Abstract
Meiotic crossing over ensures proper segregation of homologous chromosomes and generates genotypic diversity. Despite these functions, little is known about the genetic factors and population genetic forces involved in the evolution of recombination rate differences among species. The dicistronic meiosis gene, mei-217/mei-218, mediates most of the species differences in crossover rate and patterning during female meiosis between the closely related fruitfly species, Drosophila melanogaster and D. mauritiana. The MEI-218 protein is one of several meiosis-specific mini-chromosome maintenance (mei-MCM) proteins that form a multi-protein complex essential to crossover formation, whereas the BLM helicase acts as an anti-crossover protein. Here we study the molecular evolution of five genes— mei-218, the other three known members of the mei-MCM complex, and Blm— over the phylogenies of three Drosophila species groups— melanogaster, obscura, and virilis. We then use transgenic assays in D. melanogaster to test if molecular evolution at mei-218 has functional consequences for crossing over using alleles from the distantly related species D. pseudoobscura and D. virilis. Our molecular evolutionary analyses reveal recurrent positive selection at two mei-MCM genes. Our transgenic assays show that sequence divergence among mei-218 alleles from D. melanogaster, D. pseudoobscura, and D. virilis has functional consequences for crossing over. In a D. melanogaster genetic background, the D. pseudoobscura mei-218 allele nearly rescues wildtype crossover rates but alters crossover patterning, whereas the D. virilis mei-218 allele conversely rescues wildtype crossover patterning but not crossover rates. These experiments demonstrate functional divergence at mei-218 and suggest that crossover rate and patterning are separable functions.
Keywords: recombination, crossing over, evolution, positive selection, Drosophila
During the early stages of meiosis, recombination occurs between homologous chromosomes, serving two functions. First, recombination repairs programmed DNA double-strand breaks (DSBs) and ensures proper Mendelian segregation of homologous chromosomes (Baker and Hall 1976; Lindsley and Sandler 1977). Second, recombination increases the efficacy of natural selection by reducing genetic linkage and creating novel genotypes (Fisher 1930; Felsenstein 1974; Crow 1992; Barton and Charlesworth 1998). Despite these benefits, recombination has risks. Dispersed selfish repetitive DNA sequences— e.g., transposons— introduce the risk of non-homologous ectopic exchange that generates chromosomal duplications and deletions (Goldberg et al. 1983; Charlesworth et al. 1994). The rate and distribution of crossing over may therefore evolve to balance the benefits of recombination and the costs of ectopic exchange (Montgomery et al. 1987; Charlesworth and Barton 1996; Kent et al. 2017; Brand et al. 2018).
Recombination landscapes vary within and among taxa, but the genes, mechanisms, and evolutionary causes involved are still largely unknown (Ritz et al. 2017; Stapley et al. 2017). In mammals, four loci are associated with intraspecific variation in recombination rates: RNF212, CPLX1, REC8, and PRDM9 (Baudat et al. 2010; Sandor et al. 2012; Kong et al. 2014). Best studied is the gene, Prdm9, which encodes a trans-acting major determinant of recombination distribution in most mammals (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). The PRDM9 protein binds specific DNA sequence motifs and modifies local histones, initiating the formation of DSBs nearby (Baudat et al. 2010; Myers et al. 2010). These DSBs are concentrated in the immediate vicinity of the motif, creating recombination “hotspots” once repaired. In rodents and primates, Prdm9 shows signals of recurrent positive selection, particularly at sites encoding a zinc finger array that mediates DNA motif binding specificity (Oliver et al. 2009). The recurrent evolution at Prdm9 alters the genomic distribution of recombination hotspots between closely related species and can, incidentally, cause sterility in species hybrids (Ptak et al. 2005; Davies et al. 2016; Smagulova et al. 2016).
In Drosophila, variation in the rate of recombination during female meiosis (males are achiasmate) exists along the lengths of chromosomes, among individuals, and between closely related species. Along chromosomes, rates of crossing over tend to be highest in medial euchromatic regions, lowest in centromere- and telomere-proximal regions (Dobzhansky 1930; Beadle 1932; Baker and Carpenter 1972; Lindsley and Sandler 1977), and absent in heterochromatic regions where repetitive DNA sequences are abundant (Baker 1958). Among individuals, natural genetic variation in crossover rates exists and responds to artificial selection (Kidwell 1972; Charlesworth and Charlesworth 1985; Brooks and Marks 1986; Brooks 1988; Comeron et al. 2012; Hunter et al. 2016). Between species, mean crossover frequencies vary more than two-fold (Ortiz-Barrientos et al. 2006; Comeron et al. 2012; L. Hemmer et al. unpublished). Despite these observations, few genetic loci are known that contribute to the observed variation in recombination rates (Hunter et al. 2016).
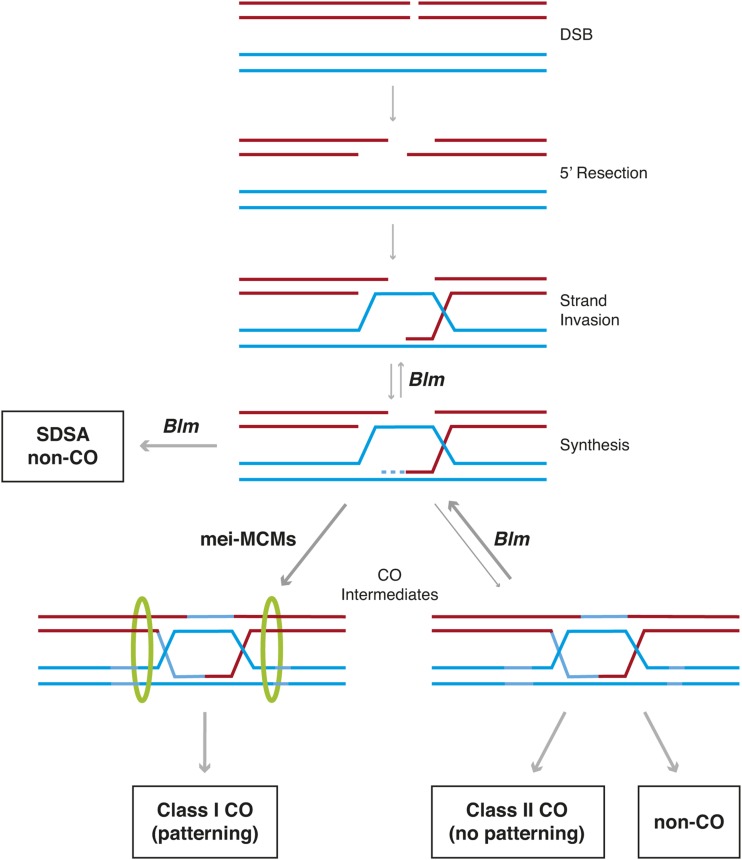
The genetic basis for recombination differs between flies and mammals. For one, many genes identified in mammals, including Prdm9, appear to be absent from Drosophila (Oliver et al. 2009; Heil and Noor 2012). Consistent with this, Drosophila lack comparably strong recombination hotspots (Comeron et al. 2012; Hunter et al. 2016). Drosophila also lack Msh4 and Msh5, the canonical proteins that promote crossover formation in most eukaryotes. Instead, flies have co-opted a meiosis-specific mini-chromosome maintenance (mei-MCM) complex encoded by the genes mei-217, mei-218, rec, and (presumably) Mcm5 to promote the formation of class I crossovers (Zalevsky et al. 1999; Kohl et al. 2012; Kohl and Sekelsky 2013). Class I crossovers are derived from heteroduplex DNA molecules (crossover intermediates) which are stabilized by the mei-MCM complex and ultimately resolved as crossover events (Figure 1; Kohl et al. 2012; Kohl and Sekelsky 2013; Hatkevich et al. 2017; Hatkevich and Sekelsky 2017). Class I crossovers are patterned by interference mechanisms that reduce the probability of a second crossover establishing nearby (Muller 1916) and by crossover suppression mechanisms that discourage crossover formation in telomere- and centromere-proximal regions (Dobzhansky 1930; Beadle 1932). Mutations in mei-MCM genes result in a >90% reduction in crossover frequency and a uniform chromosomal distribution of residual crossovers (Baker and Carpenter 1972; Carpenter and Sandler 1974; Grell 1984; Lake et al. 2007). These residual crossovers, termed class II crossovers, are uniformly distributed with chromosome length and thus lack the spatial patterning that results from crossover interference or telomere- and centromere-proximal crossover suppression (Figure 1; reviewed in Kohl and Sekelsky 2013). While the mei-MCMs promote class I crossover formation, the BLM helicase antagonizes crossover formation by dissolving heteroduplex DNA at multiple stages (Hatkevich and Sekelsky 2017). BLM unwinds the D-loops formed by strand invasion, leading to synthesis-dependent strand annealing (SDSA), the cause of most non-crossover gene conversion events (Figure 1; Allers and Lichten 2001). BLM also dissolves crossover intermediates in the class II pathway, as the mei-MCMs do not act to stabilize these (Figure 1; Kohl et al. 2012; Hatkevich et al. 2017; Hatkevich and Sekelsky 2017). Thus, BLM has anticrossover function, whereas the mei-MCMs are said to have “anti-anticrossover” function (Kohl and Sekelsky 2013). Given the antagonistic activities of the mei-MCM proteins and the BLM helicase, it seems plausible that molecular evolution at any of these proteins could contribute to phenotypic evolution of crossover rate and distribution.
Figure 1.
Meiotic recombination in Drosophila. During meiotic recombination programmed DSBs are formed, the 5′ ends are recessed and the resulting 3′ single-stranded tails invade the homologous chromosome. After synthesis off the template, the BLM Helicase can unwind the structure which is then resolved via synthesis-dependent strand annealing (SDSA) into a non-crossover gene conversion. If the invading strand synthesizes far enough, second-end capture can occur, creating a crossover intermediate. Most crossover intermediates are processed via the class I pathway in which they are stabilized by the mei-MCM complex (green rings) and resolved into interfering crossovers. A smaller fraction of crossover intermediates enters the class II pathway in which they are resolved as either non-interfering crossovers or non-crossovers with equal probability. The BLM Helicase inhibits crossover intermediate processing though the class II pathway and therefore promotes the class I pathway.
Recently, we showed that the dicistronic gene, mei-217/mei-218 (hereafter mei-217/-218), is a major contributor to evolved species differences in recombination rate and patterning between D. melanogaster and D. mauritiana (Brand et al. 2018). The total genetic map of D. mauritiana is ∼1.8-fold longer than that of D. melanogaster, and the chromosomal distribution of crossover events differs between species (True et al. 1996). This is most evident in telomere- and centromere-proximal regions where crossover formation is more suppressed in D. melanogaster than D. mauritiana (True et al. 1996). When a wildtype D. mauritiana allele of mei-217/-218 is transgenically introduced into mutant D. melanogaster females lacking mei-218 function, a largely (∼82%) D. mauritiana-like genetic map is observed (Brand et al. 2018). The D. mauritiana allele of mei-217/-218 results in weaker telomeric and centromeric suppression of crossing over as well as reduced crossover interference in medial euchromatic regions (Brand et al. 2018). Although mei-217 and mei-218 are encoded on a single transcript with two translation start sites that yield two distinct proteins (Liu et al. 2000), population genetic signals of recurrent positive selection in the D. melanogaster and D. mauritiana lineages localize exclusively to mei-218 (Brand et al. 2018). These findings imply that species differences in meiotic crossing over can be mediated by adaptive evolution at mei-218.
In this paper, we study the long-term molecular evolution of MEI-218, and its interacting proteins, and we test for further evidence of its functional divergence in other Drosophila lineages. First, we ask if recurrent positive selection at mei-218 is limited to the D. melanogaster and D. mauritiana lineages or instead extends to the broader Drosophila phylogeny, as seen with Prdm9 in rodents and primates (Oliver et al. 2009). We survey protein-coding sequence evolution at genes encoding components of the mei-MCM complex (mei-218, mei-217, rec, Mcm5) and the Blm helicase among members of the melanogaster, obscura, and virilis species groups. Our analyses reveal evidence for recurrent positive selection at two genes— mei-218 and rec— in the melanogaster and in the obscura species groups. Second, we ask if evolution at mei-218 has functional consequences for meiotic crossing over. The genetic maps of D. pseudoobscura and D. virilis are ∼2 times longer than that of D. melanogaster (Ortiz-Barrientos et al. 2006; L. Hemmer et al. unpublished). While substitutions at mei-218 might have affected crossing over, it is also possible that they were inconsequential (neutral) or mediated the evolution of some other (unknown) function (e.g., in the male germline; Chintapalli et al. 2007; Chen et al. 2014). To distinguish these possibilities, we assayed the meiotic crossover phenotypes of transgenes bearing wildtype D. pseudoobscura or D. virilis alleles in an otherwise D. melanogaster genetic background. Our transgene experiments show that both species’ alleles have functionally diverged from that of D. melanogaster: for the D. pseudoobscura allele, overall crossover rates are comparable to D. melanogaster whereas crossover patterning shifts to a more uniform distribution; for the D. virilis allele, crossover rates are aberrantly low compared to wildtype whereas crossover patterning is comparable to D. melanogaster. These observations suggest that crossover rate and crossover patterning may be separable functions.
Materials and Methods
Alignments and PAML analyses
We extracted and aligned the coding sequences of five genes, mei-217, mei-218, rec, Mcm5, and Blm from 23 species spread across the melanogaster, obscura, and virilis groups (Table S1). Coding sequences for D. affinis and the species in the virilis group were generously provided by Rob Unckless (University of Kansas) and Yasir Ahmed-Braimah (Cornell University), respectively. Once all coding sequences for each gene were compiled for each of the three species groups, we translated the coding sequence into predicted protein sequences, aligned the amino acid sequences using MUSCLE v3.8.425 (Edgar 2004), then back-translated the alignment into the original nucleotide sequences. CDS alignments were assessed and gap-adjusted by hand to retain in-frame codons (alignments available by request). Each gene alignment was fit to an NSsites model, part of the CODEML package in PAML (Yang 1997). We compared model 7 (M7), which does not allow dN/dS to exceed 1 for any codons, to model 8 (M8) which allows dN/dS > 1 for a subset of codons. We used a likelihood ratio test to determine the best fit model. Sites identified as having experienced positive selection were those found to have posterior probabilities >95% with Bayes Empirical Bayes (Yang et al. 2005).
Generating transgenic flies
To generate transgenic flies, we cloned the D. pseudoobscura and D. virilis alleles of mei-217/-218 and used the ΦC31 integrase to place the transgenes in (the same) desired chromosomal landing sites via site-specific integration (Venken et al. 2006). We amplified three sections of the D. pseudoobscura mei-217/-218 extended gene region with associated 5′ and 3′ noncoding regions from the D. pseudoobscura reference genome strain using iProof polymerase (Bio-Rad, Hercules, CA) and sequentially reconstructed the fragments within a pBluescript KS+ vector in a three-step process using standard molecular techniques. The first fragment, containing the upstream region and most of mei-217, was amplified as a 2.2kb fragment, phosphorylated, and cloned into the SpeI site of KS+ generating an intermediate plasmid KS[psefrag1]. The second fragment, containing the 3′ end of mei-217 and most of mei-218, was amplified as a 2.3kb fragment, phosphorylated, and cloned separately into the SpeI site of KS+ generating a second intermediate plasmid KS[psefrag2]. We then digested KS[psefrag1] with XbaI and NotI (New England Biolabs, Ipswich, MA), gel purified the resulting 2.2kb fragment, and cloned it upstream of KS[psefrag2] into XbaI/NotI sites. This generated a third intermediate plasmid KS[psefrag1+2]. The third fragment, containing the 3′ end of mei-218 and downstream 3′ non-coding regions, was amplified and digested with XhoI and NarI (New England Biolabs, Ipswich, MA) resulting in a 1.4kb fragment which was subsequently cloned into the XhoI/NarI sites downstream of KS[psefrag1+2] generating the final plasmid KS[psefrag1+2+3]. This final recombinant plasmid, KS[psefrag1+2+3], reconstitutes the entire D. pseudoobscura 5.7kb mei-217/-218 gene region. We confirmed the absence of introduced mutations in the cloned mei-217/-218pse allele by direct Sanger sequencing of the KS[psefrag1+2+3] plasmid. We then cut the mei-217/-218 insert from the KS+ vector with NotI and subcloned into an attB[Pacman]-ApR vector obtained from the Drosophila Genomics Resource Center (Bloomington, IN).
In an analogous manner, the D. virilis mei-217/-218 extended gene region was amplified from the D. virilis reference genome strain and sequentially reconstructed within a pBluescript KS+ vector (hereafter KS+; Stratagene, La Jolla, CA) in a three-step process using standard molecular biology techniques. All bacterial transformations were performed at room temperature to enhance plasmid stability in One Shot TOP10 chemically competent E. coli (Invitrogen, Carlsbad, CA). First, the upstream region and most of mei-217 was amplified as a 2.6 kb fragment, using a 5′ primer that contained a SphI site. The 5′ end of the resulting PCR product was digested with SphI (New England Biolabs, Ipswich, MA) while the 3′ end was made blunt using FastAp Thermosenstive Alkaline Phosphatase (Thermo Fisher, Waltham, MA). This fragment was cloned into the SphI/SpeI sites of KS+, generating intermediate plasmid KS[virfrag1]. Second, the 3′ end of the mei-217 and most of mei-218 was amplified as a 3.2kb fragment. The amplicon was phosphorylated with T4 Polynucleotide Kinase (Invitrogen, Carlsbad, CA) and ligated into the SpeI site of KS+ generating a second intermediate plasmid KS[virfrag2]. We then digested KS[virfrag2] with AatII and NotI (New England Biolabs, Ipswich, MA), gel purified the resulting 3.2kb fragment, and cloned it into the SphI/SmaI sites downstream of the virfrag1 generating a third intermediate plasmid KS[virfrag1+2]. Third, the 3′ end of mei-218 and downstream 3′ non-coding regions was amplified and digested with AatII and SalI (New England Biolabs, Ipswich, MA) resulting in a 1.1kb fragment which was subsequently cloned into the AatII/SalI sites downstream of KS[virfrag1+2] generating the final plasmid KS[virfrag1+2+3]. This final recombinant plasmid, KS[virfrag1+2+3], reconstitutes the entire D. virilis 6.9kb mei-217/-218 gene region. We then cut the mei-217/-218 insert from the KS+ vector with NotI and subcloned into an attB[Pacman]-ApR vector obtained from the Drosophila Genomics Resource Center (Bloomington, IN). We confirmed the absence of introduced mutations in the cloned mei-217/-218vir allele by Sanger sequencing.
Both D. pseudoobscura and D. virilis transgene constructs were introduced into D. melanogaster y w; PBac[y+-attP-9A]VK00005 flies, which have an attP transgene landing site at cytological position 75A10 on chromosome arm 3L, via injections performed by BestGene (Chino Hills, CA). The attB-P[w+ mei-218pse]-ApR and attB-P[w+ mei-218vir]-ApR transgenic flies (for simplicity, hereafter referred to as P[mei-217/218pse] and P[mei-217/-218vir], respectively) were then made homozygous and maintained as stocks. Following the crossing protocol in (Brand et al. 2018), we estimated crossover rates for a multiply marked second chromosome in two female genotypes:
mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218pse]/ +; and
mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218vir]/ +.
To estimate crossover frequencies, we crossed the female genotypes above to homozygous net dppd-ho dp b pr cn males and scored the progeny for all markers. (For clarity, we refer to dppd-ho throughout by its mutant synonym, ho.) We performed n = 13 and n = 14 crosses for mei-217/-218pse and mei-217/-218vir, respectively (see Table S2 for data). For each cross, we collected either ∼10 virgin mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218pse]/+ females or mei-2181; net ho dp b p cn/ + + + + + +; P[mei-217/-218vir]/+ females, aged them for three to five days, and crossed them to ∼10 net ho dp b pr cn males that were aged for at least two days. After five days, parents were discarded, and the vials were hydrated with a solution of 0.5% propionic acid. All crosses were maintained in an incubator at 24C under a 12-hour light/dark cycle on standard corn-meal media. We estimated means and standard deviations of crossover frequency among the independent, replicate crosses and compared genotypes using standard t-tests (Figure 3B,4B).
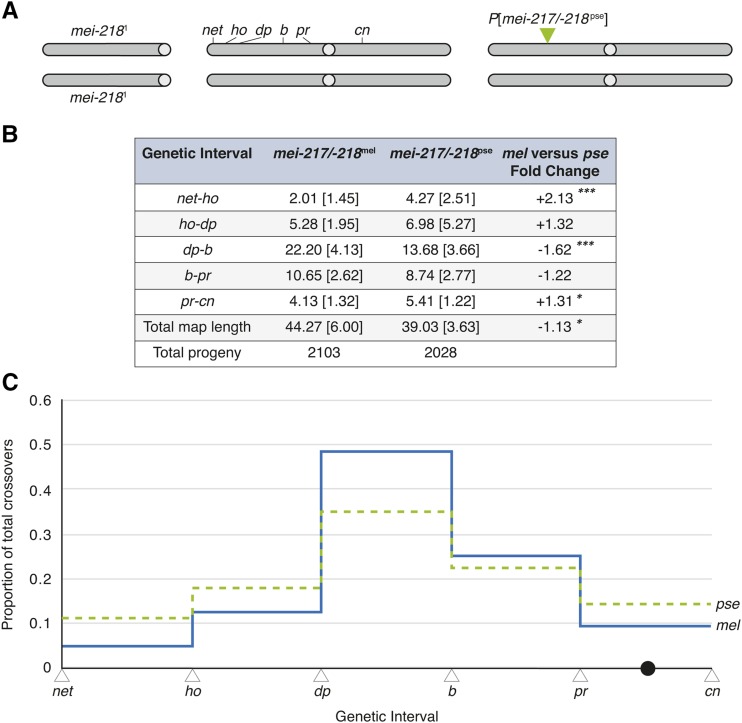
Figure 3.
The mei-217/-218 allele of D. pseudoobscura alters the rate and patterning of crossing over in D. melanogaster. (A) D. melanogaster females containing a transgene of a D. pseudoobscura mei-217/-218 allele inserted on chromosome 3L (75A10) were assayed for crossing over. The endogenous mei-2181 allele contains a nonsense mutation. Crossover frequencies were estimated among the six visible markers spanning the left arm of chromosome 2 and the centromere: net (net), decapentaplegic (ho), dumpy (dp), black (b), purple (pr), and cinnabar (cn). mei-217/-218mel data re-produced from Brand et al. (2018). (B) For each genotype, the means and standard deviations [in brackets] of crossover frequency for the five genetic intervals measured in the two transgenic genotypes. The p-values are for unpaired t-tests (*P < 0.05, **P < 0.01, ***P < 0.001). (C) The proportion of total crossovers distributed across the five intervals in the net-cn region in mei-217/-218mel (blue) and mei-217/-218pse (green) females. The total number of crossovers scored for mei-217/-218mel and mei-217/-218pse females is 956 and 786, respectively (see Table S2).
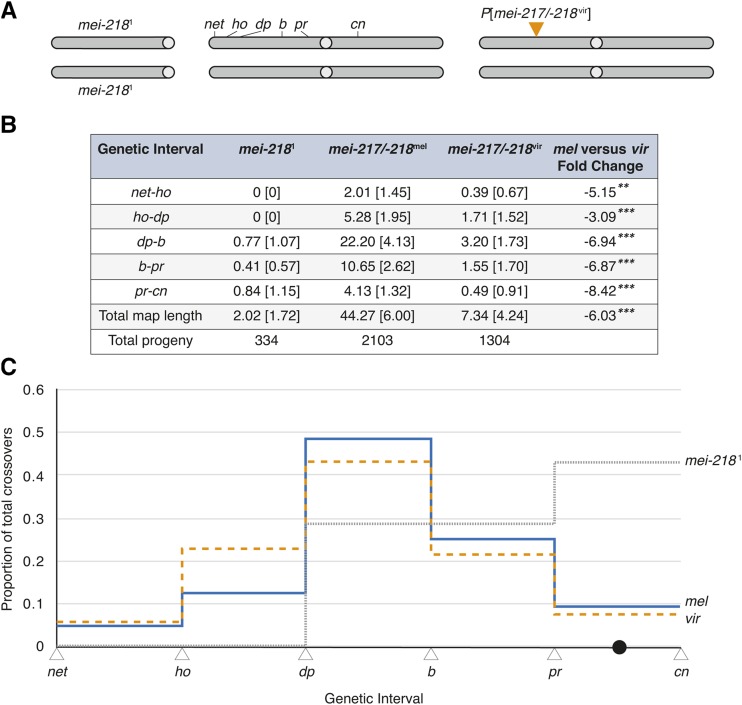
Figure 4.
The mei-217/-218 allele of D. virilis alters the rate and patterning of crossing over in D. melanogaster. (A) D. melanogaster females containing a transgene of a D. virilis mei-217/-218 allele inserted on chromosome 3L (75A10) were assayed for crossing over. The endogenous mei-2181 allele contains a nonsense mutation. Crossover frequencies were estimated among the six visible markers spanning the left arm of chromosome 2 and the centromere: net (net), decapentaplegic (ho), dumpy (dp), black (b), purple (pr), and cinnabar (cn). mei-2181 and mei-217/-218mel data re-produced from Brand et al. (2018). (B) For each genotype, the means and standard deviations [in brackets] of crossover frequency for the five genetic intervals measured in the mei-218 mutant and two transgenic genotypes. p-values are derived for unpaired t-tests. *P < 0.05, **P < 0.01, ***P < 0.001. (C) The proportion of total crossovers distributed across the five intervals in the net-cn region in mei-217/-218mel (blue), mei-217/-218vir (orange), and mei-2181 (gray) females. The total number of crossovers scored for the mei-217/-218mel, mei-217/-218vir, and mei-2181 females is 956, 93, and 7, respectively (see Table S2).
To compare the spatial distribution of crossovers across the five intervals spanning the net-cn region (Table S3) while controlling for overall crossover rate differences, we compared standardized distributions among genotypes based on the proportion of crossovers occurring in each interval using χ2 tests. We also tested for genotypic differences in crossover event distributions among tetrads by inferring the frequencies of non-, single-, double- or triple crossovers (E0, E1, E2, and E3, respectively) using the algebraic methods of Weinstein (1936). We estimated the strength of Interference (I) as 1− (observed double crossovers / expected double crossovers). All statistical analyses were performed using R (http://www.R-project.org/). The Drosophila stocks and plasmids used in this study are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Data Availability
Fly stocks and transgenic constructs are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9162431.
Results
Positive selection at meiosis genes that regulate crossover formation and patterning
We identified and extracted protein-coding sequences encoding members of the mei-MCM complex (mei-218, mei-217, rec, Mcm5) and, because it antagonizes the mei-MCMs, Blm (Kohl et al. 2012), from whole genome sequence data of 23 species (see Materials and Methods, Table S1). In some cases, we used Sanger sequencing to complement retrieved sequence data that had gaps and/or quality issues (Table S1). To investigate patterns of long-term molecular evolution, we used maximum likelihood methods to test for phylogenetic evidence of positive selection. We analyzed the melanogaster (n = 14), pseudoobscura (n = 5), and virilis (n = 4) species groups separately, as the phylogenetic distances among the three groups are so large that synonymous site divergence (dS) is saturated (Figure 2A; Larracuente et al. 2008; Stanley and Kulathinal 2016). Using the codeml program in the PAML suite, we performed likelihood ratio tests to identify genes that have elevated rates of nonsynonymous substitution relative to synonymous substitution (dN/dS) (Yang 1997). In particular, we compared the log-likelihood of a model for which the estimated values of dN/dS for individual codons is β-distributed between 0 and 1 (model 7) to that of an alternative model for which the distribution includes an additional class of codons with dN/dS > 1 (model 8). Positive selection is inferred for cases in which model 8 provides a significantly better fit to the data (Yang 1997; see Materials and Methods).
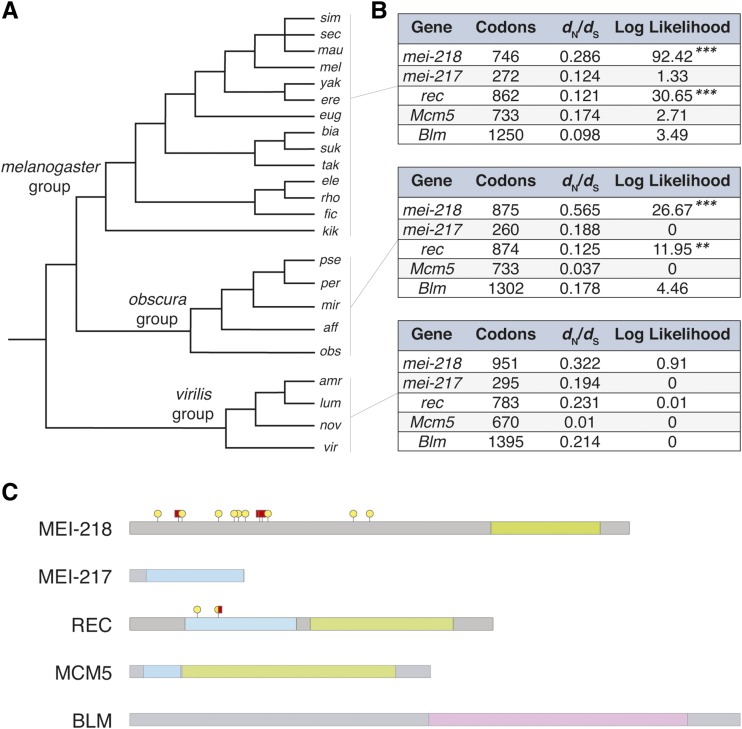
Figure 2.
Molecular evolution across the Drosophila phylogeny. (A) Phylogenetic relationship of the 23 species analyzed within the melanogaster, obscura, and virilis species group. (B) PAML analyses for the three species groups were performed separately because the phylogenetic distances among them is so large that ds is saturated. We report the log-likelihood estimates from a model 7 – model 8 comparison (*P < 0.05, **P < 0.01, ***P < 0.001). (C) A schematic of the structural domains in the five proteins analyzed. In the MEI-MCMs the AAA ATPase MCM domains are shaded in green and the MCM N-terminal domains are shaded in blue. In the BLM Helicase the RecQ DNA Helicase domain in shaded in purple. In MEI-218 and REC, the pins represent codons with evidence for positive selection in the melanogaster group (yellow circle) and the obscura group (red squares).
Two of the five genes show phylogenetic evidence of recurrent positive selection. First, we find that mei-218 has a history of positive selection in both the melanogaster and obscura species groups (Figure 2B). A Bayes Empirical Bayes (BEB) analysis (Yang et al. 2005) identified nine codons in the melanogaster group and three codons in the obscura group, all clustered in the second and third exons, with evidence of positive selection (Figure 2C; posterior probability >0.95). The quality of the local sequence alignment is low for both species groups, compromising our confidence in the BEB-identified codons. To address this problem, we performed PAML analyses on each of the five exons, separately, to broadly localize the signal of positive selection within mei-218. Consistent with the codons identified by the BEB analysis, we find that the second and third exons, which encode a disordered protein region, show evidence of positive selection in both the melanogaster and the obscura species groups. We aligned coding sequences using MUSCLE (Edgar 2004), although other alignment algorithms (i.e., ClustalW and Geneious) give qualitatively similar results. The alignment uncertainty results partly from high rates of indel evolution at mei-218. For example, compared to the D. melanogaster reference, sequences from the other species of the melanogaster group contain ≥13 indels ranging in size from 1 to 267 codons. This frequent insertion and/or deletion of codons limits the power of our PAML analysis of mei-218: of the 5,166 sites in the mei-218 melanogaster group alignment, 2,967 sites (57%) were not analyzed due to gaps in the multi-species alignment.
Among the other four genes studied, only rec shows evidence of positive selection in the melanogaster and obscura species groups (Figure 2B). BEB analyses identified two codons in the MCM N-terminal domain with histories of positive selection (Figure 2C; posterior probability >0.95): one codon experienced positive selection in the melanogaster group and the other codon experienced positive selection in both the melanogaster and obscura groups. At the positively selected codon detected in the melanogaster group, eight different amino acid states are represented among 14 species. At the positively selected codon detected in both the melanogaster and obscura groups, different amino acid states are represented in 8/14 and 4/5 species, respectively. PAML analyses find no support for positive selection at the remaining three genes (mei-217, Mcm5, or Blm). In the virilis species group, none of the five genes tested show evidence for positive selection (Figure 2B).
Functional analysis of mei-217/-218 from D. pseudoobscura
D. pseudoobscura diverged from D. melanogaster ∼30 mya, and its genetic map is ∼2-fold longer, with less expansive centromeric suppression of crossing over and a more uniform recombination landscape (Hamblin and Aquadro 1999; Ortiz-Barrientos et al. 2006; Drosophila 12 Genomes Consortium 2007). The levels of coding sequence and length divergence between D. pseudoobscura and D. melanogaster are extraordinary, and different regions of the mei-217/-218 sequence have experienced strikingly different rates of molecular evolution. Between D. pseudoobscura and D. melanogaster, pairwise amino acid identity for MEI-218 is much lower for than for MEI-217 (∼34% vs. 61%, respectively). The very low identity for MEI-218 is attributable to the N-terminal disordered region— which shares only ∼20% identity and differs in length by 183 codons— not the C-terminal AAA ATPase MCM domain which shares 70% identity (Figure 2C). Given the extraordinary protein sequence (and indel) divergence at MEI-218, the statistical evidence for recurrent positive selection at mei-218 in both species groups, and previous experimental evidence that mei-217/-218 mediates species differences in crossing over (Brand et al. 2018), we sought to test if molecular evolution at mei-217/-218 between D. pseudoobscura and D. melanogaster has functional consequences for crossing over.
To experimentally test for functional effects of molecular divergence at mei-217/-218 between D. pseudoobscura and D. melanogaster, we cloned the entire mei-217/-218 gene region, including all of the upstream and downstream noncoding regions, from D. pseudoobscura into an attB-P[acman] vector (hereafter mei-217/-218pse; see Materials and Methods). We integrated this transgene construct into an attP site on chromosome arm 3L of D. melanogaster (cytological position 75A10) and used genetic crosses to place the transgene in a mei-218 loss-of-function genetic background resulting in the D. melanogaster stock, mei-2181; P[mei-217/-218pse] (see Materials and Methods). We then estimated crossover frequencies among six visible markers (net ho dp b pr cn) that span chromosome arm 2L and the centromere for replicate crosses of mei-2181; net ho dp b pr cn/+ + + + + +; P[mei-217/-218pse]/+ females to net ho dp b pr cn males (n = 13 crosses, 2028 progeny; Figure 3, Table S2). In this genotype, the D. pseudoobscura wildtype allele is the only source of mei-218 function. As our transgenes include mei-217/-218 coding and non-coding sequence, we are unable to attribute phenotypic effects to species differences in the protein sequence vs. expression level.
These experiments have three possible outcomes. First, sequence evolution at mei-217/-218 may be of no functional consequence to crossing over: mei-217/-218pse might rescue the mei-218 mutant phenotype but produce wildtype D. melanogaster-like rates and patterning of crossing over. Second, sequence evolution at mei-217/-218 may render it incompatible between species: the mei-217/-218pse allele might be sub- or non-functional in D. melanogaster so that it cannot fully rescue the mei-218 mutant phenotype. Last, sequence evolution at mei-217/-218 may recapitulate some of the wildtype species differences in crossing over: the mei-217/-218pse allele might rescue the null mei-218 mutant phenotype but produce rates and/or patterning of crossing over that differ from D. melanogaster in a way similar to D. pseudoobscura.
We find that, in a D. melanogaster genetic background lacking mei-218 function, the mei-217/-218pse rescues crossing over: the total net-cn genetic map length of 39.03 map units is smaller (∼1.13-fold) than, but comparable to mei-217/-218mel controls (t-test, P = 0.044; Figure 3B). However, while the total lengths of the mei-217/-218pse and mei-217/-218mel genetic maps are comparable, we observe highly significant crossover rate heterogeneity among intervals between the transgenes. In mei-217/-218pse females, the medial largest interval of chromosome arm 2L (dp-b) experiences a 1.6-fold lower crossover frequency than the mei-217/-218mel control (t-test, P < 0.0001; Figure 3B) whereas the telomere-proximal (net-ho) and centromere (pr-cn) regions experience 2.13- and 1.31-fold higher crossover frequencies, respectively (t-test, P = 0.001 and P = 0.028, respectively; Figure 3B). To distinguish crossover frequency and crossover patterning, we calculated the proportion of total crossovers that occurred in each genetic interval for each transgene. These values differ from genetic map distances (the proportion of recombinant progeny), for which crossover rate and patterning are confounded, and instead provide profiles of crossover patterning that are independent of crossover rate. We find that the patterning of crossovers differs significantly between mei-217/-218mel and mei-217/-218pse (χ2 test, df = 4, P < 9.52e-28; Figure 3C, Table S3). Specifically, crossovers in mei-217/-218mel females are concentrated in the medial ho-dp-b regions and occur at lower frequencies in the telomeric net-ho and centromeric pr-cn regions, resulting in relatively larger variability in the proportion of crossovers among the genetic intervals (range between lowest net-ho and highest dp-b intervals = 0.44; Figure 3B,C; Table S3). In contrast, crossovers in mei-217/-218pse females are distributed more uniformly across the genetic intervals, with less variability in the proportion of crossovers among the genetic intervals (range between lowest net-ho and highest dp-b intervals = 0.24; Figure 3B,C; Table S3). The mei-217/-218pse transgene thus produces a total genetic map length that is nearly D. melanogaster-like but crossover patterning that differs from D. melanogaster in ways similar to D. pseudoobscura.
The distribution of the number of crossovers per recovered chromosome— the number of non-crossover (NCO), single-crossover (SCO), double-crossover (DCO) chromosomes, and so on— differs significantly between mei-217/-218pse and mei-217/-218mel transgenes (χ2 test, df = 3, P < 1.29e-16; Table S2). We used these data to infer the distribution of the number of crossovers per tetrad using the methods of Weinstein (1936). Two mechanisms constrain the distribution of the number of crossovers per meiosis: (1) crossover assurance encourages the formation of at least one obligate crossover per tetrad to guarantee proper segregation; and (2) crossover interference discourages the formation of multiple crossovers near one another (Jones and Franklin 2006; Berchowitz and Copenhaver 2010; Wang et al. 2015). As a result, the number of crossovers per tetrad is under-dispersed relative to Poisson expectations, with zero- and multiple-crossover classes under-represented in wildtype D. melanogaster (Mehrotra et al. 2007). Consistent with regulation, the inferred number of crossovers per tetrad is similarly under-dispersed relative to Poisson expectation in mei-217/-218pse females (χ2 test, df = 5, P < 1.3e−237). However, the distribution of the number of crossovers per tetrad differs between the mei-217/-218pse and mei-217/-218mel females (χ2 test, df = 3, P = 2.2e−16; Table 1). The mean number of crossovers per tetrad is reduced in mei-217/-218pse females compared to mei-217/-218mel controls (0.78 vs. 0.91). This difference is largely attributable to a 1.9-fold increase in non-crossover (E0) tetrads at the expense of single-crossovers (E1), which are reduced 1.47-fold (Table 1). Crossover assurance thus appears weaker in mei-217/-218pse females. At the same time, however, we find that multiple-crossover tetrads are more frequent in mei-217/-218pse females: double- and triple-crossover tetrads are increased 1.16- and 5-fold, respectively (Table 1). We tested whether this increase in multi-crossover tetrads is enabled by weaker crossover interference in mei-217/-218pse females. Indeed, crossover interference for the two largest adjacent intervals (ho-b-pr) is ∼28% weaker in mei-217/-218pse females, although the difference is not significant (I = 0.567 vs. 0.793, Mann-Whitney, P = 0.795; Table S2). Together, the weaker crossover assurance and crossover interference in mei-217/-218pse females increase the relative variance (variance/mean) in the number of crossovers per tetrad (0.698) compared to that for in mei-217/-218mel (0.350; Table 1).
Table 1.
The distribution of the inferred number of crossovers in the net-cn region per meiosis differs among genotypes
| Tetrad Class# | mei-2181‡ | mel‡ | pse | vir | pse-mel fold-diff* | vir-mel fold-diff* | vir-pse fold-diff* |
|---|---|---|---|---|---|---|---|
| E0 | 0.958 | 0.205 | 0.389 | 0.885 | +1.90 | +4.32 | +2.27 |
| E1 | 0.042 | 0.685 | 0.467 | 0.094 | −1.47 | −7.29 | −4.97 |
| E2 | 0.000 | 0.107 | 0.124 | 0.015 | +1.16 | −7.13 | −8.27 |
| E3 | 0.000 | 0.004 | 0.020 | 0.006 | +5 | +1.5 | −3.33 |
| Mean† | 0.042 | 0.909 | 0.775 | 0.143 | |||
| Variance† | 0.000 | 0.319 | 0.541 | 0.190 | |||
| Relative variance^ | 0.000 | 0.350 | 0.698 | 1.33 |
E0, E1, E2, E3 are the estimated frequencies of tetrads with zero, one, two and three inferred crossovers, respectively. Tetrad frequencies were estimated using Weinstein's (1936) algebraic method.
mei-2181, mei-217/-218mel data are reproduced from Brand et al. (2018).
Weinstein estimates for mei-217/-218mel, mei-217/-218pse, and mei-217/-218vir alleles differ significantly from one another (χ2 test, df = 5, P < 1.49e-130).
Mean and variance = mean and sample variance of the inferred number of crossovers per tetrad, respectively.
Relative variance = variance/mean.
These results show that crossover assurance, interference, and centromeric and telomeric suppression are all weaker in mei-217/-218pse females. These observations are qualitatively consistent with crossover patterning in wildtype D. pseudoobscura, which also shows reduced (or even possibly absent) centromeric suppression and a more uniform distribution of crossovers (Hamblin and Aquadro 1999; Ortiz-Barrientos et al. 2006; Kulathinal et al. 2008). Alternatively, it is possible that the mei-217/-218pse allele is incompatible with interactors from D. melanogaster, so that the functionally divergent mei-217/-218pse allele is unable to fully receive and/or implement endogenous crossover patterning signals from D. melanogaster. Under this incompatibility hypothesis, the shift in crossover patterning toward one that is D. pseudoobscura-like would be coincidental rather than a reflection of the wildtype properties of the D. pseudoobscura allele. As with any heterologous transgene (or interspecific genetic) experiment, formally distinguishing between these two interpretations is difficult. However, under either interpretation, the results demonstrate that the effects of D. pseudoobscura and D. melanogaster alleles on meiotic crossing over have diverged and support the notion that mei-217/-218 has two separable functions— crossover formation and crossover patterning.
Functional analysis of mei-217/-218 From D. virilis
We also assayed a mei-217/-218 wildtype allele from a species more distantly related to D. melanogaster. D. virilis diverged from D. melanogaster ∼50 mya and has a ∼2-fold longer genetic map (L. Hemmer et al. unpublished; Drosophila 12 Genomes Consortium 2007). Although our PAML analyses failed to detect evidence of positive selection at mei-218 within the virilis species group, these analyses are uninformative about the possibility of positive selection in the lineages ancestral to the virilis and melanogaster species groups. The MEI-217 protein shows ∼58% pairwise amino acid sequence identity between D. melanogaster and D. virilis. In contrast, the MEI-218 protein shares only ∼34% pairwise amino acid sequence identity overall. Divergence at mei-218 is not uniform across the protein: while the C-terminal AAA ATPase MCM domain shows ∼63% identity, the disordered N-terminal shows just ∼23% identity (Figure 2C; see also Manheim et al. 2002). The D. virilis protein is 954 amino acids long, shorter than both D. melanogaster (1186 aa) and D. pseudoobscura (1002 aa). The difference in length is driven by indel evolution largely concentrated in the disordered region of MEI-218, as the N-terminal MCM domain has remained relatively unchanged between the three species (D. melanogaster 337 aa; D. pseudoobscura 336 aa; D. virilis 334 aa; see also Kohl et al. 2012).
To functionally assay the D. virilis allele of mei-217/-218, we followed the same strategy used to create transgenic flies with mei-217/-218pse (see above) and mei-217/-218mau (Brand et al. 2018). We first cloned the entire D. virilis mei-217/-218 gene region and flanking non-coding sequences into an attB-P[acman] vector (hereafter mei-217/-218vir) and integrated it into an attP site on 3L (75A10) in D. melanogaster (see Materials and Methods). We then used crosses to place the transgene into a D. melanogaster mei-218 mutant background and measured crossover frequencies among visible markers spanning part of the second chromosome in replicate mei-2181; net ho dp b pr cn/+ + + + + +; P[mei-217/-218vir]/+ females (n = 14 crosses, 1304 progeny; Figure 4, Table S2; see Materials and Methods).
Rates of crossing over are strongly reduced in mei-217/-218vir transgene-bearing females (Figure 4B). While the control mei-217/-218mel transgene produces a total genetic map length of 44.27 across the net-cn region, the mei-217/-218vir transgene produces a genetic map that is ∼6-fold smaller (7.34; t-test, P < 0.0001; Figure 4B). Two lines of evidence, however, indicate that mei-217/-218vir does not behave like a null allele in D. melanogaster: compared to mei-2181 mutant females, mei-217/-218vir females produce longer genetic maps (2.02 map units; t-test, P = 0.001; Figure 4B) and show non-uniform spatial patterning of crossovers. In mutant mei-2181 females, ∼90% of crossovers are eliminated, and the residual crossovers fail to show crossover interference or centromeric suppression (Baker and Carpenter 1972); see also Figure 4C, Table S3). These observations suggest that most crossovers in wildtype females correspond to interfering class I crossovers, whereas residual crossovers in mei-2181 females correspond to non-interfering class II crossovers which originate via a different pathway (Figure 1; Berchowitz and Copenhaver 2010; Kohl and Sekelsky 2013). To determine if the crossovers in mei-217/-218vir behave like residual (presumed class II) crossovers of mei-2181 females, we tested whether crossover patterning across the genetic intervals is disrupted. The distribution of crossovers between mei-217/-218vir and mei-2181 is significantly different (χ2 test, df = 4, P < 1.2e-12; Figure 4C, Table S3). Specifically, mei-2181 females show a non-uniform reduction in crossover frequency, with crossing over reduced in centromere-proximal regions (pr-cn) to only 20.3% of the mei-217/-218mel value compared to 4.6% across the entire 2L (net-cn) region (see also Carpenter and Sandler 1974). In contrast, mei-217/-218vir females show comparable reductions in crossover frequency among regions, with crossing over reduced in the centromere-proximal regions (pr-cn) to 11.9% of the mei-217/-218mel value compared to 16.6% across the entire 2L (net-cn) region (Figure 4B). As a result, crossover patterning in mei-217/-218vir females is comparable to mei-217/-218mel (χ2 test, df = 4, P = 0.05; Figure 4C). In mei-217/-218vir females, then, overall crossover frequencies are reduced 83.4%, but gross crossover patterning is largely unchanged. These findings show that mei-217/-218vir is unable to support wildtype (D. melanogaster or D. virilis) rates of crossing over in a D. melanogaster genetic background but appears able to integrate patterning information specified by D. melanogaster.
The distributions of the observed number of crossover events per recovered chromosome (χ2 test, df = 3 P = 7.07e-137; Table S2) and the estimated number of crossovers per tetrad helps to explain why the total net-cn map length is so much smaller in mei-217/-218vir females than mei-217/-218mel females. The estimated number of crossovers per tetrad is under-dispersed in females bearing the mei-217/-218mel transgene (variance/mean = 0.350), which experience a mean of 0.91 crossovers per tetrad (χ2 test, df = 5, P < e-200; Table 1; Brand et al. 2018). In contrast, the estimated number of crossovers per tetrad is over-dispersed in females bearing the mei-217/-218vir transgene (variance/mean = 1.33)— with a deficit of single-crossover tetrads (E1 = 0.094) and an excess of multiple-crossover tetrads (E≥2 = 0.021; χ2 test, df = 5, P = e-20; Table 1)— which experience a mean of only 0.14 crossovers per tetrad. The reduced genetic map in mei-217/-218vir females therefore occurs because most tetrads experience no crossovers during meiosis. Crossover assurance therefore appears strongly compromised in mei-217/-218vir females implying that the mei-217/-218vir allele is unable to ensure an obligate crossover in a D. melanogaster genetic background. Such achiasmate tetrads suffer elevated rates of mis-segregation and nondisjunction leading to production of aneuploid gametes and reduced fecundity (Baker and Carpenter 1972; Bhagat et al. 2004). Consistent with nondisjunction, mei-217/-218vir females produce significantly fewer progeny (mean ± SD = 93.14 ± 17.3) than mei-217/-218mel females (161.77 ± 52.76; Figure 4B; t-test P = 0.0005).
The mei-217/-218vir allele provides additional evidence that crossover rate and patterning are separable: in D. melanogaster, mei-217/-218vir is hypomorphic with respect to crossover formation but not crossover patterning. The fact that mei-217/-218vir cannot fully complement the mei-2181-mediated loss of crossover formation in D. melanogaster suggests either of two possibilities. The wildtype function of mei-217/-218vir may differ from mei-217/-218mel: whereas mei-217/-218mel functions in crossover formation and patterning in D. melanogaster (Baker and Carpenter 1972; McKim et al. 1996; Kohl et al. 2012), mei-217/-218vir may be less essential to crossover formation but still essential to crossover patterning in D. virilis. Alternatively, mei-217/-218vir may be genetically incompatible with factors from D. melanogaster: mei-217/-218vir may fail to interact appropriately with D. melanogaster-encoded proteins such that crossover formation (but not patterning) is compromised. Under either model, the molecular divergence at mei-217/-218 between D. melanogaster and D. virilis has functional consequences for female meiosis.
Discussion
Our phylogenetic analyses revealed that two mei-MCM genes, mei-218 and rec, have histories of recurrent positive selection in the melanogaster and obscura species groups, and our transgenic assays show that the different species’ mei-217/-218 alleles have functionally diverged with respect to crossover patterning (mei-217/-218pse) and crossover formation (mei-217/-218vir). These observations are superficially reminiscent of the recurrent positive selection at Prdm9, the major trans-acting factor that controls the distribution of recombination hotspots in mammals. The forces driving the rapid molecular evolution of Prdm9 are reasonably well understood. During recombination-repair of DSBs, the DNA sequence motifs recognized by the PRDM9 zinc fingers tend to be replaced with non-motif sequence. As the number of recombination hotspots erodes over time, the overall frequency of recombination decreases to suboptimal levels, elevating the risk of chromosomal mis-segregation and/or breakage (Ségurel et al. 2011; Smagulova et al. 2016). This model explains why there is selection for PRDM9 to acquire novel zinc fingers that recognize novel DNA sequence motifs, creating a new class of recombination hotspots, and thereby reestablishing appropriate recombination frequencies. This process can quickly lead to differences in the identity and distribution of recombination hotspots between closely related species and, incidentally, to sterility in species hybrids (Ptak et al. 2005; Davies et al. 2016; Smagulova et al. 2016). For the mei-MCMs, mei-218 and rec, the causes of recurrent positive selection are unclear. In Drosophila, fine-scale heterogeneity in recombination rates exists, but recombination hotspots comparable to those in mammals do not (Comeron et al. 2012; Hunter et al. 2016). Moreover, unlike PRDM9, the mei-MCMs do not have DNA binding domains known to recognize specific motifs. It therefore seems doubtful that the positive selection we have observed involves DNA motif turnover.
The phylogenetic evidence for recurrent bouts of positive selection at mei-218 and rec are similarly consistent with adaptation to moving fitness optima but the causes of selection are unclear. For instance, despite the absence of crossing over in Drosophila males, FlyAtlas and modEncode data show that both mei-218 and rec are expressed in D. melanogaster testes (Chintapalli et al. 2007; Chen et al. 2014). It is therefore possible that the history of positive selection at these genes reflects adaptation for male reproductive functions, although the functions of mei-218 and rec in testes are unknown and mutant males are fertile. If, instead, recurrent positive selection at mei-218 and rec has occurred to modulate crossing over, then we require a model in which the optimal rate and/or distribution of crossing over has changed repeatedly. Selfish genetic elements could provide one source of such fluctuating selection. First, meiotic drive in the female germline can generate selection for modifiers of crossing over. Depending on such details as whether drive occurs in meiosis I or II, or whether drive involves the centromere or telomere(s), selection can favor modifiers that increase or decrease rates of crossing over (Brandvain and Coop 2012). Second, while crossing over provides important meiotic and evolutionary functions, it also entails the risk of ectopic non-homologous exchange between similar but dispersed sequences, like transposons. Ectopic exchange can generate deleterious duplications, deletions, and other chromosomal aberrations (Goldberg et al. 1983; Barrón et al. 2014). The optimal recombination rate should thus evolve to balance the benefits of crossing over against the costs. The requirement for at least one crossover per chromosome (arm) sets a minimum rate, whereas the risk of ectopic exchange may constrain the maximum rate. The risk of ectopic exchange depends on the abundance of dispersed repetitive DNA sequences with high similarity. In D. melanogaster, ≥2% of meioses yield aberrant chromosomes as a result of ectopic exchange between transposons (Miller et al. 2016). The rate of such ectopic exchange undoubtedly fluctuates over time, tracking with the load(s) of transposons of high sequence similarity. The typical evolutionary-demographic history of transposons involves invasion of a new host genome via horizontal transfer (or escape from suppression by the host surveillance system); a burst of proliferation; and eventual silencing upon capture by the host surveillance system (Charlesworth et al. 1994; Kidwell and Lisch 2000; Barrón et al. 2014). Under this scenario, the risk of ectopic exchange due to any particular transposon will spike with transposon proliferation, as genomes come to harbor a high number of highly similar transposon sequences, and then fade as the sequences of silenced transposons diverge from one another and degenerate. The response to fluctuating selection pressures on crossover rates could be mediated by meiosis genes like mei-218 and rec. Consistent with this hypothesis, in both the melanogaster and obscura groups, D. mauritiana and D. pseudoobscura have higher mean rates of crossing over and smaller transposon loads compared to their respective sister species, D. melanogaster and D. persimilis (Dowsett and Young 1982; True et al. 1996; Ortiz-Barrientos et al. 2006; Hill and Betancourt 2018). Similarly, within D. melanogaster, transposon densities are highest in chromosomal regions that experience little or no crossing over (reviewed in Lee and Langley 2010; Barrón et al. 2014). It is important to note, however, that strong alternative models exist in which the presence of transposons favors increased rates of crossing over (Charlesworth and Barton 1996). While distinguishing among these hypotheses will be challenging (Charlesworth 2018), it is clear that selfish genetic elements present a ubiquitous, powerful, and perhaps underappreciated source of selection on rates of recombination.
Acknowledgments
We thank Jan Spence for comments on an earlier draft of the manuscript; Rob Unckless and Yasir Ahmed-Braimah for providing sequence data; Christina Muirhead for statistical advice; and Jeff Sekelsky for sharing fly stocks. This work was supported by funds from an NSF-DDIG (DEB 1403154) to CLB, and the David and Lucile Packard Foundation, the University of Rochester, and the NIH (R01 GM111380) to DCP.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9162431.
Communicating editor: J. Comeron
Literature Cited
- Allers T., and Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. 10.1016/S0092-8674(01)00416-0 [DOI] [PubMed] [Google Scholar]
- Baker W. K., 1958. Crossing over in heterochromatin. Am. Nat. 92: 59–60. 10.1086/282010 [DOI] [Google Scholar]
- Baker B. S., and Carpenter A. T. C., 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., and Hall J. C., 1976. Meiotic mutants: genetic control of meiotic recombination and chromosome segregation, pp. 352–434 in Genetics and Biology of Drosophila, edited by Novitski E., and Ashburner M. Academic Press, New York. [Google Scholar]
- Barrón M. G., Fiston-Lavier A.-S., Petrov D. A., and González J., 2014. Population Genomics of Transposable Elements in Drosophila. Annu. Rev. Genet. 48: 561–581. 10.1146/annurev-genet-120213-092359 [DOI] [PubMed] [Google Scholar]
- Barton N. H., and Charlesworth B., 1998. Why sex and recombination? Science 281: 1986–1990. 10.1126/science.281.5385.1986 [DOI] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C. et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. Erratum 328: 690. 10.1126/science.1183439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. 10.1073/pnas.18.2.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., and Copenhaver G. P., 2010. Genetic interference: Don’t stand so close to me. Curr. Genomics 11: 91–102. 10.2174/138920210790886835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat R., Manheim E. A., Sherizen D. E., and McKim K. S., 2004. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenet. Genome Res. 107: 160–171. 10.1159/000080594 [DOI] [PubMed] [Google Scholar]
- Brand C. L., Cattani M. V., Kingan S. B., Landeen E. L., and Presgraves D. C., 2018. Molecular evolution at a meiosis gene mediates species differences in the rate and patterning of recombination. Curr. Biol. 28: 1289–1295.e4. 10.1016/j.cub.2018.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., and Coop G., 2012. Scrambling Eggs: Meiotic Drive and the Evolution of Female Recombination Rates. Genetics 190: 709–723. 10.1534/genetics.111.136721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L. D., and Marks R. W., 1986. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics 114: 525–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks L. D., 1988. The evolution of recombination rates, pp. 87–105 in The Evolution of Sex: An Examination of Current Ideas, edited by Michod R., and Levin B.. Sinauer Associates. [Google Scholar]
- Carpenter A. T. C., and Sandler L., 1974. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics 76: 453–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., and Charlesworth D., 1985. Genetic variation in recombination in Drosophila. I. Responses to selection and preliminary genetic analysis. Heredity 54: 71–83. 10.1038/hdy.1985.10 [DOI] [Google Scholar]
- Charlesworth B., Sniegowski P., and Stephan W., 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. 10.1038/371215a0 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., and Barton N. H., 1996. Recombination load associated with selection for increased recombination. Genet. Res. 67: 27–41. 10.1017/S0016672300033450 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2018. Evolution: Increased recombination caused by a single gene. Curr. Biol. 28: R342–R344. 10.1016/j.cub.2018.02.072 [DOI] [PubMed] [Google Scholar]
- Chen Z.-X., Sturgill D., Qu J., Jiang H., Park S. et al. , 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24: 1209–1223. 10.1101/gr.159384.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., and Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., and Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium et al., 2007 Evolution of genes and genomes on the Drosophila phylogeny. Nature 8: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Crow J. F., 1992. An advantage of sexual reproduction in a rapidly changing environment. J. Hered. 83: 169–173. 10.1093/oxfordjournals.jhered.a111187 [DOI] [PubMed] [Google Scholar]
- Davies B., Hatton E., Altemose N., Hussin J. G., Pratto F. et al. , 2016. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530: 171–176. 10.1038/nature16931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1930. Translocations Involving the Third and the Fourth Chromosomes of Drosophila melanogaster. Genetics 15: 347–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett A. P., and Young M. W., 1982. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc. Natl. Acad. Sci. USA 79: 4570–4574. 10.1073/pnas.79.15.4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The genetical theory of natural selection, Oxford Clarnedon Press. Oxford, UK; 10.5962/bhl.title.27468 [DOI] [Google Scholar]
- Goldberg M. L., Sheen J.-Y., Gehring W. J., and Green M. M., 1983. Unequal crossing-over associated with asymmetrical synapsis between nomadic elements in the Drosophila melanogaster genome. Proc. Natl. Acad. Sci. USA 80: 5017–5021. 10.1073/pnas.80.16.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell R. F., 1984. Time of recombination in the Drosophila melanogaster oocyte. III. Selection and characterization of temperature-sensitive and -insensitive, recombination-deficient alleles in Drosophila. Genetics 108: 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M. T., and Aquadro C. F., 1999. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: A study of the second chromosome. Genetics 153: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Kohl K. P., McMahan S., Hartmann M. A., Williams A. M. et al. , 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27: 96–102. 10.1016/j.cub.2016.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., and Sekelsky J., 2017. Bloom syndrome helicase in meiosis: Pro-crossover functions of an anti-crossover protein. BioEssays 39: 1700073 10.1002/bies.201700073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil C. S. S., and Noor M. A. F., 2012. Zinc finger binding motifs do not explain recombination rate variation within or between species of Drosophila. PLoS One 7: e45055 10.1371/journal.pone.0045055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer L. W., Dias G., Smith B., Van Vaerenberghe K., Howard A. et al. , 2019. The meiotic recombination landscape of Drosophila virilis is robust to mitotic damage during hybrid dysgenesis. bioRxiv 342824; 10.1101/342824 [DOI] [Google Scholar]
- Hill T., and Betancourt A. J., 2018 . Extensive exchange of transposable elements in the Drosophila pseudoobscuragroup. Mob. DNA 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C. M., Huang W., Mackay T. F. C., and Singh N. D., 2016. The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet. 12: e1005951 10.1371/journal.pgen.1005951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. H., and Franklin F. C. H., 2006. Meiotic crossing-over: Obligation and interference. Cell 126: 246–248. 10.1016/j.cell.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Kent T. V., Uzunović J., and Wright S. I., 2017. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160458 10.1098/rstb.2016.0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G., 1972. Genetic change of recombination value in Drosophila melanogaster. I. Artificial selection for high and low recombination and some properties of recombination-modifying genes. Genetics 70: 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G., and Lisch D. R., 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15: 95–99. 10.1016/S0169-5347(99)01817-0 [DOI] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., and Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. 10.1126/science.1228190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., and Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. 10.1534/genetics.113.150581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Frigge M. L., Masson G., Gudbjartsson D. F. et al. , 2014. Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 46: 11–16. 10.1038/ng.2833 [DOI] [PubMed] [Google Scholar]
- Kulathinal R. J., Bennett S. M., Fitzpatrick C. L., and Noor M. A. F., 2008. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc. Natl. Acad. Sci. USA 105: 10051–10056. 10.1073/pnas.0801848105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Teeter K., Page S. L., Nielsen R., and Hawley R. S., 2007. A genetic analysis of the Drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176: 2151–2163. 10.1534/genetics.107.073551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D. et al. , 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24: 114–123. 10.1016/j.tig.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Lee Y. C. G., and Langley C. H., 2010. Transposable elements in natural populations of Drosophila melanogaster. Philos. Trans. R. Soc. B Biol. Sci. 365: 1219–1228. 10.1098/rstb.2009.0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., and Sandler L., 1977. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277: 295–312. 10.1098/rstb.1977.0019 [DOI] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Graham J., Nycz K., and McKim K. S., 2000. Two genes required for meiotic recombination in Drosophila are expressed from a dicistronic message. Genetics 154: 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manheim E. A., Jang J. K., Dominic D., and McKim K. S., 2002. Cytoplasmic Localization and Evolutionary Conservation of MEI-218, a Protein Required for Meiotic Crossing-over in Drosophila. Mol. Biol. Cell 13: 84–95. 10.1091/mbc.01-06-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Dahmus J. B., and Hawley R. S., 1996. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics 144: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., Hawley R. S., and McKim K. S., 2007. Synapsis, double-strand breaks, and domains of crossover control in Drosophila females, pp. 125–151 in Recombination and Meiosis, Genome Dynamics and Stability, edited by Egel R. and Lankenau D.-H.. Springer, Berlin, Heidelberg. [Google Scholar]
- Miller D. E., Smith C. B., Kazemi N. Y., Cockrell A. J., Arvanitakis A. V. et al. , 2016. Whole-Genome Analysis of Individual Meiotic Events in Drosophila melanogaster Reveals That Noncrossover Gene Conversions Are Insensitive to Interference and the Centromere Effect. Genetics 203: 159–171. 10.1534/genetics.115.186486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E., Charlesworth B., and Langley C. H., 1987. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet. Res. 49: 31–41. 10.1017/S0016672300026707 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1916. The mechanism of crossing-over. Am. Nat. 50: 193–221. 10.1086/279534 [DOI] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C. et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. 10.1126/science.1182363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z., Roach K. C. et al. , 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5: e1000753 10.1371/journal.pgen.1000753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D., Chang A. S., and Noor M. A. F., 2006. A recombinational portrait of the Drosophila pseudoobscura genome. Genet. Res. 87: 23–31. 10.1017/S0016672306007932 [DOI] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., and Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 12: 835 10.1126/science.1181495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak S. E., Hinds D. A., Koehler K., Nickel B., Patil N. et al. , 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37: 429–434. Erratum: 445. 10.1038/ng1529 [DOI] [PubMed] [Google Scholar]
- Ritz K. R., Noor M. A. F., and Singh N. D., 2017. Variation in Recombination Rate: Adaptive or Not? Trends Genet. TIG 33: 364–374. 10.1016/j.tig.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Sandor C., Li W., Coppieters W., Druet T., Charlier C. et al. , 2012. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 8: e1002854 10.1371/journal.pgen.1002854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségurel L., Leffler E. M., and Przeworski M., 2011. The case of the fickle fingers: How the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 9: e1001211 10.1371/journal.pbio.1001211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Brick K., Pu Y., Camerini-Otero R. D., and Petukhova G. V., 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30: 266–280. 10.1101/gad.270009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley C. E., and Kulathinal R. J., 2016. flyDIVaS: A Comparative Genomics Resource for Drosophila Divergence and Selection. G3(Bethesda). 6: 2355–2363. 10.1534/g3.116.031138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J., Feulner P. G. D., Johnston S. E., Santure A. W., and Smadja C. M., 2017. Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160455 Erratum 373: 20170360. 10.1098/rstb.2016.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., and Laurie C. C., 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A., and Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Wang S., Zickler D., Kleckner N., and Zhang L., 2015. Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14: 305–314. 10.4161/15384101.2014.991185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein A., 1936. The theory of multiple-strand crossing over. Genetics 21: 155–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. CABIOS 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W. S. W., and Nielsen R., 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., and Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway rhat Is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Fly stocks and transgenic constructs are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9162431.