Abstract
The current understanding of Inflammatory Bowel Disease (IBD) pathogenesis implicates a complex interaction between host genetics, host immunity, microbiome and environmental exposures. Mechanisms gleaned from genetics and molecular pathogenesis offer clues to the critical triggers of mucosal inflammation and guide the development of therapeutic interventions. A complex network of interactions between host genetic factors, microbes, and microbial metabolites governs intestinal homeostasis, making classification and mechanistic dissection of involved pathways challenging. In this review we discuss these challenges, areas of active translation, and opportunities for development of next-generation therapies.
1. The current state of treatment: Challenges posed
Clinical and endoscopic characteristics, such as disease location, distinguish the two major types of IBD, ulcerative colitis (UC) and Crohn’s disease (CD). While UC is restricted to the colon, mucosal inflammation in CD can involve the entire depth of the tissue and is associated with complications such as strictures and fistulas. In some patients, however, IBD cannot be definitively classified as either UC or CD. Treatment regimens overlap for these conditions, and include aminosalicylates; corticosteroids; immunomodulators; janus kinase inhibitors; or ‘biologic’ therapies, including monoclonal antibodies to Tumor necrosis factor-ɑ (TNF-ɑ), interleukin (IL)-12/23, or integrins to target inflammatory/trafficking pathways. Biologic therapies have increased quality of life and reduced the risk of disease-related complications, including surgery and hospitalization. However, up to 40% of patients do not respond to initial treatment. Among initial responders, 13–46% lose response over the subsequent year, with estimates varying by therapy, disease subtype and definition of non-response. In some fistulizing CD cases, fistula recurrence after treatment is as high as 64% (Roda et al., 2016).
The lack of treatment response raises two challenges to the advancement of IBD therapy: (i) further need for mechanistic understanding of disease drivers, incorporating features of heterogeneity, host genetics and the microbiome and (ii) renewed classification systems based on clinical, molecular and artificial intelligence methods to facilitate prognostication for clinicians to identify therapeutic non-responders and guide proactive treatment algorithms. Our understanding and ability to treat IBD is limited by factors including variable disease onset, genetic and clinical heterogeneity and pharmacokinetics [Table 1]. Many non-responders have adequate circulating levels of the drug, suggesting that rapid drug clearance is not a major contributor to lack of response. Additionally, the complexity of underlying disease mechanisms which impact distinct inflammatory pathways point to an unmet need to develop therapies that target specific pathways, including those that alter intestinal barrier function, tissue metabolism, immunity, fibrosis and fistulization. While existing IBD therapies target inflammation broadly, utilizing a pathway-focused approach in individual patients may increase the likelihood of success, reduce off-target effects, and result in durable benefits and modification of disease trajectory.
Table 1:
Advancing therapy in IBD
| Factors influencing IBD therapy | Clinical observations | Opportunities for discovery and translation |
|---|---|---|
| Variable onset of disease | Interval between untreated disease/symptoms and treatment associated with treatment failure | Triggers for disease-onset at different ages, recurrence. Early stratification of therapy based on baseline clinical and molecular profiling for individualized therapy |
| Genetic heterogeneity | GWAS-identified loci, resequencing efforts | Elucidating gene-variant function relationships; genetic risk score development |
| Disease progression | Different trajectories of disease | Factors determining complications, including recurrence, fibrosis, fistula, stricture and surgery |
| Infectious modifiers | CMV, C.difficile | Shared pathways in response to infection/inflammation and off-target |
| Dietary modifiers | Exclusive enteral nutrition | Pre-, pro- and syn-biotic interventions, mechanism of disease modification, interaction with non-dietary therapy |
| Drug delivery | Entyvio integrin therapy vs. TNF | Nanotechnology, gut-on-a-chip |
| Drug turn-over | Frequency of administration | Methods to measure drug levels, antibodies |
| Side-effect / off-target effects | Immune suppression, infectious complications | Pathway-based discovery |
Current disease classification systems incorporate anatomic distribution, clinical severity, age of onset, and behavior (fibrosing, penetrating). Antibodies towards self, microbial antigens and other peptides have been proposed as biomarkers of antigen-reactivity with limited clinical utility (Prideaux et al., 2012). Classifiers incorporating genetic, microbiome and clinical parameters to differentiate responders from non-responders to TNF and anti-integrin therapy have been developed (Ananthakrishnan et al., 2017). Although promising as a proof-of-concept, such classifiers require further validation for use in routine clinical practice. Given these challenges, this review proposes specific areas to improve mechanistic understanding and integrate bench and bedside studies to advance therapeutic options.
2. IBD genetics and functional links to the microbiome
Studies of IBD genetics have identified over 200 loci that explain a modest 8–13% of disease susceptibility risk variance (Jostins et al., 2012; de Lange et al., 2017; Rivas et al., 2011), highlighting the importance of non-genetic modifiers, such as intestinal microbes, that drive the pathological immune response in a genetically susceptible host (Xavier and Podolsky, 2007). Paradoxically, this breakdown in immune tolerance can be driven by seemingly opposing host genetic vulnerabilities. Both hyperinflammatory phenotypes and immunodeficiency phenotypes have been associated with risk of IBD onset. Chronic inflammation resulting from intestinal barrier dysfunction and/or colonization of the epithelial surface can create an intestinal tissue environment that alters colonization niches in the gut. A chronic low-grade inflammatory intestinal milieu can significantly impact the microbiome by selecting against species sensitive to inflammation and promoting blooms of functionally adapted species. Despite the intimate relationship between host and microbiome, identifying confident associations between host genetics (IBD SNPs) and microbiome features remains challenging.
The dysbiotic gut microbiome, defined as having a reduced microbial diversity and richness compared to healthy controls, has been extensively characterized in IBD (Lloyd-Price et al., 2019; Vich Vila et al., 2018) and reviewed in detail (Schirmer et al., 2019). The reduced richness does not necessarily simplify the task of finding links to host genetics. Inter-personal microbiome variation remains high in IBD, with many species occurring rarely in the general population due to diet, use of medication or ecological interactions. In addition, the different physiological functions and geography of the GI tract creates ecological niches that select for distinct microbial communities, which are additionally stratified by their mucosal or lumenal location (Zmora et al., 2018). Several studies identified links between genetic variants and gut microbes. For example, NOD2 variants are associated with abundance of Faecalibacterium prausnitzii, genus Roseburia and family Enterobacteriaceae (Aschard et al., 2019; Knights et al., 2014). NOD2 is a host intracellular sensor of bacterial infection by that recognizes the prokaryotic cell wall component muramyl dipeptide (MDP) and elicits a proinflammatory cytokine response in phagocytes (Girardin et al., 2003). The ubiquity of MDP across microbiome taxa/phyla, however, likely precludes identification of functional associations. In fact, NOD2 variants associate with taxa that are also observed in microbiome case-control studies in non-IBD diseases such as liver cirrhosis (Qin et al., 2014), suggesting that NOD2-associated dysregulation of microbial sensing broadly contributes to dysbiosis. This example underscores the complexity of IBD genetics and microbiome diversity and highlights challenges in identifying functional host-microbiome connections.
Current challenges highlight the need for new computational approaches to associate microbiome traits with host genetics. For example, the fungal mycobiome, often overlooked in IBD studies, can be functionally distinguished from the bacterial microbiome based on the mechanisms by which the host innate immune system recognizes microbial ligands. CARD9 is an essential adaptor protein required for NF-κB activation downstream of fungal ligand-sensing receptors such as Dectin-1. Consistent with its role in antifungal immunity, a common CARD9 missense variant (S12N) was associated with intestinal colonization by a commensal fungus Malassezia restricta in CD (Limon et al., 2019). Furthermore, CARD9 has been linked to circulating antibodies against M. restricta, indicating that breakdown of innate immunity translates into durable adaptive immunity in CD patients (Limon et al., 2019). Similar to the impact of fungi on immunity, host genetic variation interacts with viral encounters to modulate intestinal homeostasis. For example, the IBD risk gene Atg16l1 functionally interacts with viral sensing pathways mediated by cGAS-STING to regulate intestinal inflammation in an IL-22-dependent manner (Aden et al., 2018). Atg16l1 hypomorphic mice with dextran sodium sulfate-induced epithelial injury exhibit Paneth cell granule morphology abnormalities and exacerbated colitis when infected with a murine norovirus (MNV CR6) (Cadwell et al., 2010). These examples underscore the role of non-bacterial microbiome constituents in host immunity. Further unraveling these interactions will require cohort expansion and stool sample biobanking, followed by microbiome typing of bacteria as well as fungi, small eukaryotes and vira in patients with genotypes of interest.
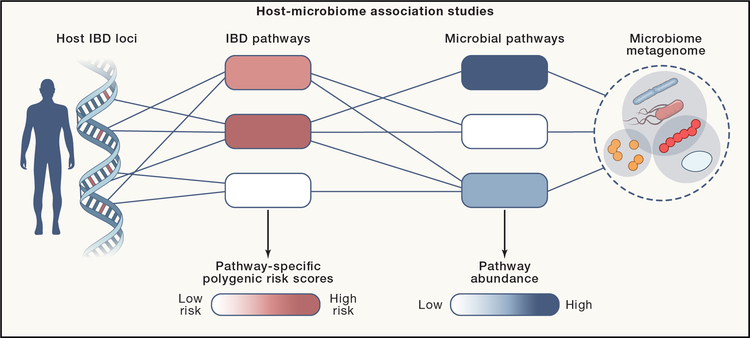
In IBD genetics, risk variants implicate a core set of functionally related pathways that represent vulnerabilities to intestinal inflammation (Momozawa et al., 2018). They encompass four broad overlapping categories: (i) microbial sensors and innate cytokine pathways, (ii) antibacterial effector mechanisms (oxidative burst, autophagy), (iii) antigen presentation and adaptive cytokine pathways, and (iv) epithelial barrier function. For example, NOD2 and CARD9 variants alter NF-κB signaling, increasing inflammatory cytokine production across microbial sensing pathways. According to this pathway-level perspective, functional connections between host and microbiome are best approached by searching for associations between host-pathway genetic risk scores and microbe-pathway metagenomic annotations (Figure 1). In essence, this strategy condenses a multitude of host genetic risk factors into functionally related pathway scores and parses many metagenomes into functionally related gene classes, thereby allowing for discovery of pathway-level associations between host genetic factors and the microbiome. This approach addresses inter-personal genetic heterogeneity, which introduces diverse vulnerabilities for microbiome dysbiosis. Encapsulating these genetic variants into separate scores or mutation loads for the major groups of IBD pathways could account for the molecular programs implicated in UC and CD and facilitate identification of common and rare variants in the same genes (Christodoulou et al., 2013). Furthermore, it would reduce the multiple testing burden when analyzing associations between IBD genetics and environmental modifiers such as the gut microbiome.
Figure 1. Genetic interactions between host and microbiome.
Associations between host genetics and microbiome features can be identified by integrating GWAS SNPs into polygenic risk scores for pathways relevant to IBD pathology and microbiome features into coherent functional pathways. The objective of this strategy is to identify statistical associations between host and microbiome pathways that reveal causal mechanisms by which the host and microbiome interact to impact IBD.
Coding variants associated with early onset IBD, including rare XIAP variants that diminish NOD2 signaling or mutations in nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex subunit genes, suggest functional interactions between the innate immune system and the microbiome (Amininejad et al., 2018; Denson et al., 2018). For example, the NADPH oxidase complex (CYBB, CYBBA, NCF1, NCF2, NCF4) is highly expressed in phagocytes, providing innate defense against microbial infection by producing toxic reactive oxygen intermediates (ROIs). Loss of function mutations in NADPH oxidase subunits result in chronic granulomatous disease (CGD), a severe primary immunodeficiency associated with recurrent bacterial and fungal infections. CGD patients are susceptible to infection with catalase-positive bacteria (Muise et al., 2012), suggesting that in IBD, hypomorphic alleles of NADPH oxidase subunits may alter host immunity to allow catalase positive gut commensals to occupy a permissive niche. This rationale suggests a possible relationship between host genotypes and abundance of microbiome genes mitigating ROI toxicity (e.g. catalase, superoxide dismutase, thioredoxin). Thus, carriers of IBD risk variants in NCF4 may have higher incidences of colonization by various facultative anaerobes in the Enterobacteriaceae family that can tolerate oxygen levels found in intestinal tissue but are normally susceptible to killing by phagocyte-derived ROI. In summary, condensing functionally related host genetic variables and microbiome traits could facilitate discovery and strengthening of associations between human genetics and the gut microbiome (Rothschild et al., 2018), particularly within the context of immune functions discussed in detail in the following sections.
3. Mechanisms of adaptive immunity towards microbiome antigens
Among the most robust genetic associations linking host immunity with microbial infection and/or autoimmunity are variants within the human leukocyte antigen (HLA) locus. Population-level HLA gene variation is concentrated within the peptide-binding groove of class I and class II major histocompatibility complexes (MHC). Variants alter MHC binding specificity and profoundly impact the spectrum of antigenic peptides presented to T cells. How HLA impacts commensal antigen presentation to T cells in IBD and how commensal-specific T cells impact intestinal pathology remains unclear. MHC class I and II (HLA) allele associations have been reported for CD (15 alleles) and UC (16 alleles, mostly class II), while fine variant mapping detected signal heterogeneity for clinical IBD phenotypes (Goyette et al., 2015). DRB1*01:03 is a strong MHC II risk allele for IBD with colonic involvement, while DRB1*07:01 in CD associates with non-colonic inflammation (Goyette et al., 2015). Sequence preferences for HLA binding are established for many alleles, however it remains unclear how the T cell pool is shaped by HLA-associated peptidomes during thymic selection and in the periphery.
The host immune system maintains tolerance to diverse immunogenic antigens produced by commensals. In this regard, the microbiome can be considered a host “self” tissue that is uniquely immune privileged. The intestinal lumen is not constitutively occupied by immune cells and is not directly surveilled by mechanisms of cellular immunity, but is subject to protection by mechanisms of humoral immunity (Bunker et al., 2017). The intestinal mucosa is rich in neutralizing commensal-specific IgA, which differs from other immunoglobulin isotypes that engage host cellular effector mechanisms, such as the complement system and Fc receptors. Accordingly, the immune system does not sterilize the lumen in an antigen-specific manner and is generally not equipped for killing lumenal microbes/pathogens. For example, adaptive immunity to C. difficile does not eradicate the bacterium from the lumen, but can instead inhibit infection of host mucosal tissue and systemic infection. Thus, coordinated T and B cell responses confer immunity to commensals, with T cells providing cytokine support for B cell functions including antibody class switch recombination, somatic hypermutation, and affinity maturation. In particular, T cell dependent anti-commensal IgG responses are associated with future diagnosis of type 1 diabetes (T1D) and depended on HLA genotype (Paun et al., 2019). Similar mechanisms may apply to IBD, as CD patients exhibit higher levels of immunoglobulins directed towards food antigens and commensal species (Lerner et al., 1989; Paun et al., 2019). These findings suggest impaired self-tolerance in T cell-dependent antibody responses. Recent studies showed that commensal-reactive B cells can evade autoreactivity by undergoing receptor editing, a process in which B cells re-rearrange light chain loci to mitigate local hyper-reactivity (Wesemann et al., 2013). While an analogous process in T cells has not been conclusively demonstrated, commensal-specific T cells have been identified that express dual T cell receptors (TCRs) with two distinct alpha chains. T cell reactivity towards segmented filamentous bacteria (SFB) is associated with dual TCR usage in lung autoimmune T cells, suggesting that dual-specificity T cells directed against microbiome antigens and autoantigens may contribute to autoimmune pathologies (Bradley et al., 2017).
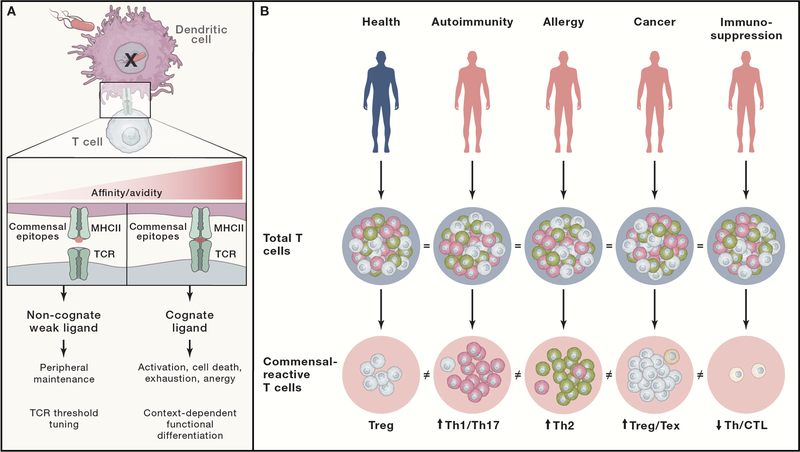
If we consider the microbiome to be a “self” tissue, how is tolerance established and maintained? Commensal-reactive T cells that develop in the thymus are not subject to the typical negative selection mechanisms that enforce central tolerance. Instead, multiple peripheral tolerance mechanisms control immune responses mediated by T cells expressing TCRs with high affinity to commensal antigens (Figure 2A). However, T cells expressing TCRs with low affinity/avidity for non-cognate commensal epitopes may function as positively selecting self ligands in the periphery that aid in T cell survival and tuning of TCR activation thresholds (Figure 2A). Recent studies demonstrated the importance of these ligands for conditioning T cell responses (Persaud et al., 2014; Wegorzewska et al., 2019). Thus, peripheral T cell maintenance may be promoted by tonic TCR signaling induced by interactions with weak self/commensal peptide ligands presented by MHCII. Taken together, commensal epitopes may function as weak self ligands that condition host T cell responses and/or strong antigenic ligands that either drive T cell effector responses or induce peripheral tolerance. Defining the functional impact of the microbiome on host adaptive immunity will require a more thorough characterization of the commensal antigen landscape and the T cell responses directed towards these antigens.
Figure 2. The dynamics of host adaptive T cell responses to commensal antigens.
The host T cell response to commensals is constant but dynamic over time such that it changes qualitatively and quantitatively according to the health status of the host. Thus, the T cell response to commensals represents a potentially powerful surrogate measure of immune health. (A) T cell activation is facilitated through TCR binding to a peptide-MHC class II (pMHCII) complex on antigen presenting cells (e.g. dendritic cells (DC)). Commensal epitopes can elicit very different outcomes in T cells depending on the context and affinity of pMHCII for TCRs. High affinity cognate interactions between pMHCII and TCR can drive T cell activation, differentiation, deletion, or exhaustion. Low affinity non-cognate interactions between pMHCII and TCR can elicit low grade signaling to promote T cell survival or fine tune TCR activation thresholds. (B) The total T cell pool is large and functionally heterogeneous, which poses challenges in monitoring changes in immune health during disease. Functionally characterizing the relationship of T cells with the microbiome can reveal the health status of an individual. For example, Tregs (blue) decrease and inflammatory Th1/Th17 cells (red) increase in autoimmunity, while Th2 cells (green) predominate over Th1 in response to allergens. Increased Tregs and hyporeactive exhausted T cells (Tex, gray) are associated with cancer. Immunosuppression can be characterized by decreased T helper (Th) cells and cytotoxic T lymphocytes (CTL) (orange). While these disease-associated perturbations in immune function can be difficult to measure at the level of the total T cell pool, they can become more clear and quantitative when focusing on a subset of microbiome-specific T cells.
Microbiome antigen discovery efforts identified several MHCII-restricted CD4 T cell epitopes in mouse models. Early efforts used serum from a mouse model that develops spontaneous colitis to identify T cell epitopes in bacterial flagellin (Cong et al., 2009). Subsequent work identified Helicobacter hepaticus-specific Treg responses (Xu et al., 2018), Th17 reactivity with specific SFB antigens (Yang et al., 2014), and Tfh reactivity with Akkermansia mucinophila (Ansaldo et al., 2019). T cell antigen discovery is more challenging in humans, given the diversity of HLA alleles and individual microbiomes. For these reasons, much of the work aimed at monitoring human adaptive immunity to commensals has focused on antibody-mediated B cell responses. Current approaches in IBD utilize serology to measure antibodies against a panel of microbial or self-antigens, including anti-Saccharomyces cerevisiae antibody (ASCA) and self perinuclear anti-neutrophil cytoplasmic antibody (pANCA), as well as more recently identified epitopes derived from E. coli OmpC or the conserved flaggelin epitope CBir1. These antibodies are observed in UC and CD patients, although at varying frequencies, thus limiting their diagnostic sensitivity (Prideaux et al., 2012). Collectively, these antigens represent a narrow view of the immunogenic landscape in IBD, and as efforts in antigen discovery advance, the field will benefit from gaining a more expansive perspective on the relationship between the microbiome and the adaptive immune system (Honda and Littman, 2016; Sefik et al., 2015).
With accumulating metagenomic datasets, predictive computational algorithms can facilitate antigen discovery and prioritization of antigenic peptides from the gut proteome (Graham et al., 2018). Once predicted epitopes are experimentally validated, it is equally important to establish immunodominance hierarchies by quantitatively monitoring antigen-specific T cell responses, TCR repertoires, and functional T cell phenotypes (Graham et al., 2018). New approaches in single cell RNA sequencing paired with targeted TCR sequencing have provided unique insights into T cell function relative to TCR antigen specificity, and high throughput TCR fingerprinting approaches have been leveraged to screen for reactivity to candidate peptide-MHCII ligands (Graham et al., 2018). Several additional approaches have been developed to facilitate high-throughput screening of MHCI-associated tumor antigens. Screens employing yeast display technology (Gee et al., 2018), DNA-barcoded tetramers (Bentzen et al., 2016), trogocytosis (Li et al., 2019) or T-cell based antigen discovery platforms (Joglekar et al., 2019) have enabled discovery of MHCI-associated antigens for cytotoxic T cells.
The clinical utility of antigen discovery holds potential for biomarker identification and antigen-specific immunotherapies. The continuous interaction of the microbiome and host adaptive immune system is dynamic and depends on factors intrinsic to immune function. Immunomodulation resulting from aging, infection or autoimmunity can qualitatively alter the T cell response to commensal epitopes by skewing cytokine profiles and/or impacting peripheral maintenance and attrition (Figure 2B). Thus, immunomonitoring technology aimed at quantifying the T cell response to commensal epitopes has direct relevance for diagnostic applications. In addition, antigen discovery efforts have the potential to enable novel therapeutic approaches based on vaccination strategies mobilizing or repurposing commensal-specific T cells to induce bystander tolerance in autoimmunity or unleash anti-tumor immunity. Similarly, cell-based therapies that employ emerging chimeric antigen receptor T cell (CAR-T) technologies may be adapted to selectively target pathogenic TCRs in the context of clonal resection therapy. Fundamental breakthroughs await decryption of the antigen-specific adaptive immune responses that underlie the host-microbiome relationship.
4. Microbiome-encoded molecular patterns condition host immunity
Antigens drive the T cell response, but cytokine context shapes the functional outcome of immunity to microbiome-derived antigens. Evidence suggests that microbe-associated accessory signals, transduced by the innate immune system, shape the intestinal cytokine milieu to polarize T cells to differentiate into Treg, Th17, or other customized phenotypes that allow adaptation to the environmental context (Honda and Littman, 2016). In addition to the antigen, the contribution of microbe-specific signals to T cell polarization has been elegantly illustrated in mouse models. SFB antigen, which typically induces Th17 cells, instead induced Th1 when heterologously expressed in Listeria monocytogenes (Yang et al., 2014), highlighting the fact that microbes produce diverse macromolecules that influence levels of cytokines such as TGF-β, TNF-ɑ or IL-6. For instance, 17 Clostridia strains have been identified that induce Treg expansion by stimulating epithelial cells to release TGF-β, which is considered necessary for Treg differentiation (Atarashi et al., 2013). Systematic study across an array of typical gut commensals has shown that immunomodulatory function is not phylogenetically conserved; rather, closely related species or strains display variable immune cell-polarizing potential. Additional research will be required to map the molecular sources of instructive signals that shape the immune cytokine milieu in mucosal tissues.
Although incompletely understood, the crucial question regarding how microbial signaling skews immune cell populations and impacts health/disease has been addressed by several landmark studies. IBD patients, primarily those with active disease, have increased numbers of inflammatory IL-17+ immune cells infiltrating the mucosa, higher serum levels of IL-17, and diminished populations of anti-inflammatory Tregs compared to healthy subjects (Fujino et al., 2003). These studies have utilized germ-free mouse models to identify changes in circulating or gut immune cell populations upon inoculation with bacterial species in the absence of confounding effects of the endogenous microbiome (Sefik et al., 2015). Specific strains or communities of commensal bacteria have been isolated and shown in gnotobiotic settings to modulate development and/or function of diverse immune cell types. These include mouse SFB and 20 human strains that promote Th17 cells (Atarashi et al., 2015; Ivanov et al., 2009), 46 mouse and 17 human Treg-promoting Clostridia (Atarashi et al., 2011, 2015), human Th1-promoting Klebsiella strains (Atarashi et al., 2017) and 11 human strains that promote IFNγ CD8+ cells (Tanoue et al., 2019). Similarly, gut microbiome transplantation from IBD individuals into germ-free mice induces colitis, promotes expansion of Th2 and Th17 populations, and leads to a concurrent decrease in Tregs (Britton et al., 2019). In contrast, transplantation of a “healthy” microbiome into germ-free mice creates a permissive environment for development of Tregs that produce anti-inflammatory IL-10, which in turn suppresses Th17 activity by inhibiting NF-κB signaling (Britton et al., 2019; Sefik et al., 2015).
The effects of genetic context on the gut microbiome and microbial signaling, as well as on immune function and disease susceptibility, can be evaluated in germ-free mice harboring IBD-associated genetic variants. One such variant, Atg16L1 T300A, which impairs xenophagy and Paneth cell function, induced a gut microbiome re-configuration in conventionally housed mice that was characterized by expansion of Bacteroidetes under normal and colitis conditions (Lavoie et al., 2019). Introduction of stool from patients with active CD, but not UC, into Atg16L1 T300A germ-free animals had a similar effect, inducing expansion of Th17, Th1 and Tregs in the colonic and ileal lamina propria relative to wild-type recipients (Lavoie et al., 2019). This work highlights the notion that immunomodulatory signals are intrinsic to the microbiome, but require a specific host genetic context and microbiome configuration to manifest. Single species or combinatorial inoculations into germ-free mice will remain an attractive way to attribute immunomodulatory function to microbes. In practice, immune screening efforts will be aided by increased strain biobanking and detailed molecular characterization of key strains in the context of healthy and diseased tissues. Hypothesis-driven mechanistic experiments can help to uncover and validate the molecular or antigenic signals that directly modulate immune function.
Early successes in identifying microbiome-derived effector molecules and host target receptors resulted from studying pathogens and their microbe-associated molecular patterns (MAMPS) that engage pattern recognition receptors (PRRs) of the innate immune system. For instance, IBD-associated Ruminococcus gnavus produces glucorhamnan polysaccharide that potently activates dendritic cells to secrete TNF-ɑ in a TLR4-dependent manner (Henke et al., 2019). Beyond MAMPS, the microbiome interacts with the host through metabolites generated by a combination of de-novo synthesis and secondary transformation of dietary and non-dietary precursors. Integrated metabolite and metagenomic profiles revealed metabolite classifiers that differentiated IBD cases from healthy controls (Franzosa et al., 2019). CD patients exhibited robust differences in global stool metabolite compositions compared to healthy controls, while UC patients straddled across both healthy and CD metabolite signatures. Specific metabolites were enriched in CD patients and correlated with markers of inflammation such as fecal calprotectin. These include sphingolipids, carboximidic acid and bile acid derivatives (Franzosa et al., 2019). The importance of sphingolipids in modulating immune responses is highlighted by the finding that colonization of germ-free mice with Bacteroides thetaiotaomicron maintained intestinal homeostasis while a mutant strain of B. thetaiotaomicron deficient in sphingolipid synthesis induced intestinal inflammation associated with elevated IL-6 and monocyte chemoattractant protein-1 (Brown et al., 2019). These findings attribute an anti-inflammatory role to sphingolipids that are differentially abundant in IBD compared to healthy controls, thus providing functional context to a clinical observation.
Our understanding of IBD dysbiosis stems largely from cross-sectional studies in treatment naive CD or UC patients (Gevers et al., 2014; Schirmer et al., 2018), as well as longitudinal studies in broader IBD populations that focus largely on stool and biopsy microbiomes and metabolomes (Lloyd-Price et al., 2019). These efforts show that IBD is characterized by an increased abundance of facultative anaerobes, many of which are autochthonous to the oral cavity and display pro-inflammatory characteristics. In the context of the microbiome, these anaerobes are associated with a decrease of bacterial metabolites, including short-chain fatty acids (SCFAs) or secondary bile acids. Substantial efforts will be required to advance the field from species-level disease associations and correlations with metabolite levels to causal mechanisms linking the microbiome to host physiology. This challenge is confounded by secondary effects of generic inflammation, exposure to medications and dietary variables. Establishing mechanistic connections between molecular effectors and microbial species will be necessary to identify metabolite producers and prioritize them as key mediators of host physiology.
5. Pharmacology of microbiome-derived metabolites and host targets.
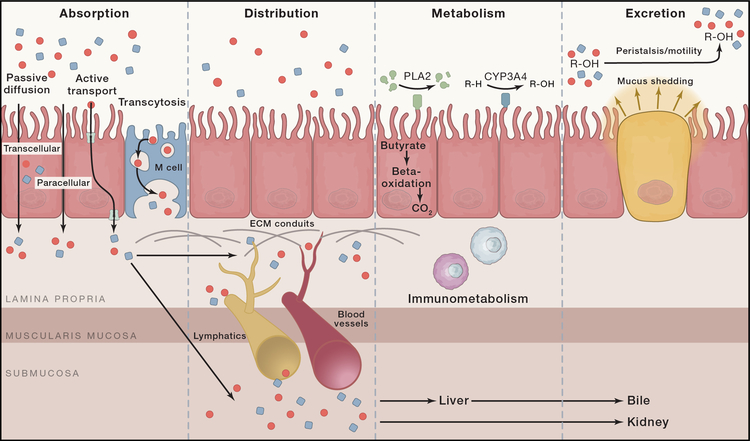
Translating microbiome-derived bioactives into novel therapeutics may benefit from viewing the challenge through the lens of pharmacology. For example, the principles of ADME (absorption, distribution, metabolism, excretion) highlight the fact that stool metabolites may or may not be the same chemical entities that directly engage host receptors/targets (Figure 3). This is exemplified in recent studies wherein mice were inoculated with C13-labelled, non-replicating E. coli to track ADME of microbe-derived metabolites in the host. Accordingly, profiling by mass spectrometry identified hundreds of metabolites across 23 host tissues (Uchimura et al., 2018). Using a gnotobiotic mouse model, researchers identified a biosynthetic pathway in Clostridium sporogenes that produces amino acid metabolites that modulate host physiology by regulating intestinal permeability (Dodd et al., 2017). This important observation establishes that microbe-derived metabolites directly modulate host physiology in vivo. Additional studies have implicated aromatic amino acids and their metabolites in engaging key cytoprotective host pathways, including AhR and Keap1/Nrf2, in IBD (Wlodarska et al., 2017). The discovery of organisms capable of producing similar compounds reinforces the potential for aromatic amino acid derivatives to promote intestinal barrier integrity and mitigate inflammation.
Figure 3. Local and systemic ADME dictate biological effects of microbial metabolites.
The pharmacokinetic principles of ADME (absorption, distribution, metabolism and excretion) determine the fate of dietary and microbiome-produced metabolites in the intestine and dictate their potential biological effects on the host. Absorption of microbial metabolites across the intestinal epithelium can occur by passive (transcellular transport, paracellular transport or diffusion) or active mechanisms (via transporters). Molecules can also be actively transported by transcytosis facilitated by M cells. Absorbed molecules are distributed locally throughout the intestinal mucosa via extracellular matrix (ECM) conduits and systemically by the circulatory and lymphatic systems. Molecules may undergo chemical transformation by means of metabolism in the lumen, within the mucosa, or in the liver. Excretion can occur through the gastrointestinal tract by peristalsis and mucus shedding, while systemic excretion is generally mediated by biliary or urinary excretion. Taken together, microbiome-derived molecules are subject to multiple biological processes that modulate or limit their ability to engage host target pathways.
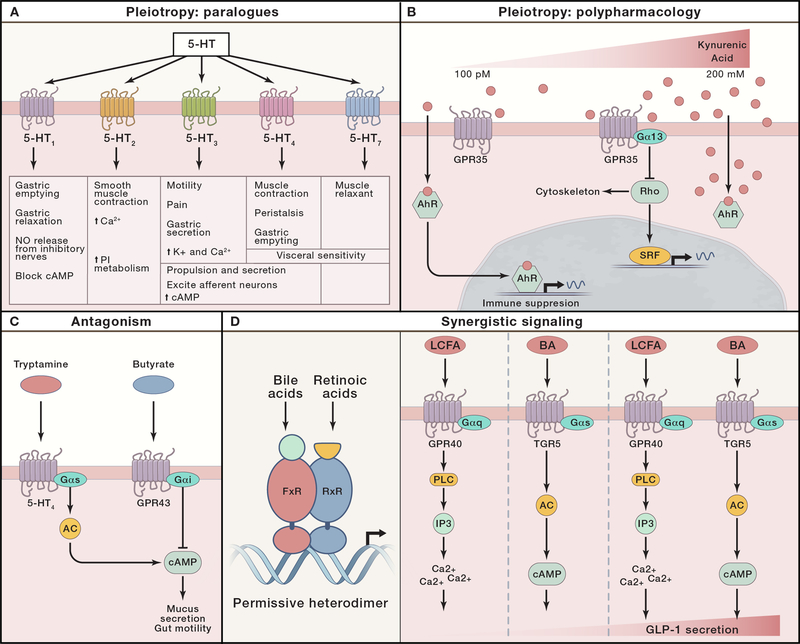
The diversity of microbiome-derived biosynthetic gene clusters and their metabolite products highlights the need to expand our view of microbe-host interactions to account for the integrated circuitry within and between host cells that coordinates adaptive responses to environmental cues in the microbiome. Specifically, pleiotropy can arise from the action of a discrete metabolite on a singular host target or paralogues that are expressed in different cell types and that induce distinct cell type-specific functions (Figure 4). Serotonin is a tryptophan metabolite that exhibits pleiotropy by binding to many closely related paralogs (serotonin signals through 14 closely related 5-hydroxytryptamine receptors (5-HTRs)) (Figure 4A). Engagement of 5-HTRs in enteric neurons induces smooth muscle contraction and gut motility (Mawe and Hoffman, 2013), promotes mucous secretion in goblet cells (Yang et al., 2013), potentiates IL-10 and inhibits IL-12/23 in myeloid cells (de las Casas-Engel et al., 2013), and induces degranulation and histamine release in mast cells (Wang et al., 2013). Thus, serotonin derived from host enterochromaffin cells or from microbiome metabolism exemplifies paralogous signaling wherein similar signaling pathways engaged in specialized cell types result in diverse biological outcomes.
Figure 4. Pharmacology of microbial metabolites and host targets.
Metabolites engage host pathways and elicit diverse signaling outputs. (A) Pleiotropy by paralogs: serotonin (5-HT) has pleiotropic effects on the host due to the fact that there are 14 different 5-HT receptors, each associated with distinct downstream effects in different cell types. (B) Pleiotropy by polypharmacology: kynurenic acid exerts pleiotropic effects on the host by engaging distinct host target proteins with varying affinities. Specifically, kynurenic acid engages aryl hydrocarbon receptor (AhR) and GPR35, thus engaging unique signaling pathways that coordinately regulate immune function (CREB, cAMP response element-binding protein). (C) Antagonism: butyrate can act through GPR43 to antagonize mucus secretion induced by tryptamine signaling through 5-HT4 receptors (AC, adenylate cyclase). (D) Synergistic signaling: heterodimer FxR/RxR transcriptional activity is augmented by concurrent binding to bile acid and retinoic acid derivatives (left panel). Secretion of GLP-1 is dramatically enhanced by concurrent engagement of GPR40 and TGR5 by long-chain fatty acids and bile acids, respectively (right panel) (PLC, phospholipase C; IP3, inositol triphosphate).
Similarly, pleiotropy can result from polypharmacology in which a singular metabolite engages different targets with distinct biological outcomes (Figure 4B). Bile acids function as agonists for the G-protein coupled receptor (GPCR) GPBAR1 and the nuclear hormone receptor FxR, resulting in pleiotropic effects on digestion, metabolism, and inflammation (Malhi and Camilleri, 2017). Recent studies identified central roles for the microbiome in host-derived bile acid metabolism and host physiology (Devlin and Fischbach, 2015; Yao et al., 2018). In another example, microbiome fermentation of dietary fiber produces butyrate, which engages multiple host targets including HDAC, GPR109a, GPR43 and GPR41 (McKenzie et al., 2017). Butyrate promotes Treg expansion by inhibiting HDAC, suppressing Th17 responses, promoting IL-6 production in macrophages, and suppressing stem cell proliferation (Kaiko et al., 2016). Thus, the physiological host response is the result of integrative polypharmacology. In this context, ligand dose may preferentially engage pathways such that a low dose induces the more sensitive target pathway but a high dose allows the less sensitive target pathway to dominate. For example, kynurenic acid engages the AhR pathway more potently (EC50 ~100nM) (Seok et al., 2018) than it does the GPR35 pathway (EC50 ~50uM) (Wang et al., 2006), suggesting that variations in intestinal concentrations of kynurenic acid may differentially engage host target pathways and elicit distinct responses.
While individual metabolites may have pleiotropic effects, multiple microbial metabolites have synergistic and/or antagonistic effects on host pathways (Figure 4C and D). In this context, differential GPCR engagement can result in synergy or antagonism. Microbiome-derived tryptamine engages 5-HT4R to induce fluid secretion and promote intestinal motility (Bhattarai et al., 2018). Agonist-induced signaling through 5-HT4R is transmitted through activation of Gɑ-s proteins, which induce the second messenger cAMP. Thus, tryptamine and serotonin can synergistically induce cAMP production downstream of 5-HT4R. Conversely, the cAMP response can be antagonized by GPCRs that signal through Gɑ-i proteins, such as the butyrate receptor GPR43 (Figure 4C). More broadly, several host receptor systems in addition to GPCRs engage in antagonistic and synergistic signaling. The bile acid receptor and nuclear hormone receptor FxR forms permissive heterodimers with RxR retinoic acid receptors (Figure 4D). Consequently, both bile acids and retinoic acid derivatives coordinate synergistic induction of FxR/RxR-mediated transcriptional programs that control cellular metabolism and bile acid synthesis (Evans and Mangelsdorf, 2014).
As our understanding of microbial metabolite pharmacology evolves, it remains a challenge to systematically define and functionally characterize the microbiome metabolome. Carefully curated hypothesis-driven approaches have generated promising leads to manipulate inflammation (reviewed in detail in Milshteyn et al., 2018), however a few important points warrant consideration for future efforts. To date, candidate bioactives have been identified from (i) metabolomics in the context of disease (Franzosa et al., 2019; Hoyles et al., 2018), (ii) metagenomics and computational prediction of bioactive biosynthetic gene clusters, (iii) synthetic biology approaches that permute bacterial genetic circuits (Woodruff et al., 2017), and (iv) monoculture and biochemical fractionation of disease-associated isolates (Cohen et al., 2017). Such approaches, which aim to curate and assemble libraries of candidate bioactive molecules, serve as valuable resources for host phenotypic screens and target-focused screens. For example, fractionation and activity-guided screening approaches have identified novel mechanisms of metabolite-directed lymphocyte homing (Lu et al., 2019). Additionally, target-focused screening has benefited from technology development enabling multiplexed screening of GPCRs in high throughput (Jones et al., 2019). Recent studies have leveraged functional and phenotypic screening to uncover novel mechanisms underlying the host-microbiome relationship, and these have been covered in greater detail in excellent reviews (Milshteyn et al., 2018; Skelly et al., 2019). Collectively, these screening efforts show great potential towards establishing mechanistic connections between the microbiome and host that impact mucosal homeostasis and IBD.
6. Microbiome-inspired therapeutics
Efforts to leverage the microbiome to advance therapeutic ends have taken several forms. Given the scope of chemical diversity encoded within and produced by the microbiome, interest is growing in mining the microbiome for bioactive compounds to derive drugs from bugs. Alternatively, live microorganisms have been used as therapeutic agents or as vectors for delivery of therapeutic agents in a strategy for deploying bugs as drugs. Finally, approaches for altering microbiome function or composition have led to the exploration of drugs for bugs. These approaches may involve decolonization to reduce harmful species or ecological engineering to restore homeostasis to the microbiome community through dietary intervention or manipulation of quorum-sensing pathways. Below, we highlight key examples of microbiome-inspired therapeutics and discuss potential applications of these approaches.
Drugs from bugs
Are bioactive microbiome metabolites good therapeutic leads? Clinically useful antibiotics, including vancomycin, streptothricin, streptomycin or tetracycline, have been discovered from soil microorganisms living in complex communities, primarily Actinobacteria (actinomycetes). Because commensals coevolved with their host, this mutualistic relationship may be particularly permissive for evolution of bioactives that modulate host physiology. For example, digestion of dietary fibers results in the production of SCFAs that nourish colonocytes and exhibit broad immunomodulatory function, such as butyrate-mediated Treg expansion. Levels of butyrate its microbial producers are lower in patients with IBD flares compared to controls, a finding that inspired numerous translational studies (Machiels et al., 2014; Sokol et al., 2009). Moreover, interventional studies in humans have shown that butyrate administration may be effective, however the sample size of these trials precludes conclusive statements. Although butyrate is beneficial with respect to host physiology, caveats to therapeutic translation include delivery challenges due to potency in the mM range, uncertainty of host target due to pleiotropic effects, and sub-optimal pharmacokinetic properties (Daniel et al., 1989). Engineering of commensals to produce therapeutic molecules offers an alternative pathology management strategy for IBD patients. For instance, Lactobacillus strains that deliver IL-10 or anti-TNF antibodies showed promise in controlling inflammation in mouse colitis models (Steidler et al., 2000; Vandenbroucke et al., 2010). Such approaches present dosing challenges and require special attention to ADME, as payload delivery to the gut lumen may or may not reach the site of inflammation or therapeutic target.
Although microbiome metabolites may lack ideal pharmacologic properties (potency, selectivity, metabolic stability), they represent powerful tools as bioactive probes to identify host targets that can be exploited for drug development. CRISPR technology has been adapted to link bioactive molecules with often elusive host targets through combinatorial use of gene expression knockdown and transcriptional activation (CRISPRi/CRISPRa) that allows filtering and prioritization of genetic signals responsible for small molecule bioactivity (Jost et al., 2017). Similar approaches can be applied to target identification for bioactive microbial metabolites. Host target identification enables traditional drug discovery campaigns to identify compounds that can be optimized by medicinal chemistry to improve potency, selectivity, and impart more favorable pharmacokinetic properties (Figure 5).
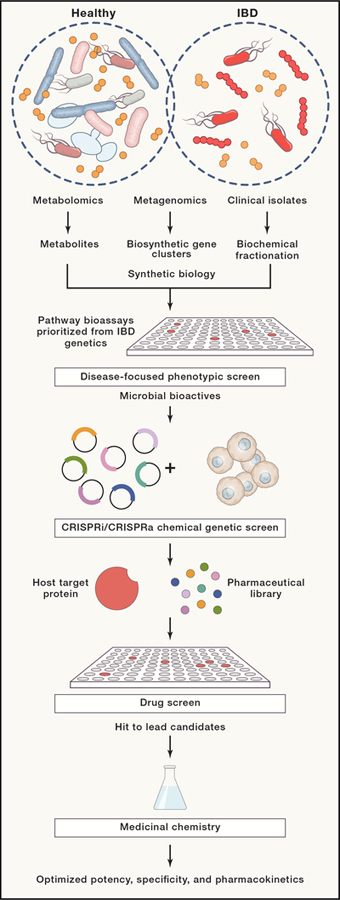
Figure 5. Translating microbiome bioactives into therapeutic agents.
Natural product molecular libraries can be designed and assembled based on microbiome metabolomics datasets. Alternatively, compounds can be directly produced by synthetic biology approaches and/or fractionated from microbial cultures. With microbiome-derived molecular libraries, compounds and molecules can be screened and prioritized based on their bioactivity in phenotypic screening assays that model biological processes and pathways associated with IBD pathology. In turn, chemical-genetic screens can facilitate identification of host target proteins through which microbiome bioactives exert their function. For example, a given microbiome bioactive is predicted to lose its biological effect in host cells that lack their specific target protein due to genetic knock-out or knock-down (CRISPR KO or CRISPRi). Conversely, a given microbiome bioactive is predicted to exhibit increased biological activity in host cells that overexpress their specific target protein (CRISPRa). Subsequently, pharmaceutical libraries with drug-like properties can be screened for activity against validated host targets. Finally, lead candidate small molecules can be further engineered by medicinal chemistry to optimize potency, selectivity, and pharmacokinetic properties.
Bugs as drugs
Even with an incomplete mechanistic understanding of how the microbiome impacts health and disease, progress has been made in therapeutic intervention by fecal microbial transplantation (FMT). Perhaps the most significant clinical benefit has been observed in the treatment of Clostridium difficile infection. Studies modeling time-course data after C. difficile infection and antibiotic treatment identified microbiome species that correlated negatively with occurrence of the disease agent (Buffie et al., 2015). This approach revealed that Clostridium scindens chemically modifies primary bile acids to inhibit C. difficile germination (Buffie et al., 2015), paving the way for design of a simplified therapy using a single metabolite or species, or a consortium of secondary bile acid metabolizers. In contrast, randomized FMT clinical trials showed modest effects on inducing remission in UC (Paramsothy et al., 2017). Evidence suggests that donor-specific effects strongly influence likelihood of remission in recipients (Moayyedi et al., 2015), and studies of transmission suggest that strain tracking could be employed to monitor engraftment and assess responses over time (Smillie et al., 2018). In addition to low efficacy and uncertainty regarding long-term durability and safety of FMT in IBD, additional challenges include a lack of procedural standardization, risk of transferring pathogenic agents, and induction of unwanted phenotypes, such as flares in UC patients.
Probiotics or designed microbial consortia offer potential alternatives for optimizing and standardizing treatments (Stein et al., 2018), but to date, these interventions have limited efficacy. Suboptimal probiotic engraftment was observed in a study that sampled the human mucosal and lumenal microbiome along the length of the gastrointestinal tract, suggesting that therapeutic efficacy may require promotion of colonization niches (Zmora et al., 2018). Accordingly, in specific-pathogen-free mice inoculated with a cocktail of 11 commensal strains that induce CD8+ IFNγ+ T cells, engraftment was only possible after antibiotic treatment (Tanoue et al., 2019). Similarly in UC patients with mild to moderate active disease, administering a capsule of proprietary spores in combination with vancomycin pre-treatment showed transient clinical and endoscopic features of remission when compared to patients receiving no pre-treatment (Misra et al., 2018). Since vancomycin itself positively affects remission in UC patients (de Chambrun et al. 2018), it is difficult to disentangle the role of antibiotics in opening new niches compared to removal of pathogenic or antigenic species. Future studies will be necessary to optimize the technical aspects of composition, delivery, and engraftment of the transplanted microbial organisms.
Drugs for bugs
The decreased microbial density in stool from IBD patients primarily affects total numbers of Firmicutes, Bacteroidetes and Actinobacteria, while Proteobacteria, which includes species of Enterobacteriaceae that are well adapted to thrive in an oxidizing environment, maintain a stable absolute population (Contijoch et al., 2019; Vandeputte et al., 2017). The elevated oxidative environment in the gut can result from dysbiosis caused by antibiotic treatment, increased oxygen tension due to intestinal bleeding, or the presence of anaerobic electron acceptors (e.g. nitrate / NO3-), some of which could be by-products of anti-microbicidal immune functions that generate ROI and nitric oxide (Reese et al., 2018; Winter et al., 2013). For instance, Escherichia coli and Salmonella typhimurium can respire on nitrate and tetrathionate (S4O62−)) respectively, and bloom in the presence of colitis-associated inflammation (Thiennimitr et al., 2011; Winter et al., 2013). This could potentially explain why the absolute abundance of Proteobacteria does not decrease in IBD patients (Contijoch et al., 2019). Overgrowth of facultative anaerobes from family Enterobacteriaceae can be controlled under anaerobic conditions by impairing their respiration on ROI by-products and inhibiting nitrate reductase using tungstate (Zhu et al., 2018). Similarly, nutrient inhibition targeting epithelia-derived ethanolamine, which increases during inflammation and promotes growth of S. typhimurium, may represent a viable strategy to control microbial outgrowth in the gut (Thiennimitr et al., 2011). The multi-gene ethanolamine utilization locus is detected across multiple intestinal Firmicutes and Proteobacteria, suggesting that ethanolamine is an important nutrient source for certain species (Kaval and Garsin, 2018).
Decolonization
Emerging strategies in microbiome therapeutics have begun to explore selective removal of specific microbes. Phage therapy holds particular promise, as it allows for precise targeting of conserved, species-specific membrane proteins for phage docking and delivery of cargo that eliminates the target organism, modifies its genome or regulates its function (Ramachandran and Bikard, 2019). The latter is possible by employing CRISPR-based nuclease activity to introduce double stranded breaks in a genome using a library of specific guide RNAs (Gomaa et al., 2014). In this context, adherent-invasive E. coli (AIEC) have been associated with IBD and may represent a viable target for such approaches. The FimH gene product promotes AIEC binding to the small bowel epithelium via the host CEACAM6 receptor, rendering AIEC resistant to clearance by the immune system (Barnich et al., 2007). Thus, it may be feasible to develop phages that exploit FimH binding for docking and delivery of CRISPR cargo that specifically targets the AIEC genome. Studies of inter-individual strain diversity suggest that microbiome strains carry genes that are unique to a given individual, a trait that could be exploited to design personalized, strain-specific CRISPR targets with minimal risk for unintended species depletion due to off-target effects.
Ecological engineering
The most basic and clinically feasible approach to altering the microbiome is through changes to human diet that modulate the nutrient pool available to gut microbes. For IBD maintenance, a treatment termed the ‘elemental diet’ has shown efficacy on par with corticosteroid therapy. The elemental diet, which is comprised of amino acids, essential fatty acids, monosaccharides, vitamins and minerals, was shown to be particularly effective in pediatric IBD where 80% of patients experienced disease remission (Penagini et al., 2016). The ability to extend treatment to adults and achieve a similar effect, particularly in the United States, has been hampered by palatability and compliance. Although it is unclear how the elemental diet impacts IBD, various studies have pointed towards augmenting mucosal barrier function by excluding substances present in typical diets (e.g. emulsifiers, carrageenan or carboxymethylcellulose) that irritate the epithelium or by excluding typical dietary antigens. It is well known that CD patients develop food-specific IgGs, for example those that are directed against milk proteins (Lerner et al., 1989). However, it remains unclear if immune responses directed against dietary antigens contribute to the pathogenesis of IBD or are predominantly a downstream consequence of pathological inflammation. An additional aspect to consider is that diet can act indirectly, by modulating the expression of microbe-encoded antigens (Wegorzewska et al., 2019). While the underlying mechanisms responsible for dietary impacts on the microbiome remain incompletely understood, several studies have begun to define the effects of diet on the microbiome, host physiology, and immune function. These include reports on broad dietary regimes (e.g. whole-grain, gluten-free diets) and targeted prebiotic supplementations in a controlled background diet (Gurry et al., 2018; Hansen et al., 2018; Roager et al., 2019). By anchoring dietary interventions on the background of the elemental diet, it is possible to define how introduction of macronutrients found in a typical western diet impacts intestinal homeostasis and immune function. Finally, identifying dietary IBD effectors could inspire personalized dietary interventions, as has been proposed for control of glycemic response (Mendes-Soares et al., 2019; Zeevi et al., 2015).
Re-establishing a healthy gut microbiome may require limiting the colonization of certain species while promoting others. In this context, promoting engraftment of desirable species has become feasible with the identification of nutrients that act as prebiotics that selectively create a niche for particular species. The recently described Bacteroides ovatus and Bacteroides plebeius strains harbor a rare gene cluster that metabolizes porphyran from a marine algae, which allows these species to thrive and engraft in an otherwise colonization-resistant endogenous microbiome (Kearney et al., 2018; Shepherd et al., 2018). Similarly, community-level metabolic activity within the microbiome can modulate nutrient pools to impact growth and engraftment of gut microbes. For example, C. diff colonization restriction by antagonistic commensal species illustrates the capability of microbiome-derived metabolic activity to affect growth and engraftment. Such species-species interactions can be deconvoluted in reductionist co-cultivation experiments, as shown for B. plebeius and Akkermansia muciniphila, which exhibit differential growth kinetics when co-cultivated on host mucins versus other carbon sources (Kearney et al., 2018). In the context of IBD, however, entire networks of typically correlated species are perturbed, and such widespread ecological disruptions are difficult to reset even with generalized introduction of an entire community by FMT (Yilmaz et al., 2019). Defining critical ecological relationships between gut microbiome constituents will require experimental and computational innovations. Metatranscriptomic analyses of gene expression networks within microbial communities have defined codependent transcriptional programs engaged by microbes in two-species systems (Mahowald et al., 2009) and in the context of higher-order gut microbiome interactions (Plichta et al., 2016). Functional interpretation of these transcriptional networks has inspired approaches for modelling the metabolic output of gut microbes to reveal which metabolites are released to the common microbial nutrient pool versus those utilized by single microbes (Zelezniak et al., 2015). Such systems biology approaches, when extended towards epithelial and immune gene expression and secreted metabolites, could reveal necessary cues for reengineering a stable microbiome ecology. To reduce the inevitable complexity of such a holistic model, experimental systems have been developed that incorporate more controlled organoid based systems with microbiome, mucosa, epithelium and immune cell compartments (Kim et al., 2016). These model systems can be leveraged to reveal the signaling cues necessary for stable coexistence of varying microbiome configurations, dietary variables, and immune mediators.
More refined therapies have been conceived to focus on identifying molecules that can redirect the behavior of a community, and in turn, promote a desired phenotype. Quorum sensing is a mechanism by which bacteria react to external stimuli, for example changes in population density or other microbes, and reactively coordinate gene expression or metabolic programs regulating growth, metabolism or inflammation. These mechanisms are implicated in bacterial biofilm formation and regulation of virulence factor expression (Rutherford and Bassler, 2012). Several groups of quorum sensing molecules have been identified, including N-acyl homoserine lactone (AHL) molecules (autoinducer-1, AI-1), modified oligopeptides (autoinducer peptides, AIP) and boron-furan derived signal molecules (autoinducer-2, AI-2) produced and detected by both Gram-negative and Gram-positive bacteria (Rutherford and Bassler, 2012). In fact, the host may also contribute to generating auto-inducers (Ismail et al., 2016). The observation that molecules produced by bacteria can direct the metabolic programs of the community highlights the need to decode the operations of complex quorum-sensing pathways and their downstream targets to purposefully direct community function.
Perspectives
Host genetic variables create a favorable background for manifesting IBD and its associated pathologies in the context of a dysbiotic microbiome, whose activity and metabolic outputs adversely impact host physiology and immune function. Substantial progress has been made towards understanding these components in isolation, and the field is starting to unravel their mechanistic interconnections to guide therapeutic strategies aimed at altering the course of IBD. Still, many outstanding questions remain. How and to what extent do multiple, genetic susceptibilities across distinct molecular pathways induce pathological shifts in microbiome composition? Does an unstable microbiome dysregulate host physiology in a progressive manner to eventually surpass a certain threshold that induces pathological inflammation? What are the dietary factors and microbiome metabolites that associate with health or disease, and on what molecular receptors and pathways do they act? These are non-trivial questions that are fundamentally important given the challenges associated with inducing long-term sustained remission in IBD patients. Pursuing solutions to these questions should help decode host-microbiome interactions and guide the development of combinatorial treatments that simultaneously restore health and homeostasis to the intestinal barrier, immune system and microbiome.
Systems biology approaches can functionally connect biological components of IBD to improve our understanding of disease and patient care. A molecular basis for individualized therapy holds promise for a new wave of context- and microbiome-specific therapies, though difficulty in identifying triggers of IBD flares will remain a hurdle for the field of microbiome therapeutics. Furthermore, deciphering disease mechanisms from human cohorts is complicated by variables related to genetics, microbiome, treatment history, and diet. Longitudinal sampling of human cohorts has created opportunities to interrogate real-time microbiome patterns in a multi-dimensional matrix. Several studies have utilized machine learning algorithms to decode associations between microbiome structure, dietary patterns and clinical metadata to successfully predict glucose response (Mendes-Soares et al., 2019; Zeevi et al., 2015). We anticipate burgeoning opportunities to synthesize patterns of disease variables and risk factors within an individual or groups of individuals over time, akin to machine learning to understand consumer preference selection among various commercial products available today. Further alluring is the possibility of utilizing algorithms to help detect patterns in clinical metrics that emerge during observation and can be interpreted to subsequently elicit interventions that promote a health outcome. Treatment-naive IBD patients in clinical trials may be subjected to serial microbiome profiling, functional immunomonitoring, and pathway-focused genotype profiling. Individualized patient data may be subsequently used to predict the likelihood of attaining sustained remission with a given therapy, or for guiding when and how to adjust therapeutic strategies. This may also help collate patterns of disease among IBD patients who are otherwise currently defined by other parameters such as disease location rather than genetic/microbiome-based stratifications. Current limitations to the application of artificial intelligence include sparsity of longitudinal data, variations in the data generation methods and ultimate biological interpretability of predictions. Utilizing functional measures of microbiome output through better mucosal immune system read-outs, as well as molecular subtyping of disease may offer more biologically actionable and interpretable predictions with direct benefit to patients.
Glossary
- Anergy
Functional inactivation of self-antigen reactive T cells
- Clonal deletion
apoptotic removal of T cells specific for self-antigens prior to their development into fully mature lymphocytes
- Bystander suppression
suppression of T cell response to an unrelated antigen
- Exhaustion
a state characterized by loss of T cell effector function due to chronic antigen exposure, as in chronic infections or cancer
- Trogocytosis
Transfer of plasma membrane fragments from antigen presenting cells to lymphocytes
Footnotes
Declaration of interests
The authors declare no competing interests.
Bibliography
- Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Häsler R, et al. (2018). ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J. Exp. Med 215, 2868–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amininejad L, Charloteaux B, Theatre E, Liefferinckx C, Dmitrieva J, Hayard P, Muls V, Maisin J-M, Schapira M, Ghislain J-M, et al. (2018). Analysis of Genes Associated With Monogenic Primary Immunodeficiency Identifies Rare Variants in XIAP in Patients With Crohn’s Disease. Gastroenterology 154, 2165–2177. [DOI] [PubMed] [Google Scholar]
- Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, and Xavier RJ. (2017). Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 21, 603–610.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, et al. (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Laville V, Tchetgen ET, Knights D, Imhann F, Seksik P, Zaitlen N, Silverberg MS, Cosnes J, Weersma RK, et al. (2019). Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLoS Genet. 15, e1008018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. (2017). Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser A-L, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel J-F, et al. (2007). CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest 117, 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, Such L, Furness AJS, McGranahan N, Rosenthal R, et al. (2016). Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol 34, 1037–1045. [DOI] [PubMed] [Google Scholar]
- Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, Linden DR, Akiba Y, Kandimalla KK, Zachos NC, et al. (2018). Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 23, 775–785.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, Huang H, Knox KS, Littman DR, and Wu H-JJ. (2017). Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 22, 697–704.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R, Li Z, Mortha A, Merad M, Das A, et al. (2019). Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 50, 212–224.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, Arthur TD, Fornelos N, Heim C, Franzosa EA, et al. (2019). Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 25, 668–680.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, and Bendelac A. (2017). Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu T-C, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, and Virgin HW. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Casas-Engel M, Domínguez-Soto A, Sierra-Filardi E, Bragado R, Nieto C, Puig-Kroger A, Samaniego R, Loza M, Corcuera MT, Gómez-Aguado F, et al. (2013). Serotonin skews human macrophage polarization through HTR2B and HTR7. J. Immunol 190, 2301–2310. [DOI] [PubMed] [Google Scholar]
- Christodoulou K, Wiskin AE, Gibson J, Tapper W, Willis C, Afzal NA, Upstill-Goddard R, Holloway JW, Simpson MA, Beattie RM, et al. (2013). Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut 62, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LJ, Esterhazy D, Kim S-H, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, et al. (2017). Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, and Elson CO. (2009). A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. U. S. A 106, 19256–19261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contijoch EJ, Britton GJ, Yang C, Mogno I, Li Z, Ng R, Llewellyn SR, Hira S, Johnson C, Rabinowitz KM, et al. (2019). Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel P, Brazier M, Cerutti I, Pieri F, Tardivel I, Desmet G, Baillet J, and Chany C. (1989). Pharmacokinetic study of butyric acid administered in vivo as sodium and arginine butyrate salts. Clin. Chim. Acta 181, 255–263. [DOI] [PubMed] [Google Scholar]
- Denson LA, Jurickova I, Karns R, Shaw KA, Cutler DJ, Okou DT, Dodd A, Quinn K, Mondal K, Aronow BJ, et al. (2018). Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn’s Disease. Gastroenterology 154, 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AS, and Fischbach MA. (2015). A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, and Mangelsdorf DJ. (2014). Nuclear Receptors, RXR, and the Big Bang. Cell 157, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, and Fujiyama Y. (2003). Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee MH, Han A, Lofgren SM, Beausang JF, Mendoza JL, Birnbaum ME, Bethune MT, Fischer S, Yang X, Gomez-Eerland R, et al. (2018). Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes. Cell 172, 549–563.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The Treatment-Naive Microbiome in New-Onset Crohn’s Disease - supplement. Cell Host Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, and Sansonetti PJ. (2003). Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem 278, 8869–8872. [DOI] [PubMed] [Google Scholar]
- Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, and Beisel CL. (2014). Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 5, e00928–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L, Huang H, Ripke S, Gusareva ES, Annese V, Hauser SL, et al. (2015). High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet 47, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DB, Luo C, O’Connell J, D, Lefkovith A, Yassour M, Varma M, Abelin G, J, Conway L, K, Jasso J, G, Matar G, C, et al. (2018). Antigen discovery and specification of immunodominance hierarchies for MHCIIrestricted epitopes. Nat. Med 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurry T, HST Microbiome Consortium*, Gibbons SM, Nguyen LTT, Kearney SM, Ananthakrishnan A, Jiang X, Duvallet C, Kassam Z, and Alm EJ. (2018). Predictability and persistence of prebiotic dietary supplementation in a healthy human cohort. Sci. Rep 8, 12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LBS, Roager HM, Søndertoft NB, Gøbel RJ, Kristensen M, Vallès-Colomer M, Vieira-Silva S, Ibrügger S, Lind MV, Mærkedahl RB, et al. (2018). A low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults. Nat. Commun 9, 4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, and Clardy J. (2019). Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed]
- Honda K, and Littman DR. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. [DOI] [PubMed] [Google Scholar]
- Hoyles L, Fernández-Real J-M, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, et al. (2018). Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nature Medicine 24, 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AS, Valastyan JS, and Bassler BL. (2016). A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe 19, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AV, Leonard MT, Jeppson JD, Swift M, Li G, Wong S, Peng S, Zaretsky JM, Heath JR, Ribas A, et al. (2019). T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods 16, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EM, Jajoo R, Cancilla D, Lubock NB, Wang J, Satyadi M, Cheung R, de March C, Bloom JS, Matsunami H, et al. (2019). A Scalable, Multiplexed Assay for Decoding GPCR-Ligand Interactions with RNA Sequencing. Cell Syst 8, 254–260.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, et al. (2017). Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Mol. Cell 68, 210–223.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, and Stappenbeck TS. (2016). The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 165, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaval KG, and Garsin DA. (2018). Ethanolamine Utilization in Bacteria. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney SM, Gibbons SM, Erdman SE, and Alm EJ. (2018). Orthogonal Dietary Niche Enables Reversible Engraftment of a Gut Bacterial Commensal. Cell Rep 24, 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Li H, Collins JJ, and Ingber DE. (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U. S. A 113, E7–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. (2014). Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 6, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji S-G, et al. (2017). Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie S, Conway KL, Lassen KG, Jijon HB, Pan H, Chun E, Michaud M, Lang JK, Gallini Comeau CA, Dreyfuss JM, et al. (2019). The Crohn’s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A, Rossi TM, Park B, Albini B, and Lebenthal E. (1989). Serum antibodies to cow’s milk proteins in pediatric inflammatory bowel disease. Crohn’s disease versus ulcerative colitis. Acta Paediatr. Scand 78, 384–389. [DOI] [PubMed] [Google Scholar]
- Li G, Bethune MT, Wong S, Joglekar AV, Leonard MT, Wang JK, Kim JT, Cheng D, Peng S, Zaretsky JM, et al. (2019). T cell antigen discovery via trogocytosis. Nat. Methods 16, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. (2019). Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 0. [DOI] [PMC free article] [PubMed]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. (2019). Multi-omics Detail the Gut Microbial Ecosystem in Inflammatory Bowel Disease. Nature. [DOI] [PMC free article] [PubMed]
- Lu E, Wolfreys FD, Muppidi JR, Xu Y, and Cyster JG. (2019). S-Geranylgeranyl-l-glutathione is a ligand for human B cell-confinement receptor P2RY8. Nature 567, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A 106, 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, and Camilleri M. (2017). Modulating bile acid pathways and TGR5 receptors for treating liver and GI diseases. Curr. Opin. Pharmacol 37, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawe GM, and Hoffman JM. (2013). Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol 10, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C, Tan J, Macia L, and Mackay CR. (2017). The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev 278, 277–295. [DOI] [PubMed] [Google Scholar]
- Mendes-Soares H, Raveh-Sadka T, Azulay S, Edens K, Ben-Shlomo Y, Cohen Y, Ofek T, Bachrach D, Stevens J, Colibaseanu D, et al. (2019). Assessment of a Personalized Approach to Predicting Postprandial Glycemic Responses to Food Among Individuals Without Diabetes. JAMA Netw Open 2, e188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milshteyn A, Colosimo DA, and Brady SF. (2018). Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe 23, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra B, Curran J, Herfarth H, Jagarlamudi K, and Trucksis M. (2018). P421 SER-287, an investigational microbiome therapeutic, induces remission and endoscopic improvement in a placebo-controlled, double-blind randomised trial in patients with active mild-to-moderate ulcerative colitis. Journal of Crohn S and Colitis 12, S317–S317. [Google Scholar]
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al. (2015). Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 149, 102–109.e6. [DOI] [PubMed] [Google Scholar]
- Momozawa Y, Dmitrieva J, Théâtre E, Deffontaine V, Rahmouni S, Charloteaux B, Crins F, Docampo E, Elansary M, Gori A-S, et al. (2018). IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun 9, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muise AM, Xu W, Guo C-H, Walters TD, Wolters VM, Fattouh R, Lam GY, Hu P, Murchie R, Sherlock M, et al. (2012). NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut 61, 1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramsothy S, Paramsothy R, Rubin DT, Kamm MA, Kaakoush NO, Mitchell HM, and Castaño-Rodríguez N. (2017). Faecal Microbiota Transplantation for Inflammatory Bowel Disease: A Systematic Review and Meta-analysis. J. Crohns. Colitis 11, 1180–1199. [DOI] [PubMed] [Google Scholar]
- Paun A, Yau C, Meshkibaf S, Daigneault MC, Marandi L, Mortin-Toth S, Bar-Or A, Allen-Vercoe E, Poussier P, and Danska JS. (2019). Association of HLA-dependent islet autoimmunity with systemic antibody responses to intestinal commensal bacteria in children. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagini F, Dilillo D, Borsani B, Cococcioni L, Galli E, Bedogni G, Zuin G, and Zuccotti GV. (2016). Nutrition in Pediatric Inflammatory Bowel Disease: From Etiology to Treatment. A Systematic Review. Nutrients 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud SP, Parker CR, Lo W-L, Weber KS, and Allen PM. (2014). Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat. Immunol 15, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, Manichanh C, Fouqueray C, Levenez F, Nielsen T, et al. (2016). Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol 1, 16152. [DOI] [PubMed] [Google Scholar]
- Prideaux L, De Cruz P, Ng SC, and Kamm MA. (2012). Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm. Bowel Dis 18, 1340–1355. [DOI] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64. [DOI] [PubMed] [Google Scholar]
- Ramachandran G, and Bikard D. (2019). Editing the microbiome the CRISPR way. Philos. Trans. R. Soc. Lond. B Biol. Sci 374, 20180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, et al. (2018). Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. (2011). Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roager HM, Vogt JK, Kristensen M, Hansen LBS, Ibrügger S, Mærkedahl RB, Bahl MI, Lind MV, Nielsen RL, Frøkiær H, et al. (2019). Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: a randomised cross-over trial. Gut 68, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda G, Jharap B, Neeraj N, and Colombel J-F. (2016). Loss of Response to Anti-TNFs: Definition, Epidemiology, and Management. Clin. Transl. Gastroenterol 7, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. [DOI] [PubMed] [Google Scholar]
- Rutherford ST, and Bassler BL. (2012). Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspectives in Medicine 2, a012427–a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Denson L, Vlamakis H, Franzosa EA, Thomas S, Gotman NM, Rufo P, Baker SS, Sauer C, Markowitz J, et al. (2018). Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host Microbe 24, 600–610.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Garner A, Vlamakis H, and Xavier RJ. (2019). Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol [DOI] [PMC free article] [PubMed]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S-H, Ma Z-X, Feltenberger JB, Chen H, Chen H, Scarlett C, Lin Z, Satyshur KA, Cortopassi M, Jefcoate CR, et al. (2018). Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J. Biol. Chem 293, 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, and Sonnenburg JL. (2018). An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly AN, Sato Y, Kearney S, and Honda K. (2019). Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol 19, 305–323. [DOI] [PubMed] [Google Scholar]
- Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, et al. (2018). Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation. Cell Host Microbe 23, 229–240.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, and Doré J. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis 15, 1183–1189. [DOI] [PubMed] [Google Scholar]
- Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, and Remaut E. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. [DOI] [PubMed] [Google Scholar]
- Stein RR, Tanoue T, Szabady RL, Bhattarai SK, Olle B, Norman JM, Suda W, Oshima K, Hattori M, Gerber GK, et al. (2018). Computer-guided design of optimal microbial consortia for immune system modulation. Elife 7, e30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et al. (2019). A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605. [DOI] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, and Bäumler AJ. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A 108, 17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y, Fuhrer T, Li H, Lawson MA, Zimmermann M, Yilmaz B, Zindel J, Ronchi F, Sorribas M, Hapfelmeier S, et al. (2018). Antibodies Set Boundaries Limiting Microbial Metabolite Penetration and the Resultant Mammalian Host Response. Immunity 49, 545–559.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, et al. (2010). Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3, 49–56. [DOI] [PubMed] [Google Scholar]
- Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. (2017). Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511. [DOI] [PubMed] [Google Scholar]
- Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, et al. (2018). Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 10. [DOI] [PubMed] [Google Scholar]
- Wang G-D, Wang X-Y, Zou F, Qu M, Liu S, Fei G, Xia Y, Needleman BJ, Mikami DJ, and Wood JD. (2013). Mast cell expression of the serotonin1A receptor in guinea pig and human intestine. Am. J. Physiol. Gastrointest. Liver Physiol 304, G855–G863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Simonavicius N, Wu X, Swaminath G, Reagan J, Tian H, and Ling L. (2006). Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem 281, 22021–22028. [DOI] [PubMed] [Google Scholar]
- Wegorzewska MM, Glowacki RWP, Hsieh SA, Donermeyer DL, Hickey CA, Horvath SC, Martens EC, Stappenbeck TS, and Allen PM. (2019). Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, Panchakshari RA, Rodig SJ, Kepler TB, and Alt FW. (2013). Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 501, 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. (2017). Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 22, 25–37.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff LBA, Gorochowski TE, Roehner N, Mikkelsen TS, Densmore D, Gordon DB, Nicol R, and Voigt CA. (2017). Registry in a tube: multiplexed pools of retrievable parts for genetic design space exploration. Nucleic Acids Res 45, 1567–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, and Podolsky DK. (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434. [DOI] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR. (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Garcia MAS, and Quinton PM. (2013). Normal mucus formation requires cAMP-dependent HCO3- secretion and Ca2+-mediated mucin exocytosis. J. Physiol 591, 4581–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, Banks AS, Bry L, and Devlin AS. (2018). A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B, Juillerat P, Øyås O, Ramon C, Bravo FD, Franc Y, Fournier N, Michetti P, Mueller C, Geuking M, et al. (2019). Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med 1. [DOI] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. (2015). Personalized Nutrition by Prediction of Glycemic Responses. Cell 163, 1079–1094. [DOI] [PubMed] [Google Scholar]
- Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, and Patil KR. (2015). Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U. S. A 112, 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB-Z, et al. (2018). Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 174, 1388–1405.e21. [DOI] [PubMed] [Google Scholar]