Abstract
Clinical text de-identification enables collaborative research while protecting patient privacy and confidentiality; however, concerns persist about the reduction in the utility of the de-identified text for information extraction and machine learning tasks. In the context of a deep learning experiment to detect altered mental status in emergency department provider notes, we tested several classifiers on clinical notes in their original form and on their automatically de-identified counterpart. We tested both traditional bag-of-words based machine learning models as well as word-embedding based deep learning models. We evaluated the models on 1,113 history of present illness notes. A total of 1,795 protected health information tokens were replaced in the de-identification process across all notes. The deep learning models had the best performance with accuracies of 95% on both original and de-identified notes. However, there was no significant difference in the performance of any of the models on the original vs. the de-identified notes.
Keywords: Machine Learning, Data Anonymization, Natural Language Processing
Introduction
The use of electronic health records (EHR) data to support research is growing in the translation research community [1]. Significant amounts of those data are trapped in free-text throughout a variety of clinical notes [2,3]. Provider notes often contain patient names, dates of services, and other types of Protected Health Information (PHI). In the United States, the Health Insurance Portability and Accountability Act (HIPAA) protects the confidentiality of patients [4], and based on the Common Rule, researchers are required to obtain either an informed consent from patients or a waiver of informed consent from an Internal Review Board (IRB) in order to use data for research [5]. As a result, automated de-identification in order to make text data more accessible for research is being investigated [6]. The impact of de-identification on the fidelity of information content as well as the utility of the data for research purposes, and information retrieval are also being evaluated [7,8]. In this paper, we examine the impact of de-identification on machine learning-based text classifiers. This experiment was conducted in the context of an automated approach for the detection of altered mental status (AMS) in emergency department (ED) physician notes for the purpose of decision support in the evaluation and management of pulmonary embolism [9–11].
Methods
This study was approved by the IRB at the Medical University of South Carolina (MUSC) under protocol # Pro00080055. We extracted ED physician notes from the MUSC Research Data Warehouse, which contains data extracted from the EHR system. The text notes span a period of 6 years, exported from one commercial EHR system. The notes were enriched with records from adult patients with concurrent International Classification of Diseases (ICD)-10 codes indicating AMS (e.g. codes under the R41 ICD-10 code hierarchy, which includes symptoms and signs involving cognitive functions and awareness) and an equal number of records from patients without AMS ICD codes as controls or negative cases. Using regular expressions to identify the section headers, a total of 8,194 clinical notes were segmented into the different components of a clinical record including the history of present illness (HPI), physical exam, assessment, etc.
Labeling
The parsed notes were imported into REDCap [12] and made available to clinical members of our research team (two ED physicians, a Pediatrician and a Doctor of Pharmacy) for review and labeling as either AMS or not AMS. The team was instructed to look for any signs or symptoms of AMS in the context of pulmonary embolism as described in the literature [9,10] within the HPI, e.g. disorientation, lethargy, stupor, somnolence, confusion, coma, loss of consciousness, or syncope. They were also asked to drop repetitive notes for patients with frequent ED visits in order to minimize bias in testing the models. Cases that were deemed not clear cut AMS by a reviewer, were labeled by consensus after consultation with other team members. The team completed the labeling on 1,113 HPI text notes from 849 patients, with 487 notes labeled as AMS, and 626 labeled as non-AMS. A sub-sample of 100 notes was labeled independently by two labelers in order to estimate the interrater reliability.
De-identification
For automated de-identification, we used a system based on ‘BoB’ (best-of-breed clinical text de-identification application) [13,14] with a previously demonstrated precision of 93% and recall of 76% on MUSC data [15]. This system combines high precision algorithms (e.g., regular expression matching) and high recall algorithms, e.g., Conditional Random Fields (CRF) and Support Vector Machines (SVM). After all models are run on a document, their individual PHI determinations are consolidated into a single coherent output using an ensemble method. Tokens tagged as PHI can be replaced with tags, (e.g. a name replaced with a single generic token “[***PHI***]” or a corresponding PHI-type or class name e.g. “[*** Patient ***]”) or “resynthesized” (i.e., PHI is replaced with realistic surrogates so a name will be replaced with a different, randomly sampled name). For the purposes of this study, we selected to use the multi-class token replacements in the de-identified output.
Text Processing
We used R version 3.5.1 [16] for constructing the machine learning pipelines, and Keras [17] and TensorFlow [18] for constructing and training the deep learning models. We ran both the original text (i.e., not de-identified) and de-identified text through several traditional bag-of-words (BOW) text classifiers, and word-embedding (WE)-based deep learning models. Text processing for the BOW models was done using the quanteda package [19] and included lower casing, punctuation removal, stop-word removal, stemming, and tokenization. The BOW word frequencies were normalized using term-frequency, inverse document frequency (tf-idf) a weighted approach for term discrimination [20]. For the WE models, we used Keras for text processing, which included lower casing, sentence segmentation, punctuation removal, and tokenization, followed by sequence padding to ensure that all sequences have the same length.
Baseline Approaches
As a baseline, we used regular expressions (regex) on lowercased notes to identify AMS key words as described in the literature in the context of pulmonary embolism [9,10] (e.g. altered, disorientation, disoriented, lethargy, stupor, confusion, syncope, etc.). The regex algorithm was refined after several iterations of testing against the labeled data to include other patterns based on the root words and abbreviations. We also examined the accuracy of ICD codes against the labeled data.
BOW-based Machine Learning Models
The traditional models included: Naïve Bayes Classifier (NBC) with uniform priors, smoothing of 1; Lasso (LASS) with a default alpha=1 and lambda=NULL [21]; Single Decision Tree (SDT) [22] with a maximum depth of 20; Random Forrest (RF) [23] with 201 trees and the number of variables randomly sampled as candidates at each split (mtry)=150; SVM Type 1 with a radial basis kernel [24]; and a Simple Multilayer Perceptron (SMP) artificial neural network with 64-node input and hidden layers.
Word Embedding - Deep Learning Models
The deep learning model was based on the architecture described by Kim [25]. However, instead of using parallel channels for the embedding layer, we used either a pre-trained layer using word2vec (W2V) [26] or word embedding without pre-training (vectors randomly initialized using a uniform distribution) with either of 50 (D50) or 200 (D200) dimensions per word. The W2V weights were derived by pre-training the W2V skip-gram model on all 8,194 HPI (original, not de-identified) notes using a window of 5 words in each direction and 200 dimensions per word. The next layer was a convolutional layer or convolutional neural network (CNN) with multi-filter sizes (3, 4 and 5), 200 nodes each, with global maxpooling, followed by a merge tensor, a fully connected 200 node layer, then a single sigmoid activation output node. A dropout rate of 0.2 was used after both the embedding layer and the last dense layer. Other deep neural network architectures were tested including Kim’s and CNN’s with larger window sizes; however, we chose the above architecture due to its superior performance in our hands for the purpose of demonstrating the impact of de-identification.
Training and Evaluation
Due to the relatively small number of labeled clinical notes, all the models were trained and evaluated using 5-fold train/test cycles, i.e. the test set in each of those runs was used as an unseen holdout set. Moreover, in order to ensure further consistency in results, the experiment was repeated 5 times by bootstrap sampling using different random seeds. Therefore, all models were run a total of 25 times on different train/test sets. In order to allow comparison between models, the same train/test sets were used during each cycle for all the models. The same random seeds were used for both the original and de-identified sets, thus ensuring identical partitioning of train/test sets and that the models trained on original notes were trained on the de-identified version of the same original notes. The performance metrics, including area under the receiver operating characteristic curve (AUC), were calculated based on the pooled predictions of the test data from the 25 runs. The caret package was used for k-fold, bootstrap sampling and calculations of the metrics [27].
Results
Dataset Statistics
The BOW matrices generated after lower casing, stop-word removal and stemming had token dimensions of 5,200 and 4,925 for the original and de-identified sets respectively. The WE sequences, which did not undergo stop-word removal and stemming, had vocabulary sizes of 7,260 and 6,923 for the original and de-identified sets respectively. The HPI note sizes ranged from 21 to 716 words with a median of 174 words for both original and identified sets. The corpus of the labeled HPI notes included a total of 207,475 tokens.
De-identification Results
The automated de-identification resulted in the replacement of 1,795 PHI tokens from a variety of types or classes of PHI (Table 1), which is less than 1% of all tokens in the corpus. The most prevalent replacements within our HPI data were related to health care unit names (such as “MUSC” or “Gastrointestinal” unit) and ages. The different PHI classes and their prevalences are listed in Table 1.
Table 1–
Breakdown of the Numbers of PHI Tokens Replaced by the De-identification System.
| PHI Token Class | n | % |
|---|---|---|
| Health care unit names | 558 | 31.1% |
| Ages greater than 89 | 512 | 28.5% |
| Dates | 360 | 20.1% |
| Provider names | 128 | 7.1% |
| Patient names | 122 | 6.8% |
| Street or City | 73 | 4.1% |
| State or Country | 16 | 0.9% |
| Phone numbers | 15 | 0.8% |
| Other organization names | 10 | 0.6% |
| Other IDs | 1 | 0.1% |
| Total | 1795 | 100.0% |
Baseline Analyses
We had a fairly high interrater reliability between labelers (Cohen’s Kappa = 0.94). Using the ICD codes listed in table 2, the accuracy of concurrent ICD codes assignments associated with the labeled clinical notes was 81.1% (precision 72.2%, recall 92.4%). The presence of any of these codes was considered as positive for AMS otherwise the note was considered negative for AMS by ICD.
Table 2–
List of ICD-9 and ICD-10 Codes Considered to be AMS in the Context of Pulmonary Embolism.
| Code Set | ICD Code | Diagnosis Name |
|---|---|---|
| ICD9 | 780.0x | Alteration of consciousness |
| ICD9 | 780.2 | Syncope and collapse |
| ICD9 | 780.97 | Altered mental status |
| ICD9 | 799.5x | Signs and symptoms involving cognition |
| ICD10 | R40.x | Somnolence, stupor and coma |
| ICD10 | R41.0 | Disorientation, unspecified |
| ICD10 | R41.8x | Other symptoms and signs involving cognitive functions and awareness |
| ICD10 | R41.9 | Unspecified symptoms and signs involving cognitive functions and awareness |
| ICD10 | R55 | Syncope and collapse |
The accuracy of classification using the refined regex patterns against the labeled notes was 88.1% (precision 81.3%, recall 94.7%).
Machine Learning Performance
There was no discernable difference in performance of any of the models between original and de-identified text, with significant overlap in the 95% confidence intervals of the AUC’s for all the models across original vs. de-identified (Figure 1). Table 3 shows the performance of the models, along with the differences in performance between original and de-identified text (Δ’s).
Figure 1.
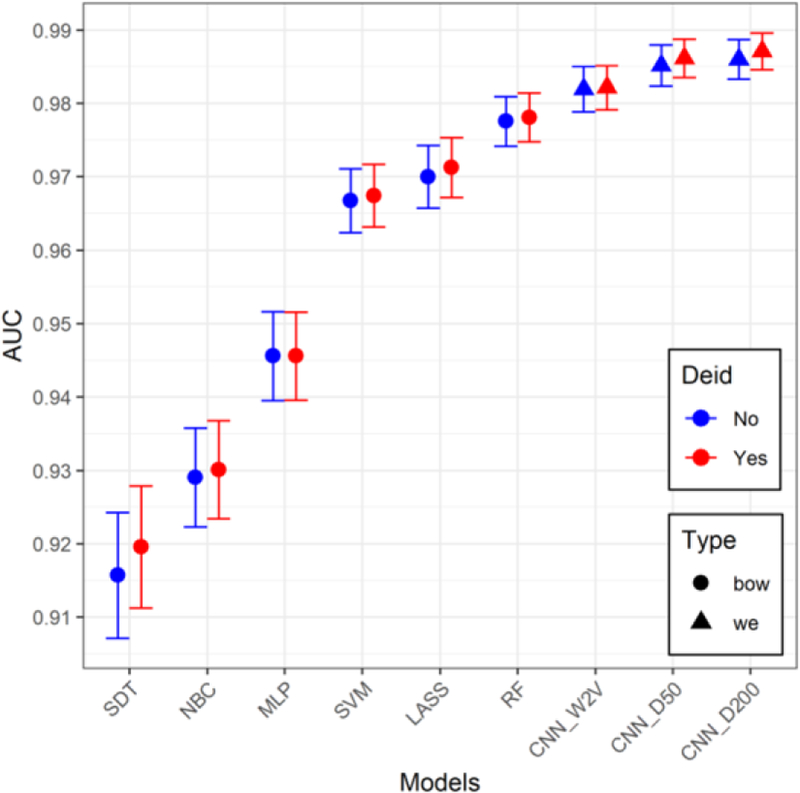
AUC values and 95% confidence intervals for all the models for both original and de-identified (Deid) data.
Table 3–
Performance of models. (Acc=accuracy, Prec=precision; other abbreviations are in the text, Δ = De-identified – Original, 95% CI=95% confidence interval)
| Original | De-identified | Δ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | AUC (95% CI) | Acc | Prec | Recall | AUC (95% CI) | Acc | Prec | Recall | AUC | Acc |
| RF | 0.978 (0.974–0.981) | 0.924 | 0.938 | 0.885 | 0.978 (0.975–0.981) | 0.928 | 0.943 | 0.890 | 0.001 | 0.004 |
| LASS | 0.970 (0.966–0.974) | 0.906 | 0.955 | 0.824 | 0.971 (0.967–0.975) | 0.907 | 0.958 | 0.823 | 0.001 | 0.001 |
| SVM | 0.967 (0.962–0.971) | 0.907 | 0.905 | 0.879 | 0.967 (0.963–0.972) | 0.908 | 0.907 | 0.880 | 0.001 | 0.001 |
| MLP | 0.946 (0.940–0.952) | 0.885 | 0.875 | 0.860 | 0.946 (0.940–0.952) | 0.883 | 0.869 | 0.863 | 0.000 | −0.002 |
| NBC | 0.929 (0.922–0.936) | 0.842 | 0.776 | 0.898 | 0.930 (0.923–0.937) | 0.848 | 0.782 | 0.903 | 0.001 | 0.006 |
| SDT | 0.916 (0.907–0.924) | 0.911 | 0.921 | 0.870 | 0.920 (0.911–0.928) | 0.913 | 0.927 | 0.870 | 0.004 | 0.002 |
| CNN D200 | 0.986 (0.983–0.989) | 0.946 | 0.946 | 0.929 | 0.987 (0.985–0.990) | 0.949 | 0.949 | 0.934 | 0.001 | 0.004 |
| CNN D50 | 0.985 (0.982–0.988) | 0.948 | 0.945 | 0.934 | 0.986 (0.984–0.989) | 0.948 | 0.947 | 0.934 | 0.001 | 0.001 |
| CNN W2V | 0.982 (0.979–0.985) | 0.939 | 0.941 | 0.918 | 0.982 (0.979–0.985) | 0.937 | 0.936 | 0.919 | 0.000 | −0.001 |
The RF model was the best performer in the BOW models with AUC’s of 0.978, in both original and de-identified texts. Both CNN’s (D50 and D200) with the randomly initialized word embeddings had the best overall performance with AUC’s near 99% and average accuracies near 95% across both original and de-identified text, exceeding those of the W2V model.
Discussion
The de-identification resulted in the replacement of 1,795 PHI tokens out of a total of over 200 thousand tokens in our sample of 1,113 HPI notes. Table 1 demonstrates the extent of de-identification that the notes were subjected to. The results show negligible difference in performance of text classifiers on original vs. de-identified HPI notes, across all types of machine learning models. The deep learning models in particular seem to perform exceedingly well in both environments.
We hypothesized that the replacement of a number of different named entities with uniform classes of tokens, which slightly reduces the vocabulary size in our corpus and therefore the number of features that the BOW and WE models have to deal with, could potentially improve machine learning performance due to reduced dimensionality and noise in the data. If such an advantage exists, it could not be definitively demonstrated in our results. Several of the models seem to have a slight advantage when applied to the de-identified set, but given the 95% confidence intervals’ overlap between original and de-identified, this difference is not significant. It is worth noting that such a result would also be expected if a PHI ‘resynthesis’ process were used (i.e., replacement of PHI with realistic surrogates), which is an option in the automatic de-identification system described above, and is based on a large and diverse database of possible surrogates (e.g., all last names found in the U.S. national census). The resynthesis might allow for conservation of diversity of the original PHI.
The fact that automated de-identification did not reduce the accuracy of machine learning performance should be of interest to the translational research community. Clinical text corpora are critical for biomedical informatics research in domains such as machine learning, ontology annotations, predictive modeling and precision medicine, to name a few. Automated de-identification should make such corpora more accessible at scale for such research. While automated de-identification technologies have matured significantly in recent years, regulatory guidelines still lag behind. Looking ahead, we envision several governance models in which de-identified text corpora could be made available with appropriate data use agreements with minimal oversight and review by ethics boards. Institutional research leaders at academic medical centers should work closely with their offices of research oversight and local informatics experts to make such resources more accessible.
Regarding the performance of deep learning vs. the traditional classifiers, the deep learning classifiers seem to significantly outperform the BOW-based classifiers with the exception of RF, which had a performance approaching that of the CNN’s. Not surprisingly, all machine learning models outperformed ICD codes on accuracy, which is consistent with reports in the literature on coder errors, such as misattribution, unbundling, and upcoding [28,29]. However, only the better performing models (MLP and above) had better accuracies than the optimized regex classifier. The accuracies of the deep learning models were particularly far superior to the regex accuracy. However, none of the models outperformed the high recall demonstrated by our regex classifier. However, the regex approach required significant fine-tuning specific to the detection of AMS keywords in order to yield this performance. As such, the regex approach is more difficult to generalize to other classification problems. Finally, it should be noted that the W2V initialized WE models, did not outperform the randomly initialized WE models. In fact they seem to have consistently lower AUC’s, but with overlapping 95% confidence intervals. The lower performance is possibly due to the relatively small amount of pre-training data, for example, compared to models trained on all of PubMed.
Limitations
Our text corpus represents data from one health system, on a single EHR system making it difficult to draw generalizations about performance in other environments. We also examined the performance of machine learning through the narrow prism of a simple text classifier to identify AMS in one type of EHR clinical text, namely the HPI. Future work should include expanding the study to other institutions and examining other types of notes and machine learning tasks, such as predictions of outcomes.
Conclusions
Despite the limitations outlined above, this simple experiment demonstrates the power of an automated de-identification pipeline, and the preservation of text features that are necessary for the performance of both deep learning models as well as the more traditional machine learning models used in text classification tasks.
Acknowledgements
This project was supported in part by the South Carolina SmartState Program, the South Carolina Clinical & Translational Research Institute, with an academic home at the Medical University of South Carolina, through the National Institutes of Health - National Center for Advancing Translational Sciences grant number UL1-TR001450, and the Delaware-CTR Accel program through the National Institute of General Medical Sciences grant number U54-GM104941.
References
- [1].Obeid JS., Beskow LM., Rape M., Gouripeddi R., Black RA., Cimino JJ., Embi PJ., Weng C., Marnocha R., and Buse JB., A survey of practices for the use of electronic health records to support research recruitment, Journal of Clinical and Translational Science 1 (2017), 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Meystre SM., Savova GK., Kipper-Schuler KC., and Hurdle JF., Extracting information from textual documents in the electronic health record: a review of recent research, Yearbook of medical informatics 17 (2008), 128–144. [PubMed] [Google Scholar]
- [3].Shivade C., Raghavan P., Fosler-Lussier E., Embi PJ., Elhadad N., Johnson SB., and Lai AM., A review of approaches to identifying patient phenotype cohorts using electronic health records, Journal of the American Medical Informatics Association 21 (2014), 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].HIPAA Privacy Rule, 45 CFR Part 160, Part 164(A,E)., U.S. Department of Health and Humans Services, 2002. [Google Scholar]
- [5].Federal Policy for the Protection of Human Subjects (‘Common Rule, HHS.Gov. (2009). https://www.hhs.gov/ohrp/regulations-and-policy/regulations/common-rule/index.html (accessed November 20, 2018).
- [6].Meystre SM., Friedlin FJ., South BR., Shen S., and Samore MH., Automatic de-identification of textual documents in the electronic health record: a review of recent research, BMC Medical Research Methodology 10 (2010), 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meystre SM., Ferrández Ó., Friedlin FJ., South BR., Shen S., and Samore MH., Text de-identification for privacy protection: a study of its impact on clinical text information content, Journal of Biomedical Informatics 50 (2014), 142–150. [DOI] [PubMed] [Google Scholar]
- [8].Deleger L., Molnar K., Savova G., Xia F., Lingren T., Li Q., Marsolo K., Jegga A., Kaiser M., Stoutenborough L., and Solti I., Large-scale evaluation of automated clinical note de-identification and its impact on information extraction, Journal of the American Medical Informatics Association 20 (2013) 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Donzé J., Le Gal G., Fine MJ., Roy P-M., Sanchez O., Verschuren F., Cornuz J., Meyer G., Perrier A., Righini M., and Aujesky D., Prospective validation of the Pulmonary Embolism Severity Index, Thrombosis and haemostasis 100 (2008), 943–948. [DOI] [PubMed] [Google Scholar]
- [10].Prandoni P., Lensing AWA., Prins MH., Ciammaichella M., Perlati M., Mumoli N., Bucherini E., Visonà A., Bova C., Imberti D., Campostrini S., Barbar S., and PESIT Investigators, Prevalence of Pulmonary Embolism among Patients Hospitalized for Syncope, New England Journal of Medicine, 375 (2016) 1524–1531. [DOI] [PubMed] [Google Scholar]
- [11].Costantino G., Ruwald MH., Quinn J., Camargo CA., Dalgaard F., Gislason G., Goto T., Hasegawa K., Kaul P., Montano N., Numé A-K., Russo A., Sheldon R., Solbiati M., Sun B., and Casazza G., Prevalence of Pulmonary Embolism in Patients With Syncope, JAMA Internal Medicine, 178 (2018), [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Harris PA., Taylor R., Thielke R., Payne J., Gonzalez N., and Conde JG., Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support, Journal of Biomedical Informatics 42 (2009), 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ferrández O., South BR., Shen S., Friedlin FJ., Samore MH., and Meystre SM., BoB, a best-of-breed automated text de-identification system for VHA clinical documents, Journal of the American Medical Informatics Association 20 (2013), 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim Y., Heider P., and Meystre SM., Ensemble-based methods to improve de-identification of electronic health record narratives., in: AMIA Annual Symposium Proceedings 2018, San Francisco, CA, n.d: pp. 663–672. [PMC free article] [PubMed] [Google Scholar]
- [15].Meystre SM., Heider P., Kim Y., Trice A., and Underwood G., Clinical text automatic de-identification to support large scale data reuse and sharing: pilot results., in: AMIA Annual Symposium Proceedings 2018, San Francisco, CA, n.d: p. 2069. [Google Scholar]
- [16].R Core Team R: A Language and Environment for Statistical Computing., (2018).
- [17].Chollet F., Keras, (2018). https://keras.io/ (accessed November 20, 2018).
- [18].Abadi Martín, Agarwal Ashish, Barham Paul, Brevdo Eugene, Chen Zhifeng, Citro Craig, Corrado Greg S., Davis Andy, Dean Jeffrey, Devin Matthieu, Ghemawat Sanjay, Goodfellow Ian, Harp Andrew, Irving Geoffrey, Isard Michael, Jia Y., Jozefowicz Rafal, Kaiser Lukasz, Kudlur Manjunath, Levenberg Josh, Mané Dandelion, Monga Rajat, Moore Sherry, Murray Derek, Olah Chris, Schuster Mike, Shlens Jonathon, Steiner Benoit, Sutskever Ilya, Talwar Kunal, Tucker Paul, Vanhoucke Vincent, Vasudevan Vijay, Viégas Fernanda, Vinyals Oriol, Warden Pete, Wattenberg Martin, Wicke Martin, Yu Yuan, and Zheng Xiaoqiang, TensorFlow: large-scale machine learning on heterogeneous systems, (2018). https://www.tensorflow.org/ (accessed November 20, 2018).
- [19].Benoit K., Watanabe K., Wang H., Nulty P., Obeng A., Müller S., and Matsuo A., quanteda: An R package for the quantitative analysis of textual data, Journal of Open Source Software. 3 (2018), 774. [Google Scholar]
- [20].Salton G., Yang CS., and Yu CT., A theory of term importance in automatic text analysis, Journal of the American Society for Information Science 26 (1975), 33–44. [Google Scholar]
- [21].Friedman J., Hastie T., and Tibshirani R., Regularization paths for generalized linear models via coordinate descent, Journal of Statistical Software 33 (2010), 1–22. [PMC free article] [PubMed] [Google Scholar]
- [22].Breiman L., Classification and regression trees, Chapman & Hall/CRC, New York, N.Y, 1984. http://lib.myilibrary.com?id=1043565 (accessed December 6, 2018). [Google Scholar]
- [23].Breiman L., Random Forests, Machine Learning 45 (2001), 5–32. [Google Scholar]
- [24].Weston J., and Watkins C., Multi-class support vector machines, Citeseer, 1998. [Google Scholar]
- [25].Kim Y., Convolutional neural networks for sentence classification, in: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Association for Computational Linguistics, Doha, Qatar, 2014: pp. 1746–1751. [Google Scholar]
- [26].Mikolov T., Chen K., Corrado G., and Dean J., Efficient Estimation of Word Representations in Vector Space, (2013). https://arxiv.org/abs/1301.3781v3 (accessed November 20, 2018).
- [27].Kuhn M., The caret Package, n.d. http://topepo.github.io/caret/index.html (accessed December 6, 2018).
- [28].O’Malley KJ., Cook KF., Price MD., Wildes KR., Hurdle JF., and Ashton CM., Measuring diagnoses: ICD code accuracy, Health Services Research 40 (2005), 1620–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wei W-Q., Leibson CL., Ransom JE., Kho AN., Caraballo PJ., Chai HS., Yawn BP., Pacheco JA., and Chute CG., Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus, Journal of the American Medical Informatics Association 19 (2012), 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]


