Abstract
Cotton is an agriculturally important crop. Because of its importance, a genome sequence of a diploid cotton species (Gossypium raimondii, D-genome) was first assembled using Sanger sequencing data in 2012. Improvements to DNA sequencing technology have improved accuracy and correctness of assembled genome sequences. Here we report a new de novo genome assembly of G. raimondii and its close relative G. turneri. The two genomes were assembled to a chromosome level using PacBio long-read technology, HiC, and Bionano optical mapping. This report corrects some minor assembly errors found in the Sanger assembly of G. raimondii. We also compare the genome sequences of these two species for gene composition, repetitive element composition, and collinearity. Most of the identified structural rearrangements between these two species are due to intra-chromosomal inversions. More inversions were found in the G. turneri genome sequence than the G. raimondii genome sequence. These findings and updates to the D-genome sequence will improve accuracy and translation of genomics to cotton breeding and genetics.
Keywords: Gossypium raimondii, Gossypium turneri, cotton, genome sequence, PacBio
In 2012, the first reference quality cotton genome was brought to fruition through a monumental, collaborative effort using a combination of next-generation sequencing technologies and targeted Sanger sequencing (Paterson et al. 2012). Gossypium raimondii, a Mesoamerican diploid species, was selected to represent the cotton genus for its small genome size and its relationship to the domesticated polyploid species (Chen et al. 2007). Subsequently, this genome has been widely used by the cotton research community, garnering ∼500 citations from a wide spectrum of research. While this genome has been a reliable resource for over 7 years, increased read lengths have improved scaffolding and assembly quality, while development of chromosome conformation capture (3C) techniques have allowed association of sequences within the interphase nucleus but separated by thousands or millions of base pairs along the linear DNA strand (de Wit and de Laat 2012; Peterson and Arick 2018).
The justification for the original G. raimondii sequence, i.e., its phylogenetic relatedness to the domesticated allopolyploid species and the recruitment of genetic factors from that subgenome during domestication, make G. raimondii and its close relatives potential genetic sources for cotton breeding. Gossypium turneri is a species from Sonora, Mexico (Fryxell 1978), that is closely related to G. raimondii (Guo et al. 2007). Like G. raimondii, fiber from G. turneri is unspinnable; however, G. turneri has phenotypic characters with agronomic potential, e.g., caducous bracts, insect resistance, and abiotic stress tolerance (Chen et al. 2018).The two species are generally similar, both having a haploid complement of 13 chromosomes and relatively small genome sizes (910 Mb vs. 880 Mb in G. turneri and G. raimondii, respectively; (Hendrix and Stewart 2005)). The two species, however, are genetically distinct, as long recognized by taxonomists and their extreme allopatry (G. raimondii is from Peru, G. turneri from Baja California), as well as by genetic and phylogenetic data (Ulloa et al. 2013), (Grover et al. 2019). Notably, a previously published draft genome suggests that gene gain and loss may be elevated in G. turneri (Grover et al. 2019).
Here we describe two de novo genome sequences, for G. raimondii (D5) and G. turneri (D10), which were assembled using newly generated PacBio, Hi-C, and Bionano (G. raimondii only) technologies. The G. raimondii genome sequence reported here represents an independent effort and identifies three significant assembly errors in the initial publication of G. raimondii, including a large assembly artifact on the original chromosome 1. We also report a high-quality sequence for G. turneri that is suitable for various comparative, genetic, and genomic analyses. Together, these genomes represent a useful resource for cotton breeding and for comparative genomics in general.
Methods & Materials
Plant material and sequencing
Leaf tissue of mature G. raimondii (accession D5-4) and G. turneri (accession D10-3) plants was collected at the Brigham Young University (BYU) greenhouse. DNA was extracted using CTAB techniques (Kidwell and Osborn 1992). DNA concentration was measured by a Qubit Fluorometer (ThermoFisher, Inc.). The sequencing library was constructed according to PacBio recommendations at the BYU DNA Sequencing Center (DNASC). Fragments >18 kb were selected for sequencing via BluePippen (Sage Science, LLC). Prior to sequencing, the size distribution of fragments in the libraries was evaluated using a Fragment Analyzer (Advanced Analytical Technologies, Inc). Eight and eleven PacBio cells were sequenced from a single library each for G. raimondii and G. turneri, respectively, on the Pacific Biosciences Sequel system. For both genomes, the raw PacBio sequencing reads were assembled using Canu V1.6 using default parameters (Koren et al. 2017).
HiC libraries were constructed from G. raimondii leaf tissue at NorthEast Normal University, China. Sequencing was performed at Annoroad Gene Technology Co., Ltd (Beijing, China). The HiC data of G. raimondii was mapped to the previous genome sequence of G. raimondii using HiC-Pro (Servant et al. 2015), and to the newly assembled CANU contigs of G. raimondii PacBio reads by PhaseGenomics. The HiC interactions were used as evidence for contig proximity and in scaffolding contig sequences. An initial draft genome sequence of pseudochromosomes (PGA assembly) was created using a custom python script from PhaseGenomics.
DNA was also extracted from young G. raimondii leaves following the Bionano Plant protocol for high-molecular weight DNA. DNA was purified, nicked, labeled, and repaired according to Bionano standard operating procedures for the Irys platform. Two optical maps of different enzymes (BspQI and BssSI) were assembled using the IrysSolve pipeline on the BYU Fulton SuperComputing cluster. The optical maps were combined into a two-enzyme composite optical map and it was aligned to the PGA assembly using an in silico labeled reference sequence. Conflicts between the Bionano maps and the PGA assembly were manually identified in the Bionano Access software by comparing the mapped Bionano contigs to the CANU contigs along the draft genome sequence. Conflicts between datasets were resolved by repositioning and reorienting CANU contigs in PGA ordering files followed by reconstruction of the fasta sequence, provided there was supporting or no-conflict evidence from the optical map ((Durand et al. 2016), Supp. Figure 1). Multiple iterations of mapping, conflict resolution, and draft sequence construction resulted in the final, new genome sequence of G. raimondii.
Leaf tissue of G. turneri was shipped to DoveTail Genomics for DNA extraction and construction of HiC sequencing libraries. These HiC sequencing libraries were sequenced on the Illumina HiSeq 2500 (PE125 bp) at the BYU DNASC. Reads were mapped to the G. raimondii (Paterson et al. 2012) reference genome, and a scaffolded assembly was created for G. turneri by Dovetail Genomics. Whole genome alignments identified in-silico assembly errors where a contiguous 25.7 Mb of Chromosome 9 (D10_09) was initially placed on D10_12, and the remainder of that chromosome was in smaller scaffolded pieces. Similar to the process above, manual iterations of scaffolding correctly assembled D10_09 and D10_11 using Juicebox (Durand et al. 2016). The final genome sequence of G. turneri was constructed using a custom python script developed by PhaseGenomics, LLC and consists of 13 assembled chromosomes.
Repeats and gene annotation
Repeats were identified using a combination of RepeatMasker (Smit et al.) and “One code to find them all” (Bailly-Bechet et al. 2014), the latter used to assemble multiple adjacent RepeatMasker hits into complete transposable element (TE) copies. RepeatMasker was run for each genome with a custom library, which combines Repbase 23.04 repeats (Bao et al. 2015) with cotton-specific repeats. Default parameters were run, except the run was “sensitive” and was set to mask only TEs (no low-complexity). Parameters are available at https://github.com/Wendellab/D5D10. “One code to find them all” was used to aggregate multiple hits into TE models using default parameters. The resulting output was aggregated and summarized in R/3.4.4 (R Development Core Team 2008) using dplyr /0.7.4 (Wickham et al. 2019). All code can be found at https://github.com/Wendellab/D5D10.
The MAKER-P pipeline (Cantarel et al. 2008) was used to annotate G. raimondii and G. turneri genomes after masking repetitive elements with RepeatMasker (Smit et al.) using a custom database that enriched for cotton-specific repeat sequences.
Gossypium raimondii was annotated using the iterative MAKER-P method previously described (Grover et al. 2017) with the following modifications: (1) assembly of RNA-seq data using Mikado (Venturini et al. 2018); (2) RNA-seq assembly provided as another prediction source instead of ESTs evidence; and (3) updated software versions. The raw RNA-seq reads are available from the SRA (PRJNA493521). The assembly and annotation quality for each genome was validated via the BUSCO (Simão et al. 2015) pipeline, which evaluates completeness by characterizing the presence, fragmentation, and/or duplication of highly conserved genes. Single-copy syntenic orthologs were inferred using MCScanX (Wang et al. 2012) with a minimum of 50 genes in a syntenic block and gap penalty of 2. Any gene belonging to two different syntenic groups was removed.
Data availability
The assembled genome sequences of G. raimondii (PRJNA493304) and G. turneri (PRJNA493521) are available in NCBI (CP032553-CP032565 and CP032571-CP032583, respectively). The raw data for G. raimondii and G. turneri are also available in NCBI (SRR6356446 and SRR7957402, respectively). Supplemental material available at FigShare: https://doi.org/10.25387/g3.9702299.
Results and Discussion
Genome assemblies
We report two de novo genome sequences for the genus Gossypium, a new and corrected assembly for G. raimondii (D5) and a new reference-quality assembly for the closely related G. turneri (D10). These new genomes integrate multiple sequencing technologies and provide a more accurate representation of each cotton genome. Notwithstanding the utility of the original G. raimondii sequence (Paterson et al. 2012), it has become evident that the genome sequence contained minor assembly errors.Our genome sequence reported here provides an improved G. raimondii assembly using PacBio long read sequencing technology and corrects some errors in the genome sequence that have been identified (Du et al. 2018; Wang et al. 2019).
The G. raimondii genome was assembled from 43.7x PacBio coverage of raw sequence reads. The assembly consisted of 187 contigs with an N50 of 6.3Mb (Table 1). The contigs were scaffolded using HiC by PhaseGenomics and the pseudomolecules were manually adjusted using JuiceBox (Durand et al. 2016). The final scaffolded assembly was independently verified using a composite optical map of two different enzymes. A comparison of assembly metrics between the previous genome sequence and our new genome sequence of G. raimondii illustrates a 45x improvement in contig length and a 97x reduction in the number of gaps. The cumulative gap length of the new assembly (17.6 kb) was reduced by 647x compared to the assembled gaps of the previous genome sequence (11,391 kb). The final genome assembly size was 14.9 Mb smaller than the previous assembly, representing 98% of previously assembled genome sequence in length.
Table 1. Assembly metrics of the G. turneri genome, the G. raimondii (our current assembly, D5), and the previous G. raimondii assembly (Paterson et al. 2012).
| G. turneri (D10) | G. raimondii (D5) | G. raimondii (2012) | |
|---|---|---|---|
| Contigs | 220 | 187 | 16,924 |
| Max Contig | 23,475,487 | 24,216,129 | 1,162,971 |
| Mean Contig | 3,432,648 | 3,929,767 | 43,597 |
| Contig N50 | 7,909,293 | 6,291,832 | 136,998 |
| Contig N90 | 1,624,019 | 2,044,991 | 32,166 |
| Total Contig Length | 755,182,540 | 734,866,495 | 737,837,083 |
| Assembly GC | 33.21 | 33.19 | 33.19 |
| Scaffolds | 13 | 13 | 13 |
| Max Scaffold | 67,704,245 | 65,701,939 | 70,713,020 |
| Mean Scaffold | 58,092,557 | 56,529,546 | 57,632,930 |
| Scaffold N50 | 60,464,062 | 58,819,159 | 62,175,169 |
| Scaffold N90 | 50,570,303 | 46,322,098 | 45,765,648 |
| Total Scaffold Length | 755,203,240 | 734,884,094 | 749,228,090 |
| Captured Gaps | 207 | 174 | 16,911 |
| Max Gap | 100 | 200 | 63,138 |
| Mean Gap | 100 | 101 | 674 |
| Gap N50 | 100 | 100 | 2,607 |
| Total Gap Length | 20,700 | 17,599 | 11,391,007 |
This is the first de novo genome sequence for G. turneri. The G. turneri genome was assembled from 73.2x PacBio of raw sequence reads. The assembly consisted of 220 contigs with an N50 of 7.9Mb (Table 1). Similar to the G. raimondii sequence, these contigs were scaffolded by Dovetail Genomics and the pseudomolecules were manually adjusted using JuiceBox. Bionano data were not collected for G. turneri. The G. raimondii Bionano data were uninformative when aligned to the G. turneri genome sequence (because the distances between labeled recognition sites were too different). After creation of the sequence assembly, the G. raimondii HiC sequence reads were also mapped to the G. turneri genome sequence (and vice versa). While the number of mapped reads was reduced significantly (29.90% and 12.67%, respectively), there were no association anomalies detected between genomes.
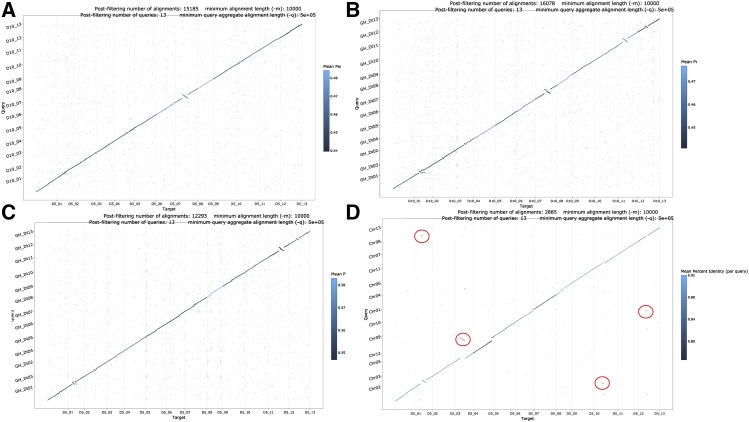
The assembled genome sequences were also verified by alignments to the DT-genome of G. hirsutum (Wang et al. 2019) and to the previous genome assembly of G. raimondii (Figure 1). The chromosomes had general agreement in their alignments between the four independently assembled sequences (old and new G. raimondii; G. turneri; DT of G. hirsutum). Such colinearity was also previously identified between cotton genomes. For example, genetic maps of G. hirsutum (e.g., (Byers et al. 2012)) were used to previously verify and sometimes establish proper scaffolding between contigs (Paterson et al. 2012).
Figure 1.
Genome comparisons between G. raimondii (D5), G. turneri (D10), G. raimondii (2012), and the DT-genome of G. hirsutum (DT). A) Genome alignment between G. turneri (D10) and G. raimondii (D5). B) Genome alignment between DT and D10. C) Genome alignment between DT and D5. D) Genome alignment between D5 (2012) and D5 (new). Red circles indicate assembly errors in the 2012 sequence as identified by these alignments and independent HiC data (e.g., D5_13 – Chr01, D5_11 – Chr03, D5_04 – Chr09, D5_02 – Chr13).
Error Correction in G. raimondii genome sequence
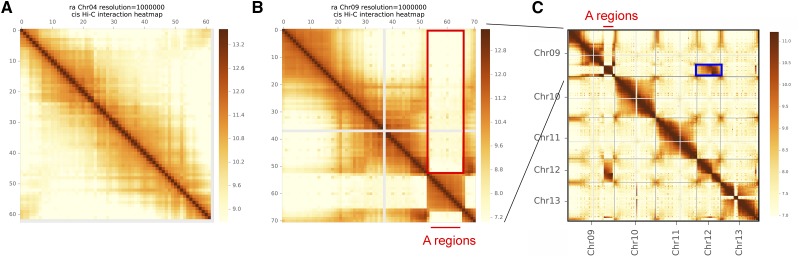
Errors were identified in the previous G. raimondii sequence (Paterson et al. 2012). In the previous genome sequence, the chromosomes were named to be consistent with previous genetic maps; however, a new chromosome naming convention has been used for diploid and allotetraploid cotton (Li et al. 2015; Zhang et al. 2015; Du et al. 2018), where homeologous chromosomes are organized in sequence pairs (e.g., AT_01 - AT_13 [Chr. 01 - Chr. 13] are homeologs of DT_01 - DT_13 [Chr. 14 – Chr. 26], respectively). We have adopted this new naming convention for the homologous chromosomes of these two genomes. Structural errors in the previously published sequence were identified by genome alignments (Figure 1) and by mapping HiC reads to the genome sequence (Figure 2, Supp Figure 1). The largest error was an assembly-derived translocation of D5_04 (previously Chr. 12) on D5_05 (previously Chr. 09) (Figure 2). Additional, smaller errors were found between Chr. 01 (now D5_07) and Chr. 13 (now D5_13); Chr. 02 (now D5_01) and Chr. 13 (now D5_13); Chr. 03 (now D5_02) and Chr. 13 (now D5_13); Chr. 02 (now D5_01) and Chr. 03 (now D5_02); Chr. 02 (now D5_01) and Chr. 07 (now D5_11); and Chr. 03 (now D5_02) and Chr. 07 (now D5_11) (Supp Figure 1). These corrections based on alignment and HiC data were also supported by the alignment of Bionano data.
Figure 2.
HiC interactions detected in the previously published G. raimondii genome sequence (Paterson et al. 2012). A) Most interaction maps of chromosome sequences suggested that the genome sequence was assembled in the correct order. B) A sequence was incorrectly assembled within Chr. 9 (now D5_05) that created a large insertion (red box). Few interactions were found between the inserted segment and the remainder of Chr. 9. C) Corresponding interactions were identified in the HiC interaction plot between Chr. 9 and Chr. 12 (now D5_04), as well as ‘pinch’ within the diagonal interaction map in Chr. 12, indicating the true position of the incorrectly assembled sequence.
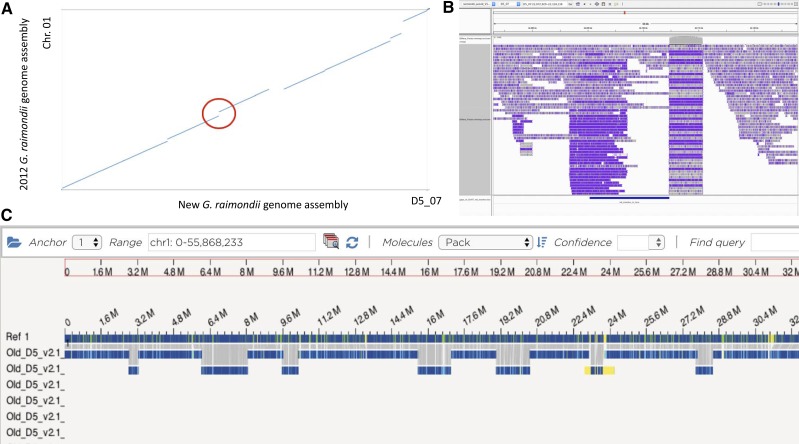
We also inspected a reported nuclear mitochondrial genome insertion (NUMT) on D5_07 (previously Chr. 1, Figure 3) located between coordinates 23.1Mb and 25Mb (Paterson et al. 2012). This region appears to have been the result of assembly error. Alignment of the two genomes (previous D5 genome vs. new D5 genome) identified a 1.26Mb segment that was inserted into the old sequence and not found in our new de novo assembly. Bionano data also indicated an insertion in the old assembly while the ‘inserted’ Bionano contig was unmapped in the new assembly of D5 (Figure 3C).
Figure 3.
Genomic assembly data of the new G. raimondii sequence suggest that the previously reported mitochondrial insertion was likely due to an assembly error. A) Genome alignments between G. raimondii Chr. 01 (Paterson et al. 2012) and our new genome sequence of D5_07. The red circle indicates the putative position of the mitochondrial genome insertion in the previous G. raimondii sequence relative to the new assembly. B) Alignment of G. raimondii PacBio reads (Track 2) to the new reference genome of G. raimondii (Track 1). The multi-colored bars represent individual PacBio reads (Track 2). The previous reference genome of G. raimondii had a mitochondrial insertion somewhere in this 14kb region indicated by the blue bar of Track 3. There are no PacBio reads that span the gap between the flanking regions of the 6,071 repeat and the repeat itself. C) Bionano data mapped to the previous reference genome sequence of G. raimondii (Paterson et al. 2012) also suggest an insertion of a sequence that is non-contiguous in the flanking regions. The Ref1 track reference to the originally published genome sequence of G. raimondii with a mitochondrial insertion between ∼23Mb and ∼24Mb. Independently constructed Bionano contigs were aligned to the 2012 reference sequence. A Bionano contig matched the reference sequence in the mitochondria insertion region, but the flanking regions of the Bionano contig (yellow) did not match flanking Bionano contigs or the reference sequence.
Since NUMTs evolve more quickly than do functional mitochondrial gene sequences, we also inspected the sequence similarity of the NUMT to the mitochondrial genome sequence of G. raimondii (Chen et al. 2017). The NUMT exhibited high similarity to the published G. raimondii mitochondrial genome (99.8% PID over 94% of region between Chr01:23,100,000-25,000,000). On an individual gene basis, over half of the genes contained within the putative NUMT were over 99% identical to the published sequence in the G. raimondii mitochondrial genome, with an average of 95% similarity. Considering the D-genome alignments and Bionano data presented above, the NUMT was more likely an assembly artifact than a recent insertion event in the G. raimondii genome.
Structural Variations Between the D-genomes
Comparisons between G. raimondii and G. turneri revealed several structural differences between the two genomes (Supp Figure 2). The genomes were largely colinear and no significant duplicated segments (relative to the genome alignments) were found in either genome (Figure 1). The assembled sequence of the G. turneri genome was 20.3 Mb longer than the G. raimondii genome and the gene content was similar (see below). The largest number of structural variants between the two genomes were chromosomal inversions. We identified several relative inversions between the two de novo genomes (Table 2). Inversions were manually identified in the genome alignment output file. A total of 64 Mb genome sequence had an inverted order between these two genomes.These regions included a total of 2,592 genes (∼6.4% of gene total number). The largest structural variant was an inversion on D10_08 (Figure 1, Supp Figure 3 - 15). This inversion could have been the result of misassembly, but the putative break points had clear overlapping, individual PacBio reads in G. raimondii (Supp Figures 6 & 9) and in G. turneri (Supp Figures 10 & 13). In addition, both genomes had consistent HiC patterns for the D8 chromosome where an inversion of ∼16 Mb would have been clearly identified had it been the result of assembly error in one of the two genome sequences (Supp Figure 1).
Table 2. Inversions between the de novo genome assemblies of G. turneri and G. raimondii.
| Chromosome | Inv. number | Total Length | Gene number |
|---|---|---|---|
| 1 | 9 | 4,856,224 | 132 |
| 2 | 9 | 7,086,444 | 114 |
| 3 | 5 | 5,569,613 | 431 |
| 4 | 4 | 2,192,874 | 60 |
| 5 | 5 | 4,213,508 | 179 |
| 6 | 6 | 1,597,287 | 164 |
| 7 | 3 | 2,453,735 | 159 |
| 8 | 7 | 16,167,439 | 345 |
| 9 | 8 | 5,741,456 | 267 |
| 10 | 2 | 417,545 | 9 |
| 11 | 9 | 7,708,678 | 501 |
| 12 | 4 | 1,771,113 | 44 |
| 13 | 10 | 4,944,400 | 187 |
| Total | 81 | 64,720,316 | 2592 |
We also compared the G. turneri and G. raimondii genome sequence to other Gossypium genomes (Supp Figure 16 & 17, (Du et al. 2018; Wang et al. 2019)). If a large inversion between G. turneri and G. raimondii was 1) also present in the genome alignment between G. arboreum and G. turneri and 2) was not present in the genome alignment between G. raimondii and G. arboreum then it was considered as an inversion derived during the natural evolutionary history of the G. turneri genome (similar logic for inversions derived in G. raimondii or G. arboreum). The inversions need to be large (>2 Mb) and present in only one genome to be confident about its description without further investigation. The largest inversions on chromosomes D10_03, D10_05, D10_07, and D10_08 appear to be specific to G. turneri (36% of the length of total inversions). Chromosome rearrangements (inversions and other events) specific for G. arboreum were found on A2_01, A2_02, A2_03, A2_07, and A2_11. Only one inversion (D5_13, 2.6 Mb) was found to be specific to the G. raimondii genome. Perhaps, these inversions were part of the speciation process between the different Gossypium genomes.
Gene annotations
Similar numbers of genes were found in the annotation of each genome. Annotation of the genomes of G. turneri and G. raimondii identified 38,489 and 40,743 gene models respectively (Table 3). BUSCO analysis reported >90% completeness scores for both G. turneri and G. raimondii genome assemblies, indicating that the evolutionarily-conserved core gene set was present in both de novo assemblies (Supp. Figure 18). Using MCScanX, we were able to identify 23,499 syntenic orthologs shared between the two species, indicating that the gene order and gene compliment are largely conserved between these two species. Genes in 34 of these syntenic orthologs were inferred to have more than one syntenic ortholog; these genes were removed from the dataset, resulting in 23,465 high-confidence syntenic orthologs (Supp. File 1). While not every gene was categorized into syntenic relationships, this is not surprising given that genes present in tandem arrays were excluded from this analysis (a default setting of MCScanX), gene loss has likely occurred in both species since they last shared a common ancestor, and subtle differences in gene annotation in the two genome assemblies likely lead to slight differences in overall gene content.
Table 3. Each of the de novo genome assemblies were annotated for gene content using Maker-P.
| Predicted Features | G. turneri (D10) | G. raimondii (D5) | G. raimondii (2012) |
|---|---|---|---|
| CDS | 205,333 | 235,836 | 486,043 |
| exon | 200,384 | 236,559 | 527,563 |
| gene | 38,489 | 40,743 | 37,505 |
| mRNA | 39,553 | 41,030 | 77,267 |
Repeats
Transposable element content was predicted for both de novo genomes and compared to the existing G. raimondii reference sequence (Paterson et al. 2012). As expected, the de novo G. raimondii genome had nearly identical predicted TE content with the previous G. raimondii genome sequence (Table 4). This difference is not significant and can be attributed to slight differences in assembly of repetitive regions. Consistent with the larger size of G. turneri than G. raimondii (910 Mb vs. 880 Mb), the G. turneri genome assembled an additional 8.5 Mb and 10.6 Mb of repetitive sequence, relative to the previous and new de novo G. raimondii genome sequences, respectively. Generally, the G. turneri genome sequence has slightly fewer DNA TEs and more LTR retrotransposons than the two G. raimondii genomes, both with respect to absolute content and percent of genome (Table 4). No non-LTR retrotransposons (e.g., LINE/SINE) were detected. For all three genome assemblies, retrotransposons comprise approximately 36% of the genome sequence, whereas all DNA elements combined comprise just under 3% in each. These results are consistent with a previous analysis of low-coverage sequencing results of these two genomes (Grover et al. 2019).
Table 4. Repetitive content of the newly sequenced G. turneri and G. raimondii genomes, and the previously published G. raimondii (Paterson et al. 2012). No LINE or SINE elements were detected. The genome size of G. turneri is 910 Mb and G. raimondii is 880 Mb.
| G. turneri | G. raimondii | G. raimondii (Paterson et al. 2012) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Family | Fragments | Copies | Total (Mb) | Fragments | Copies | Total (Mb) | Fragments | Copies | Total (Mb) |
| DNA | 20,199 | 12,453 | 18.28 | 22,503 | 13,764 | 20.63 | 23,474 | 13,969 | 20.27 |
| CMC/EnSpm | 2 | 1 | 0.00 | 2 | 2 | 0.00 | 14 | 9 | 0.00 |
| EnSpm/CACTA | 2,443 | 1,385 | 3.92 | 3,172 | 1,864 | 5.24 | 3,648 | 1,878 | 4.87 |
| Harbinger | 30 | 22 | 0.01 | 58 | 41 | 0.03 | 42 | 28 | 0.02 |
| hAT | 2,725 | 1,712 | 1.01 | 3,079 | 1,895 | 1.01 | 3,209 | 1,966 | 1.03 |
| L1 | 1,255 | 638 | 1.56 | 1,256 | 618 | 1.49 | 1,290 | 633 | 1.54 |
| Mariner/Tc1 | 98 | 51 | 0.07 | 76 | 40 | 0.06 | 84 | 43 | 0.06 |
| MuDR | 13,590 | 8,592 | 11.71 | 14,828 | 9,280 | 12.79 | 15,145 | 9,381 | 12.73 |
| MULE-MuDR | 52 | 50 | 0.01 | 21 | 19 | 0.00 | 25 | 23 | 0.00 |
| PIF-Harbinger | 4 | 2 | 0.00 | 11 | 5 | 0.00 | 17 | 8 | 0.01 |
| LTR | 338,644 | 199,672 | 277.72 | 325,760 | 190,122 | 264.75 | 336,908 | 196,564 | 267.24 |
| LTR | 224 | 216 | 0.02 | 214 | 206 | 0.02 | 311 | 304 | 0.03 |
| Copia | 48,098 | 28,294 | 45.51 | 48,911 | 29,032 | 45.29 | 50,993 | 29,965 | 45.72 |
| Gypsy | 290,322 | 171,162 | 232.19 | 276,635 | 160,884 | 219.44 | 285,604 | 166,295 | 221.49 |
| Total | 358,843 | 212,125 | 296.00 | 348,263 | 203,886 | 285.38 | 360,382 | 210,533 | 287.51 |
Conclusion
Genome sequences of many plants have been recently published, and in fact are too numerous to cite here. Many of these previously reported genome sequences are being revisited with long-read technology of PacBio or Oxford Nanopore. In this report, we present new de novo genome sequences for G. raimondii and G. turneri based on PacBio long-read sequence technology. Both of these genomes are closely related to the DT-genome of cultivated tetraploid cotton. These sequences provide an evolutionary perspective for comparative genomics of the Gossypium clades as well as providing useful resources for the genetic improvement of cotton. Because of the economic relevance of the Gossypium genus, additional genome sequences of related Gossypium species will continue to be studied and revised in the future.
Acknowledgments
We thank the National Science Foundation Plant Genome Research Program (Grant #1339412) and Cotton Inc. for their financial support. This research was funded, in part, through USDA ARS Agreements 58-6066-6-046 and 58-6066-6-059.We thank BYU Fulton SuperComputer lab for their resources and generous support. We also thank ResearchIT for computational support at Iowa State University. We thank Rise Services for office accommodations in Orem, UT.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9702299.
Communicating editor: P. Morrell
Literature Cited
- Bailly-Bechet M., Haudry A., and Lerat E., 2014. “One code to find them all”: a perl tool to conveniently parse RepeatMasker output files. Mob. DNA 5: 13 10.1186/1759-8753-5-13 [DOI] [Google Scholar]
- Bao W., Kojima K. K., and Kohany O., 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers R. L., Harker D. B., Yourstone S. M., Maughan P. J., and Udall J. A., 2012. Development and mapping of SNP assays in allotetraploid cotton. Theor. Appl. Genet. 124: 1201–1214. 10.1007/s00122-011-1780-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Korf I., Robb S. M. C., Parra G., Ross E. et al. , 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen Y., Feng S., Zhao T., and Zhou B., 2018. Overcoming obstacles to interspecific hybridization between Gossypium hirsutum and G. turneri. Euphytica 214: 35 10.1007/s10681-018-2118-2 [DOI] [Google Scholar]
- Chen Z., Nie H., Grover C. E., Wang Y., Li P. et al. , 2017. Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species. Plant Biol. 19: 484–493. 10.1111/plb.12536 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., Scheffler B. E., Dennis E., B. a Triplett, T. Zhang et al, 2007. Toward Sequencing Cotton (Gossypium) Genomes. PLANT Physiol. 145: 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., and de Laat W., 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26: 11–24. 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Huang G., He S., Yang Z., Sun G. et al. , 2018. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50: 796–802. 10.1038/s41588-018-0116-x [DOI] [PubMed] [Google Scholar]
- Durand N. C., Robinson J. T., Shamim M. S., Machol I., Mesirov J. P. et al. , 2016. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 3: 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryxell P. A., 1978. Gossypium turneri (Malvaceae), A New Species From Sonora, Mexico. Madrono 25: 155–159. [Google Scholar]
- Grover C. E., Arick M. A., Conover J. L., Thrash A., Hu G. et al. , 2017. Comparative Genomics of an Unusual Biogeographic Disjunction in the Cotton Tribe (Gossypieae) Yields Insights into Genome Downsizing. Genome Biol. Evol. 9: 3328–3344. 10.1093/gbe/evx248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. E., Arick M. A., Thrash A., Conover J. L., Sanders W. S. et al. , 2019. Insights into the Evolution of the New World Diploid Cottons (Gossypium, Subgenus Houzingenia) Based on Genome Sequencing. Genome Biol. Evol. 11: 53–71. 10.1093/gbe/evy256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. Z., Sang Z. Q., Zhou B. L., and Zhang T. Z., 2007. Genetic relationships of D-genome species based on two types of EST-SSR markers derived from G. arboreum and G. raimondii in Gossypium. Plant Sci. 172: 808–814. 10.1016/j.plantsci.2006.12.012 [DOI] [Google Scholar]
- Hendrix B., and Stewart J. M., 2005. Estimation of the nuclear DNA content of Gossypium species. Ann. Bot. 95: 789–797. 10.1093/aob/mci078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell K. K., and Osborn T. C., 1992. Simple plant DNA isolation procedures, pp. 1–13 in Plant Genomes: Methods for Genetic and Physical Mapping. Springer, Dordrecht, The Netherlands. [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. et al. , 2017. Canu: Scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Fan G., Lu C., Xiao G., Zou C. et al. , 2015. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33: 524–530. 10.1038/nbt.3208 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J. et al. , 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Peterson D. G., and Arick M., 2018. Sequencing Plant Genomes, pp. 109–193 in Progress in Botany, edited by Cánovas F., Lüttge U., Matyssek R., and Pretzsch H.. Springer, Berlin, Germany. [Google Scholar]
- R Development Core Team , 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna. [Google Scholar]
- Servant N., Varoquaux N., Lajoie B. R., Viara E., Chen C.-J. et al. , 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16: 259 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., and Green P., 2019. RepeatMasker Open-4.0. http://www.repeatmasker.org
- Ulloa M., Abdurakhmonov I. Y., Perez-M C., Percy R., and Stewart J. M., 2013. Genetic diversity and population structure of cotton (Gossypium spp.) of the New World assessed by SSR markers. Botany 91: 251–259. 10.1139/cjb-2012-0192 [DOI] [Google Scholar]
- Venturini L., Caim S., Kaithakottil G. G., Mapleson D. L., and Swarbreck D., 2018. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 7: 1–15. 10.1093/gigascience/giy093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J. D., Tan X., Li J. et al. , 2012. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tu L., Yuan D., Zhu D., Shen C. et al. , 2019. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51: 224–229. 10.1038/s41588-018-0282-x [DOI] [PubMed] [Google Scholar]
- Wickham H., Francois R., Henry L., and Muller K., 2019. dplyr: A Grammar of Data Manipulation. R package version 0.7.4. https://CRAN.R-project.org/package=dplyr
- Zhang T., Hu Y., Jiang W., Fang L., Guan X. et al. , 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33: 531–537. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled genome sequences of G. raimondii (PRJNA493304) and G. turneri (PRJNA493521) are available in NCBI (CP032553-CP032565 and CP032571-CP032583, respectively). The raw data for G. raimondii and G. turneri are also available in NCBI (SRR6356446 and SRR7957402, respectively). Supplemental material available at FigShare: https://doi.org/10.25387/g3.9702299.