Abstract
Oral squamous cell carcinoma (OSCC) is a major type of malignant tumor of the oral cavity. Despite marked advances in the management and diagnosis of OSCC, the associated overall survival ratio has only exhibited a modest increase in recent years. The present study aimed to identify potential crucial genes associated with clinical features and prognosis for OSCC, and to provide a basis for further investigation. RNA-sequencing data and corresponding clinical information were downloaded from The Cancer Genome Atlas database and differentially expressed mRNAs (DEmRNAs) were identified using the edgeR package. Bioinformatics analysis was performed to identify differentially expressed clinical features-associated mRNAs (CFmRNAs) and enhance the current knowledge of the function of them. Functional enrichment analysis and protein-protein interplay (PPI) network analysis were then performed to better understand CFmRNAs. Survival-associated genes were analyzed with Kaplan-Meier survival curves and the log-rank test. A total of 2,013 DEmRNAs between OSCC samples and normal tissues were identified, 180 of which were associated with clinical features. A total of 17 GO terms and 4 KEGG pathways were significantly enriched in functional enrichment analysis. A total of 4 hub genes (albumin, statherin, neurotensin and mucin 7) were identified in the PPI network analysis. A total of 6 genes (DDB1 and CUL4 associated factor 4 like 2, opiorphin prepropeptide, R3H domain containing like, transmembrane phosphatase with tensin homology, actin like 8 and protocadherin α 11) were observed to have an influence on survival. The DEmRNAs identified may have a crucial role in the genesis and development of OSCC and may be further developed for diagnostic, therapeutic and prognostic applications for OSCC in the future.
Keywords: oral squamous cell carcinoma, differentially expressed mRNAs, biomarkers, prognosis
Introduction
Oral squamous cell carcinoma (OSCC) is the sixth most common malignant tumor type worldwide, accounting for nearly 529,500 new cases and 292,300 deaths per year (1). Despite the advances in surgery, radiation therapy and chemotherapy, the long-term survival rate for OSCC has only improved by a small amount in the last three decades (1–3). Most OSCC patients are diagnosed at an advanced clinical stage, which is usually associated with poor prognosis (4,5). Therefore, it is essential and urgent to diagnose OSCC at an earlier stage and explore potential therapeutic targets.
Increasing evidence has demonstrated that multiple genetic and epigenetic alterations are responsible for the genesis and progression of OSCC. A study has indicated that α-protein kinase 1 (ALPK1) is associated with lymph node metastases and tumor development in OSCC, probably via regulation of cell growth, migration and invasion (6). Similarly, tumor-associated calcium signal transducer 2 (TROP2) was also determined to be involved in cell differentiation, TNM stage and vascular invasion (7). Although certain studies have reported on several genetic or epigenetic alterations contributing to OSCC, the molecular mechanisms of OSCC requires further elucidation.
Recent additions to the Cancer Genome Atlas (TCGA) provide abundant expression profiles from patient samples, as well as the associated clinicopathological data for >30 human cancer types (8,9). With the bioinformatics methods developed, integrated data analyses may be performed to further assist oncology research in several ways, including primary identification of dysregulated genes. In the present study, 2,013 differentially expressed (DE) mRNAs were identified from TCGA data. To further clarify the potential association between dysregulated mRNAs and clinical outcomes, 180 DEmRNAs were filtered according to the clinical features of patients [pathological N stage, T stage and pathological stage based on TNM staging system (10)], 26 of which were aberrantly expressed regarding all 3 different types of subgroup. Finally, 6 survival-associated genes [DDB1 and CUL4 associated factor 4 like 2 (DCAF4L2), opiorphin prepropeptide (OPRPN), R3H domain containing like (R3HDML), transmembrane phosphatase with tensin homology (TPTE), actin like 8 (ACTL8) and protocadherin α 11 (PCDHA11)] were identified from Kaplan-Meier survival curves with the log-rank test.
Materials and methods
Patients and samples
Original The Cancer Genome Atlas (TCGA) sequencing data and corresponding clinical information were obtained from TCGA database (https://portal.gdc.cancer.gov/) in October 2018. After excluding the patients without specific clinicopathological features (Tx and Nx), a total of 307 patients and 30 normal controls provided by TCGA dataset for OSCC were included. The clinical and pathological characteristics of the OSCC patients included are presented in Table I. No paired test was used to check whether the groups were age/sex-matched. Ethical approval was not required for the present study, since the data were downloaded from a publically available and open-ended TCGA database and did not involve primary data from human subjects.
Table I.
Clinicopathological characteristics of 307 patients with oral squamous cell carcinoma.
| Subtype | N (%) |
|---|---|
| Age (years) | |
| <60 | 132 (43) |
| ≥60 | 175 (57) |
| Sex | |
| Male | 207 (67.4) |
| Female | 100 (32.6) |
| Ethnicity | |
| White | 266 (86.6) |
| Black or African American | 21 (6.8) |
| Asian | 9 (2.9) |
| American Indian or Alaska native | 1 (0.3) |
| Not available | 10 (3.3) |
| Pathologic stage | |
| I | 19 (6.2) |
| II | 51 (16.6) |
| III | 54 (17.6) |
| IV | 160 (52.1) |
| Not available | 23 (7.5) |
| Pathologic T-stage | |
| T1 | 29 (9.4) |
| T2 | 94 (30.6) |
| T3 | 57 (18.6) |
| T4 | 109 (35.5) |
| TX | 9 (2.9) |
| Not available | 9 (2.9) |
| Pathologic N-stage | |
| N0 | 115 (37.5) |
| N1 | 46 (15) |
| N2 | 100 (32.6) |
| N3 | 2 (0.7) |
| NX | 34 (11.1) |
| Not available | 10 (3.3) |
| Pathologic M-stage | |
| M0 | 116 (37.8) |
| MX | 33 (10.7) |
| Not available | 158 (51.5) |
| Survival status | |
| Alive | 166 (54.1) |
| Dead | 141 (45.9) |
Analysis of differential expression profiles
DEmRNAs between OSCC and normal tissues were identified using the edgeR package from Bioconductor analysis tools (https://bioconductor.org/packages/release/bioc/html/edgeR.html). For multiple testing corrections, a false discovery rate (FDR) was applied to all P-values. The cut-off criteria were set as absolute |log2fold change (FC)≥2| and FDR<0.01.
Associations between DEmRNA and clinical features
The selected DEmRNAs were analyzed to determine whether they were differentially expressed in different clinical feature categories, including pathological T stage (T3 + T4 vs. T1 + T2), pathological N stage (N2 + N3 vs. N0 + N1) and pathological stage (stage III + IV vs. stage I + II). DEmRNAs with |log2FC|≥1.5 and FDR<0.05 were regarded as being significantly linked to the relevant clinical feature.
Functional enrichment analysis
Functional enrichment analysis was performed for clinical feature-associated DEmRNAs (CFmRNAs) to reveal the functional role of these mRNAs in the oncogenesis of OSCC. The Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/) was employed to perform such functional and pathway enrichment analyses. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were applied to determine the potential biological functions and pathways of the CFmRNAs (P<0.05).
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online database (http://string-db.org) is a search tool for identifying and describing PPIs via review of recorded interactions. This tool was used to analyze the PPIs of CFmRNAs. In addition, Cytoscape software (version 3.6.1; http://www.cytoscape.org/) was then utilized to visualize the PPI network of the CFmRNAs.
Survival analysis
For each CFmRNA, the OSCC patients were classified into either a high expression group or a low expression group. The median expression value of the respective CFmRNA was used as the cut-off value. The differences in survival time between the groups were then evaluated by Kaplan-Meier survival curves and the log-rank test to identify CFmRNAs that were significantly associated with survival in OSCC. The threshold for statistical significance was set as P<0.05.
Results
DEmRNAs in OSCC
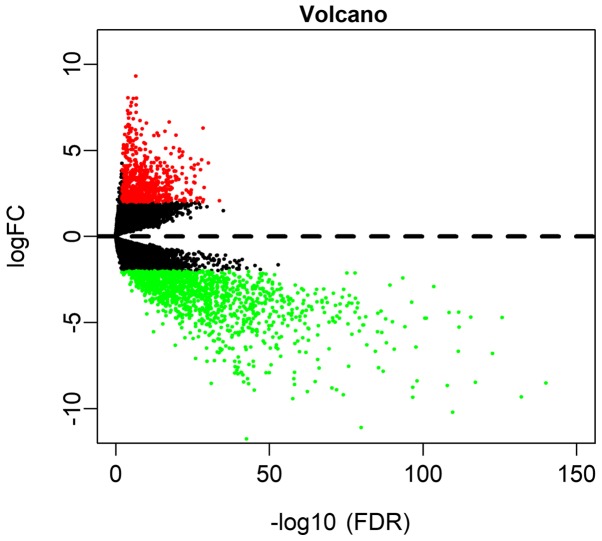
Based on the screening conditions as |log2FC|≥2 and FDR<0.01, a total of 675 (33.53%) upregulated and 1,338 (66.47%) downregulated genes were identified in OSCC (Table SI). A volcano plot was drawn to visualize the distribution of DEmRNAs between OSCC and normal controls (Fig. 1).
Figure 1.
The volcano plot of differentially expressed genes in oral squamous cell carcinoma. FDR, false discovery rate; FC, fold change.
DEmRNAs in association with clinical parameters
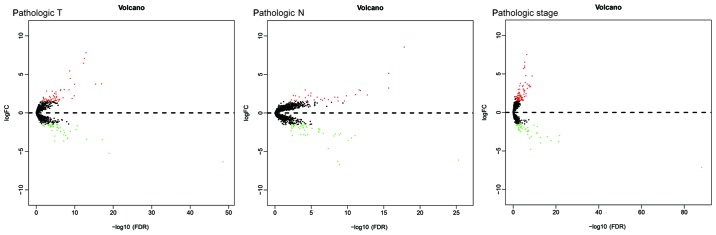
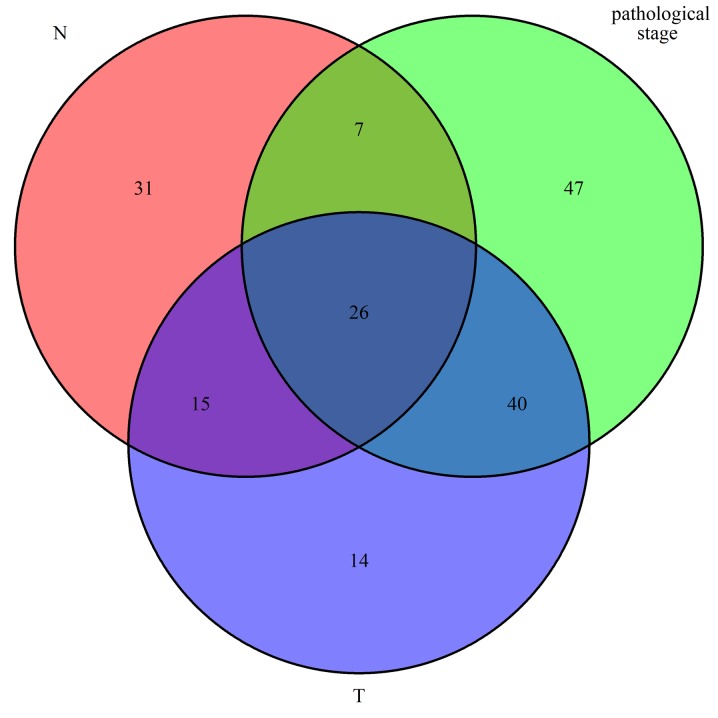
In order to further understand the association between dysregulated mRNAs and patient outcomes, associations between DEmRNAs and clinical characteristics, including pathologic T, pathologic N and pathologic stage, were analyzed. The expression levels of 180 DEmRNAs were significantly different in the clinical feature comparisons (|log2FC|≥1.5 and FDR<0.05). Among them, 95 mRNAs were differentially expressed between the pathologic T subgroups, 79 mRNAs were differentially expressed between the pathologic N subgroups and 120 mRNAs were differentially expressed between the pathologic stage subgroups (Table SII). The corresponding volcano plots are provided in Fig. 2. Finally, 26 mRNAs were screened out for all types of subgroups (Fig. 3).
Figure 2.
Volcano plot of differentially expressed genes in association with pathologic T stage, pathologic N stage and pathological stage in oral squamous cell carcinoma. FDR, false discovery rate; FC, fold change.
Figure 3.
Common differentially expressed genes in relation to clinical parameters in oral squamous cell carcinoma.
GO and pathway enrichment analysis
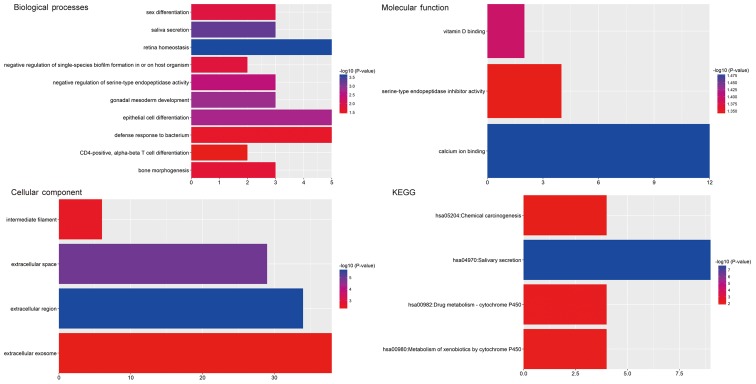
GO and pathway enrichment analyses of the identified CFmRNAs were performed using DAVID (Fig. 4). A total of 17 GO terms significantly enriched by the CFmRNAs were identified (Table II). In the GO category Biological Process, the dysregulated genes were particularly enriched in terms including retina homeostasis, saliva secretion, gonadal mesoderm development, epithelial cell differentiation and negative regulation of serine-type endopeptidase activity. In the Cellular Component category, the genes were enriched in the terms extracellular region, extracellular space, intermediate filament and extracellular exosome. In the GO category Molecular Function, the CFmRNAs were mainly enriched in calcium ion binding, vitamin D binding and serine-type endopeptidase inhibitor activity.
Figure 4.
Significantly enriched Gene Ontology terms in the categories Biological Process, Molecular Function and Cellular Component, as well as KEGG pathways regulated by differentially expressed genes in association with clinical parameters in oral squamous cell carcinoma. KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Table II.
Gene ontology analysis of differentially expressed genes in association with clinical parameters in oral squamous cell carcinoma.
| Category/term | N (%) | P-value | Genes |
|---|---|---|---|
| GOTERM_CC_DIRECT | |||
| GO:0005576~extracellular region | 34 (19.1) | 2.12×10−6 | GC, DCD, C7, BPIFB6, STATH, GAST, HTN3, TAC3, SPINK7, SPINK9, TTR, ALB, GP2, PIP, LTF, FGF3, PRB2, CRISP3, BPIFA2, SCUBE3, BPIFA1, OPRPN, R3HDML, DHRS7C, MUC6, IL22, PLG, NTS, C4ORF26, CST5, WIF1, SCGB3A2, SMR3A, DMBT1 |
| GO:0005615~extracellular space | 29 (16.3) | 1.12×10−5 | GC, DCD, BPIFB2, PRH2, MSMB, C6ORF58, SPINK1, GAST, OSTN, SCGB1A1, TAC3, ZG16B, TTR, ALB, PIP, LTF, TFF1, FGFBP2, CRISP3, BPIFA1, OPRPN, IL22, PLG, CST5, SCGB3A1, EPYC, MUC5B, DMBT1, SMR3B |
| GO:0005882~intermediate filament | 6 (3.4) | 0.002961 | KRT36, KRT27, KRT40, KRTAP17-1, KRTAP19-5, KRTAP13-2 |
| GO:0070062~extracellular exosome | 38 (21.3) | 0.004723 | GC, DCD, C7, BPIFB2, PKHD1, C6ORF58, ALDOB, AQP5, SPINK1, CRNN, CALB1, SCGB1A1, TAC3, ZG16B, TTR, KRT27, ALB, UPK1A, GP2, PIP, LTF, TGM3, KRT84, CRISP3, GSTA2, BPIFA2, VIL1, ARMC3, MUC7, PLG, LRRC26, KRT36, PYGM, CST5, SCGB3A1, MUC5B, DMBT1, SMR3B |
| GOTERM_BP_DIRECT | |||
| GO:0001895~retina homeostasis | 5 (2.8) | 2.38×10−4 | ZG16B, ALB, PIP, LTF, OPRPN |
| GO:0046541~saliva secretion | 3 (1.7) | 5.68×10−4 | CHRM1, STATH, AQP5 |
| GO:0007506~gonadal mesoderm development | 3 (1.7) | 0.001567 | TSPY2, TSPY1, TSPY10 |
| GO:0030855~epithelial cell differentiation | 5 (2.8) | 0.002003 | GSTA2, UPK1A, VIL1, DMBT1, ACTL8 |
| GO:1900004~negative regulation of serine-type endopeptidase activity | 3 (1.7) | 0.00362 | SPINK9, SPINK1, SPINK7 |
| GO:0007548~sex differentiation | 3 (1.7) | 0.01426 | TSPY1, DMRT1, TSPY10 |
| GO:1900229~negative regulation of single-species biofilm formation in or on host organism | 2 (1.1) | 0.015188 | BPIFA1, LTF |
| GO:0060349~bone morphogenesis | 3 (1.7) | 0.017868 | LTF, IFITM5, PAX1 |
| GO:0042742~defense response to bacterium | 5 (2.8) | 0.02499 | DCD, BPIFA2, STATH, HTN3, MUC5B |
| GO:0043367~CD4-positive, alpha-beta T cell differentiation | 2 (1.1) | 0.037541 | PAX1, NKX2-3 |
| GOTERM_MF_DIRECT | |||
| GO:0005509~calcium ion binding | 12 (6.7) | 0.033232 | MYL7, SCUBE3, ANXA10, MYL3, CAPSL, RPTN, VIL1, TGM3, PCDHA11, CRNN, CALB1, NECAB2 |
| GO:0005499~vitamin D binding | 2 (1.1) | 0.040216 | GC, CALB1 |
| GO:0004867~serine-type endopeptidase inhibitor activity | 4 (2.2) | 0.045216 | SPINK9, SPINK1, TFPI2, SPINK7 |
GO, Gene Ontology.
The results of the KEGG pathway enrichment analysis are displayed in Fig. 4. The most significant pathways associated with CFmRNAs were salivary secretion, drug metabolism-cytochrome P450, metabolism of xenobiotics by cytochrome P450 and chemical carcinogenesis.
Key candidate genes identified from the CFmRNA PPI network
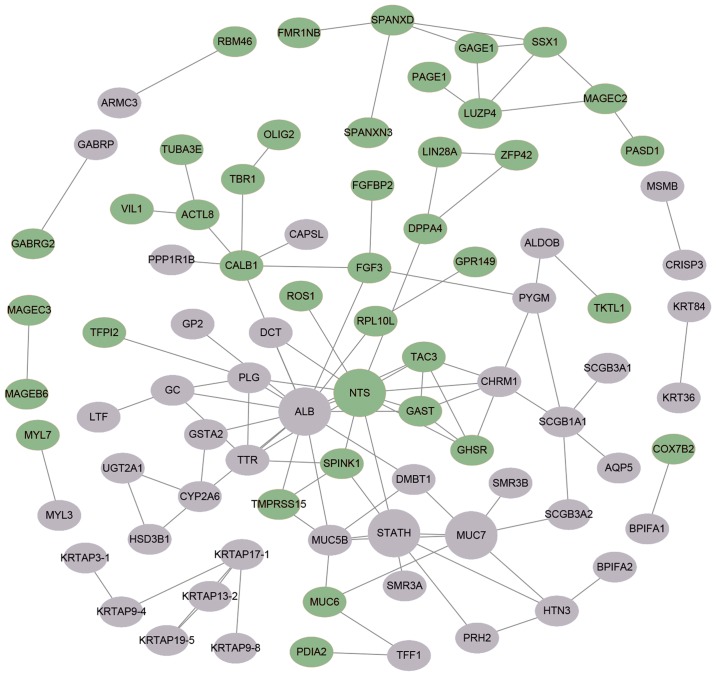
The STRING online database was used to further investigate the potential links between abnormally expressed genes. A PPI network was visualized by Cytoscape and is provided in Fig. 5. A total of 4 central node genes (>7 connections/interactions) were identified, including albumin (ALB), statherin (STATH), neurotensin (NTS) and mucin 7, secreted (MUC7).
Figure 5.
Protein-protein interaction network of differentially expressed genes in association with clinical parameters in oral squamous cell carcinoma constructed with the Search Tool for the Retrieval of Interacting Genes/Proteins database. Upregulated genes are represented as green ellipses and downregulated genes as gray ellipses. Hub genes are provided as larger circles.
CFmRNAs associated with survival
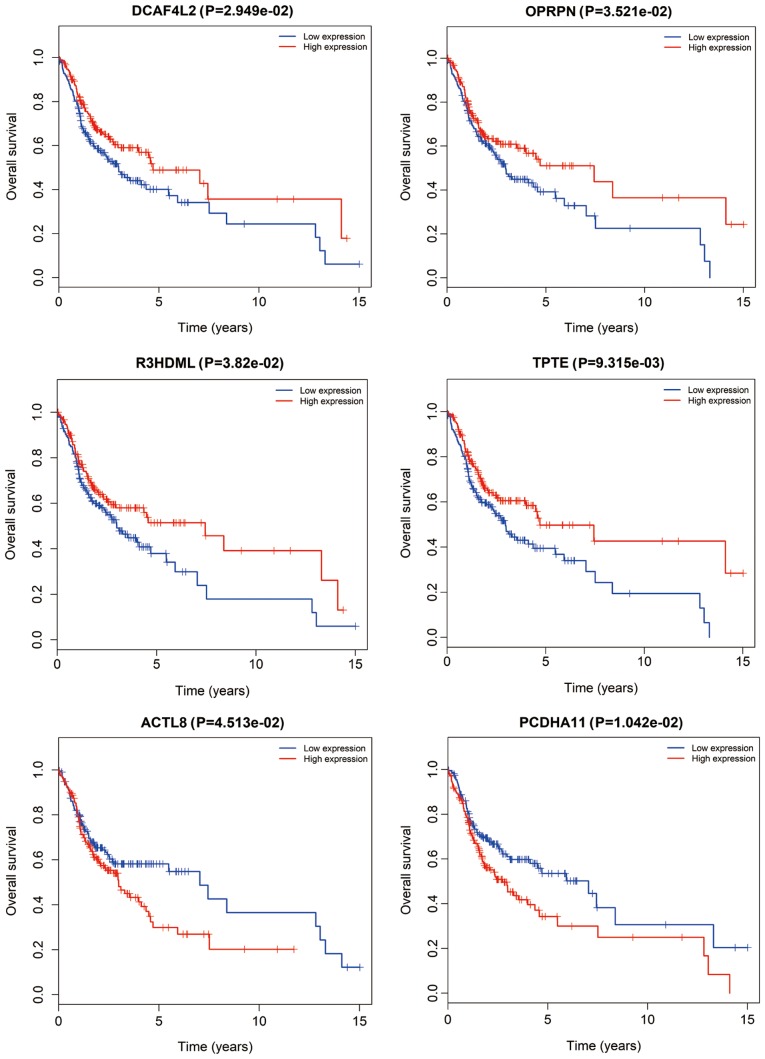
CFmRNAs associated with survival were detected by Kaplan-Meier survival curves and log-rank tests. A total of 6 CFmRNAs were identified to have a significant influence on the overall survival rate (P<0.05). Of these, 4 (DCAF4L2, OPRPN, R3HDML and TPTE) were associated with a favorable prognosis regarding overall survival (Fig. 6). Patients with higher expression of these mRNAs had a tendency for longer survival, compared with patients with lower expression (P<0.05). ACTL8 and PCDHA11 were associated with an elevated risk, since their higher expression was associated with shorter overall survival of OSCC patients.
Figure 6.
Kaplan-Meier survival curves of six genes associated with overall survival in oral squamous cell carcinoma. The horizontal axis represents overall survival time (years) and the vertical axis represents the survival function. DCAF4L2, DDB1 and CUL4 associated factor 4 like 2; OPRPN, opiorphin prepropeptide; R3HDML, R3H domain containing like; TPTE, transmembrane phosphatase with tensin homology; ACTL8, actin like 8; PCDHA11, protocadherin alpha 11.
Discussion
In spite of diagnostic and therapeutic advances in the last decades, OSCC remains a huge threat to human health. With the rapidly developed technology for gene analysis, in-depth exploration of the molecular characteristics and identification of potential valuable prognostic biomarkers for OSCC may be performed (11). The multi-step progression of carcinogenesis in the oral cavity, including gene amplification or dysregulation, has an important role in OSCC (7). In the present study, 675 upregulated and 1,338 downregulated mRNAs between OSCC and normal samples were initially identified. Of these DEmRNAs, 180 were screened out according to their association with clinical features. Furthermore, 6 survival-associated genes were identified (DCAF4L2, OPRPN, R3HDML, TPTE, ACTL8 and PCDHA11).
In the GO and KEGG analyses, 17 enriched terms and 4 signaling pathways were identified. They were involved in several relevant processes, particularly in saliva secretion and metabolism by cytochrome P450. The epithelium of the oral cavity is continuously immersed in saliva and its associated compositions, and thus, changes in the saliva composition may particularly affect the state of oral tissues (12).
The PPI network of the CFmRNAs comprised four hub genes (ALB, STATH, NTS and MUC7). STATH is derived from a secretory calcium-binding phosphoprotein gene cluster and encodes the protein statherin. It was reported to be responsible for preventing calcium phosphate precipitation in saliva, leading to persistent high calcium and phosphate levels (13,14). The levels of statherin in saliva are significantly reduced in cancerous and pre-cancerous lesions compared with those in healthy controls (15). Decreased expression of STATH means less free calcium, perhaps resulting in initiation of sustained proliferation and reduction of desmosomes, as well as alterations in calcium-associated cellular functions. Dysfunction of this gene may be a possible mechanism of carcinogenesis in OSCC.
Overexpression of NTS and its affinity receptor NTSR1 are frequently detected in cancerous tissues (16,17). Numerous studies have reported that NTS is associated with tumor progression in several solid tumor types, including breast cancer, lung cancer and hepatocellular carcinoma (HCC) (18–20). Recent studies on HCC demonstrated that NTS/NTSR1 co-expression promotes tumor invasion and epithelial-mesenchymal transition via Wnt/β-catenin signaling and the NTS/interleukin-8 pathway (21,22). However, further evidence is necessary to prove the role of NTS in the progression of OSCC.
Mucins are multifunctional glycoproteins, which may be divided into membrane-bound and secreted protein (23). MUC7 is one of these secreted mucins identified in the PPI network of the screened CFmRNAs. Higher expression of MUC7 was previously reported to be associated with poor overall survival in clear-cell renal cell carcinoma (24). In the oral cavity, MUC7 acts as an innate immunity component, interacting with several oral microorganisms (25). MUC7 is typically distributed in normal salivary glands, while MUC7 is rarely expressed in malignant transformations into mucoepidermoid carcinoma (23,26). The secreted proteins MUC5B and MUC6 were identified to be dysregulated among the pathological T, N and stage subgroups in the present study. A previous study suggested that MUC5B may be associated with poor prognosis regarding survival in lung cancer (27). The also associated MUC1 belongs to the transmembrane mucins. It has been indicated that MUC1 is a reliable biomarker to predict the metastatic/invasive potential of OSCC (28,29). However, further research is required to elucidate the detailed function of MUC7 in OSCC.
From the 180 CFmRNAs, six mRNAs were identified that were able to predict the survival probability. DCAF4L2, OPRPN, R3HDML and TPTE were regarded as protective, as their expression was associated with consistently increased survival rates. DCAF4L2 is a member of the WD-repeat proteins, which commonly mediates protein-protein interlay (30), and its expression is a potential cause of cleft lip with or without cleft palate (31). In the present study, DCAF4L2 expression was elevated in tumor tissues, but was also associated with an increased survival rate. This may due to increases in the expression of DCAF4L2, which promotes polyubiquitination of protein phosphatase, Mg2+/Mn2+-dependent 1B (PPM1B) and then decreases in the totality of PPM1B (32). Reduced PPM1B in U2OS cells combined with anti-cancer drugs suppresses cellular proliferation of these cells and lowers the threshold of cell death (33). In colorectal cancer, upregulated DCAF4L2 induces neoplastic cell invasion and metastasis (32). OPRPN (also known as PROL1) localizes on chromosome 4q13.3, encoding basic proline-rich lacrimal protein (34,35). In addition, dysregulated OPRPN was reported to be associated with invasion in breast cancer (36).
TPTE and ACTL8 belong to the cancer-testis antigens (CTAs). In the present study, elevated TPTE indicated longer overall survival, while higher expression of ACTL8 predicted a worse prognosis. Similarly, prolonged survival of TPTE-seropositive lung cancer patients was reported in a previous study (37). CTAs consist of tumor-associated antigens that are widely distributed in tumors, and are prospective targets for cancer immunotherapy (38). Previous studies have already proved that CT genes are more frequently expressed in invasive head and neck squamous cell carcinoma (HNSCC) and are associated with an unfavorable phenotype (39,40). Furthermore, the proliferation and migration of HNSCC PCI-12 cells were significantly inhibited by knockdown of ACTL8 (41). On the contrary, co-expression of ACTL8, CCCTC-binding factor like, Opa interacting protein 5 and X antigen family member 3 may be associated with increased overall survival in glioblastoma patients (42). Taken together, these results suggest the prospect of CT genes as prognostic biomarkers that may be useful in the development of vaccine treatments based on CTAs (43).
At present, little is known regarding the role of dysregulated R3HDML and PCDHA11 in various diseases, including OSCC, and further research is required to investigate their potential molecular mechanisms. Of note, the present study has certain limitations. For instance, it lacks corresponding experiments to validate the in silico results; therefore, further experiments, including quantitative PCR analysis, are required to confirm the association of the genes identified with OSCC in future studies. Furthermore, a paired-test was not performed to assess whether the groups were age/sex-matched in the current study, which needs to be taken in to account in further studies.
In conclusion, six differently expressed genes linked to oncogenesis and development were identified to be associated different clinical features in the present study. These genes (DCAF4L2, OPRPN, R3HDML, TPTE, ACTL8 and PCDHA11) may serve as potential prognostic markers for OSCC. Although further experimental verification is imperative, these results may provide a basis for in-depth study regarding the diagnosis, therapy and prognosis of OSCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QW, XX and RC designed the study and wrote the manuscript. RC collected and analyzed the data. JC helped to interpret the results. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shimomura H, Sasahira T, Nakashima C, Shimomura-Kurihara M, Kirita T. Downregulation of DHRS9 is associated with poor prognosis in oral squamous cell carcinoma. Pathology. 2018;50:642–647. doi: 10.1016/j.pathol.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Sandulache VC, Michikawa C, Kataria P, Gleber-Netto FO, Bell D, Trivedi S, Rao X, Wang J, Zhao M, Jasser S, et al. High-risk TP53 mutations are associated with extranodal extension in oral cavity squamous cell carcinoma. Clin Cancer Res. 2018;24:1727–1733. doi: 10.1158/1078-0432.CCR-17-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng Q, Deng Z, Pan H, Gu L, Liu O, Tang Z. Mitogen-activated protein kinase signaling pathway in oral cancer (Review) Oncol Lett. 2018;15:1379–1388. doi: 10.3892/ol.2017.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol. 2015;6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Sun S, Zeng X, Cui L. Expression profiles analysis identifies a novel three-mRNA signature to predict overall survival in oral squamous cell carcinoma. Am J Cancer Res. 2018;8:450–461. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen PK, Hua CH, Hsu HT, Kuo TM, Chung CM, Lee CP, Tsai MH, Yeh KT, Ko YC. ALPK1 expression is associated with lymph node metastasis and tumor growth in oral squamous cell carcinoma patients. Am J Pathol. 2019;189:190–199. doi: 10.1016/j.ajpath.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Tang G, Tang Q, Jia L, Xia S, Li J, Chen Y, Li H, Ding X, Wang F, Hou D, et al. High expression of TROP2 is correlated with poor prognosis of oral squamous cell carcinoma. Pathol Res Pract. 2018;214:1606–1612. doi: 10.1016/j.prp.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Liu J, Fu X, Yang A. Identification of key genes and pathways in tongue squamous cell carcinoma using bioinformatics analysis. Med Sci Monit. 2017;23:5924–5932. doi: 10.12659/MSM.905035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Niu L, Jiang S, Zhai J, Wang P, Kong F, Jin X. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7:86174–86185. doi: 10.18632/oncotarget.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, Loomis AM, Shah JP. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:122–137. doi: 10.3322/caac.21389. [DOI] [PubMed] [Google Scholar]
- 11.Gao C, Zhuang J, Zhou C, Ma K, Zhao M, Liu C, Liu L, Li H, Feng F, Sun C. Prognostic value of aberrantly expressed methylation gene profiles in lung squamous cell carcinoma: A study based on The cancer genome atlas. J Cell Physiol. 2019;234:6519–6528. doi: 10.1002/jcp.27389. [DOI] [PubMed] [Google Scholar]
- 12.Sarode SC, Sarode GS, Patil S. Role of statherin in oral carcinogenesis. Oral Oncol. 2014;50:e55–e56. doi: 10.1016/j.oraloncology.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 13.de Sousa-Pereira P, Amado F, Abrantes J, Ferreira R, Esteves PJ, Vitorino R. An evolutionary perspective of mammal salivary peptide families: Cystatins, histatins, statherin and PRPs. Arch Oral Biol. 2013;58:451–458. doi: 10.1016/j.archoralbio.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Sabatini LM, Carlock LR, Johnson GW, Azen EA. cDNA cloning and chromosomal localization (4q11-13) of a gene for statherin, a regulator of calcium in saliva. Am J Hum Genet. 1987;41:1048–1060. [PMC free article] [PubMed] [Google Scholar]
- 15.Contucci AM, Inzitari R, Agostino S, Vitali A, Fiorita A, Cabras T, Scarano E, Messana I. Statherin levels in saliva of patients with precancerous and cancerous lesions of the oral cavity: A preliminary report. Oral Dis. 2005;11:95–99. doi: 10.1111/j.1601-0825.2004.01057.x. [DOI] [PubMed] [Google Scholar]
- 16.Souazé F, Dupouy S, Viardot-Foucault V, Bruyneel E, Attoub S, Gespach C, Gompel A, Forgez P. Expression of neurotensin and NT1 receptor in human breast cancer: A potential role in tumor progression. Cancer Res. 2006;66:6243–6249. doi: 10.1158/0008-5472.CAN-06-0450. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Galmiche A, Liu J, Stadler N, Wendum D, Segal-Bendirdjian E, Paradis V, Forgez P. Neurotensin regulation induces overexpression and activation of EGFR in HCC and restores response to erlotinib and sorafenib. Cancer Lett. 2017;388:73–84. doi: 10.1016/j.canlet.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 18.Morgat C, Mishra AK, Varshney R, Allard M, Fernandez P, Hindié E. Targeting neuropeptide receptors for cancer imaging and therapy: Perspectives with bombesin, neurotensin, and neuropeptide-Y receptors. J Nucl Med. 2014;55:1650–1657. doi: 10.2967/jnumed.114.142000. [DOI] [PubMed] [Google Scholar]
- 19.Dupouy S, Viardot-Foucault V, Alifano M, Souazé F, Plu-Bureau G, Chaouat M, Lavaur A, Hugol D, Gespach C, Gompel A, Forgez P. The neurotensin receptor-1 pathway contributes to human ductal breast cancer progression. PLoS One. 2009;4:e4223. doi: 10.1371/journal.pone.0004223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Younes M, Wu Z, Dupouy S, Lupo AM, Mourra N, Takahashi T, Fléjou JF, Trédaniel J, Régnard JF, Damotte D, et al. Neurotensin (NTS) and its receptor (NTSR1) causes EGFR, HER2 and HER3 over-expression and their autocrine/paracrine activation in lung tumors, confirming responsiveness to erlotinib. Oncotarget. 2014;5:8252–8269. doi: 10.18632/oncotarget.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y, Long X, Zhang L, Chen J, Liu P, Li H, Wei F, Yu W, Ren X, Yu J. NTS/NTR1 co-expression enhances epithelial-to-mesenchymal transition and promotes tumor metastasis by activating the Wnt/β-catenin signaling pathway in hepatocellular carcinoma. Oncotarget. 2016;7:70303–70322. doi: 10.18632/oncotarget.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao P, Long X, Zhang L, Ye Y, Guo J, Liu P, Zhang R, Ning J, Yu W, Wei F, Yu J. Neurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of Tumor-Associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncoimmunology. 2018;7:e1440166. doi: 10.1080/2162402X.2018.1440166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alos L, Lujan B, Castillo M, Nadal A, Carreras M, Caballero M, de Bolos C, Cardesa A. Expression of membrane-bound mucins (MUC1 and MUC4) and secreted mucins (MUC2, MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of salivary glands. Am J Surg Pathol. 2005;29:806–813. doi: 10.1097/01.pas.0000155856.84553.c9. [DOI] [PubMed] [Google Scholar]
- 24.NguyenHoang S, Liu Y, Xu L, Chang Y, Zhou L, Liu Z, Lin Z, Xu J. High mucin-7 expression is an independent predictor of adverse clinical outcomes in patients with clear-cell renal cell carcinoma. Tumour Biol. 2016;37:15193–15201. doi: 10.1007/s13277-016-5375-5. [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Pavlidis P, Thamadilok S, Redwood E, Fox S, Blekhman R, Ruhl S, Gokcumen O. Recent evolution of the salivary mucin MUC7. Sci Rep. 2016;6:31791. doi: 10.1038/srep31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahomed F. Recent advances in mucin immunohistochemistry in salivary gland tumors and head and neck squamous cell carcinoma. Oral Oncol. 2011;47:797–803. doi: 10.1016/j.oraloncology.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Nagashio R, Ueda J, Ryuge S, Nakashima H, Jiang SX, Kobayashi M, Yanagita K, Katono K, Satoh Y, Masuda N, et al. Diagnostic and prognostic significances of MUC5B and TTF-1 expressions in resected non-small cell lung cancer. Sci Rep. 2015;5:8649. doi: 10.1038/srep08649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar MH, Sanjai K, Kumarswamy J, Keshavaiah R, Papaiah L, Divya S. Expression of MUC1 mucin in potentially malignant disorders, oral squamous cell carcinoma and normal oral mucosa: An immunohistochemical study. J Oral Maxillofac Pathol. 2016;20:214–218. doi: 10.4103/0973-029X.185916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakur A, Tupkari JV, Joy T, Kende PP, Siwach P, Ahire MS. Expression of mucin-1 in oral squamous cell carcinoma and normal oral mucosa: An immunohistochemical study. J Oral Maxillofac Pathol. 2018;22:210–215. doi: 10.4103/jomfp.JOMFP_188_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: Novel targets for anticancer therapies. J Clin Oncol. 2013;31:1231–1238. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–781. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Chen Y, Han J, Meng Q, Xi Q, Wu G, Zhang B. DCAF4L2 promotes colorectal cancer invasion and metastasis via mediating degradation of NFκB negative regulator PPM1B. Am J Transl Res. 2016;8:405–418. [PMC free article] [PubMed] [Google Scholar]
- 33.Miller RE, Uwamahoro N, Park JH. PPM1B depletion in U2OS cells supresses cell growth through RB1-E2F1 pathway and stimulates bleomycin-induced cell death. Biochem Biophys Res Commun. 2018;500:391–397. doi: 10.1016/j.bbrc.2018.04.084. [DOI] [PubMed] [Google Scholar]
- 34.Saitoh E, Taniguchi M, Ochiai A, Kato T, Imai A, Isemura S. Bioactive peptides hidden in human salivary proteins. J Oral Biosciences. 2017;59:71–79. doi: 10.1016/j.job.2016.11.005. [DOI] [Google Scholar]
- 35.Saitoh E, Sega T, Imai A, Isemura S, Kato T, Ochiai A, Taniguchi M. The PBII gene of the human salivary proline-rich protein P-B produces another protein, Q504X8, with an opiorphin homolog, QRGPR. Arch Oral Biol. 2018;88:10–18. doi: 10.1016/j.archoralbio.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Lang Z, Wu Y, Pan X, Qu G, Zhang T. Study of differential gene expression between invasive multifocal/multicentric and unifocal breast cancer. J BUON. 2018;23:134–142. [PubMed] [Google Scholar]
- 37.Kuemmel A, Simon P, Breitkreuz A, Röhlig J, Luxemburger U, Elsässer A, Schmidt LH, Sebastian M, Sahin U, Türeci Ö, Buhl R. Humoral immune responses of lung cancer patients against the Transmembrane phosphatase with TEnsin homology (TPTE) Lung Cancer. 2015;90:334–341. doi: 10.1016/j.lungcan.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Yao J, Caballero OL, Yung WK, Weinstein JN, Riggins GJ, Strausberg RL, Zhao Q. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res. 2014;2:371–379. doi: 10.1158/2326-6066.CIR-13-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuffel C, Rivals JP, Zaugg Y, Salvi S, Seelentag W, Speiser DE, Liénard D, Monnier P, Romero P, Bron L, Rimoldi D. Pattern and clinical significance of cancer-testis gene expression in head and neck squamous cell carcinoma. Int J Cancer. 2011;128:2625–2634. doi: 10.1002/ijc.25607. [DOI] [PubMed] [Google Scholar]
- 40.Piotti KC, Scognamiglio T, Chiu R, Chen YT. Expression of cancer/testis (CT) antigens in squamous cell carcinoma of the head and neck: Evaluation as markers of squamous dysplasia. Pathol Res Pract. 2013;209:721–726. doi: 10.1016/j.prp.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Zhu J, Meng L. High expression of ACTL8 is poor prognosis and accelerates cell progression in head and neck squamous cell carcinoma. Mol Med Rep. 2019;19:877–884. doi: 10.3892/mmr.2018.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freitas M, Malheiros S, Stávale JN, Biassi TP, Zamunér FT, de Souza Begnami M, Soares FA, Vettore AL. Expression of cancer/testis antigens is correlated with improved survival in glioblastoma. Oncotarget. 2013;4:636–646. doi: 10.18632/oncotarget.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoud AM. Cancer testis antigens as immunogenic and oncogenic targets in breast cancer. Immunotherapy. 2018;10:769–778. doi: 10.2217/imt-2017-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.