Abstract
Chromosomal inversion polymorphisms play an important role in adaptation to environmental heterogeneities. For mosquito species in the Anopheles gambiae complex that are significant vectors of human malaria, paracentric inversion polymorphisms are abundant and are associated with ecologically and epidemiologically important phenotypes. Improved understanding of these traits relies on determining mosquito karyotype, which currently depends upon laborious cytogenetic methods whose application is limited both by the requirement for specialized expertise and for properly preserved adult females at specific gonotrophic stages. To overcome this limitation, we developed sets of tag single nucleotide polymorphisms (SNPs) inside inversions whose biallelic genotype is strongly correlated with inversion genotype. We leveraged 1,347 fully sequenced An. gambiae and Anopheles coluzzii genomes in the Ag1000G database of natural variation. Beginning with principal components analysis (PCA) of population samples, applied to windows of the genome containing individual chromosomal rearrangements, we classified samples into three inversion genotypes, distinguishing homozygous inverted and homozygous uninverted groups by inclusion of the small subset of specimens in Ag1000G that are associated with cytogenetic metadata. We then assessed the correlation between candidate tag SNP genotypes and PCA-based inversion genotypes in our training sets, selecting those candidates with >80% agreement. Our initial tests both in held-back validation samples from Ag1000G and in data independent of Ag1000G suggest that when used for in silico inversion genotyping of sequenced mosquitoes, these tags perform better than traditional cytogenetics, even for specimens where only a small subset of the tag SNPs can be successfully ascertained.
Keywords: Anopheles gambiae, chromosomal inversion polymorphism, genomics, inversion genotyping, karyotype analysis, malaria vector, tag SNP
A chromosomal inversion originates when a chromosome segment reverses end to end. Inversions maintained in plant and animal populations as structural polymorphisms tend to be large (several megabases) and contain hundreds of genes (reviewed in Wellenreuther and Bernatchez 2018). Long-term balancing selection can maintain these polymorphisms through millions of generations and multiple species radiations (Wellenreuther and Bernatchez 2018). Because recombination is greatly reduced between opposite orientations in inversion heterozygotes, inversions preserve selectively advantageous combinations of alleles despite homogenizing gene flow in collinear regions. Theory and mounting evidence implicate inversions in local adaptation, adaptive divergence, and range expansion, though the precise molecular mechanisms are rarely known (Hoffmann et al. 2004; Kirkpatrick and Barton 2006; Hoffmann and Rieseberg 2008; Schaeffer 2008; Kirkpatrick 2010; Lowry and Willis 2010; Joron et al. 2011; Jones et al. 2012; Kirkpatrick and Barrett 2015; Twyford and Friedman 2015; Kapun et al. 2016; Ayala et al. 2017; Fuller et al. 2017; Wellenreuther et al. 2017; Wellenreuther and Bernatchez 2018). Importantly, because of occasional double-crossovers and gene conversion, the suppression of gene flux is not absolute. As long as inversion heterozygotes are formed in populations, any significant association between an inversion and an allele within its boundaries is subject to eventual erosion unless gene flux is countered by selection (Navarro et al. 1997; Andolfatto et al. 2001).
The Anopheles gambiae complex is a medically important group of at least eight closely related and morphologically indistinguishable mosquito sibling species from sub-Saharan Africa (White et al. 2011; Coetzee et al. 2013). Three members of the complex (the eponymous Anopheles gambiae, Anopheles coluzzii, and Anopheles arabiensis) are among the most significant malaria vectors globally, responsible for a majority of the 435,000 malaria deaths in 2017 (World Health Organization 2018). The ecological plasticity of these three species contributes greatly to their status as major human malaria vectors (Coluzzi et al. 2002). In contrast to the other five, these three species have wide distributions across diverse biomes of tropical Africa. Not coincidentally, they also segregate strikingly high numbers of paracentric inversion polymorphisms, which are implicated in adaptation to seasonal and spatial environmental heterogeneities related both to climatic variables and anthropogenic alterations of the landscape (Coluzzi et al. 1979; Bryan et al. 1982; Coluzzi et al. 1985; Toure et al. 1998; Manoukis et al. 2008; Costantini et al. 2009; Simard et al. 2009; Cheng et al. 2012; Ayala et al. 2014; Caputo et al. 2014; Ayala et al. 2017; Cheng et al. 2018). Some of these inversions also have been associated with ecologically relevant phenotypes, including desiccation and thermal tolerance (Gray et al. 2009; Rocca et al. 2009; Cassone et al. 2011; Fouet et al. 2012; Ayala et al. 2019; Cheng et al. 2018).
The sister taxa An. gambiae and An. coluzzii, the focus of the present investigation, are the most closely related species in the An. gambiae complex, sharing extensive nucleotide variation through both recent common ancestry and introgression (Fontaine et al. 2015; Hanemaaijer et al. 2018), while maintaining characteristic differences in ecology and behavior (Costantini et al. 2009; Diabaté et al. 2009; Simard et al. 2009; Gimonneau et al. 2010; Gimonneau et al. 2012a; Gimonneau et al. 2012b; Dabire et al. 2013; Tene Fossog et al. 2015; Ayala et al. 2017). They also share four of six common chromosomal inversion polymorphisms on chromosomal arm 2R (b, c, d, u) and the only inversion polymorphism on chromosomal arm 2L (a) (Figure 1) (della Torre et al. 2005). These inversions range in size from ∼4Mb to 22Mb, and together span thousands of genes and a sizeable fraction of chromosome 2: ∼61% of 2R and ∼38% of 2L polytene (euchromatic) content (Coluzzi et al. 2002). Inversions 2La and 2Rb are found in populations throughout tropical Africa and are therefore cosmopolitan, while three other inversions on 2R (c, d, and u) are widespread in West, very rare in Central Africa, and absent from East Africa. The remaining two inversions, 2Rj and 2Rk, have more restricted geographic distributions (Coluzzi et al. 2002; Ayala et al. 2017).
Figure 1.
Diagrammatic representation of the common polymorphic inversions (labeled brackets) on chromosome 2 in An. gambiae. Polytene chromosome map modified from Figure 1 and poster in Coluzzi et al. (2002). CT, centromere.
Early cytogenetic studies of An. gambiae and An. coluzzii, presumed at the time to be a single heterogenous species, uncovered genetic discontinuities that led to the designation of five presumed assortatively-mating ‘chromosomal forms’: FOREST, SAVANNA, MOPTI, BAMAKO, and BISSAU (Coluzzi et al. 1985; Toure et al. 1998; Coluzzi et al. 2002; della Torre et al. 2005). They were delineated based on stable non-random associations of different sets of chromosome 2R inversions in co-occurring populations, and differed in larval ecology. Subsequent DNA-based studies identified fixed differences in the ribosomal DNA (rDNA), located in the pericentromeric region of the X chromosome, leading to the definition of two assortatively mating M and S ‘molecular forms’ of An. gambiae (della Torre et al. 2001). The molecular forms, which were eventually given specific status as An. coluzzii (formerly M) and An. gambiae sensu stricto (formerly S) (Coetzee et al. 2013), are incongruent with the chromosomal forms. Nearly all inversion associations segregate in both species albeit at different frequencies, and likely play similar roles in ecological specialization and adaptation in both An. gambiae s.s. (hereafter, An. gambiae) and An. coluzzii (della Torre et al. 2005; Costantini et al. 2009; Simard et al. 2009; Ayala et al. 2017). Hence, inversion associations are indicative of environmental heterogeneities more so than intrinsic reproductive boundaries.
Beyond a role in ecological specialization, inversions in the An. gambiae complex are also associated with vector traits affecting malaria transmission intensity and control: biting and resting behavior (Coluzzi et al. 1979; Riehle et al. 2017), seasonality (Rishikesh et al. 1985), morphometric variation (Petrarca et al. 1990), and Plasmodium infection rates (Petrarca and Beier 1992; Riehle et al. 2017). Although a robust molecular assay is available for genotyping inversion 2La in natural populations (White et al. 2007), 2R inversions with characterized breakpoint sequences (j, b, c, and u) (Coulibaly et al. 2007a; Sangare 2007; Lobo et al. 2010) proved difficult to genotype molecularly at the breakpoints (Coulibaly et al. 2007b; Lobo et al. 2010), owing to extensive tracts of flanking repetitive DNA. The 2Rk breakpoints have yet to be characterized, but recent localization of the 2Rd breakpoints in the reference genome assembly using proximity-ligation sequencing (Corbett-Detig et al. 2019) also revealed high repeat content, suggesting that repetitive DNA at inversion breakpoints will pose a significant challenge both for breakpoint characterization and for molecular genotyping assays targeting breakpoint regions in these species.
Failure to account for the presence of inversions is a barrier to a more comprehensive understanding of epidemiologically relevant mosquito behavior and physiology. Inversion-blind analysis of population data can mislead population genetic inference, and create spurious associations in genome-wide association studies (Seich Al Basatena et al. 2013; Houle and Marquez 2015). Powerful genomic resources exist for An. gambiae, including a high-quality reference genome assembly (Holt et al. 2002) and a database of genomic variation (Ag1000G) based on deep genome re-sequencing of thousands of mosquitoes from natural populations across Africa (The Anopheles gambiae 1000 Genomes Consortium 2017). Unfortunately, inversion genotypes are not automatically revealed by genome re-sequencing, as reads are mapped to their position in the reference genome assembly, not their position in the re-sequenced mosquito genome. Despite advancing genome technology, the only method currently available to determine the An. gambiae karyotype is a method perfected half a century ago (Coluzzi 1968) involving cytological analysis of ovarian nurse cell polytene chromosomes (Coluzzi et al. 2002; Pombi et al. 2008). At best, such cytological analysis is severely rate-limiting because it is laborious and requires highly specialized training. At worst, it is prohibitive because it requires proper preservation of chromosomes harvested only from ovaries of adult females at a specific gonotrophic stage; suitable polytenization is absent at other gonotrophic stages as well as in males (della Torre 1997). While salivary glands of late fourth instar larvae also contain chromosomes with an adequate degree of polytenization, and the banding patterns of salivary and ovarian chromosomes are homologous in principle, most bands are difficult to homologize due to a different pattern of chromosome ‘puffing’ (della Torre 1997), rendering this alternative impractical. To overcome these impediments, our goal is to develop broadly accessible computational and molecular methods of genotyping chromosomal inversions in individual specimens of An. gambiae and An. coluzzii.
Here, we exploit the Ag1000G database and leverage the subset of cytologically karyotyped specimens within that database to develop a computational approach for karyotyping applicable to whole genome sequence data. We identify multiple tag single nucleotide polymorphisms (SNPs) significantly associated with inversions across geography that collectively predict with high confidence the genotypes of six common polymorphic inversions on chromosome 2 (a, j, b, c, d, u) in individually sequenced genomes of An. coluzzii and An. gambiae. We then apply this approach to data generated independently of Ag1000G to show that our approach has wider utility, even for specimens where only a small subset of the tag SNPs can be successfully ascertained.
Materials and Methods
Mosquito genotype data
Variant call data used for the discovery of inversion tag SNPs were accessed from Ag1000G (The Anopheles gambiae 1000 Genomes Consortium 2017) and Vector Observatory (VOBS; Table S1), projects of the Malaria Genomic Epidemiology Network (MalariaGEN; https://www.malariagen.net/) that provide catalogs of genomic sequence variation based on individual wild-collected An. gambiae and An. coluzzii mosquitoes sampled from multiple African countries and the Mayotte archipelago. With the exception of four atypical samples (see next section), we verified species identifications as reported in Ag1000G and VOBS using principal component analysis (PCA) of biallelic SNPs on the X chromosome. We excluded any specimens with more than 50,000 missing genotypes on chromosomal arm 2R (N = 9), and any specimens subjected to whole genome amplification (WGA) prior to genomic sequencing (N = 44), as PCA revealed strong biases associated with WGA. After filtering, we retained variant call data from 1,347 mosquitoes (Table S2).
Karyotype imputation by local PCA
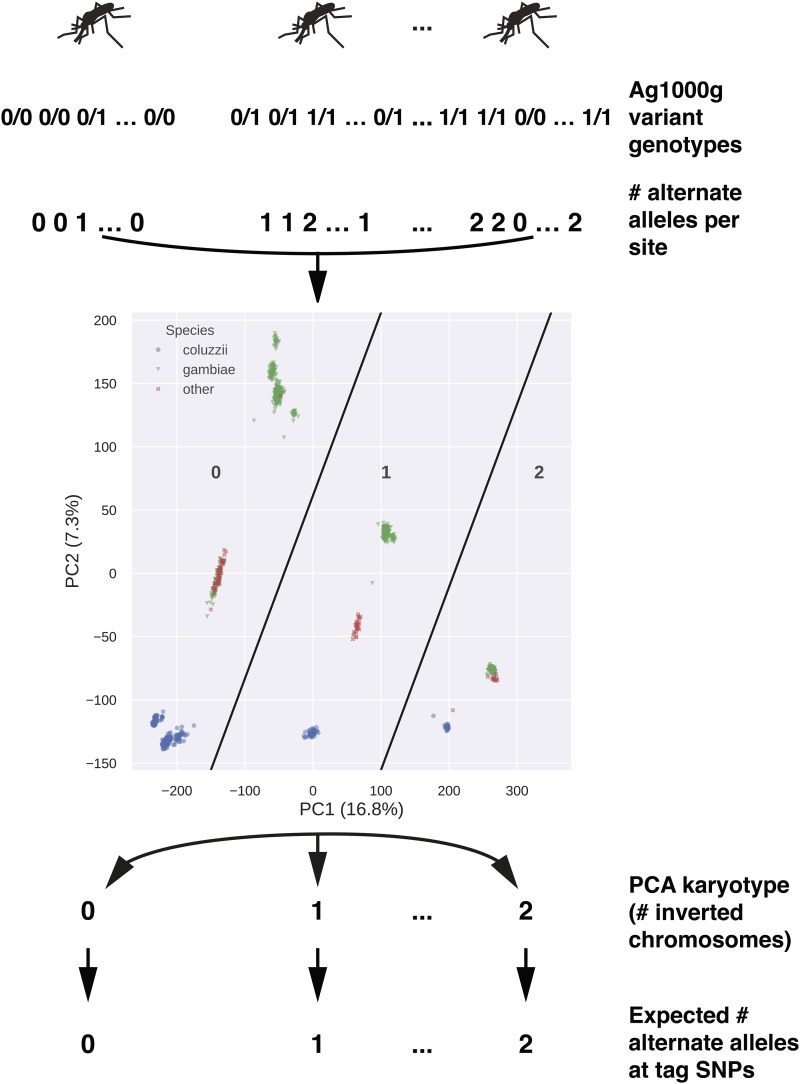
Cytological karyotype information derived from phase contrast microscopy of ovarian polytene chromosomes (della Torre 1997) was available only for a relatively small subset of specimens (N = 373) in Ag1000G/VOBS (hereafter, Ag1000G for brevity). Thus, as a first step toward discovering SNPs putatively predictive of inversion status (tag SNPs), we imputed karyotypes computationally at each of six focal inversions (Figure 1), using local PCA (where ‘local’ refers to windows of the genome corresponding to chromosomal rearrangements). Ma and Amos (2012) showed that applying PCA to SNP genotypes in a window of the genome containing an inversion polymorphic in population genomic data (an approach that we call ‘PCA karyotyping’) produces a pattern of three equidistant clusters (stripes) in a plot of the first two principal components, assuming adequate numbers of each of three possible inversion genotypes: inverted and uninverted (standard) homokaryotypes, and heterokaryotypes. The two flanking stripes represent alternative homokaryotypes, and the middle stripe represents the inversion heterokaryotype, a 1:1 “admixture” between the two homokaryotype classes (Ma and Amos 2012).
To apply this approach, we combined specimens from both species (An. gambiae and An. coluzzii) and different geographic localities into a single metapopulation sample of 1,347 mosquitoes (Tables S1, S2). We identified a set of biallelic SNPs within inversion boundaries (Table S3) with potentially informative levels of polymorphism [minor allele count ≥3 and minimum alternate allele frequency (MAF) ≥0.15 for all inversions except 2Rd, for which the MAF threshold was reduced to 0.03]. As 2Rd overlaps 2Ru in the genome (Figure 1), we limited consideration to only those SNPs found outside (proximal to) 2Ru for PCA karyotyping of 2Rd (Table S3). Next, we converted mosquito genotypes at these SNPs to a count of the number of alternate alleles (‘0’ if both matched the reference allele, ‘1’ or ‘2’ if one or both matched the alternate allele, respectively). Using the scikit-allel Python package v1.1.9 (Miles and Harding 2017), we then applied PCA to the resulting matrix of alternate allele counts, and represented the output as a scatter plot of the first two principal components for each mosquito in the population sample. The correct genotype corresponding to the two homokaryotype stripes was determined based on the inclusion in a given stripe of mosquitoes with cytologically determined karyotype. Based on this classification, mosquitoes without cytologically determined karyotypes were assigned a PCA karyotype.
The distinction between stripes was not always sharp; the stripes could be diffuse and oblique rather than tightly clustered. In extreme cases, stripes were not initially discernable. Through an iterative process of ‘leave one population sample out’ followed by PCA, we determined that absence of a clear three-stripe pattern was attributable to some or all of the same four atypical source populations, in particular, those from Kenya, Mayotte, The Gambia, and Guinea Bissau. The Kenyan sample has been found to display signs of extreme inbreeding (The Anopheles gambiae 1000 Genomes Consortium 2017), and Mayotte is an island whose mosquito population is plausibly subject both to inbreeding and a degree of isolation from mainland samples. The Gambia and Guinea Bissau are localities with unusually high degrees of hybridization and introgression between An. gambiae and An. coluzzii (Caputo et al. 2008; Oliveira et al. 2008; Caputo et al. 2011; Marsden et al. 2011; Weetman et al. 2012; Nwakanma et al. 2013). Where necessary, we removed these population samples, as well as two An. gambiae-An. coluzzii hybrid specimens from Burkina Faso and Guinea Conakry, and repeated the PCA. In addition, successful PCA karyotyping of 2Rd and 2Rj required the removal of all An. coluzzii specimens owing to taxonomic structuring of variation. Accordingly, PCA karyotyping was successful on all (2La) or subsets (all 2R inversions) of the 1,347 specimens (Table S4).
Discovery of SNPs predictive of inversion orientation
The PEST reference genome assembly for An. gambiae (AgamP4; Giraldo-Calderón et al. 2015) was derived from a colony whose karyotype was homozygous standard with respect to all common chromosomal inversions in this species. We therefore had the general expectation that an individual SNP might be a good predictor of chromosomal inversion orientation if the reference allele is strongly associated with the standard arrangement and the alternate allele is strongly associated with the inverted arrangement within and across population samples. As shown in Figure 2 in overview, we assessed SNP genotype-inversion genotype concordance for each inversion in individual mosquitoes, limiting our assessment to potentially more informative, higher frequency biallelic SNPs inside inversion boundaries (i.e., those with MAF ≥ 5%). We converted both the SNP genotype and the corresponding mosquito’s PCA-based inversion genotype to single numbers, representing the count of alternate alleles (0, 1, or 2) in the case of SNP genotype, and the count of inverted chromosomes (0, 1, or 2) in the case of inversion genotype. Successfully performing tags are expected to have a SNP genotype that correlates strongly with the PCA-based inversion genotype.
Figure 2.
Assessment of correspondence between SNP and inversion genotype in each mosquito. For each chromosomal rearrangement and mosquito, biallelic SNP genotypes inside rearrangement boundaries were converted to a number representing the count of alternative alleles (relative to the AgamP4 reference). We applied PCA to the resulting matrix to assign each individual mosquito an inversion genotype. The expectation is that the PCA-based genotype, expressed as the number of inverted chromosomes at the focal rearrangement, should match the number of alternative alleles at SNPs predictive of inversion status (tag SNPs).
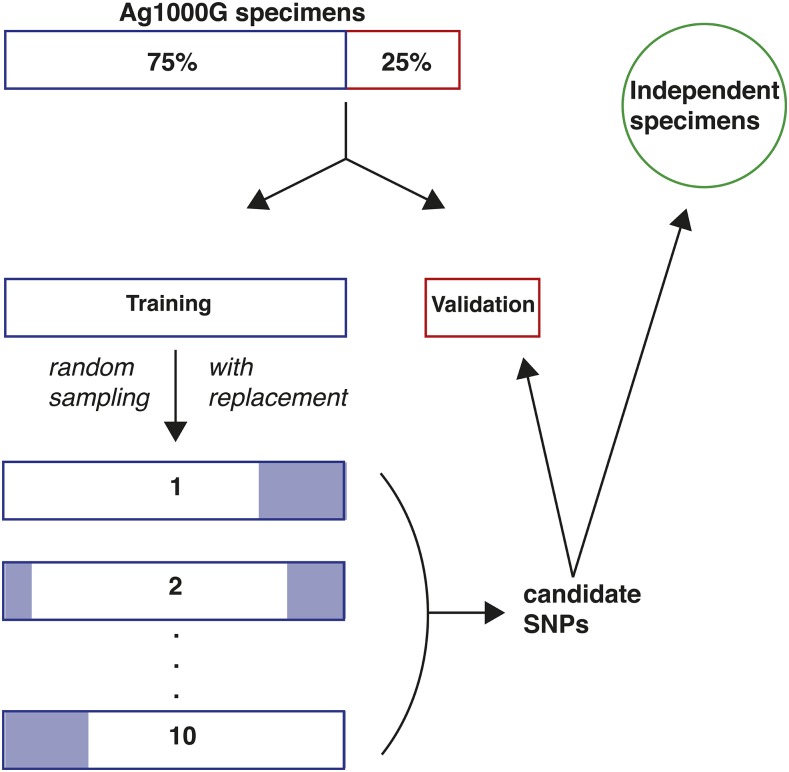
More formally, we sought to identify candidate tag SNPs using the procedure illustrated in Figure 3 (applied separately for each inversion). Specimens assigned a PCA-based karyotype for a focal inversion were divided into a training sample used for tag SNP discovery (75%) and a validation sample that was held in reserve until a later time (25%), using the model_selection module of the scikit-learn Python package (v0.19.2) (Pedregosa et al. 2011). We ensured that both partitions were balanced with regard to inversion genotypes but randomized in all other respects. For robust identification of candidate tag SNPs within the training sample, we masked all SNP genotypes inside the inversion boundaries with a genotype quality (GQ) below 20.
Figure 3.
Overview of experimental design. For each inversion, the appropriate Ag1000G sample of mosquitoes that had been successfully karyotyped by PCA was partitioned into a training set (75%) and a validation set (25%). Ten bootstrap replicates of the training set were created from a random sample of 75% of the full training set. For each bootstrap replicate and each mosquito, higher frequency biallelic SNPs within inversion breakpoints were interrogated for genotypic concordance with the PCA-based genotype. Results were summarized across the ten replicates to create a set of candidate tag SNPs with concordance rates exceeding 80%. Candidate tags were used to genotype the held-out validation set, and the computational karyotype score computed across tags was compared to the PCA-based karyotype. Candidate tags were also used to interrogate mosquitoes sequenced independently of Ag1000G, and the computational karyotype score was compared to the associated cytogenetically determined karyotype.
Next, we created ten bootstrap replicates of the training sample (Figure 3). Each of the ten replicates consisted of sub-samples of 75% of the full training sample, chosen at random with respect to all variables except inversion genotype balance. For each bootstrap replicate at each interrogated SNP (biallelic, MAF ≥ 5%), we calculated the SNP genotype-inversion genotype concordance for each mosquito in the sample, as described above (Figure 2). Genotypic concordance at each SNP interrogated in a given bootstrap replicate was expressed as the percentage of mosquitoes for which the number of alternate SNP alleles matched the number of inverted chromosomes. Because an imbalance among inversion genotypes could lead to false-positive tag SNPs, we calculated concordance separately for the three inversion genotypes in each of the ten bootstrap replicates. We then averaged the concordance scores across the ten replicates, by inversion genotype. To generate a single, conservative tag SNP concordance statistic, we used the minimum of the three mean values. Note that because the mosquito composition differed among bootstrap replicates, some SNPs were not evaluated in all ten, if they did not pass our filters in one or more iterations. Finally, to eliminate SNP positions with high levels of missing genotypes, we also calculated for each inversion genotype in each bootstrap replicate the percentage of mosquitoes with SNP genotype calls at the candidate tag (the ‘call rate’), and averaged across the ten replicates.
The procedure just described returned from 99 to 349 candidate tag SNPs for five inversions, but only two for 2Rc (Table 1). We therefore adopted a modified approach to control for suspected population structure. One possible source of structure was the haplotype configuration of 2Rc with respect to the flanking inversions (2Rb and 2Ru) (Figure 1). The inverted orientation of 2Rc is in almost perfect linkage disequilibrium with the inverted orientation of either 2Rb (as haplotype ‘2Rbc’) or 2Ru (as haplotype ‘2Rcu’). In a ∼50-year cytogenetic database compiled from samples collected in many parts of sub-Saharan Africa (described in Pombi et al. 2008), only four specimens were ever recorded as carrying the inverted orientation of 2Rc unaccompanied by either 2Rb or 2Ru (V. Petrarca, unpublished data). A second source, not mutually exclusive, was population structure between An. coluzzii, An. gambiae, and the BAMAKO chromosomal form that is subsumed taxonomically within An. gambiae but is at least partially reproductively isolated and genetically differentiated (Manoukis et al. 2008; Love et al. 2016). Although 2Rc occurs in all three taxa, there is a strong karyotype imbalance among them in natural populations and in Ag1000G. For example, of 70 An. coluzzii with 2Rc in Ag1000G, at least 49 (70%) carried the 2Rbc haplotype (haplotypes of the other specimens could not be inferred unambiguously). Similarly, of 64 non-BAMAKO An. gambiae with 2Rc, 62 (97%) carried the 2Rbc haplotype. On the other hand, all 45 BAMAKO, by definition, carried 2Rcu. We initially partitioned our sample by species, but the inclusion of BAMAKO in the An. gambiae partition resulted in very few candidate tags concordant with inversion genotype (N = 17). Ultimately, we retained two data partitions (An. coluzzii and non-BAMAKO An. gambiae), eliminating a third BAMAKO partition due to the fixation of 2Rc in this taxon (Coluzzi et al. 1985). From the non-BAMAKO An. gambiae partition (hereafter, An. gambiae for brevity), we omitted two of only three specimens carrying 2Ru (AZ0267-C from Mali and AV0043 C from Guinea), guided by PCA. As described above, both data partitions were split into training (75%) and validation (25%) sets, and ten bootstrap replicates of each training set were analyzed.
Table 1. Candidate tag SNPs predictive of inversion genotype in Ag1000G data.
| Inversion | Concordance Threshold | No. Tags |
|---|---|---|
| 2La | >0.995 | 209 |
| 2Rj-gambiae | >0.8 | 99 |
| 2Rb | >0.8 | 349 |
| 2Ru | >0.8 | 177 |
| 2Rd-gambiae | >0.8 | 147 |
| 2Rc | >0.8 | 2 |
| 2Rc-coluzzii | >0.8 | 57 |
| 2Rc-gambiae | >0.8 | 49 |
Ultimately, the candidate tag SNPs chosen (Table 1) met the following three criteria: they were (i) analyzed in at least eight of the ten bootstrap replicates; (ii) called at a rate greater than 90% within each karyotype class; and (iii) concordant with karyotype more than 80% of the time within each karyotype class (99.5% for 2La). Their approximate physical position relative to the span of each inversion is illustrated in Figure S1.
Validation of candidate tag SNPs in Ag1000G
We interrogated the candidate tag SNPs in the validation samples from Ag1000G that had been held aside during the discovery phase (Figure 3). For each mosquito in the validation set, we masked genotypes inside the focal inversion with GQ scores less than 20. Next, among the retained SNPs, we identified those corresponding to candidate tags and converted their diploid genotypes to a count of the number of alternate alleles. Finally, the number of alternate alleles at each tag SNP was summed across tags and averaged to provide an overall computational karyotype score. We compared this mean score to the PCA-based karyotype.
Testing tag SNPs in data independent of the Ag1000G pipeline
We also explored the efficacy of our tag SNPs for computational karyotyping in wild-caught mosquitoes subject to whole genome sequencing and variant calling by individual investigators, for which corresponding cytological karyotypes had been determined through phase microscopy (Figure 3). We used specimens originating from southern Mali, 8 An. gambiae BAMAKO chromosomal form (Fontaine et al. 2015; Love et al. 2016) and 17 An. coluzzii (Main et al. 2015), whose variant calls and cytogenetic metadata are publicly accessible (Tables S5, S6). These data include specimens sequenced to much lower coverage than the standard adhered to by Ag1000G. We followed the same procedure described for the Ag1000G validation set to computationally karyotype these specimens, and compared their computational and cytologically determined karyotypes.
Genetic distance trees to assess inversion history
We compared patterns of relatedness near the breakpoints of all six inversions using unrooted neighbor-joining (NJ) trees. For each inversion, we used biallelic SNPs with a MAF of 0.01 found within 5 kb upstream and downstream of the distal and proximal breakpoints (15 kb for 2Rd). Total numbers of SNPs for each inversion were: 2La, 596; 2Rj, 909; 2Rb, 428; 2Rc, 2141; 2Rd, 955; 2Ru, 1110. Using the python package anhima, we converted the number of alternate alleles at these SNPs into a Euclidean distance matrix, and then constructed neighbor-joining trees using all 1,347 specimens. To assess support for the nodes of the 2Rc tree, we used the transfer bootstrap estimate (TBE; Lemoine et al. 2018), a statistic that measures the number of taxa that must be transferred to make a given branch of a reference tree match the closest equivalent branch in a bootstrap tree. To calculate this statistic, we imported the matrix of alternate allele counts into R (v. 3.5.1, “Feather Spray”; R Core Team 2018) and used the dist() function of base R to construct the Euclidean distance matrix. We then used the nj() function in the ape package (v. 5.2) to construct the neighbor joining tree, and the boot.phylo() function to generate 1,000 bootstrap trees. We used these trees as input to booster (Lemoine et al. 2018), which calculates the TBE for each node.
Code and data availability
All genomic sequence data and variant call files used in this study are located in open data repositories as specified in Tables S1 and S2. Phases 1 and 2 of Ag1000G are available from https://www.malariagen.net/data; variant calls and metadata for Ag1000G phase 3 and Vector Observatory data are available from https://figshare.com/projects/Data_for_In_silico_karyotyping_of_chromosomally_polymorphic_malaria_mosquitoes_in_the_Anopheles_gambiae_complex_/65522 under the Ag1000G terms of use: https://www.malariagen.net/data/terms-use/ag1000g-terms-use.
The An. gambiae AgamP4 reference assembly is available through VectorBase (https://www.vectorbase.org). All custom code necessary to reproduce this analysis can be found at https://github.com/rrlove/comp_karyo_notebooks and https://github.com/rrlove/ingenos. Software packages used, and their versions, are summarized in Table S7. The complete set of tag SNPs, together with a custom script for computational karyotyping, which calculates the mean inversion genotype across the relevant tag SNPs, can be found at https://github.com/rrlove/compkaryo. Supplemental material available at Figshare: https://doi.org/10.25387/g3.9159479.
Results
After filtering, we retained the genotype data from 1,347 individually sequenced An. coluzzii and An. gambiae mosquitoes from the Ag1000G repository of natural genomic sequence variation, representing population samples from 13 West, Central, and East African countries and the island of Mayotte (Tables S1, S2).
Patterns of genetic variation at inversion breakpoints
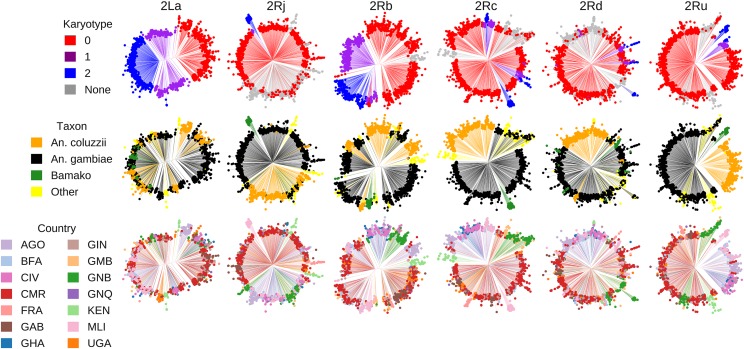
To gain insight into the relative roles of inversion history, taxonomic status, and geographic location in structuring genetic variation for each inversion, we reconstructed neighbor-joining trees based on SNPs in the immediate vicinity of the breakpoints (Figure 4). The resulting dendrograms, color-coded by inversion genotype, taxon and African country, indicate little clustering on the basis of geographic location; outlier population samples are those with a history of inbreeding or hybridization (see Methods). On the other hand, with the notable exception of 2La, taxonomic status is an important factor structuring inversion variation between An. gambiae and An. coluzzii. Moreover, BAMAKO specimens appear to comprise a differentiated outlier clade within the larger An. gambiae cluster. It is interesting to note that for inversion 2Rc, taxonomic status appears to be a more decisive factor than inversion genotype. All three 2Rc inversion genotypes cluster within their respective species (supported by bootstrap at 90%, or 98% if dendrograms are constructed after removing outlier samples from The Gambia, Guinea-Bissau and Kenya; not shown). Further investigation is required to determine whether this pattern results from a monophyletic inversion that subsequently differentiated between taxa, or from independent inversion events in the two taxa.
Figure 4.
Neighbor-joining dendrograms reconstructed from 1,347 An. gambiae and An. coluzzii mosquitoes from Ag1000G, using biallelic SNPs within 5 kb of inversion breakpoints (15 kb for 2Rd) having a minimum minor allele frequency of 0.01. Columns represent the same inversion dendrogram, alternately color-coded by inversion genotype as determined from PCA (first row), taxon (second row), or geographic source (third row). Some specimens that could not be karyotyped by PCA for inversions 2Rc, 2Rd, 2Rj, and 2Ru had cytogenetically determined karyotypes, which were used in place of PCA for color-coding the inversion genotype. ‘None’ refers to mosquitoes that were not assigned an inversion genotype either by PCA or cytogenetically; ‘Other’ refers to mosquitoes that were not identified taxonomically. Countries: AGO, Angola; BFA, Burkina Faso; CIV, Cote d’Ivoire; CMR, Cameroon; GAB, Gabon; GHA, Ghana; GIN, Guinea; GMB, The Gambia; GNB, Guinea-Bissau; GNQ, Equatorial Guinea; KEN, Kenya; MLI, Mali; FRA, France (Mayotte Archipelago); UGA, Uganda.
Inversion karyotype imputation by PCA
Only 373 of the 1,347 mosquitoes were associated with metadata that included cytologically determined inversion karyotypes. As discovery of candidate tag SNPs requires provisional inversion genotype assignments, we applied local PCA to assign genotypes for individual inversions on chromosome 2, following Ma and Amos (2012). A recognized limitation to this population-level approach, beyond the fact that it cannot be applied to individual mosquitoes, is that its success depends upon the presence of all three inversion genotypes in the sample under study. For this reason, and with the goal of finding the most flexible solution to inversion genotyping across geography and taxa, we began with PCA based on the entire set of 1,347 mosquitoes, under the simplifying assumption that the expected ‘three-stripe’ signal on a PCA plot would not be overwhelmed by geographic or population structure. Only in the case of 2La could genotype assignments be confidently inferred from the combination of all 1,347 specimens. For inversions on 2R, from one to four admixed (An. gambiae-An. coluzzii) or geographic outlier populations (highly inbred or island samples) had to be removed from analysis before a three-genotype pattern could be discerned on the PCA plot (Tables S2, S4; see Methods). Additionally, for 2Rd and 2Rj, An. gambiae-An. coluzzii population structure dominated the PCA. Taken together with the fact that 2Rj has yet to be found in An. coluzzii (Coluzzi et al. 2002; della Torre et al. 2005), we removed all 341 An. coluzzii specimens (Tables S2, S4) prior to PCA karyotyping of 2Rd and 2Rj in An. gambiae. Ultimately, PCA karyotypes were imputed for 780-1,347 mosquitoes, depending upon the inversion (Table S4).
Tag SNP ascertainment and validation in Ag1000G
Dividing the Ag1000G samples into training (75%) and validation (25%) sets for each inversion, and working within the training sets using a bootstrapping procedure, we screened for candidate tag SNPs in the five 2R inversions and 2La (see Methods for details). Candidate tag SNPs were those whose genotypes were concordant with the corresponding PCA genotypes, when averaged across ten bootstrap replicates, for more than 80% of the specimens that were scored (99.5% for 2La). The number of candidate tags ranged from 99 (2Rj) to 349 (2Rb) excluding 2Rc, in which only two candidates were found due to population structure between An. gambiae and An. coluzzii (Figure 4; Table 1). Partitioning the 2Rc sample by taxon (and omitting BAMAKO; see Methods) resulted in 49 and 57 tags for An. gambiae and An. coluzzii, respectively (Table 1). Notably, there was no overlap between the two sets of tags.
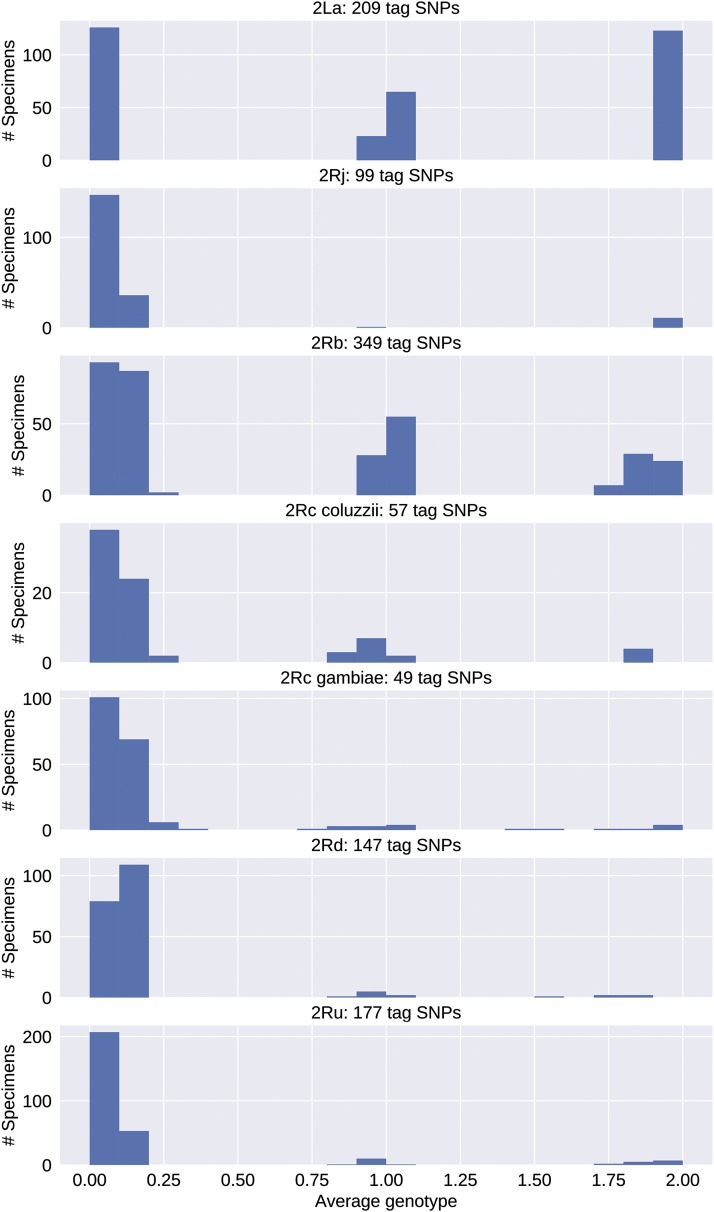
To assess the performance of these candidate tags, we used them to predict karyotypes in the held-out validation sets of Ag1000G specimens. For each inversion and specimen, we calculated a computational karyotype score representing the average genotype inferred across all candidate tag SNPs ascertained (see Methods). Histograms of resulting computational karyotype scores generally showed tight clustering around the three theoretical genotypic optima (0, 1, 2), reflecting close agreement among specimens (Figure 5). For each specimen in a validation set, we then compared the computational karyotype score to its PCA karyotype, and tallied the number of disagreements (Table 2). All except one specimen had matching PCA and computational karyotype scores. This exception, one of 254 (0.4%) assignments for 2Rc in An. gambiae, involved a specimen carrying 2Ru (AZ0267-C) already noted as an outlier (see Methods).
Figure 5.
Histograms of computational karyotyping scores calculated by interrogating tag SNPs in An. gambiae and An. coluzzii mosquitoes from the Ag1000G validation sets. Mean scores cluster around 0, 1, and 2. Note differences in Y-axis scale.
Table 2. Mismatches between PCA and computational karyotypes in the Ag1000G validation sets.
| Matching karyotypes | Mismatched karyotypes | |||||
|---|---|---|---|---|---|---|
| Inversion | Total specimens | No. tags scored | No. specimens | % tags supporting score | No. Specimens | % tags supporting score |
| 2La | 337 | 168-203 | 337 | 93.6-100 | 0 | – |
| 2Rj-gambiae | 195 | 94-99 | 195 | 83.8-100 | 0 | – |
| 2Rb | 325 | 304-349 | 325 | 77.5-97.7 | 0 | – |
| 2Rc-coluzzii | 80 | 55-57 | 80 | 78.9-100 | 0 | – |
| 2Rc-gambiae | 196 | 45-49 | 195 | 59.6-100 | 1 | 67.3 |
| 2Rd-gambiae | 201 | 128-147 | 201 | 55.2a-95.9 | 0 | – |
| 2Ru | 286 | 124-177 | 286 | 76.6-100 | 0 | – |
Next highest value is 70.1%.
Performance of tag SNPs in resequencing data independent of Ag1000G
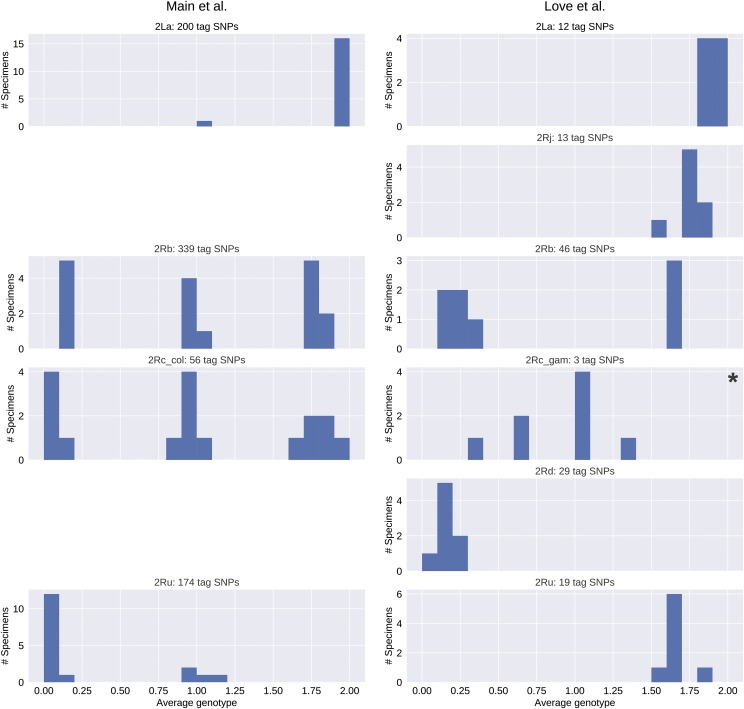
Previous studies re-sequenced An. gambiae or An. coluzzii mosquitoes from Mali whose karyotypes had been determined from the polytene chromosome banding pattern (Main et al. 2015; Love et al. 2016). Although sample size is limited, these data allow a direct comparison of cytogenetic and in silico karyotyping under less ideal conditions—lower sequencing depth, with variant calls made independently of the Ag1000G pipeline. For each specimen and inversion, we calculated computational karyotype scores (averaged across all tag SNPs that could be ascertained in a specimen) (Tables S5, S6). Histograms of these scores by inversion, similar to those based on Ag1000G validation sets, reveal clustering of scores around the three genotypic optima provided that taxon-specific tags (2Rc-coluzzii and 2Rc-gambiae) are applied to the conspecific taxon, and heterospecific applications (including use of 2Rc-gambiae tags to genotype BAMAKO) are avoided (Figure 6, Figure S2).
Figure 6.
Histograms of computational karyotyping scores calculated by interrogating tag SNPs in An. gambiae and An. coluzzii mosquitoes re-sequenced independently of the Ag1000G pipeline, often at lower sequencing depth. Scores cluster near 0, 1, and 2 with little dispersion except when taxon-specific tag SNPs are applied to a different taxon (indicated by an asterisk).
In the BAMAKO sample of Love et al. (2016) where mean sequencing depth ranged from 9-10x, there was concordance in karyotype assignments between cytogenetic and computational methods for five inversions including 2La, even though only 10-12 2La tags were ascertained (Tables S5, S6). However, as expected for BAMAKO, the An. gambiae 2Rc tags failed. Due to the extreme geospatial restriction of BAMAKO, this specific problem is limited in scope.
In the An. coluzzii sample of Main et al. (2015), mean sequencing coverage varied widely (4-66x; Table S5). The impact of very low sequencing coverage on the success of computational karyotyping is illustrated by specimens 04SEL021 and 04SEL02 (4x and 5x, respectively). For 04SEL021, there is no apparent disagreement between the cytogenetic and mean computational genotype scores for any of the six inversions. Nevertheless, for those inversions classified as heterozygotes both cytogenetically and computationally (2La, 2Rb, 2Rc), the proportion of tags whose genotype matches the mean computational score drops drastically to ∼30% (Table S5), likely because true heterozygous sites are often scored as homozygous either for the reference or alternate allele (0 or 2) due to low sequencing coverage. (Indeed, using chromosome 3L, we confirmed the expected drop in the rate of heterozygosity with decreasing coverage in these 17 specimens; data not shown). Low coverage alone is less likely to bias computational scores toward zero or two. For 04SEL02 (5x coverage), where cytogenetic vs. computational discrepancies occur at 2Rb and 2Ru, the computational karyotype is supported respectively by 81% of 208 tags and >94% of 57 tags, favoring the computational genotype by weight of evidence. The remaining six specimens with discordant inversion genotypes were sequenced to at least 10x coverage. In these cases, when the computational genotype score signaled ‘1’ in contradiction to a homokaryotypic cytogenetic genotype (02SEL85, 02SEL006, 02SEL009, 01Osel134), the proportion of tags supporting the computational genotype ranged from 65 to >92%. For other types of genotypic disagreements between methods, the computational score was supported by >80% of tags scored. Overall, these results suggest that computational karyotyping using tag SNPs can be successful in data derived independently of Ag1000G (Tables S5, S6), though care should be taken when this approach is applied to very low coverage samples.
Performance of tag SNPs against cytogenetically karyotyped Ag1000G specimens
We compared the cytogenetic karyotype assignments for 373 specimens in Ag1000G to their corresponding computational karyotype assignments (Table 3). Conflicts were few overall, and for every inversion, all but one conflict (involving specimen AZ0267-C, the exceptional An. gambiae carrier of the 2Ru inversion) could be attributed to errors in the cytogenetically assigned scores, as genotypes imputed from both PCA and tag SNPs contradict the cytogenetic assignment. Visual reference back to the PCA plots clearly confirmed that for specimens whose cytogenetic and tag SNP assignments differed and for whom PCA karyotypes could be determined, their locations on the plot strongly agreed with the tag SNP genotype (Figure S3). Considering that we ascertained tens or hundreds of tags per specimen, and that the proportion of tags whose SNP genotype matched the computational score was greater than 83% in all except the unusual specimen AZ0267-C (Table 3), the computational scores more confidently predict the true inversion genotype than traditional cytogenetics for these five inversions. The most dramatic example is with respect to inversion 2Ru, where we noted an unusually large number of erroneous cytogenetic genotypes of ‘1’ (N = 18/29) conflicting with both PCA and computational assignments of ‘0’. It is not immediately clear what could lead to such an elevated rate of cytogenetic error (which otherwise is ∼4%), but it is possible that the 2Ru heterozygous loop was mistaken either for a loop created by a rare inversion (sensu Pombi et al. 2008) in the same chromosomal region, or for a 2Rd loop in samples from regions where 2Ru is rare (as supported by the fact all 11 cytogenetic errors in An. gambiae were found in samples from the same small region in Cameroon, six of which were scored computationally as ‘1’ for 2Rd).
Table 3. Discrepancies between cytogenetic and computational karyotypes in Ag1000G mosquitoes analyzed.
| Specimens with discrepancies | ||||||
|---|---|---|---|---|---|---|
| Inversion Tags | Partition | CYT | Specimens (N) | Mismatch CYT-TAG (%) | Match TAG-PCA (%) | No. tag SNPs scored (% matching TAG) |
| 2La | 0 | 117 | 5 (4.3) | 5 (100) | 200-203 (99.5-100) | |
| 1 | 68 | 5 (7.4) | 5 (100) | 193-203 (100) | ||
| 2 | 160 | 2 (1.3) | 2 (100) | 201-203 (99.5-100) | ||
| 2Rj-gambiae | gambiae | 0 | 236 | 0 (0) | – | – |
| 1 | 4 | 0 (0) | – | – | ||
| 2 | 45 | 0 (0) | – | – | ||
| 2Rb | 0 | 127 | 2 (1.6) | 2 (100) | 348 (85.3-87.6) | |
| 1 | 124 | 4 (3.2) | 4 (100) | 346-349 (86.8-93.7) | ||
| 2 | 121 | 6 (5.0) | 6 (100) | 331-348 (88.8-91.6) | ||
| 2Rc-gambiae | gambiaea | 0 | 184 | 7 (3.8) | 7 (100) | 48-49 (83.7-98.0) |
| 1 | 32 | 3 (9.4) | 2 (66.7) | 47-49 (42.9b-91.5) | ||
| 2 | 24 | 2 (8.3) | 2 (100) | 48-49 (90.0-91.8) | ||
| 2Rc-coluzzi | coluzzii | 0 | 13 | 1 (7.7) | 1 (100) | 56 (87.5) |
| 1 | 25 | 0 (0) | – | – | ||
| 2 | 16 | 0 (0) | – | – | ||
| 2Rd-gambiae | gambiae | 0 | 234 | 9 (3.8) | 9 (100) | 143-147 (84.9-96.6) |
| 1 | 28 | 4 (14.3) | 4 (100) | 146-147 (88.4-93.9) | ||
| 2 | 22 | 3 (13.6) | 3 (100) | 146-147 (89.1-91.8) | ||
| 2Ru | col+gam | 0 | 263 | 1 (0.38) | 1 (100) | 176 (85.2) |
| 1 | 29 | 18 (62.1) | 18 (100) | 170-177 (88.7-99.4) | ||
| 2 | 47 | 1 (2.1) | 1 (100) | 176 (97.2) | ||
CYT, cytogenetic genotype; TAG, computational genotype; PCA, genotype inferred by PCA.
An. gambiae excluding BAMAKO.
This value corresponds to one of three non-BAMAKO An. gambiae carriers of the 2Ru inversion, AZ0267-C. The next highest value is 85.7.
Our results also highlight the pitfalls of using taxon-specific tags to genotype other taxa, or populations with high levels of admixture between taxa (Table S8; Figure S4 summarizes the correct use of taxon-specific tags). As expected, we find elevated numbers of cytogenetic-computational disagreements when (i) 2Rc-gambiae tags are applied to BAMAKO (60% of the 45 specimens), (ii) 2Rd-gambiae tags are used to genotype An. coluzzii, and (iii) 2Rd-gambiae tags are applied to admixed An. gambiae-An. coluzzii populations such as those from Guinea Bissau. These disagreements involve specimens carrying inverted arrangements according to cytogenetic analysis which are not tagged as inverted computationally, due to the lack of correlation between tags and the inverted orientation in the heterospecific genetic background.
Discussion
In recent years, numerous approaches have been developed for the discovery and detection of chromosomal inversions aside from traditional cytogenetics, including optical mapping, Hi-C, read cloud sequencing, and identification of discordantly mapping sequencing reads. Our method is not intended to substitute for these largely discovery-based approaches. Here, we are concerned with specific inversions that segregate at relatively high frequencies in natural populations, and have a long history of cytogenetic study. Our approach benefits from the fact that the chromosomal coordinates of their breakpoints already are known precisely or to a good approximation. The goal was to overcome the substantial limitations of cytogenetic analysis by identifying SNPs inside these rearrangements whose allelic state is predictive of inversion status, allowing for rapid in silico or molecular karyotyping of hundreds or thousands of specimens.
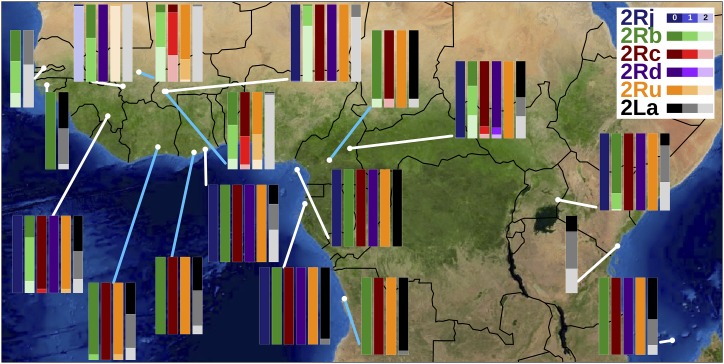
Analysis of the Ag1000G database allowed us to develop the first standardized computational karyotyping of the six main polymorphic chromosomal inversions in the major malaria vectors An. coluzzii and An. gambiae, despite the fact that only a small subset of specimens in the database had cytogenetic karyotype assignments (Figure 7). Direct comparison of computational karyotype scores with the cytogenetic assignment for the same specimen in Ag1000G suggests that computational karyotyping outperforms traditional cytogenetics in terms of accuracy, given that assignments are based on tens or hundreds of individual tags. Preliminary testing on specimens sequenced and computationally processed by individual laboratories outside of Ag1000G standards suggests that our tag SNPs have the potential to perform well, even on specimens sequenced to much lower depth. Our approach not only has a lower error rate compared to classical cytogenetics, but also is more widely applicable (regardless of mosquito gender, physiological status, or method of preservation), more widely accessible to those without specialized expertise, higher throughput, and therefore, ultimately cheaper to implement at scale. This method can now be used to predict inversion genotypes in previously sequenced data sets for which ecological and behavioral data may already be available. Even more important, easy large-scale adoption of this approach allows for new and properly powered association studies to be performed on ecologically and epidemiologically relevant mosquito phenotypes, studies that that would have been prohibitively ambitious when relying on cytogenetic karyotyping. In addition, this method can now facilitate sequencing experiments for which inversion karyotype is relevant at scale. Expanding the possibilities further, molecular assays based on these results that will allow inversion genotyping without whole genome sequencing are under development.
Figure 7.
Map of the study area with the frequency of An. gambiae and An. coluzzii inversion genotypes inferred for up to six polymorphic chromosome 2 inversions summarized by country (and the island of Mayotte; see Table S2). Blue connecting lines point to An. coluzzii samples, while white connecting lines point to An. gambiae and hybrid/outlier populations. Image: Visible Earth, NASA. Produced with cartopy v.0.17.1.
However, some important limitations exist. Computational karyotyping is strictly dependent upon tag SNPs that are strongly correlated with inversion status, a contingency that depends upon representative sampling. Although Ag1000G is populated by samples derived from multiple countries in West, Central and East Africa, An. coluzzii is underrepresented, as is southern Africa (The Anopheles gambiae 1000 Genomes Consortium 2017). Even more importantly, with the exception of the cosmopolitan inversions 2La and 2Rb, the inverted orientation of other rearrangements (2Rj, 2Rc, 2Rd, and 2Ru) is underrepresented in the Ag1000G data that was available at the time of our analysis. It is clear that population structure is an especially important factor in the application and further development of tags for 2Rc and 2Rd. The current taxon-specific tags should not be used to genotype heterospecific specimens (including BAMAKO) or samples from areas where high rates of interspecific hybridization are known. The presence of strong population structure means that correlations between tags and the inverted orientation characteristic of the target taxon cannot be assumed in a different taxon. The absence of correlation should downwardly bias the computational score, resulting in false negatives when genotyping true inverted homozygotes and heterozygotes. Finally, our inversion breakpoint dendrograms raise the possibility that at least one cytologically-recognized inversion, 2Rc, may have arisen repeatedly at the molecular level, a result that requires further investigation beyond the scope of this study. With the exception of 2Rc, 2Rd, and 2Rj, for which we developed taxon-specific tags, our approach implicitly assumed that inversions shared by An. gambiae and An. coluzzii are monophyletic, and may yield unexpected results if this assumption is violated. Accordingly, these tools should be applied with caution, and there is ample room for improvement as more data become available. Fortunately, our standardized approach makes it easy to accommodate improvements. The success of our method thus far suggests that the general approach may be suitable for studying inversions more broadly, in additional malaria vectors as well as other systems where inversions are implicated in local adaptation.
Nearly twenty years ago, Coluzzi and colleagues predicted that the then-newly-assembled An. gambiae reference genome would facilitate our analyses of polymorphic chromosomal inversions in the An. gambiae complex (Coluzzi et al. 2002). Our work continues the realization of that prediction by providing, for the first time, cross-continent diagnostics for multiple inversions. These computational diagnostics, and the molecular diagnostics that they leverage, take us one step closer to understanding the contribution of chromosomal inversions to the deadly facility of An. gambiae and An. coluzzii for vectoring malaria.
Acknowledgments
We thank the Notre Dame Center for Research Computing for technical support, and C. Liu, C. Sweet, and J. Young for helpful discussions. We thank M. Kern and R. Montañez-Gonzalez for assistance with DNA extraction. This work was supported by the National Institutes of Health (R01 AI125360 awarded to NJB). During this work, NJB was supported by Target Malaria, which receives core funding from the Bill & Melinda Gates Foundation and from the Open Philanthropy Project Fund, an advised fund of Silicon Valley Community Foundation.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.9159479.
Communicating editor: R. Anholt
Literature Cited
- Andolfatto P., Depaulis F., and Navarro A., 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. 10.1017/S0016672301004955 [DOI] [PubMed] [Google Scholar]
- The Anopheles gambiae 1000 Genomes Consortium , 2017. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552: 96–100. 10.1038/nature24995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Acevedo P., Pombi M., Dia I., Boccolini D. et al. , 2017. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution 71: 686–701. 10.1111/evo.13176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Ullastres A., and Gonzalez J., 2014. Adaptation through chromosomal inversions in Anopheles. Front. Genet. 5: 129 10.3389/fgene.2014.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D., Zhang S., Chateau M., Fouet C., Morlais I. et al. , 2019. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol. Ecol. 28: 1333–1342. 10.1111/mec.14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J. H., Di Deco M. A., Petrarca V., and Coluzzi M., 1982. Inversion polymorphism and incipient speciation in Anopheles gambiae s. str. in The Gambia, West Africa. Genetica 59: 167–176. 10.1007/BF00056539 [DOI] [Google Scholar]
- Caputo B., Nwakanma D., Caputo F. P., Jawara M., Oriero E. C. et al. , 2014. Prominent intra-specific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol. Ecol. 23: 4574–4589. 10.1111/mec.12866 [DOI] [PubMed] [Google Scholar]
- Caputo B., Nwakanma D., Jawara M., Adiamoh M., Dia I. et al. , 2008. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar. J. 7: 182 10.1186/1475-2875-7-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B., Santolamazza F., Vicente J. L., Nwakanma D. C., Jawara M. et al. , 2011. The “far-west” of Anopheles gambiae molecular forms. PLoS One 6: e16415 10.1371/journal.pone.0016415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone B. J., Molloy M. J., Cheng C., Tan J. C., Hahn M. W. et al. , 2011. Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Mol. Ecol. 20: 2567–2580. 10.1111/j.1365-294X.2011.05114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Tan J. C., Hahn M. W., and Besansky N. J., 2018. A systems genetic analysis of inversion polymorphisms in the malaria mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 115: E7005–E7014. 10.1073/pnas.1806760115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., White B. J., Kamdem C., Mockaitis K., Costantini C. et al. , 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics 190: 1417–1432. 10.1534/genetics.111.137794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M., Hunt R. H., Wilkerson R., della Torre A., Coulibaly M. B. et al. , 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619: 246–274. 10.11646/zootaxa.3619.3.2 [DOI] [PubMed] [Google Scholar]
- Coluzzi M., 1968. Cromosomi politenici delle cellule nutrici ovariche nel complesso gambiae del genere Anopheles. Parassitologia 10: 179–183. [Google Scholar]
- Coluzzi M., Petrarca V., and DiDeco M. A., 1985. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll. Zool. 52: 45–63. 10.1080/11250008509440343 [DOI] [Google Scholar]
- Coluzzi M., Sabatini A., della Torre A., Di Deco M. A., and Petrarca V., 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. 10.1126/science.1077769 [DOI] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., Petrarca V., and Di Deco M. A., 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans. R. Soc. Trop. Med. Hyg. 73: 483–497. 10.1016/0035-9203(79)90036-1 [DOI] [PubMed] [Google Scholar]
- Corbett-Detig R., Said I., Calzetta M., Genetti M., McBroome J. et al. , 2019. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. bioRxiv. 10.1101/662114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C., Ayala D., Guelbeogo W. M., Pombi M., Some C. Y. et al. , 2009. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 9: 16 10.1186/1472-6785-9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly M. B., Lobo N. F., Fitzpatrick M. C., Kern M., Grushko O. et al. , 2007a Segmental duplication implicated in the genesis of inversion 2Rj of Anopheles gambiae. PLoS One 2: e849 10.1371/journal.pone.0000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly M. B., Pombi M., Caputo B., Nwakanma D., Jawara M. et al. , 2007b PCR-based karyotyping of Anopheles gambiae inversion 2Rj identifies the BAMAKO chromosomal form. Malar. J. 6: 133 10.1186/1475-2875-6-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabire K. R., Sawadodgo S., Diabate A., Toe K. H., Kengne P. et al. , 2013. Assortative mating in mixed swarms of the mosquito Anopheles gambiae s.s. M and S molecular forms, in Burkina Faso, West Africa. Med. Vet. Entomol. 27: 298–312. 10.1111/j.1365-2915.2012.01049.x [DOI] [PubMed] [Google Scholar]
- della Torre A., 1997. Polytene chromosome preparation from anopheline mosquitoes, pp. 329–336 in Molecular Biology of Disease Vectors: A Methods Manual, edited by Crampton J. M., Beard C. B., and Louis C.. Chapman & Hall, London: 10.1007/978-94-009-1535-0_28 [DOI] [Google Scholar]
- della Torre A., Fanello C., Akogbeto M., Dossou-yovo J., Favia G. et al. , 2001. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol. Biol. 10: 9–18. 10.1046/j.1365-2583.2001.00235.x [DOI] [PubMed] [Google Scholar]
- della Torre A., Tu Z., and Petrarca V., 2005. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem. Mol. Biol. 35: 755–769. 10.1016/j.ibmb.2005.02.006 [DOI] [PubMed] [Google Scholar]
- Diabaté A., Dao A., Yaro A. S., Adamou A., Gonzalez R. et al. , 2009. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae. Proc. Biol. Sci. 276: 4215–4222. 10.1098/rspb.2009.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine M. C., Pease J. B., Steele A., Waterhouse R. M., Neafsey D. E. et al. , 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347: 1258524 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C., Gray E., Besansky N. J., and Costantini C., 2012. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One 7: e34841 10.1371/journal.pone.0034841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller Z. L., Haynes G. D., Richards S., and Schaeffer S. W., 2017. Genomics of natural populations: Evolutionary forces that establish and maintain gene arrangements in Drosophila pseudoobscura. Mol. Ecol. 26: 6539–6562. 10.1111/mec.14381 [DOI] [PubMed] [Google Scholar]
- Gimonneau G., Bouyer J., Morand S., Besansky N. J., Diabate A. et al. , 2010. A behavioral mechanism underlying ecological divergence in the malaria mosquito Anopheles gambiae. Behav. Ecol. 21: 1087–1092. 10.1093/beheco/arq114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G., Pombi M., Choisy M., Morand S., Dabire R. K. et al. , 2012a Larval habitat segregation between the molecular forms of the mosquito, Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Med. Vet. Entomol. 26: 9–17. 10.1111/j.1365-2915.2011.00957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G., Pombi M., Dabire R. K., Diabate A., Morand S. et al. , 2012b Behavioural responses of Anopheles gambiae sensu stricto M and S molecular form larvae to an aquatic predator in Burkina Faso. Parasit. Vectors 5: 65 10.1186/1756-3305-5-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón G. I., Emrich S. J., MacCallum R. M., Maslen G., Dialynas E. et al. , 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43: D707–D713. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E. M., Rocca K. A., Costantini C., and Besansky N. J., 2009. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar. J. 8: 215 10.1186/1475-2875-8-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanemaaijer M. J., Collier T. C., Chang A., Shott C. C., Houston P. D. et al. , 2018. The fate of genes that cross species boundaries after a major hybridization event in a natural mosquito population. Mol. Ecol. 27: 4978–4990. 10.1111/mec.14947 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., and Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42. 10.1146/annurev.ecolsys.39.110707.173532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Sgro C. M., and Weeks A. R., 2004. Chromosomal inversion polymorphisms and adaptation. Trends Ecol. Evol. 19: 482–488. 10.1016/j.tree.2004.06.013 [DOI] [PubMed] [Google Scholar]
- Holt R. A., Subramanian G. M., Halpern A., Sutton G. G., Charlab R. et al. , 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- Houle D., and Marquez E. J., 2015. Linkage disequilibrium and inversion-typing of the Drosophila melanogaster genome reference panel. G3 (Bethesda) 5: 1695–1701. 10.1534/g3.115.019554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E. et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M., Frezal L., Jones R. T., Chamberlain N. L., Lee S. F. et al. , 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Fabian D. K., Goudet J., and Flatt T., 2016. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol. Biol. Evol. 33: 1317–1336. 10.1093/molbev/msw016 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: e1000501 10.1371/journal.pbio.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., and Barrett B., 2015. Chromosome inversions, adaptive cassettes and the evolution of species’ ranges. Mol. Ecol. 24: 2046–2055. 10.1111/mec.13074 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., and Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F., Domelevo Entfellner J. B., Wilkinson E., Correia D., Davila Felipe M. et al. , 2018. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556: 452–456. 10.1038/s41586-018-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo N. F., Sangare D. M., Regier A. A., Reidenbach K. R., Bretz D. A. et al. , 2010. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar. J. 9: 293 10.1186/1475-2875-9-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love R. R., Steele A. M., Coulibaly M. B., Traore S. F., Emrich S. J. et al. , 2016. Chromosomal inversions and ecotypic differentiation in Anopheles gambiae: the perspective from whole-genome sequencing. Mol. Ecol. 25: 5889–5906. 10.1111/mec.13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., and Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500 (erratum: PLoS Biol. 10: 10.1371/annotation/caa1b7dd-9b6d-44db-b6ce-666954903625). 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., and Amos C. I., 2012. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One 7: e40224 10.1371/journal.pone.0040224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main B. J., Lee Y., Collier T. C., Norris L. C., Brisco K. et al. , 2015. Complex genome evolution in Anopheles coluzzii associated with increased insecticide usage in Mali. Mol. Ecol. 24: 5145–5157. 10.1111/mec.13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukis N. C., Powell J. R., Touré M. B., Sacko A., Edillo F. E. et al. , 2008. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 105: 2940–2945. 10.1073/pnas.0709806105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden C., Lee Y., Neimen C., Sanford M., Dinis J. et al. , 2011. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol. Ecol. 20: 4983–4994. 10.1111/j.1365-294X.2011.05339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles A., and Harding N. J., 2017. scikit-allel: A Python package for exploring and analysing genetic variation data. http://github.com/cggh/scikit-allel. 10.5281/zenodo.597309 [DOI]
- Navarro A., Betran E., Barbadilla A., and Ruiz A., 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwakanma D. C., Neafsey D. E., Jawara M., Adiamoh M., Lund E. et al. , 2013. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics 193: 1221–1231. 10.1534/genetics.112.148718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E., Salgueiro P., Palsson K., Vicente J. L., Arez A. P. et al. , 2008. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J. Med. Entomol. 45: 1057–1063. 10.1603/0022-2585(2008)45[1057:HLOHBM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B. et al. , 2011. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12: 2825–2830. [Google Scholar]
- Petrarca V., and Beier J. C., 1992. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am. J. Trop. Med. Hyg. 46: 229–237. 10.4269/ajtmh.1992.46.229 [DOI] [PubMed] [Google Scholar]
- Petrarca V., Sabatinelli G., Di Deco M. A., and Papakay M., 1990. The Anopheles gambiae complex in the Federal Islamic Republic of Comoros (Indian Ocean): some cytogenetic and biometric data. Parassitologia 32: 371–380. [PubMed] [Google Scholar]
- Pombi M., Caputo B., Simard F., Di Deco M. A., Coluzzi M. et al. , 2008. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol. Biol. 8: 309 10.1186/1471-2148-8-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2018. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Riehle M. M., Bukhari T., Gneme A., Guelbeogo W. M., Coulibaly B. et al. , 2017. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. eLife 6 10.7554/eLife.25813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishikesh N., Di Deco M. A., Petrarca V., and Coluzzi M., 1985. Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Trop. 42: 165–170. [PubMed] [Google Scholar]
- Rocca K. A., Gray E. M., Costantini C., and Besansky N. J., 2009. 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar. J. 8: 147 10.1186/1475-2875-8-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangare, D. M., 2007 Breakpoint analysis of the Anopheles gambiae s.s chromosome 2Rb, 2Rc, and 2Ru inversions in PhD Thesis, Graduate Program in Biological Sciences, University of Notre Dame. University of Notre Dame, Notre Dame, IN. [Google Scholar]
- Schaeffer S. W., 2008. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution 62: 3082–3099. 10.1111/j.1558-5646.2008.00504.x [DOI] [PubMed] [Google Scholar]
- Seich Al Basatena N. K., Hoggart C. J., Coin L. J., and O’Reilly P. F., 2013. The effect of genomic inversions on estimation of population genetic parameters from SNP data. Genetics 193: 243–253. 10.1534/genetics.112.145599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F., Ayala D., Kamdem G. C., Pombi M., Etouna J., et al. , 2009. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 9: 17 10.1186/1472-6785-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B., Ayala D., Acevedo P., Kengne P., Ngomo Abeso Mebuy I. et al. , 2015. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol. Appl. 8: 326–345. 10.1111/eva.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure Y. T., Petrarca V., Traore S. F., Coulibaly A., Maiga H. M. et al. , 1998. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia 40: 477–511. [PubMed] [Google Scholar]
- Twyford A. D., and Friedman J., 2015. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution 69: 1476–1486. 10.1111/evo.12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., Wilding C. S., Steen K., Pinto J., and Donnelly M. J., 2012. Gene flow-dependent genomic divergence between Anopheles gambiae M and S forms. Mol. Biol. Evol. 29: 279–291. 10.1093/molbev/msr199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M., and Bernatchez L., 2018. Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol. Evol. 33: 427–440. 10.1016/j.tree.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Wellenreuther M., Rosenquist H., Jaksons P., and Larson K. W., 2017. Local adaptation along an environmental cline in a species with an inversion polymorphism. J. Evol. Biol. 30: 1068–1077. 10.1111/jeb.13064 [DOI] [PubMed] [Google Scholar]
- White B. J., Collins F. H., and Besansky N. J., 2011. Evolution of Anopheles gambiae in relation to humans and malaria. Annu. Rev. Ecol. Evol. Syst. 42: 111–132. 10.1146/annurev-ecolsys-102710-145028 [DOI] [Google Scholar]
- White B. J., Santolamazza F., Kamau L., Pombi M., Grushko O. et al. , 2007. Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am. J. Trop. Med. Hyg. 76: 334–339. 10.4269/ajtmh.2007.76.334 [DOI] [PubMed] [Google Scholar]
- World Health Organization , 2018. World Malaria Report: 2018, https://www.who.int/malaria/publications/world-malaria-report-2018/report/en/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genomic sequence data and variant call files used in this study are located in open data repositories as specified in Tables S1 and S2. Phases 1 and 2 of Ag1000G are available from https://www.malariagen.net/data; variant calls and metadata for Ag1000G phase 3 and Vector Observatory data are available from https://figshare.com/projects/Data_for_In_silico_karyotyping_of_chromosomally_polymorphic_malaria_mosquitoes_in_the_Anopheles_gambiae_complex_/65522 under the Ag1000G terms of use: https://www.malariagen.net/data/terms-use/ag1000g-terms-use.
The An. gambiae AgamP4 reference assembly is available through VectorBase (https://www.vectorbase.org). All custom code necessary to reproduce this analysis can be found at https://github.com/rrlove/comp_karyo_notebooks and https://github.com/rrlove/ingenos. Software packages used, and their versions, are summarized in Table S7. The complete set of tag SNPs, together with a custom script for computational karyotyping, which calculates the mean inversion genotype across the relevant tag SNPs, can be found at https://github.com/rrlove/compkaryo. Supplemental material available at Figshare: https://doi.org/10.25387/g3.9159479.