Abstract
Background:
Exosomes are nanovesicles that have been shown to mediate carcinogenesis in pancreatic ductal adenocarcinoma (PDAC). Given the direct communication of pancreatic duct fluid with the tumor and its relative accessibility, we aimed to determine the feasibility of isolating and characterizing exosomes from pancreatic duct fluid.
Methods:
Pancreatic duct fluid was collected from 26 patients with PDAC (n=13), intraductal papillary mucinous neoplasm (IPMN) (n=8) and other benign pancreatic diseases (n=5) at resection. Exosomes were isolated by serial ultracentrifugation, proteins were identified by mass spectrometry, and their expression was evaluated by immunohistochemistry.
Results:
Exosomes were isolated from all specimens with a mean concentration of 5.9 +/− 1 x 108 particles/mL and most frequent size of 138 +/− 9nm. Among the top 35 proteins that were significantly associated with PDAC, multiple carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) and extracellular matrix (ECM) proteins were identified. Interestingly, CEACAM 1/5 expression by immunohistochemistry was seen only on tumor epithelia whereas tenascin C positivity was restricted to stroma, suggesting that both tumor and stromal cells contributed to exosomes.
Conclusions:
This is the first study showing that exosome isolation is feasible from pancreatic duct fluid, and that exosomal proteins may be utilized to diagnose patients with PDAC.
Keywords: Exosomes, extracellular vesicles, proteomics, pancreatic ductal adenocarcinoma, intraductal papillary mucinous neoplasm, CEACAM, tenascin C
INTRODUCTION
Exosomes are lipid-bilayer extracellular nanovesicles containing proteins, DNAs, and microRNAs.(1, 2) Secreted from many normal and cancerous cells, exosomes have emerged as an important component of the intercellular communication between cancer cells and their microenvironment.(3, 4) Cancer-derived exosomes have been shown to mediate tumorigenesis and metastasis in melanoma, colon cancer, and pancreatic ductal adenocarcinoma (PDAC).(5–8)
PDAC develops as a stepwise progression from precursor and premalignant lesions such as intraductal papillary mucinous neoplasm (IPMN) and pancreatic intraepithelial neoplasia (PanIN).(9, 10) Molecular alterations in KRAS, p16, and p53 have revealed some understanding of this progression, but much of its biological behavior and time to progression remain unknown.(9, 11) Not only is it challenging to detect invasive component in pre-existing IPMN and PanIN, it is also difficult to differentiate PDAC from benign pancreas lesions.(9) In studying PDAC derived exosomes, glypican-1 was identified to distinguish PDAC from normal pancreas, and macrophage migration inhibitory factor was found to stimulate liver metastasis.(7, 8, 12)
Prior studies involving PDAC-related exosome interrogation have primarily utilized peripheral blood.(7, 8, 13) Although peripheral blood based proteins are more appealing due to easy access, pancreatic duct fluid has the inherent advantage of potential enrichment of proteins given the direct contact with the tumor epithelia and stroma.(9) Exosomes from primary tumor and stroma secreted into pancreatic duct fluid would provide PDAC-specific information and its unique exosomal protein composition may be useful to distinguish patients with PDAC from other pathology. Endoscopic ultrasound guided fine needle aspiration or core needle biopsies are often used in patients presenting with suspicious pancreatic lesions to confirm diagnosis and dictate course of treatment.(14, 15) Although specificity is high, the sensitivity of pancreatic duct fluid cytopathology may vary due to the evaluation of suspicious, atypical, or nondiagnostic cells.(14) We rationalized that pancreatic duct fluid would be an ideal medium to study PDAC, as it may contain a high concentration of PDAC-specific exosomes that are relatively accessible such as via endoscopic approach and would reflect contribution of exosomes from tumor and stroma without invasive biopsies or excision.
Since pancreatic duct fluid exosomes have not been characterized, the goal of this pilot study was to evaluate the feasibility of exosome isolation from pancreatic duct fluid of patients with PDAC as well as IPMN and other benign pancreatic diseases, and to compare their protein composition in order to distinguish PDAC from other pancreatic diagnoses.
METHODS
Study patients
With the approval of the Memorial Sloan Kettering Cancer Center Institutional Review Board (IRB), pancreatic duct fluid was prospectively collected from patients during pancreaticoduodenectomy or distal pancreatectomy in 2010 to 2014. At the time of pancreatic parenchymal transection, 1mL of pancreatic duct fluid was aspirated from the both sides of the transected main pancreatic duct using a 19-gauge needle. Patients included in the study had PDAC, IPMN with or without microinvasive cancer, or other benign pancreatic diseases serving as the controls. Patients were excluded from the study if less than 1mL of pancreatic duct fluid was collected at the time of the surgery.
In addition to patients enrolled for exosome analysis, we obtained non-matched pancreatic tumor from additional patients with PDAC and IPMN for immunohistochemistry. These patients were selected from a prospective biospecimen banking protocol approved by IRB based on the absence of neoadjuvant chemoradiation and availability of adequate formalin-fixed paraffin-embedded (FFPE) tumor blocks from tumors resected during 2003 to 2016. None of these patients had concurrent malignancy of another type and all patients either developed recurrence or had at least 3 years of recurrence-free follow-up.
Pancreatic duct fluid analysis
Pancreatic duct fluid was snap-frozen in liquid nitrogen and stored at −80°C until analysis. One mL of pancreatic duct fluid from each patient was used for analysis. Serial ultracentrifugation protocol was used to remove cell debris and isolate exosomes, as previously described protocol for plasma(8). In brief, pancreatic duct fluid was centrifuged at 500xg for 10 minutes at 4C. The supernatant then was centrifuged at 3,000xg for 20 minutes, followed by 12,000xg for 20 minutes, and then 100,000xg for 70 minutes. The supernatant of the last centrifugation was removed and the pellet was resuspended in 2mL of phosphate-buffer saline (PBS), centrifuged at 100,000xg for 70 minutes, and the pellet was resuspended in 100 uL of PBS.
We quantified exosome size and concentration using the DS500 nanoparticle characterization system (NanoSight, Malvern Instruments) equipped with a blue laser (405 nm). Presence and morphology of exosomes and characteristic size of approximately 100nm were visually confirmed by transmission electron microscopy. Exosomal proteins were identified and analyzed using liquid chromatography-mass spectrometry (LC-MS), as previously described. (8, 16) Levels of expressions of exosomal proteins were quantified as presence or absence as well as by their relative expression levels using Log2 transformed label-free quantification (LFQ) intensities.(16, 17)
Pancreatic tumor immunohistochemistry
The expression of selected exosomal proteins was evaluated on non-matched primary pancreatic tumors by IHC. Representative FFPE tumor blocks were obtained for each patient and the IHC studies were performed with a Leica Bond RX (Leica Biosystems) automated stainer on 5-μm tissue sections using the standard avidin-biotin peroxidase method. Pancreatic tumors were immunostained with anti-CEACAM1 + CEACAM 5 antibody (Abcam #190718) and anti-tenascin C antibody (Abcam #ab108930). To ensure antibody specificity, mouse and rabbit immunoglobulins were used as negative controls for CEACAM 1/5 and tenascin C, respectively. After staining, the sections were mounted with Permount for digital scanning with Pannoramic Confocal (3dHistech). Stained tissue slides were scored by 2 pathologists (A.G. and B.O.) blinded to clinical information. The epithelial tumor cells and adjacent stromal components were both evaluated for immuno-labeling. The staining pattern was scored as negative (0), focal (1) (labeling in 10-25% of cells), and diffuse (labeling in ≥ 25% of cells) (2), whereas staining intensity was scored as weak (1), moderate (2), or strong (3). The expression for CEACAM 1/5 and tenascin C was accepted as positive if a total score of ≥ 4 was obtained from combined staining intensity and pattern. For CEACAM, expression pattern was further categorized as being in the membranous or luminal part of the tumor epithelial cells.
Statistical analysis
Categorical variables were expressed as frequency and percentage and were compared using Fisher’s exact test. Continuous variables were expressed as median and range and were compared using Mann-Whitney. P values < 0.05 from two-sided tests were considered significant. Statistical analyses were performed using GraphPad Prism (GraphPad Software, CA) and SPSS version 22 (IBM Corp., Armonk, NY). Venn’s diagram was plotted using Venny 2.1 (BioinfoGP Service, Spain, http://bioinfogp.cnb.csic.es/tools/venny/).
RESULTS
Patients
Exosomes were isolated and characterized from the pancreatic duct fluid of patients with PDAC (n=13), IPMN with microinvasive cancer (n=4), IPMN without invasive component (n=4), or other benign pancreatic diseases, including 3 with chronic pancreatitis, 1 with retention cyst, and 1 with duodenal adenoma. Whole tissue sections of primary pancreas tumors were obtained from additional patients with PDAC (n=27) or IPMN (n=12) for IHC. These included 6 patients with PDAC and 1 patient with IPMN from the original exosome isolation cohort. Clinical and pathological characteristics of all study patients are shown (Table 1).
Table 1.
Study patients.
| Exosome Quantification Cohort | Immunohistochemistry Cohort | |||||
|---|---|---|---|---|---|---|
| PDAC (n=13) | IPMNca (n=4) | IPMN (n=4) | Benign (n=5) | PDAC (n=27) | IPMN (n=12) | |
| Age | 66 (54-86) | 67 (54-76) | 77 (61-81) | 76 (57-92) | 66 (45-87) | 71 (50-81) |
| Male | 9 (69%) | 2 (50%) | 1 (25%) | 2 (40%) | 17 (63%) | 7 (58%) |
| CA 19-9, unit/mL | 176 (16-921) | 14 (13-42) | 19 | 29 (16-69) | 251 (21-9337) | 45 (4-89) |
| Biliary stent | 5 (38%) | 1 (25%) | 0 (0%) | 1 (20%) | 14 (52%) | 0 (0%) |
| Whipple | 12 (92%) | 4 (100%) | 2 (50%) | 4 (80%) | 27 (100%) | 12 (100%) |
| Pathologic diagnosis | 13 (100%) PDAC | 1 (25%) BD 3 (75%) MD |
1 (25%) BD 3 (75%) MD |
3 (60%) pancreatitis 1 (20%) adenoma 1 (20%) cyst |
27 (100%) PDAC | 3 (25%) BD 9 (75%) MD |
| TNM stage | -- | |||||
| IA | 2 (15%) | 2 (50%) | -- | -- | 0 (0%) | |
| IIA | 3 (23%) | 1 (25%) | 7 (26%) | |||
| IIB | 8 (62%) | 1 (25%) | 20 (74%) | |||
| Tumor size, cm | 2.5 (0.6-4) | 0.8 (0.3-4) | 2.6 (1.3-3.2) | -- | 2.4 (1.3-5) | 1.9 (1.2-3.5) |
| Differentiation/dysplasia | ||||||
| Moderate/intermediate | 5 (38%) | 1 (25%) | 3 (75%) | -- | 13 (48%) | 3 (25%) |
| Poor/high | 8 (62%) | 3 (75%) | 1 (25%) | -- | 13 (48%) | 6 (50%) |
| PanIN | ||||||
| 1-2 | 2 (15%) | 1 (25%) | 2 (50%) | 4 (80%) | 6 (22%) | 3 (25%) |
| 3 | 9 (69%) | 0 (0%) | 0 (0%) | 0 (0%) | 17 (63%) | 0 (0%) |
| Lymphovascular invasion | 11 (85%) | 0 (0%) | -- | -- | 20 (74%) | -- |
| Perineural invasion | 12 (92%) | 0 (0%) | -- | -- | 24 (89%) | -- |
PDAC, pancreatic ductal adenocarcinoma; IPMN, intraductal papillary mucinous neoplasm; IPMNca, IPMN with microinvasive cancer; BD, branch duct IPMN; MD, main duct IPMN; TNM, tumor node metastasis; PanIN, pancreatic intraepithelial neoplasia
Categorical variables are expressed as frequency (percentage) and continuous variables are expressed as median (range).
Exosome quantification
Using Nanosight characterization, the mean concentration of exosomes isolated was 5.9 +/− 1 x 108 particles/mL. The most frequent particle size was 138 +/− 9nm with the mean size of 188 +/− 5nm. No significant differences were detected in size distribution or concentration of exosomes when patients were stratified by diagnosis (Supplementary Table 1). Isolated exosomes were visualized by transmission electron microscopy (Fig. 1A). Characteristic exosomal surface markers including CD9, CD81, and TSG101 were identified using Data Dependent Acquisition-based liquid chromatography-mass spectrometry (LC-MS/MS) as described previously. (2, 16, 18) As expected, cellular organelle markers such as Golgi marker 130 (GM130) and mitochondrial marker cytochrome C1 (CYC1) were not detected on the exosomes.(18)
Fig. 1.
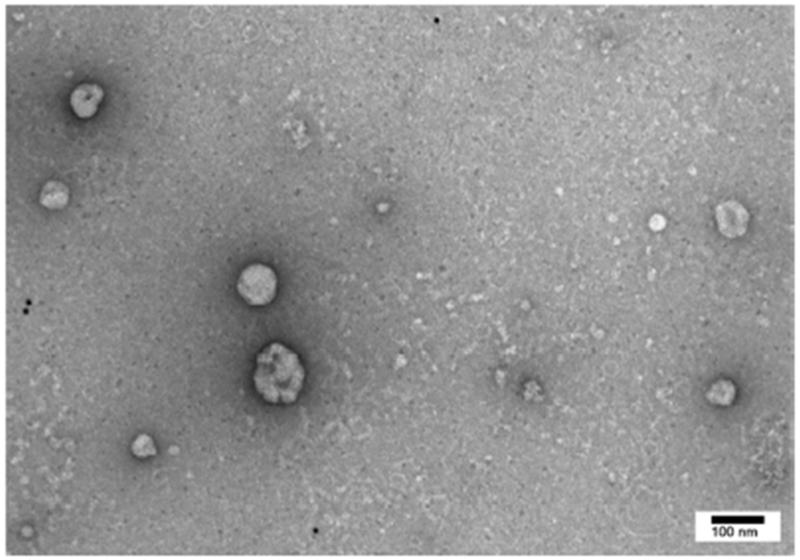
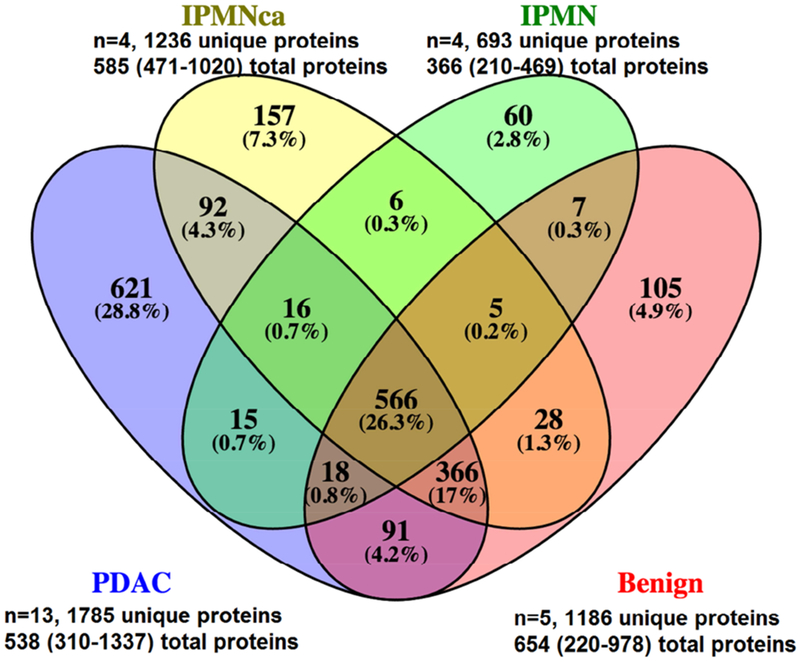
Representative exosome as visualized on transmission electronic microscopy (Fig. 1A). Exosomal proteins profiled using liquid chromatography - mass spectrometry (Fig. 1B). Total proteins are expressed as median (range).
Exosomal proteins
A total of 2153 unique exosomal proteins in the pancreatic duct fluid were identified using LC-MS/MS, with 693 unique proteins identified in patients with IPMN to 1785 in patients with PDAC (Fig. 1B). More than 26% of the proteins were present in all patients irrespective of the diagnosis, while 29% of proteins were unique to patients with PDAC. When patients were stratified by diagnosis, no significant differences were detected in the total numbers of proteins identified (p=0.293). Multiple carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) and extracellular matrix (ECM) proteins were identified among those top 35 proteins that distinguished PDAC (Supplementary Table 2). These 35 proteins were present in most patients with PDAC (n=13 total) and present in no more than 1 patient without cancer (n=9 total, including n=4 IPMN without microinvasive component or n=5 other benign diseases). The median expression levels using LFQ intensities were also shown to be higher among patients with PDAC compared to those without cancer. Selected CEACAM and ECM exosomal protein expression data from the top 35 proteins are shown (Table 2).
Table 2.
Multiple extracellular matrix (ECM) proteins and carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) were identified among the top 35 most abundant exosomal proteins from patients with PDAC compared to IPMN without microinvasive cancer and other benign diseases.
| Proteins | Genes | PDAC (n=13) | IPMNca (n=4) | IPMN (n=4) | Benign (n=5) | p-value | PDAC (LFQ*) | IPMNca (LFQ*) | IPMN (LFQ*) | Benign (LFQ*) |
|---|---|---|---|---|---|---|---|---|---|---|
| Carcinoembryonic antigen-related cell adhesion molecule 1 | CEACAM1 | 12 | 0 | 0 | 1 | <0.001 | 28.7 | 0 | 0 | 27.2 |
| Tenascin C | TNC | 11 | 1 | 0 | 1 | 0.002 | 28.9 | 29.9 | 0 | 26.5 |
| Matrix metalloproteinase-7 | MMP7 | 9 | 0 | 0 | 0 | 0.002 | 28.2 | 0 | 0 | 0 |
| Laminin subunit beta-3 | LAMB3 | 8 | 1 | 0 | 0 | 0.006 | 28.5 | 26.9 | 0 | 0 |
| Laminin subunit gamma-2 | LAMC2 | 8 | 1 | 0 | 0 | 0.006 | 29.6 | 28.5 | 0 | 0 |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | CEACAM5 | 9 | 1 | 1 | 0 | 0.012 | 28.8 | 27.9 | 27.9 | 0 |
PDAC, pancreatic ductal adenocarcinoma; IPMN, intraductal papillary mucinous neoplasm; IPMNca, IPMN with microinvasive cancer; LFQ, label-free quantification
median LFQ was reported only for those present in patients of each cohort.
CEACAM1/5 and tenascin C appeared to be among the most discriminating proteins between malignant and benign diagnoses. CEACAM1 was identified in 12 (92%) of patients with PDAC compared to zero patients with IPMN without microinvasive component and 1 patient with chronic pancreatitis and PanIN 1-2. Of those patients with CEACAM 1 identified, the median expression level of CEACAM1 among patients with PDAC was 2-fold higher than that of the benign patient. Tenascin C was identified in 11 (85%) of patients with PDAC compared to zero patients with IPMN without microinvasive component and 1 patient with chronic pancreatitis and PanIN 1. Of those patients with tenascin C present in their pancreatic duct fluid exosome, the median expression level of tenascin C among patients with PDAC was 4-fold higher than the tenascin C level of the benign patient.
Immunohistochemistry
CEACAM1/5 and tenascin C were selected for further evaluation by IHC, using primary pancreatic tumors from 27 patients with PDAC and 12 patients with IPMN without microinvasive cancer. When examining CEACAM 1/5 staining on pancreatic tumors, patients with PDAC had stronger membranous staining in the tumor epithelial cells compared to patients with IPMN (Table 3, p=0.020). In both PDAC and IPMN tumors, tenascin C only stained stroma and did not stain any tumor epithelial cells. Tenascin C staining was stronger and more diffuse in the stroma of PDAC as compared to IPMN (p=0.002). Representative immunostaining of CEACAM 1/5 and tenascin C are shown (Fig. 2).
Table 3.
CEACAM 1/5 and tenascin C staining intensity and pattern differentiates patients with PDAC from IPMN without microinvasive cancer.
| PDAC (n=27) | IPMN (n=12) | p-value | |
|---|---|---|---|
| CEACAM 1/5 staining on tumor epithelial cells | |||
| Staining intensity | |||
| Weak (1) | 0 (0%) | 0 (0%) | 0.401 |
| Moderate (2) | 7 (26%) | 2 (17%) | |
| Strong (3) | 20 (74%) | 10 (83%) | |
| Staining pattern | |||
| Membranous + luminal | 23 (0 focal, 23 diffuse) | 6 (1 focal, 5 diffuse) | 0.026 |
| Membranous only | 1 (1 focal, 0 diffuse) | 0 (0 focal, 0 diffuse) | |
| Luminal only | 3 (2 focal, 1 diffuse) | 6 (4 focal, 2 diffuse) | |
| Total score on membranous part of epithelial tumor cells | |||
| < 4 | 4 (15%) | 6 (50%) | 0.020 |
| ≥ 4 | 23 (85%) | 6 (50%) | |
| Tenascin C staining on stroma | |||
| Staining intensity | |||
| Weak (1) | 0 (0%) | 4 (33%) | 0.002 |
| Moderate (2) | 10 (37%) | 6 (50%) | |
| Strong (3) | 17 (63%) | 2 (17%) | |
| Staining pattern | |||
| Focal (1) | 13 (48%) | 12 (100%) | 0.002 |
| Diffuse (2) | 14 (52%) | 0 (0%) | |
| Total score | |||
| < 4 | 8 (30%) | 10 (83%) | 0.002 |
| ≥ 4 | 19 (70%) | 2 (17%) | |
PDAC, pancreatic ductal adenocarcinoma; IPMN, intraductal papillary mucinous neoplasm The staining pattern was scored as negative (0), focal (1), and diffuse (2), while staining intensity was scored as weak (1), moderate (2), or strong (3). The expression for CEACAM 1/5 and tenascin C was accepted as positive if a total score of ≥ 4 was obtained from combined staining intensity and pattern.
Fig. 2.
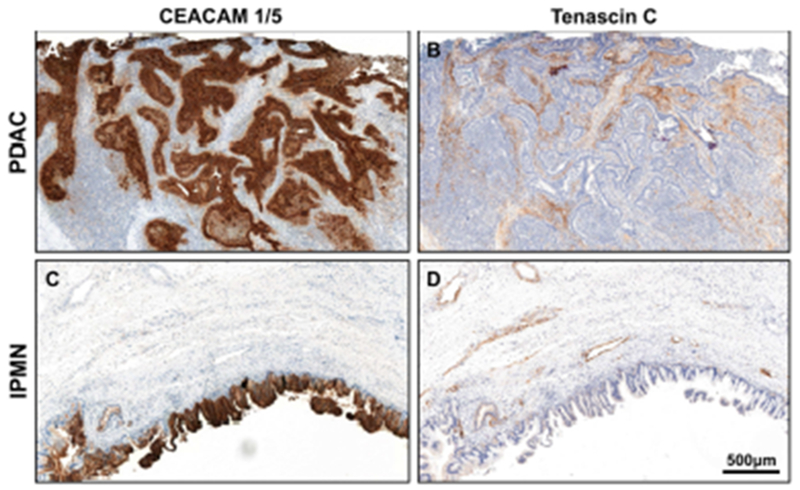
A representative PDAC with CEACAM 1/5 staining that is strong and diffuse in the tumor epithelial cells (A), whereas tenascin C staining that is strong and diffuse in the stroma (B). A representative IPMN with CEACAM 1/5 staining that is strong and diffuse in the tumor epithelial cells (C), whereas tenascin C staining that is moderate and focal in the stroma (D).
DISCUSSION
In this study, we have documented the feasibility of using pancreatic duct fluid to isolate tumor specific exosomes. Importantly, we adapted a protocol that is readily reproducible by any group with access to pancreatic duct fluid and a high-speed centrifuge. In undertaking this study, we have shown the potential for biomarker discovery in discriminating pancreatic pathology. We have also demonstrated that tumor and stroma contribute to the exosomal protein composition, which may be exploited to further investigate the role of exosomes in the biology of primary pancreatic tumors.
CEACAM 1/5 and tenascin C were highly abundant proteins in pancreatic duct fluid exosomes among patients with PDAC. CEACAM 1 and 5 are members of the glycosylphosphatidylinositol (GPI)-linked immunoglobulin family, and they function in cell adhesion as well as intracellular and intercellular signaling.(19, 20) CEACAM 5, also known as the carcinoembryonic antigen (CEA), is a well-known biomarker in many cancers, including colorectal cancer and PDAC.(19) In patients with PDAC, CEACAM 1 and 5 were overexpressed on the neoplastic epithelial cells and they have demonstrated to be diagnostic and prognostic in prior studies.(21, 22) Tenascin C is a large ECM glycoprotein that binds to other members of ECM proteins and cell surface receptors and is crucial in tissue remodeling.(23, 24) Tenascin C has been implicated in PDAC cell growth and migration, increased in expression from PanIN-1 lesions to PDAC, and correlates with vascular and lymph node invasion as well as liver metastasis.(23–25) It was suggested that factors released from pancreas tumor stimulated pancreatic stellate cells to produce tenascin C, revealing the importance of communication and contribution by both tumor and stromal cells in the microenvironment.(23) This cross-talk between tumor and stromal cells have shown to be vital in inducing transcriptional and metabolic changes modulating PDAC progression.(26) Interestingly, while tenascin C positivity was limited to stroma by immunohistochemistry in our study, whereas CEACAM 1/5 expression was limited to tumor epithelia, suggesting that both tumor and stroma contributed to the exosomal microenvironment.
Although CEACAM1/5 and tenascin C appeared to be among the most discriminating proteins between patients with PDAC and patients with IPMN and other benign diagnoses, patients with chronic pancreatitis may cause false positive result. CEACAM 1 was detected in pancreatic duct fluid exosomes of a patient with chronic pancreatitis and tenascin C was found in an another patient with chronic pancreatitis. However, the expressions of CEACAM 1 and tenascin C were 2-fold and 4-fold lower, respectively, in patients with chronic pancreatitis than in patients with PDAC. Thus, it is important to evaluate both presence and expression level of these exosomal proteins.
In addition to tenascin C, other highly abundant ECM proteins in patients with PDAC included matrix metalloproteinases 7, laminin β3 and laminin γ2. Matrix metalloproteinases 7, a member of zinc-dependent endopeptidases, has also been implicated in PDAC progression and associated with metastatic disease and worse survival.(27–29) Laminin β3 and laminin γ2 are two chains of laminin-5, which are components of basement membrane and are involved in cell adhesion, differentiation, migration, angiogenesis, and metastasis.(30, 31) In PDAC, laminin β3 and laminin C2 have been implicated in nerve invasion, and its strong membraneous overexpression occurred as early as high grade PanIN lesions and in the majority of PDAC.(31, 32) The addition of laminin C2 to CA 19-9 has been shown to improve the diagnostic performance of detecting early stage PDAC.(30)
Identification and analysis of exosomal proteins from pancreatic duct fluid is a novel tool to study PDAC biology. Although pancreatic fluid was obtained intraoperatively in this study, it could also be obtained preoperatively by endoscopic retrograde cholangio-pancreatography.(14,33)A limitation of this study is having small sample size, as mass spectrometry can be expensive and time consuming to perform(17). Mass spectrometry based proteomic analysis can quantify the expression of hundreds of proteins in each sample, but it also has its inherent limitations.(17,33) For example, absence of protein may be due to identification of peptides below the detection level, not because the protein was truly absent.(17, 35) Although mass spectrometry based proteomic analysis is complex, the potential biomarker identified in the pancreatic duct fluid exosomes can be validated in the peripheral blood using available antibodies.(9, 34, 36, 37) In addition, pancreatic duct fluid may have high concentration of PDAC-specific proteins given its proximity to the primary tumor, whereas potential biomarkers in the systemic circulation might be diluted and masked by other abundant proteins.(38) The only routinely use PDAC biomarker from the systemic circulation is CA 19-9, but its high false positive rate in patients with chronic pancreatitis limits its use in preoperative decision making.(9) Another limitation of our study is that although our samples include exosomes given visual confirmation on transmission electronic microscopy and identification of the characteristic exosomal surface markers, the average size of nanovesicles isolated was slightly larger than typical exosomes (50-150nm) although the most frequent size was characteristic of exosome size.(1, 4) We utilized serial ultracentrifugation protocol for this study, and although it is likely optimal for pancreatic duct fluid in general, the more mucinous or viscous fluid found in patients with IPMN represents a technical challenge. Optimization may however not be possible given the variable viscosities encountered and thus the same isolation protocol was utilized to minimize experimental variability. However, we are continuing to experiment with different protocols to optimize exosome isolation, such as using a sucrose gradient to further purify exosomes from other larger nanovesicles but the use of sucrose also has its own limitations as well.(39)
In conclusion, this is the first study to show that exosome isolation is feasible from pancreatic duct fluid. The exosomal protein composition reflects contribution from both tumor epithelia and stroma, as we have demonstrated in this study with CEACAMs and ECM proteins. The study of pancreatic duct fluid proteins has the potential to enhance our understanding of the crosstalk between cancer cell and their microenvironment, while also identifying potential biomarkers of patients with different pancreatic diagnoses.
Supplementary Material
Synopsis:
Pancreas cancer specific exosomes can be isolated from pancreatic duct fluid. High abundance of exosomal proteins, including extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules, distinguished pancreatic cancer from benign and pre-neoplastic diseases.
ACKNOWLEDGEMENT
Funding: This work was supported in part by NIH/NCI P30 CA008748 Cancer Center Support Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Meeting presentation: Some findings of this study were presented at Academic Surgical Congress as oral presentation in February 2017 in Las Vegas, NV.
Disclosure: The authors report no conflicts of interest in this study.
REFERENCES
- 1.Patel GK, Patton MC, Singh S, Khushman M, Singh AP. Pancreatic Cancer Exosomes: Shedding Off for a Meaningful Journey. Pancreat Disord Ther. 2016;6(2):e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochimica et biophysica acta. 2014;1841(1):108–20. [DOI] [PubMed] [Google Scholar]
- 3.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. The Journal of clinical investigation. 2016;126(4): 1139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S ELA, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–57. [DOI] [PubMed] [Google Scholar]
- 5.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nature communications. 2017;8:14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature cell biology. 2015;17(6):816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remmers N, Bailey JM, Mohr AM, Hollingsworth MA. Molecular pathology of early pancreatic cancer. Cancer Biomark. 2010;9(1-6):421–40. [DOI] [PubMed] [Google Scholar]
- 10.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63(5):318–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosoda W, Chianchiano P, Griffin JF, Pittman ME, Brosens LA, Noe M, et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J Pathol. 2017;242(1):16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu H, Niu F, Liu F, Gao J, Sun Y, Zhao X. Elevated glypican-1 expression is associated with an unfavorable prognosis in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6(6):1181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer letters. 2017;393:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellizzi AM, Stelow EB. Pancreatic cytopathology: a practical approach and review.Arch Pathol Lab Med. 2009;133(3):388–404. [DOI] [PubMed] [Google Scholar]
- 15.Vera R, Dotor E, Feliu J, Gonzalez E, Laquente B, Macarulla T, et al. SEOM Clinical Guideline for the treatment of pancreatic cancer (2016). Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2016;18(12):1172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(3-4):643–71. [DOI] [PubMed] [Google Scholar]
- 20.Tchoupa AK, Schuhmacher T, Hauck CR. Signaling by epithelial members of the CEACAM family - mucosal docking sites for pathogenic bacteria. Cell Commun Signal. 2014;12:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebauer F, Wicklein D, Horst J, Sundermann P, Maar H, Streichert T, et al. Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PloS one. 2014;9(11):e113023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeone DM, Ji B, Banerjee M, Arumugam T, Li D, Anderson MA, et al. CEACAM1, a novel serum biomarker for pancreatic cancer. Pancreas. 2007;34(4):436–43. [DOI] [PubMed] [Google Scholar]
- 23.Esposito I, Penzel R, Chaib-Harrireche M, Barcena U, Bergmann F, Riedl S, et al. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J Pathol. 2006;208(5):673–85. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Li Z, Jiang P, Wu G, Chen K, Zhang X, et al. The co-expression of MMP-9 and Tenascin-C is significantly associated with the progression and prognosis of pancreatic cancer. Diagn Pathol. 2015;10:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paron I, Berchtold S, Voros J, Shamarla M, Erkan M, Hofler H, et al. Tenascin-C enhances pancreatic cancer cell growth and motility and affects cell adhesion through activation of the integrin pathway. PloS one. 2011;6(6):e21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu S, Truitt ML, et al. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(5): 1129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamahashi U, Kumagai J, Takizawa T, Sekine M, Eishi Y. Expression and intracellular localization of matrix metalloproteinases in intraductal papillary mucinous neoplasms of the pancreas. Virchows Arch. 2008;453(1):79–87. [DOI] [PubMed] [Google Scholar]
- 29.Wang SC, Parekh JR, Porembka MR, Nathan H, D’Angelica MI, DeMatteo RP, et al. A Pilot Study Evaluating Serum MMP7 as a Preoperative Prognostic Marker for Pancreatic Ductal Adenocarcinoma Patients. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2016;20(5):899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan A, Prassas I, Dimitromanolakis A, Brand RE, Serra S, Diamandis EP, et al. Validation of biomarkers that complement CA19.9 in detecting early pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(22):5787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian J, Niu J, Li M, Chiao PJ, Tsao MS. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer research. 2005;65(12):5045–53. [DOI] [PubMed] [Google Scholar]
- 32.Mitsunaga S, Fujii S, Ishii G, Kinoshita T, Hasebe T, Aoyagi K, et al. Nerve invasion distance is dependent on laminin gamma2 in tumors of pancreatic cancer. International journal of cancer Journal international du cancer. 2010;127(4):805–19. [DOI] [PubMed] [Google Scholar]
- 33.Adler DG, Baron TH, Davila RE, Egan J, Hirota WK, Leighton JA, et al. ASGE guideline: the role of ERCP in diseases of the biliary tract and the pancreas. Gastrointest Endosc. 2005;62(1):1–8. [DOI] [PubMed] [Google Scholar]
- 34.Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016;2(3):185–95. [DOI] [PubMed] [Google Scholar]
- 35.Listgarten J, Neal RM, Roweis ST, Wong P, Emili A. Difference detection in LC-MS data for protein biomarker discovery. Bioinformatics. 2007;23(2):e198–204. [DOI] [PubMed] [Google Scholar]
- 36.Porterfield M, Zhao P, Han H, Cunningham J, Aoki K, Von Hoff DD, et al. Discrimination between adenocarcinoma and normal pancreatic ductal fluid by proteomic and glycomic analysis. J Proteome Res. 2014;13(2):395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157–71. [DOI] [PubMed] [Google Scholar]
- 38.Makawita S, Smith C, Batruch I, Zheng Y, Ruckert F, Grutzmann R, et al. Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol Cell Proteomics. 2011;10(10):M111 008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


