig 4A–B; [49,52]). 3) The bristle cell’s prominent stereotypic organization (Fig 4A–B; [49,52]) promotes detailed cytoskeletal, membranous, and organelle resolution and characterization (including the ability to measure actin and microtubule dynamics in real-time). 4) The bristle cell is an in vivo model (Fig 4A–B; [49,52]), and thus more emblematic of in situ (native) conditions than cell/tissue culturing. 5) The bristle cell and its process are large in size (Fig 4A–B; [49,52]), which simplifies analysis and enables high-throughput visualization/delineation, including with a dissecting stereomicroscope. 6) The bristle cell extends its process outside of the fly’s body (Fig 4A–B; [49,52]), which allows a range of high-resolution and time-lapse imaging approaches without requiring tissue processing/sectioning. 7) The bristle cell’s presence in Drosophila (Fig 4A–B; [49,52]) makes it amenable to efficient and extensive gene-editing/manipulation approaches (including tracking specific proteins over time and increasing or decreasing their levels in a bristle cell-specific/single cell-specific manner). 8) The bristle cell’s isolated nature (Fig 4A–B; [49,52]) allows for pharmacological manipulations and other selective alterations such as single-cell culturing. In the same way, available studies reveal that the molecules that shape the bristle process are similar to those used to shape axons and dendrites. In particular, mutations affecting Drosophila bristle shape, extension, and orientation have been known for over 100 years [53] – many of which have turned out to mutate specific actin and microtubule regulatory proteins that are also critical for axon growth and guidance such as bundlers, assemblers, and disassemblers [31]. Indeed, the bristle has long served as an in vivo model to characterize the molecules and mechanisms that regulate the cytoskeleton [49–52]. Thus, the bristle cell provides a precise high-resolution tractable tool for defining the cell and molecular biology that builds a membranous process.
Proof of Principle: gaining insights from simple cellular processes
This idea of using as a model the simplest of cellular processes – such as the bristle – has also arisen from our results using simple model systems to investigate the mechanisms of axon guidance. Specifically, multiple labs have been using the Drosophila model system to investigate how one of the largest families of axon guidance cues, the Semaphorins (with over 20 members conserved from invertebrates to humans; [54–56]), exert their effects (e.g., [29,57–64]). Semaphorins (Semas) are best known for their inhibitory/repulsive effects on extending axons [52,65] – but they also regulate the motility of cells throughout the body including those involved in immunity, angiogenesis, cardiovascular development, and cancer [66–69]. Semas elicit destabilizing effects on actin filaments (F-actin) that include a loss of F-actin, the decreased ability to polymerize new F-actin, a decrease in the number of F-actin bundles, and the regulation of F-actin-rich filopodia/spines/branches [52]. Yet, despite significant progress in the identification of Sema receptors (e.g., Plexins) and their signaling pathways [18,56,70,71], the molecules and mechanisms linking them to the control of the cytoskeleton have remained poorly understood.
To further understand these molecular and biochemical mechanisms, multiple groups have been searching for molecules that work together with Semas/Plexins. For example, we have been studying a large cytosolic protein, Mical, which has multiple protein interaction domains and is a member of a new family of Plexin-interacting proteins (the MICALs) [72]. MICALs are conserved from flies to humans and direct Sema-mediated repulsive axon guidance [72,73]. Yet, a major question that emerged early-on was: what is the specific role of MICAL proteins in axon guidance? MICALs showed similarity to oxidoreductase (Redox) enzymes [72], thereby suggesting the intriguing hypothesis that oxidation-reduction (Redox) signaling mechanisms might play a role in axon guidance [72]. Interestingly, as we tested this possibility in axons, we noticed that surviving Mical mutants exhibited defects in the shape, length, and orientation of their bristle cell processes (Fig 4C, compare middle and left; [74]). Likewise, elevating Mical levels dramatically rearranged bristle processes in a Redox-dependent manner – “transforming” unbranched bristles into branched structures (Fig 4C, right; Fig 4D, bottom [74]) with a similar degree of morphological complexity to that observed in navigating growth cones (Fig 4D, top). Harnessing this high-resolution, single cell model revealed that Mical does not alter bristle cell proliferation, differentiation, or survival – but specifically co-localizes with and controls the organization of F-actin during development [74]. Moreover, developing the bristle cell as a model has been instrumental in defining the effects of this Mical Redox enzyme on F-actin, revealing: 1) that Mical uses F-actin as a direct substrate – which it post-translationally oxidizes to simultaneously dismantle filaments and prevent polymerization [74,75], 2) that Mical is counteracted by a specific reductase enzyme called SelR/MsrB [76], 3) that MICALs do not function in an isolated manner but integrate with other well-known actin regulatory proteins and signaling pathways to drive cytoskeletal remodeling [29,77], and 4) that mammalian MICALs work in a similar manner biochemically and in vivo to Drosophila Mical [78]. Importantly, our findings using the bristle have now also been confirmed in neurons and their axonal and dendritic processes, as well as in in vitro systems and other cells including muscle, skin, and immune cells (e.g., [63,74,76,79–86]) – indicating that these important mechanisms have commonality in process extension among simple and complex cells and further validating the use of the bristle process as a model.
Thus, the bristle system has been an instrument for discovery of new molecular and biochemical mechanisms controlling the actin cytoskeleton. Likewise, the bristle system has provided a new understanding of the cellular mechanisms that generate and shape membranous processes. In particular, the repellent Sema is present in the dendrite that extends alongside the developing bristle (Fig 4E; [74]) and Sema activates Plexin/Mical signaling in the bristle to locally disassemble F-actin (Fig 4F; [74]). This F-actin disassembly leads to an increased complexity of the bristle process, with extending filopodia/branches (Fig 4F; [74]). The response of the bristle process to Sema/Plexin/Mical is therefore similar to the effect that cellular repellents including Semas, Slits, and Ephrins have on axons – triggering an initial disassembly of the actin cytoskeleton and collapse of the growth cone, but ultimately inducing the growth cone to reform in a more complex and branched organization (reviewed in [52]). Employing the cell biological resolution of the bristle cell process indicated that this branching results from a Mical-induced “transformation” of parallel-arranged bundles of F-actin, which are a hallmark of bristles and growth cone filopodia, into branched meshwork arrays of F-actin that are reminiscent of that seen in lamellipodia (Fig 4F; [74]). These and other observations have allowed the formulation of a model that repellents such as Semas disassemble (prune back) the actin network in vivo, and this pruning process initiates secondary events that serve to enhance cellular complexity/plasticity [52,74]. Strikingly, this Sema/Plexin/Mical-triggered bristle morphology is similar in appearance to what has been described when an elongating axon contacts a repellent in vivo (compare Fig 4G and 4H; arrows; [87]): the axonal growth cone “collapses” and then sends out a “back” branch that allows the axon to navigate away in a new direction (Fig 4G; [87]). Thus, a simple membranous process-containing cell, such as the bristle cell, is one such reductionist system that can provide new insights into the cellular, molecular, and biochemical mechanisms that generate membranous extensions.
Conclusions
Stimulating membranous process formation, extension, and navigation – in the diseased or damaged brain or spinal cord, for example – is crucial for curing many devastating pathologies. Yet, how can we hope to get cellular extensions such as axons to regrow if we do not know how they form, grow, and navigate? The observations highlighted in this review support the view that we can learn a great deal from simple systems, which can be applied back to more complex ones. Specifically, we propose that it will be advantageous to harness the attributes of simpler membranous process-containing cells and use them as a prototype for neurons. We suggest that simple model processes, such as the bristle process, provide a means to determine how a cellular process is built, thereby answering fundamental cell biological questions that are highly relevant to neuroscience, including defining 1) the membrane and organelle biology of a membranous process, 2) the cytoskeletal biology of a membranous process, 3) the cell-cell interactions specifying a process, 4) the membrane, organelle, and cytoskeletal biology of branch formation, 5) the gene expression of a process-containing cell, and 6) the genes that build a process. Of course, as new molecules and mechanisms are discovered using these simple but powerful model processes, they should continue to be tested in neurons using available approaches. Nevertheless, even if only basic mechanisms of formation and extension are translatable, understanding how one simple membranous process is built will provide a framework for manipulations of all types of membranous process-containing cells, including different types of neurons. In conclusion, it is our current opinion that this strategy provides a complementary approach to work being done in neurons, with the goal of significantly advancing the understanding, diagnosis, and treatment of diseases not only affecting the nervous system, but also many other tissues.
HIGHLIGHTS.
Technical hurdles and complexity have hindered our knowledge of neuronal extension
Other types of cells also extend similar axonal/dendritic-like membranous processes
These other types of process-containing cells can serve as simple models for neurons
These simple cells have uncovered mechanisms that form, extend, and navigate axons
ACKNOWLEDGMENTS
We thank Carla Green, Ege Kavalali, Helmut Krämer, and members of the Terman lab for comments. Supported by NIH (MH085923) and Welch Foundation (I-1749) grants to J.R.T.
REFERENCES
- 1.Cajal S: Degeneration and regeneration of the nervous system New York: Hafner; 1928. [Google Scholar]
- 2.Sperry RW: The growth of nerve circuits. Sci Am 1959, 201:68–75. [DOI] [PubMed] [Google Scholar]
- 3.Tessier-Lavigne M, Goodman CS: The molecular biology of axon guidance. Science 1996, 274:1123–1133. [DOI] [PubMed] [Google Scholar]
- 4.Pasterkamp RJ, Kolodkin AL: SnapShot: Axon Guidance. Cell 2013, 153:494, 494e491–492. [DOI] [PubMed] [Google Scholar]
- 5.Kolodkin AL, Pasterkamp RJ: SnapShot: Axon guidance II. Cell 2013, 153:722 e721. [DOI] [PubMed] [Google Scholar]
- 6.Lin C-H, Thompson CA, Forscher P: Cytoskeletal reorganization underlying growth cone motility. Curr. Op. Neurobiol 1994, 4:640–647. [DOI] [PubMed] [Google Scholar]
- 7.Dickson BJ: Molecular mechanisms of axon guidance. Science 2002, 298:1959–1964. [DOI] [PubMed] [Google Scholar]
- 8.Dent EW, Gertler FB: Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 2003, 40:209–227. [DOI] [PubMed] [Google Scholar]
- 9.Huber AB, Kolodkin AL, Ginty DD, Cloutier JF: Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci 2003, 26:509–563. [DOI] [PubMed] [Google Scholar]
- 10.Parrish JZ, Emoto K, Kim MD, Jan YN: Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 2007, 30:399–423. [DOI] [PubMed] [Google Scholar]
- 11.Lowery LA, Van Vactor D: The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol 2009, 10:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raper J, Mason C: Cellular strategies of axonal pathfinding. Cold Spring Harb Perspect Biol 2010, 2:a001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashaw GJ, Klein R: Signaling from axon guidance receptors. Cold Spring Harb Perspect Biol 2010, 2:a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dent EW, Gupton SL, Gertler FB: The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol 2011, 3:a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vitriol EA, Zheng JQ: Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron 2012, 73:1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shigeoka T, Lu B, Holt CE: Cell biology in neuroscience: RNA-based mechanisms underlying axon guidance. J Cell Biol 2013, 202:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez TM, Letourneau PC: Actin dynamics in growth cone motility and navigation. J Neurochem 2014, 129:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiradake E, Jones EY, Klein R: Structural Perspectives on Axon Guidance. Annu Rev Cell Dev Biol 2016, 32:577–608. [DOI] [PubMed] [Google Scholar]
- 19.Menon S, Gupton SL: Building Blocks of Functioning Brain: Cytoskeletal Dynamics in Neuronal Development. Int Rev Cell Mol Biol 2016, 322:183–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales D, Kania A: Cooperation and crosstalk in axon guidance cue integration: Additivity, synergy, and fine-tuning in combinatorial signaling. Dev Neurobiol 2017, 77:891–904. [DOI] [PubMed] [Google Scholar]
- 21.Stoeckli ET: Understanding axon guidance: are we nearly there yet? Development 2018, 145.*The author reviews the recent progress and revisions made in the axon guidance field and speaks to areas in the field that are still in their infancy.
- 22.Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, Hiesinger PR: The Developmental Rules of Neural Superposition in Drosophila. Cell 2015, 162:120–133.**The authors’ use of multi-photon microscopy in intact fly pupae to follow photoreceptor axons over time and anatomical mapping along with meticulous, large-scale quantification of growth cone filopodia dynamics, led them to define a set of simple rules that are sufficient for wiring specificity of these neurons; suggesting fundamentally important mechanisms of growth cone guidance that have previously been missed from studies of fixed tissue.
- 23.Ozel MN, Langen M, Hassan BA, Hiesinger PR: Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. Elife 2015, 4:e10721.**Using continuous, fast, and high-resolution imaging over long periods to examine growth cone dynamics in cultured developing Drosophila brains, the authors observed differences in growth cone dynamics from what had previously been understood from studies using still imaging, including differentiating effects on stabilizing growth cones versus “guiding” growth cones to their targets. Additionally, the authors find that in contrast to previous thought, N-cadherin plays a role in the stabilization of R7 photoreceptor growth cones and not in their targeting.
- 24.McConnell RE, Edward van Veen J, Vidaki M, Kwiatkowski AV, Meyer AS, Gertler FB: A requirement for filopodia extension toward Slit during Robo-mediated axon repulsion. J Cell Biol 2016, 213:261–274.**The authors find that contrary to traditional thought that repellents induce actin disassembly, the repulsive guidance cue Slit stimulates the formation and elongation of actin-based filopodia that are required for subsequent axonal repulsion away from Slit.
- 25.Akin O, Zipursky SL: Frazzled promotes growth cone attachment at the source of a Netrin gradient in the Drosophila visual system. Elife 2016, 5:e20762.**See also Refs. 27 and 28, using live imaging in intact Drosophila pupae the authors find that R8 photoreceptor growth cones reach and recognize their targets without Netrin signaling, but do not remain attached to them; revealing that contrary to traditional thought, at least in some contexts a graded ligand distribution of netrin along a guidance path is not used for chemoattraction but for target adhesion.
- 26.Long H, Yoshikawa S, Thomas JB: Equivalent Activities of Repulsive Axon Guidance Receptors. J Neurosci 2016, 36:1140–1150.*Using genetics and the high-resolution Drosophila midline model, the authors surprisingly find that there is no qualitative difference between the effects induced by chimeric receptors composed of three different repulsive guidance receptors, indicating they converge on a common repulsive pathway.
- 27.Dominici C, Moreno-Bravo JA, Puiggros SR, Rappeneau Q, Rama N, Vieugue P, Bernet A, Mehlen P, Chedotal A: Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature 2017, 545:350–354.**Similar to Ref. 28, and see also Ref 25, the authors use specific genetic approaches to find that the traditional view that Netrin’s action on commissural axons is mediated by a chemoattractant gradient of floorplate-derived netrin-1 is incomplete, and that Netrin acts locally by promoting growth cone adhesion and haptotaxis.
- 28.Varadarajan SG, Kong JH, Phan KD, Kao TJ, Panaitof SC, Cardin J, Eltzschig H, Kania A, Novitch BG, Butler SJ: Netrin1 Produced by Neural Progenitors, Not Floor Plate Cells, Is Required for Axon Guidance in the Spinal Cord. Neuron 2017, 94:790–799.**Similar to Ref 27, and see also Ref 25, the authors find that the long-held model regarding the critical role for long-range chemoattraction for commissural axons by netrin is incomplete, and that Netrin supplied by neural progenitors guides commissural axons in the developing spinal cord by depositing it on the pial surface, which ventrally directs axonal growth by haptotaxis.
- 29.Yoon J, Kim SB, Ahmed G, Shay JW, Terman JR: Amplification of F-Actin Disassembly and Cellular Repulsion by Growth Factor Signaling. Dev Cell 2017, 42:117–129.**Cellular behaviors are controlled by extracellular cues that have been historically classified into two independently acting and antagonistic groups – growth-promoting/attractants and growth-preventing/repellents – but here the authors critically amend long-held beliefs regarding the antagonistic action of these opposing groups of cues by finding that a recognized growth-promoting signaling pathway amplifies the F-actin disassembly and repulsive effects of a growth-preventing pathway.
- 30.Liao CP, Li H, Lee HH, Chien CT, Pan CL: Cell-Autonomous Regulation of Dendrite Self-Avoidance by the Wnt Secretory Factor MIG-14/Wntless. Neuron 2018, 98:320–334 e326.*The authors’ findings surprisingly support a model that dendrite self-avoidance occurs at least in some contexts by orchestrating F-actin assembly at dendrite tips coincident with dendrite contact and retraction.
- 31.Svitkina T: The Actin Cytoskeleton and Actin-Based Motility. Cold Spring Harb Perspect Biol 2018, 10:a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borisy G, Heald R, Howard J, Janke C, Musacchio A, Nogales E: Microtubules: 50 years on from the discovery of tubulin. Nat Rev Mol Cell Biol 2016, 17:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahn R, Lang T, Sudhof TC: Membrane fusion. Cell 2003, 112:519–533. [DOI] [PubMed] [Google Scholar]
- 34.Morecraft RJ, Ugolini G, Lanciego JL, Wouterlood FG, Pandya DN: Classic and contemporary neural tract-tracing techniques. In Diffusion MRI: from quantitative measurement to in vivo neuroanatomy, edn 2nd Edited by Johansen-Berg H, Behrens TEJ: Elsevier; 2014:359–399. [Google Scholar]
- 35.Wolpert L, Gingell D: Cell surface membrane and amoeboid movement. Symp Soc Exp Biol 1968, 22:169–198. [PubMed] [Google Scholar]
- 36.Vasiliev JM, Gelfand IM: Mechanisms of morphogenesis in cell cultures. Int Rev Cytol 1977, 50:159–274. [DOI] [PubMed] [Google Scholar]
- 37.Trinkaus JP: Cells into organs–the forces that shape the embryo edn Second Englewood Cliffs, NJ: Prentice-Hall Inc.; 1984. [Google Scholar]
- 38.Bray D: Cell Movements New York & London: Garland Publishing, Inc; 1992. [Google Scholar]
- 39.Condeelis J: Life at the leading edge: the formation of cell protrusions. Annu Rev Cell Biol 1993, 9:411–444. [DOI] [PubMed] [Google Scholar]
- 40.Stossel TP: On the Crawling of Animal-Cells. Science 1993, 260:1086–1094. [DOI] [PubMed] [Google Scholar]
- 41.Lauffenburger DA, Horwitz AF: Cell migration: a physically integrated molecular process. Cell 1996, 84:359–369. [DOI] [PubMed] [Google Scholar]
- 42.Rafelski SM, Theriot JA: Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu Rev Biochem 2004, 73:209–239. [DOI] [PubMed] [Google Scholar]
- 43.Chhabra ES, Higgs HN: The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol 2007, 9:1110–1121. [DOI] [PubMed] [Google Scholar]
- 44.Ridley AJ: Life at the leading edge. Cell 2011, 145:1012–1022. [DOI] [PubMed] [Google Scholar]
- 45.Carmeliet P, Tessier-Lavigne M: Common mechanisms of nerve and blood vessel wiring. Nature 2005, 436:193–200. [DOI] [PubMed] [Google Scholar]
- 46.Adams RH, Eichmann A: Axon guidance molecules in vascular patterning. Cold Spring Harb Perspect Biol 2010, 2:a001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chauvet S, Burk K, Mann F: Navigation rules for vessels and neurons: cooperative signaling between VEGF and neural guidance cues. Cell Mol Life Sci 2013, 70:1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Botstein D, Fink GR: Yeast: an experimental organism for 21st Century biology. Genetics 2011, 189:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilney LG, DeRosier DJ: How to make a curved Drosophila bristle using straight actin bundles. Proc Natl Acad Sci U S A 2005, 102:18785–18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutherland JD, Witke W: Molecular genetic approaches to understanding the actin cytoskeleton. Curr Opin Cell Biol 1999, 11:142–151. [DOI] [PubMed] [Google Scholar]
- 51.Bitan A, Guild G, Abdu U: The highly elongated Drosophila mechanosensory bristle--a new model for studying polarized microtubule function. Fly (Austin) 2010, 4:246–248. [DOI] [PubMed] [Google Scholar]
- 52.Hung R- J, Terman JR: Extracellular inhibitors, repellents, and Semaphorin/Plexin/MICAL-mediated actin filament disassembly. Cytoskeleton 2011, 68:415–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan TH, Bridges CB: Sex-linked inheritance in Drosophila. Publs. Carnegie Instn 1916, 237:1–88. [Google Scholar]
- 54.Kolodkin AL, Matthes DJ, O’Connor TP, Patel NH, Admon A, Bentley D, Goodman CS: Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron 1992, 9:831–845. [DOI] [PubMed] [Google Scholar]
- 55.Kolodkin AL, Matthes DJ, Goodman CS: The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 1993, 75:1389–1399. [DOI] [PubMed] [Google Scholar]
- 56.Alto LT, Terman JR: Semaphorins and their Signaling Mechanisms. Methods Mol Biol 2017, 1493:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo SK, Pascoe HG, Pereira T, Kondo S, Jacinto A, Zhang X, Hariharan IK: Plexins function in epithelial repair in both Drosophila and zebrafish. Nat Commun 2016, 7:12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Syed DS, Gowda SB, Reddy OV, Reichert H, VijayRaghavan K: Glial and neuronal Semaphorin signaling instruct the development of a functional myotopic map for Drosophila walking. Elife 2016, 5:e11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasse S, Klambt C: Repulsive Epithelial Cues Direct Glial Migration along the Nerve. Dev Cell 2016, 39:696–707. [DOI] [PubMed] [Google Scholar]
- 60.Meltzer S, Yadav S, Lee J, Soba P, Younger SH, Jin P, Zhang W, Parrish J, Jan LY, Jan YN: Epidermis-Derived Semaphorin Promotes Dendrite Self-Avoidance by Regulating Dendrite-Substrate Adhesion in Drosophila Sensory Neurons. Neuron 2016, 89:741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen HC, Chu SY, Hsu TC, Wang CH, Lin IY, Yu HH: Semaphorin-1a prevents Drosophila olfactory projection neuron dendrites from mis-targeting into select antennal lobe regions. PLoS Genet 2017, 13:e1006751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez-Fleming M, Rohrbach EW, Bashaw GJ: Sema-1a Reverse Signaling Promotes Midline Crossing in Response to Secreted Semaphorins. Cell Rep 2017, 18:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orr BO, Fetter RD, Davis GW: Retrograde semaphorin-plexin signalling drives homeostatic synaptic plasticity. Nature 2017, 550:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeong S, Yang DS, Hong YG, Mitchell SP, Brown MP, Kolodkin AL: Varicose and cheerio collaborate with pebble to mediate semaphorin-1a reverse signaling in Drosophila. Proc Natl Acad Sci U S A 2017, 114:E8254–E8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo Y, Raible D, Raper JA: Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 1993, 75:217–227. [DOI] [PubMed] [Google Scholar]
- 66.Gu C, Giraudo E: The role of semaphorins and their receptors in vascular development and cancer. Exp Cell Res 2013, 319:1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumanogoh A, Kikutani H: Immunological functions of the neuropilins and plexins as receptors for semaphorins. Nat Rev Immunol 2013, 13:802–814. [DOI] [PubMed] [Google Scholar]
- 68.Worzfeld T, Offermanns S: Semaphorins and plexins as therapeutic targets. Nat Rev Drug Discov 2014, 13:603–621. [DOI] [PubMed] [Google Scholar]
- 69.Epstein JA, Aghajanian H, Singh MK: Semaphorin signaling in cardiovascular development. Cell Metab 2015, 21:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascoe HG, Wang Y, Zhang X: Structural mechanisms of plexin signaling. Prog Biophys Mol Biol 2015, 118:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battistini C, Tamagnone L: Transmembrane semaphorins, forward and reverse signaling: have a look both ways. Cell Mol Life Sci 2016, 73:1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL: MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell 2002, 109:887–900. [DOI] [PubMed] [Google Scholar]
- 73.Alto LT, Terman JR: MICALs. Curr Biol 2018, 28:R538–R541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR: Mical links semaphorins to F-actin disassembly. Nature 2010, 463:823–827.**Initial observation that Semaphorin-Plexin-Mical signaling affects the morphology of the membranous processes of simple (bristle) cells.
- 75.Hung RJ, Pak CW, Terman JR: Direct redox regulation of F-actin assembly and disassembly by Mical. Science 2011, 334:1710–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR: SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol 2013, 15:1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grintsevich EE, Yesilyurt HG, Rich SK, Hung RJ, Terman JR, Reisler E: F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat Cell Biol 2016, 18:876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu H, Yesilyurt HG, Yoon J, Terman JR: The MICALs are a Family of F-actin Dismantling Oxidoreductases Conserved from Drosophila to Humans. Sci Rep 2018, 8:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giridharan SS, Rohn JL, Naslavsky N, Caplan S: Differential regulation of actin microfilaments by human MICAL proteins. J Cell Sci 2012, 125:614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee BC, Peterfi Z, Hoffmann FW, Moore RE, Kaya A, Avanesov A, Tarrago L, Zhou Y, Weerapana E, Fomenko DE, et al. : MsrB1 and MICALs Regulate Actin Assembly and Macrophage Function via Reversible Stereoselective Methionine Oxidation. Mol Cell 2013, 51:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lundquist MR, Storaska AJ, Liu TC, Larsen SD, Evans T, Neubig RR, Jaffrey SR: Redox Modification of Nuclear Actin by MICAL-2 Regulates SRF Signaling. Cell 2014, 156:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Battum EY, Gunput RA, Lemstra S, Groen EJ, Yu KL, Adolfs Y, Zhou Y, Hoogenraad CC, Yoshida Y, Schachner M, et al. : The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat Commun 2014, 5:4317. [DOI] [PubMed] [Google Scholar]
- 83.Fremont S, Hammich H, Bai J, Wioland H, Klinkert K, Rocancourt M, Kikuti C, Stroebel D, Romet-Lemonne G, Pylypenko O, et al. : Oxidation of F-actin controls the terminal steps of cytokinesis. Nat Commun 2017, 8:14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grintsevich EE, Ge P, Sawaya MR, Yesilyurt HG, Terman JR, Zhou ZH, Reisler E: Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun 2017, 8:2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vitali T, Maffioli E, Tedeschi G, Vanoni MA: Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and modulation by its CH, LIM and C-terminal domains. Arch Biochem Biophys 2016, 593:24–37. [DOI] [PubMed] [Google Scholar]
- 86.McDonald CA, Liu YY, Palfey BA: Actin stimulates reduction of the MICAL-2 monooxygenase domain. Biochemistry 2013, 52:6076–6084. [DOI] [PubMed] [Google Scholar]
- 87.Godement P, Wang LC, Mason CA: Retinal axon divergence in the optic chiasm: dynamics of growth cone behavior at the midline. J Neurosci 1994, 14:7024–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jabr F: Know your neurons: the discovery and naming of the neuron. Scientific American 2012.
- 89.Yuste R: Dendritic spines and distributed circuits. Neuron 2011, 71:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Smet F, Segura I, De Bock K, Hohensinner PJ, Carmeliet P: Mechanisms of vessel branching: filopodia on endothelial tip cells lead the way. Arterioscler Thromb Vasc Biol 2009, 29:639–649. [DOI] [PubMed] [Google Scholar]
- 91.Tilney LG, Tilney MS, Guild GM: F actin bundles in Drosophila bristles. I. Two filament cross-links are involved in bundling. J Cell Biol 1995, 130:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.The Open University, Hearing: the structure and function of the inner ear. An OpenLearn chunk reworked by permission of The Open University Copyright 1999–2018 (https://creativecommons.org/licenses/by-nc-sa/4.0/deed.en_GB). 2016.
- 93.Gschmeissner S: Metastasis of a cancerous cell, SEM. Science Source Images
- 94.Holloway I, Kayser M, Lee DA, Bader DL, Bentley G, Knight MM: Increased presence of cells with multiple elongated processes in osteoarthritic femoral head cartilage. Osteoarthritis Cartilage 2004, 12:17–24. [DOI] [PubMed] [Google Scholar]
- 95.Mescher AL: Connective Tissue. In Junqueira’s Baic Histology Edited by: McGraw-Hill; 2013. [Google Scholar]
- 96.Gray H, Lewis WH: Anatomy of the human body edn 20th Philadelphia and New York: Lea & Febiger; 1918. [Google Scholar]
- 97.Cummings B: Epidermal Cell Types Pearson Education; 2006. [Google Scholar]
- 98.Humoral immunity. Antibody Structure. Antibody diversity (http://slideplayer.com/slide/10422023/).
- 99.SlideShare (https://www.slideshare.net/Medical_PPT_Images/neuroglial-cells-astrocyte-medical-images-for-power-point).
- 100.Rezaie P, Male D: Mesoglia & microglia--a historical review of the concept of mononuclear phagocytes within the central nervous system. J Hist Neurosci 2002, 11:325–374. [DOI] [PubMed] [Google Scholar]
- 101.Assinger A: Platelets and infection - an emerging role of platelets in viral infection. Front Immunol 2014, 5:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robbins AH: Biological perspectives on human pigmentation Cambridge, UK: Cambridge University Press; 1991. [Google Scholar]
- 103.Abramsson A, Lindblom P, Betsholtz C: Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J Clin Invest 2003, 112:1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chodniewicz D, Klemke RL: Guiding cell migration through directed extension and stabilization of pseudopodia. Exp Cell Res 2004, 301:31–37. [DOI] [PubMed] [Google Scholar]
- 105.Weiss P: Cell contact. Int Rev Cytol 1958, 7:391–423. [Google Scholar]
- 106.Steinel MC, Whitington PM: The atypical cadherin Flamingo is required for sensory axon advance beyond intermediate target cells. Dev Biol 2009, 327:447–457. [DOI] [PubMed] [Google Scholar]
- 107.Yamada KM, Spooner BS, Wessells NK: Axon Growth: Roles of microfilaments and microtubules. Proc Natl Acad Sci U S A 1970, 66:1206–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]


