Abstract
Lateral meningocele syndrome (LMS), a genetic disorder characterized by meningoceles and skeletal abnormalities, is associated with NOTCH3 mutations. We created a mouse model of LMS (Notch3tm1.1Ecan) by introducing a tandem termination codon in the Notch3 locus upstream of the PEST domain. Microcomputed tomography demonstrated that Notch3tm1.1Ecan mice exhibit osteopenia. The cancellous bone osteopenia was no longer observed following the intraperitoneal administration of antibodies directed to the negative regulatory region (NRR) of Notch3. The anti-Notch3 NRR antibody suppressed the expression of Hes1, Hey1 and Hey2 (Notch target genes), and decreased Tnfsf11 (RANKL) mRNA in Notch3tm1.1Ecan osteoblast cultures. Bone marrow-derived macrophages (BMM) from Notch3tm1.1Ecan mutants exhibited enhanced osteoclastogenesis in culture; this was increased in co-cultures with Notch3tm1.1Ecan osteoblasts. Osteoclastogenesis was suppressed by anti-Notch3 NRR antibodies in Notch3tm1.1Ecan osteoblasts/BMM co-cultures. In conclusion, the cancellous bone osteopenia of Notch3tm1.1Ecan mutants is reversed by anti-Notch3 NRR antibodies.
Keywords: Lateral Meningocele Syndrome, Notch, Notch antibodies, bone remodeling, genetic disorders
INTRODUCTION
Notch receptors (Notch1 to 4) are transmembrane proteins that define cellular fate in multiple tissues including bone, where they influence skeletal development and bone homeostasis (Canalis, 2018; Fortini, 2009; Siebel & Lendahl, 2017; Zanotti & Canalis, 2016). Following interactions with ligands of the Jagged and Delta-like families, Notch receptors are activated. The extracellular domain is the site of Notch interacting with its ligands, and at the junction of the extracellular and the transmembrane domain rests the negative regulatory region (NRR), which is the site of cleavage necessary for the activation of Notch (Sanchez-Irizarry et al., 2004). Notch ligand interactions lead to the unfolding of the NRR making it accessible to ADAM metalloproteases and the γ-secretase complex for proteolytic cleavage freeing the Notch intracellular domain (NICD) (Gordon et al., 2015). The NICD translocates to the nucleus, and there it interacts with recombination signal-binding protein for Ig of κ (RBPJκ) and mastermind-like (MAML) to induce target gene transcription (Kovall, 2008; Nam, Sliz, Song, Aster, & Blacklow, 2006; Schroeter, Kisslinger, & Kopan, 1998; Wilson & Kovall, 2006). Genes induced by this canonical pathway include Hairy Enhancer of Split (Hes) and Hes-related with YRPW motif (Hey) (Iso, Kedes, & Hamamori, 2003; Kobayashi & Kageyama, 2014).
Notch1, 2 and 3 and low levels of Notch 4 mRNA are expressed by skeletal cells (Bai et al., 2008; Canalis, 2018; Zanotti & Canalis, 2017). Notch1 and Notch2 are detected in the osteoblast and osteoclast lineages, whereas Notch3 is present in the osteoblast but not in the osteoclast lineage. Although there is a degree of overlap in the function of Notch receptors, each Notch receptor has specific cellular patterns of expression and plays a unique role in skeletal physiology (Canalis, 2018). Notch1 suppresses the differentiation of osteoblasts and osteoclasts, Notch2 suppresses osteoblast differentiation but enhances osteoclast differentiation and Notch3 induces osteoclastogenesis by indirect mechanisms, demonstrating specific actions of Notch receptors in skeletal cells (Bai et al., 2008; Canalis, Schilling, Yee, Lee, & Zanotti, 2016; Canalis, Yu, Schilling, Yee, & Zanotti, 2018; Fukushima et al., 2008). Reaffirming the distinct function of each Notch receptor is the fact that loss- or gain-of-function mutations of the various Notch receptors are associated with distinct genetic diseases (Canalis, 2018; Zanotti & Canalis, 2016).
Lateral Meningocele Syndrome (LMS) or Lehman Syndrome (Online Mendelian Inheritance in Man 130720) is a rare genetic disorder characterized by craniofacial and skeletal abnormalities, meningoceles and neuromuscular dysfunction (Avela, Valanne, Helenius, & Makitie, 2011; Gripp et al., 1997; Lehman, Stears, Wesenberg, & Nusbaum, 1977). LMS is associated with short deletions or point mutations in exon 33 of NOTCH3 resulting in the premature termination of the protein product upstream of the proline (P), glutamic acid (E), serine (S) and threonine (T) (PEST) domain. This is necessary for the degradation of the NOTCH3 NICD, and the absence of the PEST domain results in the stabilization of the NOTCH3 protein (Gripp et al., 2015). Although the mutations extend the half-life of the NOTCH3 NICD, they do not activate Notch3 on their own since activation requires the proteolytic cleavage of the NRR. However, Notch3 activation is complex. Whereas ligand binding triggers unfolding of the NRR, and thus proteolytic cleavage of the Notch3 NRR domain, there is a degree of ligand-independent signaling, possibly leading to a gain-of-NOTCH3 function in LMS (Canalis et al., 2018; Choy et al., 2017; Siebel & Lendahl, 2017; Tiyanont, Wales, Siebel, Engen, & Blacklow, 2013; Xu et al., 2015).
We created a mouse model reproducing the functional aspects of mutations found in subjects afflicted by LMS (Canalis et al., 2018). In this model, termed Notch3tm1.1Ecan, a tandem termination codon was introduced into exon 33 of Notch3 causing the translation of a truncated NOTCH3 of 2230 amino acids lacking the PEST domain. Notch3tm1.1Ecan mice do not manifest the neuromuscular complications of the human disease, but heterozygous mice exhibit an increase in osteoclast number leading to a state of enhanced bone remodeling and osteopenia (Canalis et al., 2018).
In the present work, we attempted to answer the question as to whether the skeletal manifestations of Notch3tm1.1Ecan mutant mice could be reversed by intervention. For this purpose, Notch3tm1.1Ecan mice were treated with a novel antibody, anti-Notch3 NRR, that selectively inhibits signaling through the Notch3 receptor using the previously described mechanism in which antibody binding stabilizes the quiescent conformation of the NRR (Wu et al., 2010). Thus, our studies here aim to determine whether persistent Notch3 signaling is necessary to maintain the Notch3tm1.1Ecan phenotype and whether an anti-Notch3 NRR antibody could serve as a therapeutic modality in an experimental model of LMS. To establish the effect of the anti-Notch3 NRR antibody, Notch3tm1.1Ecan and control littermates were administered anti-Notch3 NRR or a non-targeting isotype control antibody (anti-ragweed) and characterized by bone microarchitectural analysis.
MATERIALS AND METHODS
Notch3tm1.1Ecan Mutant Mice—
Notch3tm1.1Ecan mutant mice have been described previously (Canalis et al., 2018). Briefly, the termination codon ACCAAG>TAATGA was inserted into the Notch3 locus at 6691–6696. The introduction of the tandem termination codon results in the translation of a truncated protein product of 2230 amino acids devoid of the PEST domain. Heterozygous Notch3tm1.1Ecan mutant mice in a C57BL/6J genetic background were mated with wild type C57BL/6 mice to create heterozygous Notch3tm1.1Ecan and littermate sex-matched controls for study. Genotypes were established by polymerase chain reaction (PCR) analysis of DNA in tail extracts using forward primer 5’-GTGCTCAGCTTTGGTCTGCTC-3’ and reverse primer 5’-CGCAGGAAGCGCCTCATTA-3’ for the Notch3tm1.1Ecan or 5’-CGCAGGAAGCGGGCCTTGG-3’ for the wild type allele (Integrated DNA Technologies (IDT), Coralville, IA), as described (Canalis et al., 2018). Notch3tm1.1Ecan mutant and control littermate male mice of 1 month of age were administered anti-Notch3 NRR or anti-ragweed antibody (Genentech, South San Francisco, CA), both suspended in phosphate buffered saline (PBS), intraperitoneally (IP) at a dose of 20 mg/Kg twice a week for a total of 8 doses and sacrificed at 2 months of age.
Anti-Notch3 NRR Antibody)—
Human antibodies targeting the human and mouse Notch3 NRR domain were generated via phage display technologies as described previously for Notch1 and Notch2 (Wu et al., 2010). To facilitate studies in mice, the human IgG1 antibody backbone was swapped for the mouse IgG2a backbone using standard DNA cloning techniques. Anti-Notch3 NRR (anti-NRR3.b21) was characterized in vitro and shown to selectively inhibit ligand-induced signaling from Notch3 but not Notch1 or Notch2. The control antibody is a nonbinding isotype control (as used by Wu et al., 2010) that carries the same mouse IgG2a backbone and targets the ragweed protein instead of Notch3.
Microcomputed Tomography (µCT)—
Microarchitectural analysis of femurs was conducted using a Scanco µCT 40 instrument (Scanco Medical AG, Bassersdorf, Switzerland), which underwent periodic calibrations using a manufacturer-provided phantom (Scanco Medical AG) (Bouxsein et al., 2010; Canalis, Kranz, & Zanotti, 2014; Glatt, Canalis, Stadmeyer, & Bouxsein, 2007; Yu, Zanotti, Schilling, & Canalis, 2018). Femoral bones were scanned in 70% ethanol at high resolution, energy level of 55 kVp, intensity of 145 µA, and integration time of 200 ms, as reported (Yu et al., 2018). One hundred slices at midshaft and 160 slices at the distal metaphysis were acquired at a thickness of 6 µm and isotropic voxel size of 216 µm3, and selected for analysis. Cancellous bone volume fraction (bone volume/total volume) and microarchitectural properties were assessed starting about 1.0 mm proximal from both femoral condyles. Contours were drawn manually at a 10 slice interval to define the region of interest for analysis, and the remaining slice contours were iterated automatically. Total volume, bone volume, number and thickness of trabeculae, connectivity density, structure model index (SMI) and material density were measured in cancellous bone using a Gaussian filter (σ = 0.8) and user defined thresholds (Bouxsein et al., 2010; Glatt et al., 2007). To analyze cortical bone, contours were iterated across 100 slices along the cortical shell at the midshaft of the femur, as reported (Canalis & Zanotti, 2017). Analysis of bone volume/total volume, cortical porosity and thickness, total cross sectional, marrow and cortical bone area, periosteal and endosteal perimeter and material density were carried out using a Gaussian filter (σ = 0.8, support = 1) with thresholds defined by the operator, as previously described (Canalis & Zanotti, 2017) .
Calvarial Osteoblast-enriched Cell Cultures—
Parietal bones from Notch3tm1.1Ecan mice and control littermates were obtained at 3 to 5 days of age and treated with Liberase TL 1.2 U/ml (Sigma-Aldrich, St. Louis, MO) for 20 min at 37°C and cells obtained in 5 sequential reactions, as reported (Canalis et al., 2018; Yesil et al., 2009). Cells from digestions 3 to 5 were pooled and seeded at 10,000 cells/cm2 density, as reported (Canalis et al., 2018; Canalis, Zanotti, & Smerdel-Ramoya, 2014). Osteoblast-enriched cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with non-essential amino acids (both from Thermo Fisher Scientific, Waltham, MA), 20 mM HEPES, 100 µg/ml ascorbic acid (both from Sigma-Aldrich) and 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA) in a 5% CO2 incubator at 37°C, as reported (Zanotti, Yu, Adhikari, & Canalis, 2018). Anti-Notch3 NRR antibody or control anti-ragweed antibody were tested at a concentration of 20 μg/ml of culture medium.
Bone Marrow-derived Macrophages (BMMs) Cultures—
BMMs were isolated by flushing the marrow from Notch3tm1.1Ecan and littermate mice with a 26 gauge needle, as described previously (Canalis, Sanjay, Yu, & Zanotti, 2017). Erythrocytes were lysed in 150 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA (pH 7.4) and cells were separated by centrifugation and suspended in α-minimum essential medium (α-MEM) (Thermo Fisher Scientific) in the presence of 10% FBS and 30 ng/ml of human macrophage colony stimulating factor (M-CSF). M-CSF was purified as described, and M-CSF cDNA and expression vector were provided by D. Fremont (Washington University, St. Louis, MO) (Lee et al., 2006). Cells were plated at a density of 300,000 cells/cm2 on uncoated plastic petri dishes and cultured in the presence of M-CSF for 3 days, as described (Canalis et al., 2018; Yu et al., 2018). The cell layer was then treated with 0.25% trypsin/EDTA for 5 min and cells recovered and seeded at a density of 47,000 cells/cm2 on tissue culture plates in α-MEM with 10% FBS, 30 ng/ml of M-CSF and 10 ng/ml of murine receptor activator of NF Kappa B ligand (RANKL) and anti-Notch3 NRR or control antibodies at 20 μg/ml. Tnfsf11 cDNA and expression vector were provided by from M. Glogauer (Toronto, Canada), and GST-tagged RANKL was expressed and purified as described (Wang et al., 2008).
To explore whether factors derived from the osteoblast contributed to osteoclastogenesis, cells enriched in osteoblasts and obtained from either Notch3tm1.1Ecan or littermate controls were seeded at a 15,700 cells/cm2 density in α-MEM with BMMs from either genotype seeded at 47,000 cells/cm2 density and cultured with 1,25 dihydroxyvitamin D3 (Enzo Life Science, Farmingdale, NY) at 10 nM with anti-Notch3 NRR or control antibodies at 20 μg/ml, as described (Canalis et al., 2018). Cultures were conducted until multinucleated tartrate resistant acid phosphatase (TRAP)-positive cells were formed. Enzyme histochemical analysis for TRAP was performed using a commercial kit (Sigma-Aldrich), according to manufacturer’s instructions. Cells containing 3 or more nuclei staining positive for TRAP were considered osteoclasts.
Quantitative Reverse Transcription-PCR—
RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) in accordance with instructions from the manufacturer (Nazarenko, Lowe, et al., 2002; Nazarenko, Pires, Lowe, Obaidy, & Rashtchian, 2002). The iScript RT-PCR kit was used to reverse transcribe equal amounts of RNA (BioRad, Hercules, CA) and products were amplified in the presence of specific primers (IDT) (Table 1) with SsoAdvanced Universal SYBR Green Supermix (BioRad) at 60°C for 40 cycles. Copy number was determined by comparing test samples to serial dilutions of Hes1 (from American Type Culture Collection (ATCC), Manassas, VA), Hey1, Hey2 (from T. Iso, Gunma University, Gunma, Japan), or Tnfsf11 cDNA (from Source Bioscience, Nuttingham, UK) (Iso et al., 2001; Nakagawa, Nakagawa, Richardson, Olson, & Srivastava, 1999). To estimate the copy number for Notch3tm1.1Ecan transcripts, samples were compared to serial dilutions of a 90 bp DNA fragment (IDT) surrounding the Notch3 6691–6696 ACCAAG>TAATGA mutation, and cloned into pcDNA3.1(−) (Thermo Fisher Scientific) by isothermal single reaction assembly (New England Biolabs, Ipswich, MA), as reported (Canalis et al., 2018; Gibson et al., 2009). Amplification reactions were carried out in a CFX96 real-time PCR detection system (BioRad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are reported as copy number, corrected for Rpl38 (from ATCC) (Kouadjo, Nishida, Cadrin-Girard, Yoshioka, & St-Amand, 2007).
Table 1.
Primers used for qRT-PCR determinations. GenBank accession numbers identify transcript recognized by primer pairs.
| Gene | Strand | Sequence 5'–3' | GenBank Accession Number |
|---|---|---|---|
| Hes1 | Forward Reverse |
5'-ACCAAAGACGGCCTCTGAGCACAGAAAGT-3' 5'-ATTCTTGCCCTTCGCCTCTT-3' |
NM_008235 |
| Hey1 | Forward Reverse |
5'-ATCTCAACAACTACGCATCCCAGC-3' 5'-GTGTGGGTGATGTCCGAAGG-3' |
NM_010423 |
| Hey2 | Forward Reverse |
5'-AGCGAGAACAATTACCCTGGGCAC-3' 5'-GGTAGTTGTCGGTGAATTGGACCT-3' |
NM_013904 |
| Notch3 | Forward Reverse |
5'-CCGATTCTCCTGTCGTTGTCTCC-3' 5'-TGAACACAGGGCCTGCTGAC-3' |
NM_008716 |
| Notch36691-TAATGA | Forward Reverse |
5'-AACCCGCAGTAGCCCCTAATG-3' 5'-ATAAGGATGCTCGCTGGGAACC-3' |
Not applicable |
| Rpl38 | Forward Reverse |
5'-AGAACAAGGATAATGTGAAGTTCAAGGTTC-3' 5'-CTGCTTCAGCTTCTCTGCCTTT-3' |
NM_001048058; ; NM_023372 |
| Tnfsf11 | Forward Reverse |
5'-TATAGAATCCTGAGACTCCATGAAAAC-3' 5'-CCCTGAAAGGCTTGTTTCATCC-3' |
NM_011613 |
Statistics
Values are reported as means ± SD. In vivo data represent biological replicates, and in vitro data represent technical replicates. qRT-PCR values represent 2 technical replicates of biological or technical replicates as stated in figure legends. Analysis of variance (ANOVA) for multiple comparisons with Holm-Šídák post-hoc analysis was used to establish statistical differences.
RESULTS
Notch3tm1.1Ecan Mice—
Although Notch3tm1.1Ecan mice of both sexes exhibit osteopenia defined as decreased cancellous bone volume, this persists into adulthood in male mice (Canalis et al., 2018). As a consequence, the present studies were conducted in male Notch3tm1.1Ecan mice. Heterozygous male Notch3tm1.1Ecan mice were compared to wild type littermates matched for sex in a C57BL/6J background following crosses of heterozygous Notch3tm1.1Ecan mice with wild type mice. Consistent with prior observations, the body weight and the femoral length of 2 month old Notch3tm1.1Ecan heterozygous mice were equivalent to those of control mice (Figure 1) (Canalis et al., 2018). The administration of anti-Notch3 NRR or control antibody at 20 mg/Kg IP twice a week for 4 weeks did not cause obvious unwanted effects; mice appeared healthy and their body weight and femoral length were not altered by anti-Notch3 NRR antibodies (Figure 1).
Figure 1.
Weight and femoral length of male Notch3tm1.1Ecan mutant mice (black bars) and sex-matched littermate wild type controls (white bars) treated with anti-Notch3 NRR (N3NRR) or anti-ragweed control antibody (Ctrl) twice a week for 4 weeks. Values are means ± SD; n = 7 to 10.
Skeletal Phenotype—
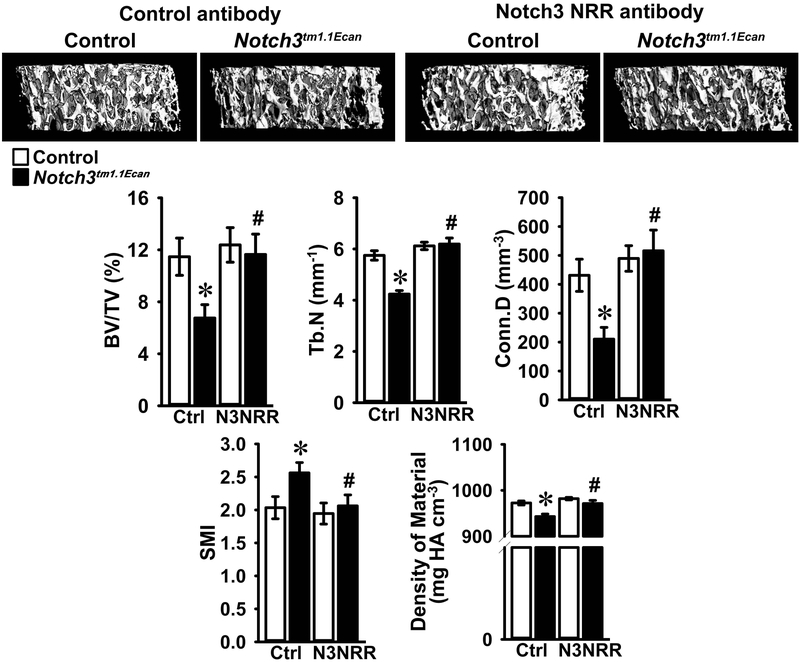
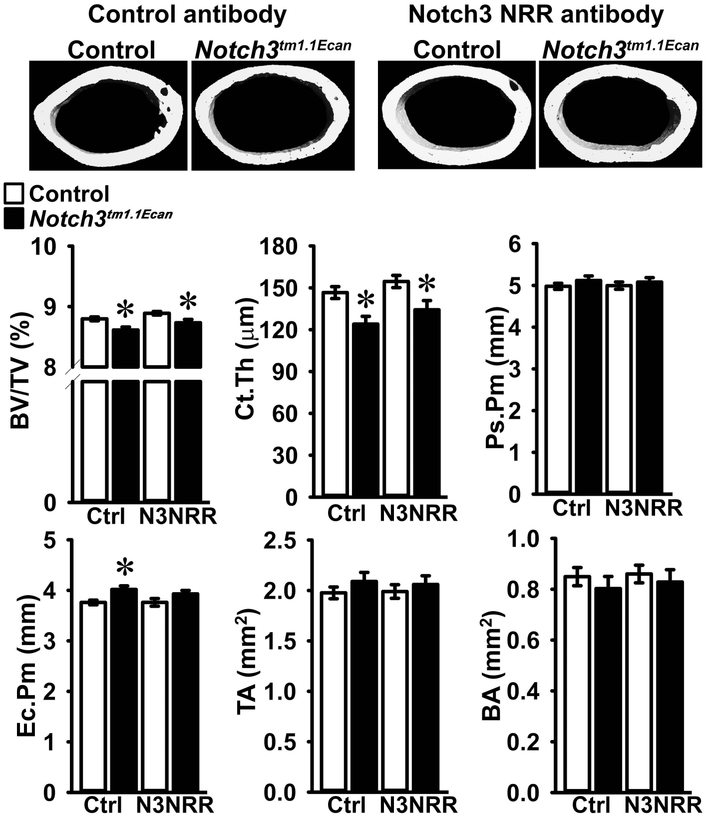
In accordance with previous work, μCT of the distal femur demonstrated that 2 month old male Notch3tm1.1Ecan mice displayed a 40% reduction in cancellous bone volume together with a decrease in the number of trabeculae, connectivity density and density of material and an increase in SMI (Figure 2) (Canalis et al., 2018). Administration of anti-Notch3 NRR antibody did not alter the cancellous bone volume or number of trabeculae of control mice. However, the osteopenia of Notch3tm1.1Ecan mice was no longer observed following the administration of anti-Notch3 NRR antibodies (Figure 2). As a consequence, bone volume/total volume, trabecular number, connectivity and density of material of Notch3tm1.1Ecan mice administered anti-Notch3 NRR antibodies were significantly higher and SMI significantly lower than placebo-treated Notch3tm1.1Ecan mice and not different from the values observed in control mice administered anti-Notch3 NRR antibodies (Figure 2). The results demonstrate that anti-Notch3 NRR antibodies can reverse the cancellous bone osteopenia of Notch3tm1.1Ecan mice. µCT of cortical bone revealed modest alterations in the cortical structure of Notch3tm1.1Ecan mice; bone volume and cortical bone thickness were decreased. Endocortical perimeter was greater in Notch3tm1.1Ecan than in control mice suggesting increased cortical remodeling (Figure 3). Anti-Notch3 NRR antibodies did not modify the decrease in cortical bone volume or in cortical thickness of Notch3tm1.1Ecan mice, and these parameters remained significantly decreased when compared to control mice treated with anti-Notch3 NRR antibodies indicating no resolution of the cortical osteopenia (Figure 3).
Figure 2.
Cancellous bone microarchitecture assessed by μCT of the distal femur from 2 month old Notch3tm1.1Ecan mutant male mice (black bars) and sex-matched littermate controls (white bars) treated with anti-Notch3 NRR (N3NRR; n = 8 to 9) or anti-ragweed control antibody (Ctrl; n = 7 to 10), both at 20 mg/Kg, twice a week for 4 weeks prior to sacrifice. Parameters shown are: bone volume/tissue volume (BV/TV); trabecular number (Tb.N); connectivity density (Conn.D); structure module index (SMI) and density of material. Values are means ± SD. *Significantly different between Notch3tm1.1Ecan and control mice, p < 0.05. #Significantly different between anti-Notch3 NRR and control antibody, p < 0.05. A representative image shows cancellous bone osteopenia in Notch3tm1.1Ecan mutant mice and its reversal by anti-Notch3 NRR antibodies.
Figure 3.
Cortical bone microarchitecture assessed by μCT of the femoral mid-shaft from 2 month old Notch3tm1.1Ecan mutant male mice and sex-matched littermate controls treated with anti-Notch3 NRR (N3NRR; n = 8 to 9) or anti-ragweed control antibody (Ctrl; n = 7 to 10), both at 20 mg/Kg, twice a week for 4 weeks prior to sacrifice. Parameters shown are: bone volume/tissue volume (BV/TV); cortical thickness (Ct.Th); periosteal (Ps.Pm) and endocortical perimeter (Ec.Pm); total (TA) and bone area (BA). Values are means ± SD. *Significantly different between Notch3tm1.1Ecan and control mice, p < 0.05. A representative image shows cortical thinning in Notch3tm1.1Ecan mutant mice and no reversal by anti-Notch3 NRR antibody.
Calvarial Osteoblast-enriched Cell Cultures—
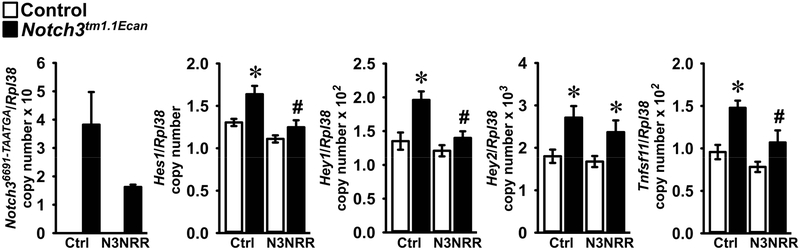
Notch36691-TAATGA transcripts were present in cells from Notch3tm1.1Ecan mutant mice but not in control cultures; and Hes1, Hey1 and Hey2 transcripts were increased in Notch3tm1.1Ecan osteoblasts revealing that Notch signaling was activated (Figure 4). In accordance with prior observations, Tnfsf11, encoding RANKL was induced in Notch3tm1.1Ecan osteoblasts. Anti-Notch3 NRR antibodies added to the culture medium for 1 week opposed the induction of Hes1 and Hey1 and of Tnfsf11, but not of Hey2 mRNA in cells from Notch3tm1.1Ecan mice (Figure 4). Notch36991-TAATGA transcripts were not affected significantly by the anti-Notch3 NRR antibody. In an alternate experiment, where Hey2 mRNA was induced in osteoblast cultures from (means ± SD; n = 4) 1.0 ± 0.1 in control cells to 1.7 ± 0.3 in Notch3tm1.1Ecan osteoblasts (p < 0.05), treatment with anti-Notch3 NRR antibody for 2 weeks reduced Hey2 mRNA to 1.2 ± 0.2 in Notch3tm1.1Ecan cells (p < 0.05 vs. ragweed antibody treated Notch3tm1.1Ecan cells). The results indicate a prevention of Notch3 activation and a reversal of the Tnfsf11 (RANKL) induction by anti-Notch3 NRR antibodies.
Figure 4.
Calvarial osteoblast-enriched cells from Notch3tm1.1Ecan mutant (black bars) and wild type (white bars) littermate controls were isolated and cultured in the presence of anti-Notch3 NRR (N3NRR) or anti-ragweed control antibody (Ctrl) at 20 μg/ml for 1 week. Total RNA was extracted, and gene expression determined by qRT-PCR in the presence of specific primers. Data are expressed as Notch36691-TAATGA mutant, Hes1, Hey1, Hey2 and Tnfsf11 copy number corrected for Rpl38. Values are means ± SD; n = 4. *Significantly different between Notch3tm1.1Ecan mutant and wild type control cells, p < 0.05. #Significantly different between anti-Notch3 NRR and control antibody, p < 0.05.
BMM Cultures and Osteoclast Formation—
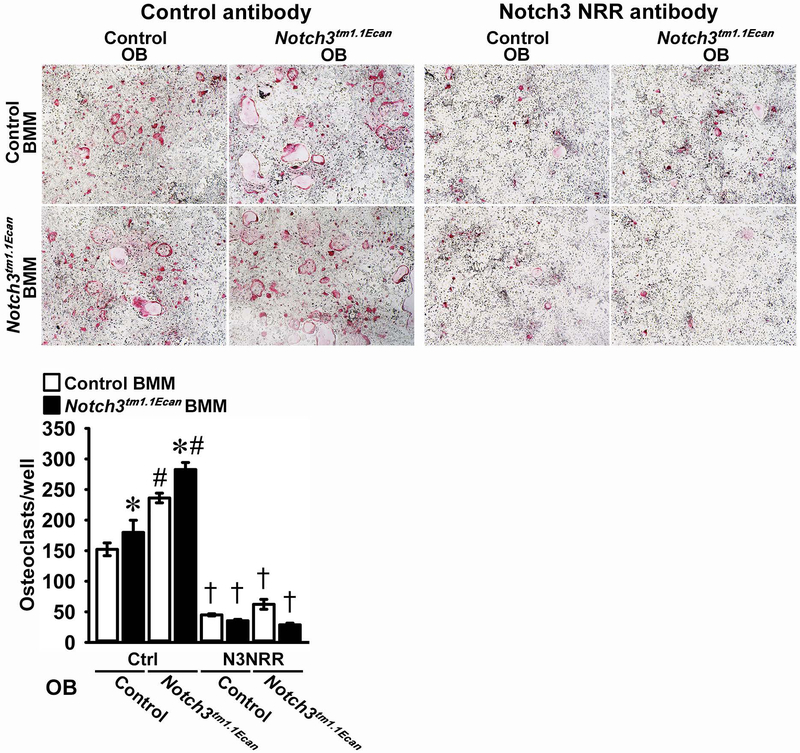
BMMs from Notch3tm1.1Ecan mutants and control littermates were incubated with M-CSF at 30 ng/ml for 3 days followed by the subsequent addition of RANKL at 10 ng/ml in conjunction with M-CSF at 30 ng/ml, as reported (Canalis et al., 2018; Yu et al., 2018). The number of osteoclasts, defined as multinucleated cells that were positive for TRAP, was augmented by ~30% in Notch3tm1.1Ecan cultures (Figure 5). However, Notch36691-TAATGA transcripts were not detected in Notch3tm1.1Ecan BMMs, and Notch3 mRNA was not detected in either mutant or wild type BMMs (data not shown). Consequently, the anti-Notch3 NRR antibody did not prevent the enhanced osteoclastogenesis observed in Notch3tm1.1Ecan BMM cultures (Figure 5). This suggests that the phenotype observed in BMM cultures is secondary to events that occurred in vivo. To determine whether the osteoblast is the cell accountable for the increased osteoclastogenesis, Notch3tm1.1Ecan and control BMMs were co-cultured with osteoblasts from either control or Notch3tm1.1Ecan mice. Osteoclast number was increased in Notch3tm1.1Ecan BMMs whether the BMMs were cultured in the presence of control or Notch3tm1.1Ecan osteoblasts (Figure 6). Osteoblasts from Notch3tm1.1Ecan mice increased osteoclast formation in both wild type and Notch3tm1.1Ecan mutant BMMs, and the effect was reversed by anti-Notch3 NRR antibodies (Figure 6). This is in line with the inhibitory effect of anti-Notch3 NRR antibodies on RANKL expression by Notch3tm1.1Ecan osteoblasts.
Figure 5.
Bone marrow cells, harvested from long bones of Notch3tm1.1Ecan mutant (black bars) and wild type littermate controls (white bars) were grown for 72 hours in the presence of M-CSF at 30 ng/ml and then seeded in the presence of M-CSF 30 ng/ml and RANKL at 10 ng/ml in the presence of anti-Notch3 NRR (N3NRR) or anti-ragweed control antibody (Ctrl) at 20 μg/ml for osteoclast formation. Representative images of TRAP-stained multinucleated cells are shown to the left. Data are expressed as total number of TRAP-positive multinucleated cells/well. Values are means ± SD; n = 4 technical replicates. *Significantly different between Notch3tm1.1Ecan mutant and wild type control cells, p < 0.05.
Figure 6.
Bone marrow cells harvested from long bones of Notch3tm1.1Ecan mutants (black bars) and wild type littermate controls (white bars) were cultured for 72 hours in the presence of M-CSF at 30 ng/ml. BMMs from control and Notch3tm1.1Ecan mutant mice were seeded on culture dishes in the presence of osteoblasts (OB) from control or Notch3tm1.1Ecan mutant mice with 1,25 dihydroxyvitamin D3 at 10 nM in the presence of anti-Notch3 NRR (N3NRR) or anti-ragweed control antibody (Ctrl) at 20 μg/ml, and assessed for the appearance of TRAP-positive multinucleated cells. Representative images of TRAP-stained multinucleated are shown in the upper panels. Data are expressed as total number of TRAP-positive multinucleated cells/well. Values are means ± SD; n = 4 technical replicates. *Significantly different between Notch3tm1.1Ecan and control BMM, p < 0.05. #Significantly different between Notch3tm1.1Ecan and control calvarial osteoblast-enriched cells, p < 0.05. †Significantly different between anti-Notch3 NRR and control antibody, p < 0.05.
DISCUSSION
The present findings confirm that the introduction of a mutation into the mouse genome replicating the one reported in LMS results in osteopenia of the cancellous and cortical bone compartments. Whereas the osteopenic phenotype is present in male and female young mice, it persists in mature male mice. Consequently, at 1 month of age male mice were treated with anti-Notch3 NRR antibodies for 4 weeks. The anti-Notch3 NRR antibody was effective in reversing the cancellous bone osteopenia of Notch3tm1.1Ecan male mice. The results do not necessarily apply to female mice since male mice were studied. An additional limitation of the present experimental design is that the same mouse could not be evaluated prior to and following the administration of anti-Notch3 NRR antibodies, because the evaluation would have required the sacrifice of mice at baseline and prior to treatment initiation.
The Notch3tm1.1Ecan mouse recapitulates selected aspects of LMS, but Notch3tm1.1Ecan mice do not manifest the neurological manifestations of the human disease. There is no obvious explanation for the difference in the phenotype observed between humans and mice. It is possible that additional phenotypic traits might appear as Notch3tm1.1Ecan mice age, since so far we have examined only relatively young adult mice.
Although cells of the osteoclast lineage do not express Notch3 mRNA, the maturation of Notch3tm1.1Ecan osteoclast precursors as multinucleated osteoclasts in response to RANKL was enhanced. This suggests that the increased osteoclast differentiation observed in BMM cultures from Notch3tm1.1Ecan mice was secondary to prior events occurring in vivo. In accordance with the lack of Notch3 expression by BMMs, the anti-Notch3 NRR antibody did not affect the enhanced osteoclastogenesis of Notch3tm1.1Ecan BMMs in vitro. In contrast, the anti-Notch3 NRR antibody prevented the enhanced osteoclastogenesis observed when BMMs were co-cultured with osteoblasts from Notch3tm1.1Ecan mice suggesting that the increased osteoclastogenesis was dependent on an event occurring in cells of the osteoblast lineage. This is in agreement with the enhanced RANKL expression in osteoblasts from Notch3tm1.1Ecan mice, which is likely responsible for the enhanced osteoclastogenesis observed and was reversed by the anti-Notch3 NRR antibody (Canalis et al., 2018). The suppression of RANKL expression explains the resolution of the osteopenia following the administration of antibodies targeting the Notch3 NRR.
In contrast to the actions of Notch1 and Notch2 on osteoclast differentiation, Notch3tm1.1Ecan mice have distinct effects in the myeloid lineage. Notch1 inhibits osteoclast maturation by direct and indirect mechanisms, whereas Notch2 induces osteoclast differentiation by direct mechanisms and by the induction of RANKL by osteoblasts (Bai et al., 2008; Canalis et al., 2016; Fukushima et al., 2008; Zanotti et al., 2017). In contrast, Notch3 induces osteoclastogenesis only by indirect mechanisms enhancing the expression of RANKL by the osteoblast and osteocyte, since BMMs do not express Notch3 mRNA (Canalis et al., 2018).
Notch signal downregulation can be achieved by diverse approaches including the utilization of biochemical inhibitors, antibodies to nicastrin or to Notch receptors or their ligands, and the use of small molecules that interfere with the formation of a NICD/RBPJκ/MAML ternary complex (Ryeom, 2011). Inhibitors of γ-secretase are often used to prevent the cleavage of Notch receptors by Presenilins (De Strooper et al., 1999). However, inhibitors of γ-secretase affect many substrates and lack specificity (Duggan & McCarthy, 2016). An alternative is the use of anti-nicastrin antibodies since nicastrin forms part of the γ-secretase complex (Siebel & Lendahl, 2017). Thapsigargin inhibits the sarco/endoplasmic reticulum Ca2+-ATPase and as such prevents Notch maturation and folding, and Notch effects (Ilagan & Kopan, 2013; Largaespada & Ratner, 2013). Thapsigargin and γ-secretase inhibitors are limited by the fact that they do not discriminate among Notch receptors when preventing their activation. Stapled peptides that preclude the assembly of a Notch transcriptional complex have been employed to inhibit Notch receptors, although their efficacy is not fully established (Moellering et al., 2009).
Individual Notch receptors can be targeted specifically by the use of antibodies to the NRR, and these have been developed to target Notch1, Notch2 and Notch3 (Li et al., 2008; Wu et al., 2010). Targeting the NRR prevents cleavage and, therefore, activation of Notch receptors, making it ideal for the specific neutralization of each Notch isoform. This is the reason why anti-Notch3 NRR antibodies were chosen to resolve the osteopenia of Notch3tm1.1Ecan mice. One should be cautious and not extrapolate the present results to the human disease since knowledge on the suppression of Notch3 activity in humans is scarce. The homozygous NOTCH3 null mutation presents with cerebrovascular abnormalities and leukoencephalopathy demonstrating that long-term deficiency of Notch3 can have negative vascular consequences (Pippucci et al., 2015). However, it is not known whether prolonged Notch3 neutralization results in unwanted events, as described for other Notch receptors (Ridgway et al., 2006; Yan et al., 2010).
In conclusion, Notch3tm1.1Ecan mice, a murine model of LMS, present with cancellous bone osteopenia, and this is reversed following their treatment with anti-Notch3 NRR antibodies.
ACKNOWLEDGMENTS
We acknowledge Genentech for providing anti-Notch3 NRR and ragweed antibodies, D. Fremont for M-CSF cDNA, M. Glogauer for RANKL cDNA, T. Iso for Hey1 and Hey2 cDNA, T. Eller for technical assistance and Mary Yurczak for secretarial support.
FUNDING STATEMENT
This work was supported by grants AR063049 and AR072987 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
The abbreviations used are:
- αMEM
α-minimum essential medium
- BMM
bone marrow-derived macrophage
- BA
bone area
- BV/TV
bone volume/tissue volume
- Ct.Th
cortical thickness
- Conn.D
connectivity density
- DMEM
Dulbecco’s modified Eagle’s medium
- Ec.Pm
endocortical perimeter
- FBS
fetal bovine serum
- Hes
Hairy Enhancer of Split
- Hey
Hes-related with YRPW motif
- M-CSF
macrophage colony stimulating factor
- MAML
mastermind-like
- μCT
microcomputed tomography
- NRR
negative regulatory region
- NICD
Notch intracellular domain
- OB
osteoblasts
- PBS
phosphate buffered saline
- PEST
proline (P), glutamic acid (E), serine (S) and threonine (T)
- Ps.Pm
periosteal perimeter
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse transcription-PCR
- RANKL
receptor activator of NF Kappa B ligand
- RBPJκ
recombination signal-binding protein for Ig of κ
- SMI
structure model index
- TRAP
tartrate-resistant acid phosphatase
- TA
total area
- Tb.N.
trabecular number
Footnotes
Disclosure Statement: Christian W. Siebel is employed by Genentech. The other authors have nothing to disclose.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CONFLICT OF INTEREST
JY, LS and EC declare no conflicts of interest with the contents of this article. CS is employed by Genentech.
REFERENCES
- Avela K, Valanne L, Helenius I, & Makitie O (2011). Hajdu-Cheney syndrome with severe dural ectasia. American Journal of Medical Genetics Part A, 155A(3), 595–598. doi: 10.1002/ajmg.a.33510 [doi] [DOI] [PubMed] [Google Scholar]
- Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, . . . Teitelbaum SL (2008). NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. Journal of Biological Chemistry, 283(10), 6509–6518. [DOI] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, & Muller R (2010). Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of Bone and Mineral Research, 25(7), 1468–1486. [DOI] [PubMed] [Google Scholar]
- Canalis E (2018). Notch in skeletal physiology and disease. Osteoporosis International, 29(12), 2611–2621. doi: 10.1007/s00198-018-4694-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Kranz L, & Zanotti S (2014). Nemo-like kinase regulates postnatal skeletal homeostasis. Journal of Cellular Physiology, 229(11), 1736–1743. doi: 10.1002/jcp.24625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Sanjay A, Yu J, & Zanotti S (2017). An Antibody to Notch2 Reverses the Osteopenic Phenotype of Hajdu-Cheney Mutant Male Mice. Endocrinology, 158(4), 730–742. doi: 10.1210/en.2016-1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Schilling L, Yee SP, Lee SK, & Zanotti S (2016). Hajdu Cheney Mouse Mutants Exhibit Osteopenia, Increased Osteoclastogenesis and Bone Resorption. Journal of Biological Chemistry, 291, 1538–1551. doi: 10.1074/jbc.M115.685453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Yu J, Schilling L, Yee SP, & Zanotti S (2018). The lateral meningocele syndrome mutation causes marked osteopenia in mice. Journal of Biological Chemistry, 293(36), 14165–14177. doi: 10.1074/jbc.RA118.004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, & Zanotti S (2017). Hairy and Enhancer of Split-Related With YRPW Motif-Like (HeyL) Is Dispensable for Bone Remodeling in Mice. Journal of Cellular Biochemistry, 118(7), 1819–1826. doi: 10.1002/jcb.25859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalis E, Zanotti S, & Smerdel-Ramoya A (2014). Connective Tissue Growth Factor is a Target of Notch Signaling in Cells of the Osteoblastic Lineage. Bone, 64, 273–280. doi:S8756–3282(14)00163-X [pii]; 10.1016/j.bone.2014.04.028 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L, Hagenbeek TJ, Solon M, French D, Finkle D, Shelton A, . . . Siebel CW (2017). Constitutive NOTCH3 Signaling Promotes the Growth of Basal Breast Cancers. Cancer Research, 77(6), 1439–1452. doi: 10.1158/0008-5472.CAN-16-1022 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, . . . Kopan R (1999). A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature, 398(6727), 518–522. doi: 10.1038/19083 [DOI] [PubMed] [Google Scholar]
- Duggan SP, & McCarthy JV (2016). Beyond gamma-secretase activity: The multifunctional nature of presenilins in cell signalling pathways. Cellular Signalling, 28(1), 1–11. doi: 10.1016/j.cellsig.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Fortini ME (2009). Notch signaling: the core pathway and its posttranslational regulation. Developmental Cell, 16(5), 633–647. [DOI] [PubMed] [Google Scholar]
- Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, . . . Okabe K (2008). The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol. Cell Biol, 28(20), 6402–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, & Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6(5), 343–345. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Glatt V, Canalis E, Stadmeyer L, & Bouxsein ML (2007). Age-Related Changes in Trabecular Architecture Differ in Female and Male C57BL/6J Mice. Journal of Bone and Mineral Research, 22(8), 1197–1207. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, . . . Blacklow SC (2015). Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev Cell, 33(6), 729–736. doi: 10.1016/j.devcel.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Robbins KM, Sobreira NL, Witmer PD, Bird LM, Avela K, . . . Sol-Church K (2015). Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. American Journal of Medical Genetics Part A, 167A(2), 271–281. doi: 10.1002/ajmg.a.36863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Scott CI Jr., Hughes HE, Wallerstein R, Nicholson L, States L, . . . Zackai EH (1997). Lateral meningocele syndrome: three new patients and review of the literature. American Journal of Medical Genetics, 70(3), 229–239. [PubMed] [Google Scholar]
- Ilagan MX, & Kopan R (2013). Selective blockade of transport via SERCA inhibition: the answer for oncogenic forms of Notch? Cancer Cell, 23(3), 267–269. doi: 10.1016/j.ccr.2013.02.020 [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, & Hamamori Y (2003). HES and HERP families: multiple effectors of the Notch signaling pathway. Journal of Cellular Physiology, 194(3), 237–255. doi: 10.1002/jcp.10208 [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, & Hamamori Y (2001). HERP, a new primary target of Notch regulated by ligand binding. Molecular and Cellular Biology, 21(17), 6071–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, & Kageyama R (2014). Expression dynamics and functions of Hes factors in development and diseases. Curr Top Dev Biol, 110, 263–283. doi: 10.1016/B978-0-12-405943-6.00007-5 [DOI] [PubMed] [Google Scholar]
- Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, & St-Amand J (2007). Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics, 8, 127 doi: 10.1186/1471-2164-8-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA (2008). More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene, 27(38), 5099–5109. [DOI] [PubMed] [Google Scholar]
- Largaespada D, & Ratner N (2013). Interweaving the strands: beta-catenin, an HIV co-receptor, and Schwann cell tumors. Cancer Cell, 23(3), 269–271. doi: 10.1016/j.ccr.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, . . . Choi Y (2006). v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med, 12(12), 1403–1409. doi: 10.1038/nm1514 [DOI] [PubMed] [Google Scholar]
- Lehman RA, Stears JC, Wesenberg RL, & Nusbaum ED (1977). Familial osteosclerosis with abnormalities of the nervous system and meninges. Journal of Pediatircs, 90(1), 49–54. [DOI] [PubMed] [Google Scholar]
- Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, . . . Zhou BB (2008). Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. Journal of Biological Chemistry, 283(12), 8046–8054. doi: 10.1074/jbc.M800170200 [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del BC, Aster JC, Blacklow SC, . . . Bradner JE (2009). Direct inhibition of the NOTCH transcription factor complex. Nature, 462(7270), 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, & Srivastava D (1999). HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Developmental Biology, 216(1), 72–84. doi: 10.1006/dbio.1999.9454 [doi];S0012–1606(99)99454-X [pii] [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, & Blacklow SC (2006). Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell, 124(5), 973–983. [DOI] [PubMed] [Google Scholar]
- Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, & Rashtchian A (2002). Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Research, 30(9), e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Pires R, Lowe B, Obaidy M, & Rashtchian A (2002). Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Research, 30(9), 2089–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippucci T, Maresca A, Magini P, Cenacchi G, Donadio V, Palombo F, . . . Seri M (2015). Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol Med, 7(6), 848–858. doi: 10.15252/emmm.201404399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, . . . Yan M (2006). Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature, 444(7122), 1083–1087. doi: 10.1038/nature05313 [DOI] [PubMed] [Google Scholar]
- Ryeom SW (2011). The cautionary tale of side effects of chronic Notch1 inhibition. The Journal of clinical investigation, 121(2), 508–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, & Blacklow SC (2004). Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Molecular and Cellular Biology, 24(21), 9265–9273. doi: 10.1128/mcb.24.21.9265-9273.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, & Kopan R (1998). Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393(6683), 382–386. [DOI] [PubMed] [Google Scholar]
- Siebel C, & Lendahl U (2017). Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiological Reviews, 97(4), 1235–1294. doi: 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- Tiyanont K, Wales TE, Siebel CW, Engen JR, & Blacklow SC (2013). Insights into Notch3 activation and inhibition mediated by antibodies directed against its negative regulatory region. Journal of Molecular Biology, 425(17), 3192–3204. doi: 10.1016/j.jmb.2013.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, & Glogauer M (2008). Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. Journal of Bone and Mineral Research, 23(2), 260–270. doi: 10.1359/jbmr.071013 [DOI] [PubMed] [Google Scholar]
- Wilson JJ, & Kovall RA (2006). Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell, 124(5), 985–996. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, . . . Siebel CW (2010). Therapeutic antibody targeting of individual Notch receptors. Nature, 464(7291), 1052–1057. doi: 10.1038/nature08878 [DOI] [PubMed] [Google Scholar]
- Xu X, Choi SH, Hu T, Tiyanont K, Habets R, Groot AJ, . . . Blacklow SC (2015). Insights into Autoregulation of Notch3 from Structural and Functional Studies of Its Negative Regulatory Region. Structure, 23(7), 1227–1235. doi: 10.1016/j.str.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, . . . Plowman GD (2010). Chronic DLL4 blockade induces vascular neoplasms. Nature, 463(7282), E6–7. doi: 10.1038/nature08751 [DOI] [PubMed] [Google Scholar]
- Yesil P, Michel M, Chwalek K, Pedack S, Jany C, Ludwig B, . . . Lammert E (2009). A new collagenase blend increases the number of islets isolated from mouse pancreas. Islets, 1(3), 185–190. doi: 10.4161/isl.1.3.9556 [DOI] [PubMed] [Google Scholar]
- Yu J, Zanotti S, Schilling L, & Canalis E (2018). Nuclear factor of activated T cells 2 is required for osteoclast differentiation and function in vitro but not in vivo. Journal of Cellular Biochemistry, 119(11), 9334–9345. doi: 10.1002/jcb.27212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, & Canalis E (2016). Notch Signaling and the Skeleton. Endocrine Reviews, 37(3), 223–253. doi: 10.1210/er.2016-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, & Canalis E (2017). Parathyroid hormone inhibits Notch signaling in osteoblasts and osteocytes. Bone, 103, 159–167. doi: 10.1016/j.bone.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Yu J, Adhikari S, & Canalis E (2018). Glucocorticoids inhibit notch target gene expression in osteoblasts. Journal of Cellular Biochemistry, 119(7), 6016–6023. doi: 10.1002/jcb.26798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Yu J, Sanjay A, Schilling L, Schoenherr C, Economides AN, & Canalis E (2017). Sustained Notch2 signaling in osteoblasts, but not in osteoclasts, is linked to osteopenia in a mouse model of Hajdu-Cheney syndrome. Journal of Biological Chemistry, 292(29), 12232–12244. doi: 10.1074/jbc.M117.786129 [DOI] [PMC free article] [PubMed] [Google Scholar]