Abstract
Epitranscriptomic modification of mRNA affects its metabolism and has recently been shown to regulate brain development. Two studies in this issue of Neuron, Koranda et al. (2018) and Engel et al. (2018), uncover dynamic and critical roles of m6A/m RNA modifications in the adult mammalian brain in regulating physiological and stress-induced behaviors.
The major functions of our brain, such as learning, memory, emotion, cognition, and motor control, depend on the ability of neurons to modify their functional properties or their connections in an activity-dependent manner. Experience-driven neuronal activity induces a complex program of gene expression, which facilitates changes to neural circuits by modulating synaptic development and connectivity. Thus, knowledge of regulatory mechanisms that control gene expression is crucial to understand the dynamic nature of brain functions. For example, neuronal activity can reshape the epigenetic landscape through modifications of DNA and histones to alter the responsiveness of neurons to environmental stimuli (Cholewa-Waclaw et al., 2016). In addition to transcriptional regulation by epigenetic mechanisms, recent evidence suggests that various chemical modifications on messenger RNA (mRNA) can affect almost every aspect of mRNA metabolism at the post-transcriptional level, including decay, transport, splicing, and translation (Zhao et al., 2017). New high-throughput sequencing approaches have begun to reveal a dynamic epitranscriptome landscape for many mRNA modifications in various organisms, such as pseudouridine (ψ), 2’-O-methylation (2’OMe), 5-methylcytidine (m5C), and N6-methyladenosine (m6A). Among these, m6A is the most abundant internal modification in mRNAs of eukaryotic cells. Until now, several studies have highlighted epitranscriptomic regulation of neurodevelopment (Yoon et al., 2018), but little was known about its potential role in modulating synaptic function and behavior in adult animals. Two studies in this issue of Neuron reveal the crucial role of m6A/m RNA modifications in physiological brain functions and stress-induced responses and behavior in vivo (Figure 1) (Koranda et al., 2018; Engel et al., 2018).
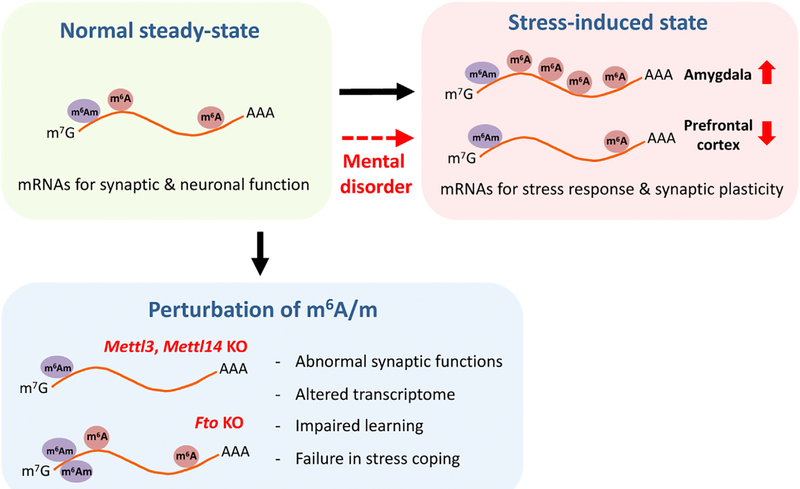
Figure 1. Dynamic m6A/m Epitranscriptomes in the Adult Mammalian Brain.
The landscape of the m6A/m epitranscriptome is actively modified during the stress response, which is compromised in mental disorders. Genetic manipulations of m6A writers and an eraser lead to dysfunctions in synaptic activities and animal behaviors, revealing the in vivo role of m6A RNA modification in the adult mammalian.
In mammals, m6A is installed by the methyltransferase complex consisting of Mettl3, Mettl14, WTAP, KIAA1429, and RBM15/RBM15B (writers) and is removed by demethylases FTO and ALKBH5 (erasers) (Zhao et al., 2017). FTO also facilitates demethylation of N6, 2’-O-di-methyladenosine (m6Am) with a higher affinity than m6A in vitro (Mauer et al., 2017). These genes are expressed both during brain development and in the adult. In both studies (Koranda et al., 2018; Engel et al., 2018), the authors mapped the steady-state m6A/m profiles in the adult mouse cortex and striatum using the m6A-seq method, which detects both m6A and m6Am modifications by using modification-specific antibodies (collectively referred to as m6A/m). m6A/m peaks are preferentially located to the 5’ UTR and around the stop codon with an enrichment of transcripts related to neuronal and synaptic regulation, such as the neuronal activity-induced gene Arc. Furthermore, Engel et al. (2018) investigated the effect of acute stress on the m6A/m landscape in the adult mouse brain. Acute restraint stress changes the global level of m6A/m, potentially by altering expression of its readers and writers. Interestingly, the stress-induced changes in the m6A/m epitranscriptome are brain region specific, since the global level of m6A/m is decreased in the prefrontal cortex but increased in the amygdala. Moreover, similar global levels of m6A/m changes occur in these brain regions upon ectopic stimulation with corticosteroid, a major mediator of the stress response. Quantitative assessment of m6A/m modifications at the individual transcripts (using m6A/m-RIP-qPCR) revealed progressive changes of stress-related and synaptic plasticity-related transcripts after acute stress. Together, these results suggest that the m6A/m epitranscriptome is dynamic and responds to external environmental stimuli in a brain-region- and gene-specific manner.
These two studies provide key insignt into the in vivo physiological roles of m6A/m RNA modifications by analyzing animal physiology and behavior using sophisticated loss-of-function models. Koranda et al. (2018) showed that conditional deletion of a m6A writer Mettl14 in striato-nigral neurons impaires striatalmediated learning and alters dopamine signaling. Mettl14-deficient striato-nigral neurons also exhibit altered neuronal excitability but no change in the number and morphology of neurons in the striatum. Engel et al. (2018) utilized inducible or adult excitatory neuron-specific knockout models of a m6A writer Mettl3 and a m6A/m eraser Fto, focusing on stress-induced responses. Pertubation of the m6A/m epitranscriptome after the conditional ablation of these genes in the adult resulted in abnormal stress-coping behaviors, such as increased fear memory during memory extinction. The Fto knockout mice also exhibited impaired synaptic plasticity. In both studies, transcriptome analyses in these animal models identified significant and specific gene expression changes in the steady-state or stress-induced responses. Collectively, these results reveal the physiological significance of the dynamic m6A/m epitranscriptome in synaptic function and behavior at the organism level.
Engel et al. (2018) also explored the potential association between m6A/m epitranscriptome and stress-related mental disorders. It has been previously suggested that m6A biology might be linked to mental disorders (Yoon et al., 2017). Engel et al. (2018) tested human patient blood samples as a peripheral indicator of the central nervous system response to stress. Interestingly, the global level of m6A/m in the blood is downregulated after acute treatment of corticosteroid. They then examined the m6A/m epitranscriptome of patients with major depressive disorder (MDD). Although the steady-state level of m6A/m was similar to healthy controls, corticosteroid treatment did not reduce the m6A/m content in blood samples from MDD patients. Next, B lymphocyte cell lines isolated from healthy control and MDD patients subjected to m6A-seq revealed the specific signature of m6A/m dysregulation with an enrichment of genes related to stress responses. While analyses were performed with a limited number of patient samples, these intriguing results raise the possibility that disturbed epitranscriptomic regulation may underlie the development of some psychiatric disorders. Future studies, such as those using induced pluripotent stem cells from mental disorder patients, could provide mechanistic information about how dysregulation of epitranscriptomes may cause various human psychiatric disorders.
While we are still in the early stages of trying to understand the nature, function, and mechanism of dynamic epitranscriptomic regulation, several recent studies, including the two in this issue, have begun to reveal several domains of epitranscriptomic influence in the nervous system. A number of studies have identified a critical role of m6A signaling in regulating neurogenesis via controlling mRNA decay during brain development (Yoon et al., 2018). In the mature nervous system, local protein translation contributes to synaptic function under both physiological and pathological conditions, and fine-tuning of gene expression at synapses in a post-transcriptional manner is an effective and rapid way to modulate neural circuitry activity. Indeed, m6A-tagged mRNAs are abundantly present in the synaptic compartment with enrichment in synaptic pathways. Such synaptic m6A epitranscriptome is interpreted by dendritically localized m6A readers to regulate dendrite development and synaptic transmission (Merkurjev et al., 2018). Therefore, specific m6A tagging may control a set of synaptic transcripts collectively and flexibly upon neuronal activity. On the other hand, the m6A epitranscriptome dynamically changes its landscape in response to pathological stimuli in the adult nervous system, similar to what was shown in Engel et al (2018). For example, nerve injury acutely elevates m6A-tagged mRNAs related to regeneration-association genes to enhance the global efficiency of protein translation, which is essential for functional axon regeneration (Weng et al., 2018). Taken together, m6A/m RNA modification-mediated transcriptome plasticity may confer more flexible and finely tuned responses of the nervous system upon physiological and pathological stimuli.
In the emerging field of epitranscriptomics, we are still limited by available technologies. Epitranscriptome profling used in both of the studies was based on the enrichment of RNA fragments by modified nucleotide-specific antibodies, which actually detect both m6A and m6Am (Koranda et al., 2018; Engel et al., 2018). Because sequencing reads the entire RNA fragments, the peaks from this technique are broad and overlapped, therefore making it difficult to distinguish m6A from m6Am. More advanced techniques, such as miCLIP-seq or third-generation sequencing, will help to identify the exact sites of m6A/m modifications with a single base pair resolution in the adult brain (Grozhik and Jaffrey, 2018). Given that the epitranscriptomic landscape differs across brain regions (Engel et al., 2018) and even subcellular compartments (Merkurjev et al., 2018), more sensitive epitranscriptome profling with region, cell-type, subcellular specificity will be required to further understand the diversity of epitranscriptomic regulations involved in brain functions. The current studies also raise many interesting questions for the future. How does the stress response alter the epitranscriptome and how is transcript specificity achieved? What are the downstream mechanisms responsible for m6A/m-mediated regulation on physiological and stress-induced functions of the brain? How do its reader proteins intepret m6A/m-tagged transcripts upon neuronal activation and eventually modulate synaptic functions and behaviors? Do m6A and m6Am RNA modifications have differential roles in brain functions? Future studies will shed light on the dynamic epitranscriptome that enables flexible and spatiotemporally coordinated regulation of gene expression in our brains.
REFERENCES
- Cholewa-Waclaw J, Bird A, von Schimmelmann M, Schaefer A, Yu H, Song H, Madabhushi R, and Tsai LH (2016). The role of epigenetic mechanisms in the regulation of gene expression in the nervous system. J. Neurosci. 36, 11427–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haffner M, et al. (2018). The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, this issue, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozhik AV, and Jaffrey SR (2018). Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 14, 215–225. [DOI] [PubMed] [Google Scholar]
- Koranda JL, Dore L, Shi H, Patel MJ, Vaasjo LO, Rao MN, Chen K, Lu Z, Yi Y, Chi W, et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99, this issue, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vas-seur JJ, Chen Q, et al. (2017). Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkurjev D, Hong WT, lida K, Oomoto I, Goldie BJ, Yamaguti H, Ohara T, Kawaguchi SY, Hirano T, Martin KC, et al. (2018). Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. (2018). Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Vissers C, Ming GL, and Song H (2018). Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. J. Cell Biol. 217, 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, and He C (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]