Abstract
Introduction
Chagas disease affects 8–10 million people worldwide, mainly in Latin America. The current therapy for Chagas disease is limited to nifurtimox and benznidazole, which are effective in treating only the acute phase of the disease but with severe side effects. Therefore, there is an unmet need for new drugs and for the exploration of innovative approaches which may lead to the discovery of new effective and safe drugs for its treatment.
Areas covered
The authors report and discuss recent approaches including structure-based design that have led to the discovery of new promising small molecule candidates for Chagas disease which affect prime targets that intervene in the sterol pathway of T. cruzi. Other trypanosome targets, phenotypic screening, the use of artificial intelligence and the challenges with Chagas disease drug discovery are also discussed.
Expert opinion
The application of recent scientific innovations to the field of Chagas disease have led to the discovery of new promising drug candidates for Chagas disease. Phenotypic screening brought new hits and opportunities for drug discovery. Artificial intelligence also has the potential to accelerate drug discovery in Chagas disease and further research into this is warranted.
Keywords: Trypanosoma cruzi sterol pathway, new drug candidates, trypanosome targets, phenotypic screening, artificial intelligence, challenges in Chagas drug discovery
1. Introduction
Trypanosoma cruzi is the protozoan parasite that causes Chagas disease, also known as American trypanosomiasis. T. cruzi parasites are predominantly transmitted to humans as metacyclic trypomastigote forms (non-dividing) in the feces of infected hematophagous triatomine bugs at the bite site. Entry is either through the wound or transfer to neighboring mucosa. T. cruzi transmission can also occur congenitally, via organ transplantation, blood transfusion, or orally by ingestion of parasite-contaminated food and drink [1]. Infective trypomastigotes invade cells and differentiate into intracellular amastigotes, which multiply by binary fission to differentiate into trypomastigotes to further be released into the circulation as bloodstream trypomastigotes to infect cells again or to be ingested by another vector. The ingested blood trypomastigotes transform into epimastigotes in the vector’s midgut to multiply and then differentiate into infective metacyclic trypomastigotes. The infective trypomastigotes and intracellular replicative amastigotes are the clinically relevant life-cycle stages of the parasite that are targets for drug intervention.
Chagas disease is endemic in 21 Latin American countries. Due to globalization, this disease has spread across European countries like Austria, Belgium, France, Germany, Italy, Netherlands, Portugal, Spain, Sweden, Switzerland, and the United Kingdom, as well as Australia, Japan, Canada and the southern United States bordering with Mexico [2,3]. In the State of Texas in the USA, Chagas disease is now endemic [4]. Chagas disease causes significant mortality and morbidity, presenting dramatic pathology in the chronic phase, involving cardiomyopathies leading to cardiac arrest followed by death as well as severe megacolon and megaesophagus [1]. The World Health Organization (WHO) estimates that 8 million people are infected with T. cruzi and more than 10,000 die every year on account of the infection [1].
In the seven southernmost American countries, the disease causes the loss of about 752,000 working days because of premature deaths and $1.2 billion in productivity [5]. The estimated annual global burden of disease is $627.46 million in health-care costs and 806,170 disability-adjusted life-years; 10% of this burden affects non-endemic countries [6]. Migration and specific modes of transmission have led to Chagas disease spreading beyond its natural geographical boundaries and becoming a global health problem [7–9].
Despite the fact that the disease was described over a century ago [10] only two drugs are available for Chagas disease treatment, benznidazole and nifurtimox, which were developed over 40 years ago. Both drugs produce significant side effects and low efficacy for the chronic phase of the disease (and extremely poor tolerability). Since the introduction of benznidazole and nifurtimox, only allopurinol and a few triazoles (ergosterol biosynthesis inhibitors) have been studied in clinical trials, observational studies, and case reports. Monotherapy with ravuconazole or posaconazole has not proven to be efficacious for the treatment of chronic T. cruzi infection [11] and the combination of posaconazole and benznidazole did not provide any further efficacy or safety advantages over benznidazole monotherapy [12]. The most plausible explanation for the failure of posaconazole in both the CHAGAZOL and STOP-CHAGAS trials, in contrast with its remarkable anti-T. cruzi activity in vitro and in animal models, is that the dose used in these studies (400 mg twice daily), leads to systemic exposures that are just 10% to 20% of those measured in mice at the curative anti-T. cruzi dose. Thus, the patients in these studies were underdosed with posaconazole for the T. cruzi application [13–15]. In the case of the E1224 trial, the parasite load in the high-dose E1224 group remained significantly lower than in the placebo group, with no difference from the benznidazole group on adjusted post-hoc comparison, and pharmacokinetic/pharmacodynamic (PK/PD) models indicated that an increase in high-dose treatment duration would significantly reduce the probability of relapse from 61% within 8 weeks to 44% within 12 weeks [16]. The reasons for these failures are not attributable to a lack of intrinsic capacity of the tested drugs to control the parasite burden on the patients, but rather to flawed clinical study designs. In this context, Chatelain [17] cites that one of the reasons could be that the patients enrolled in the trials would be infected with T. cruzi strains resistant to the azoles. This hypothesis is supported by data from Moraes et al. [18] derived from High-Content Phenotypic Screenings, that show that some T. cruzi genotypes (DTUs) are partially resistant to four ergosterol biosynthesis inhibitors (posaconazole, ravuconazole, EPL-BS967, and EPL-BS1246). Coincidentally, these genotypes are prevalent in patients from Bolivia, who were the major targets of the clinical trials [19]. Moraes et al. [18] reported that the Tulahuen strain, is sensitive to azoles different than VNI and VFV, that inhibit ergosterol biosynthesis [18]. Interestingly, as we refer in Section 2.1, VNI and VFV are effective against the drug-susceptible strain Tulahuen [20,21]. VNI and VFV present anti-trypanosomal activity in mice infected with the drug-resistant strain Y [22], VNI is effective but less potent than VFV against the drug-resistant Y strain [23], and VFV completely reduced parasitemia in mice infected with the Y strain [24]. Furthermore, VNI has shown high anti-parasitic efficacy against the drug-resistant strains Colombiana and Y [24].
Thus, new effective, safe and more potent drugs for the treatment of Chagas disease are desperately needed and drug discovery for Chagas disease is been pursued using several approaches.
Here we review and discuss the recent scientific innovations in the field of Chagas disease that have led to the discovery of new promising drug candidates for Chagas disease. We also review the status of drug targets and phenotypic screening that brought new hits and opportunities for drug discovery. Finally, we propose the use of artificial intelligence to accelerate drug discovery in Chagas disease and critically review and discuss the current challenges that need to be overcome in Chagas disease drug discovery.
2. Targeting the ergosterol pathway of T. cruzi for drug discovery
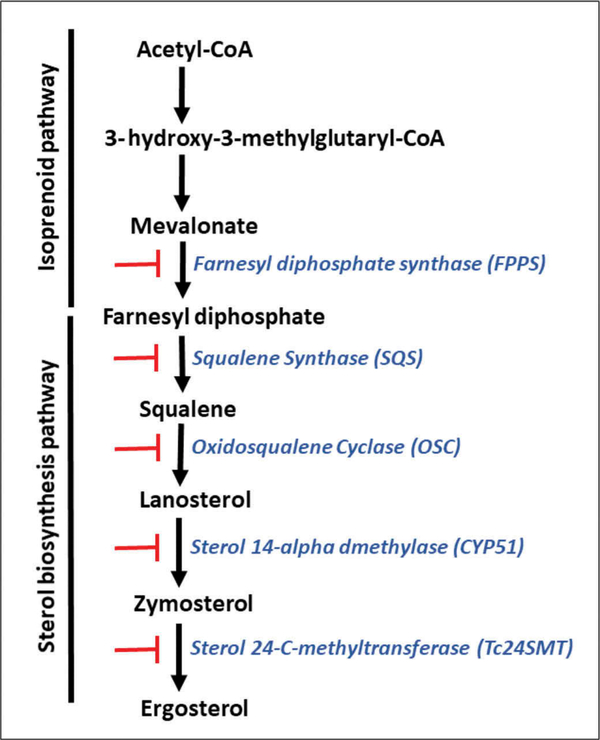
In trypanosomatids, the main sterol component of the parasite is ergosterol, which is essential for providing structure and function of trypanosome membranes and is required for parasite multiplication [25]. Ergosterol is synthesized through a biosynthetic pathway that involves two-step pathways, the isoprenoid pathway (from Acetyl-CoA to farnesyl diphosphate) and the sterol pathway (from farnesyl-diphosphate to sterols) (Figure 1). Sterol biosynthesis in trypanosomes differs from that of mammalian hosts since the final product is ergosterol instead of cholesterol, the main sterol present in the mammals. As T. cruzi is entirely dependent on endogenously produced sterols for survival and proliferation, the sterol biosynthetic pathway constitutes an attractive target for drug development [26].
Figure 1. Simplified ergosterol biosynthesis pathway in T. cruzi.
Blocking bars (red color) refer to the inhibitors listed in section 2. that block critical enzymes (blue color) of the ergosterol pathway.
The set of critical enzymes that participate in the synthesis of ergosterol in T. cruzi are targets for drug discovery. Studies in this area may facilitate in the near future the discovery of a set of small molecules that can be used as multi-drug therapy specifically targeting all critical enzymes of the T. cruzi ergosterol pathway as protease inhibitors are used for HIV/AIDS treatment.
The squalene synthase (SQS), squalene epoxidase (SQLE), oxidosqualene cyclase (OSC), lanosterol 14 α-demethylase (CYP51) and sterol 24-C-methyltransferase (24SMT) are the major enzymes targeted for inhibition in the sterol pathway (Figure 1). OSC has been less studied, but its inhibition has been shown to affect ergosterol synthesis in T. cruzi [27].
The first-committed step for sterol biosynthesis begins with the head-to-head condensation of two molecules of farnesyl diphosphate to produce squalene, a two-step reaction catalyzed by squalene synthase. The complete set of genes encoding the enzymes involved in both step pathways has been identified in the T. cruzi genome.
2.1. Sterol 14-alpha demethylase (CYP51)
The T. cruzi sterol 14-alpha demethylase (CYP51) is the only one gene of this pathway whose requirement for the parasite viability has been evaluated and found to be essential for the parasite survival as evidenced by CRISPR-Cas9 gene editing and posaconazole inhibition of the enzyme leading to arrest of T. cruzi growth [28].
The sterol14α-demethylase is by far the enzyme of the ergosterol pathway that has been studied in more detail as a therapeutic target for T. cruzi. This enzyme uses lanosterol or related compounds as substrates to produce zymosterol, which is the precursor of ergosterol. Azoles are highly effective antifungal drugs that work by inhibiting the activity of sterol 14α-demethylases. Several novel azoles have shown strong anti-T. cruzi activity in vitro and in vivo [20,21,29], so they have been considered a priority for drug development. The mechanism of action of these compounds against the T. cruzi 14α-demethylase has been studied at the structural level [29–32].
Advances in T. cruzi 14 α-demethylase and the structural basis for the enzyme druggability have been reported. Accordingly, the T. cruzi sterol 14 α-demethylase was characterized and its structure was solved in a ligand-free form and in complexes with different inhibitors such as fluconazole, posaconazole, imidazoles, triazoles, pyridines, tetrazoles, and the substrate analogue MCP [29–38]. Similarly, the structures of sterol 14 α-demethylases from the fungal organisms Aspergillus fumigatus [39,40] and Candida albicans [41], and from humans [42] were characterized. Also, the sterol 14 α-demethylase of Trypanosoma brucei and Leishmania infantum were characterized and their structures were solved in a ligand-free form and in complexes with different inhibitors and their structures were compared with the counterpart enzyme in T. cruzi and fungal organisms [43–45].
The comparative structural analysis of 14 α-demethylases revealed that sterol 14 α-demethylase enzymes preserve their conserved catalytic function regardless of the extremely low amino acid sequence identity across phylogeny by maintaining strict similarity at the secondary and tertiary structural levels [46]. All the inhibitors, including the substrate analogue MCP, bind in the sterol 14 α-demethylases active site without causing any substantial rearrangements in the protein backbone, as if ‘freezing’ the enzyme in its substrate-free conformation. The inhibitors simply occupy the available space and acquire the shape of the sterol 14 α-demethylases substrate-binding cavity.
Studies on the 14 α-demethylases structure-based drug design revealed that no conformational changes in the backbone of the sterol 14 α-demethylases substrate-binding cavity are involved in inhibitor binding, making this enzyme an attractive target for structure-based drug design. VNI is the first experimental drug for treating Chagas disease that was complexed with the active site of the T. cruzi sterol 14 α-demethylase structure [26]. Using direct testing of inhibition of protozoan sterol 14 α-demethylase orthologues, a highly potent compound was identified, VNI [(R)-N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl) ethyl)-4- (5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)], whose inhibitory effect was functionally irreversible, completely blocking the T. cruzi and T. brucei 14 α-demethylase activity [47]. VNI exhibits no toxicity to mouse cells and cures the acute and chronic forms of Chagas disease in mice caused by the Tulahuen strain of T. cruzi, with 100% survival and no observable side effects [19]. Low cost at <$0.10/mg [48], oral bioavailability, favorable pharmacokinetics, and low toxicity make this compound an exceptional candidate for clinical trials.
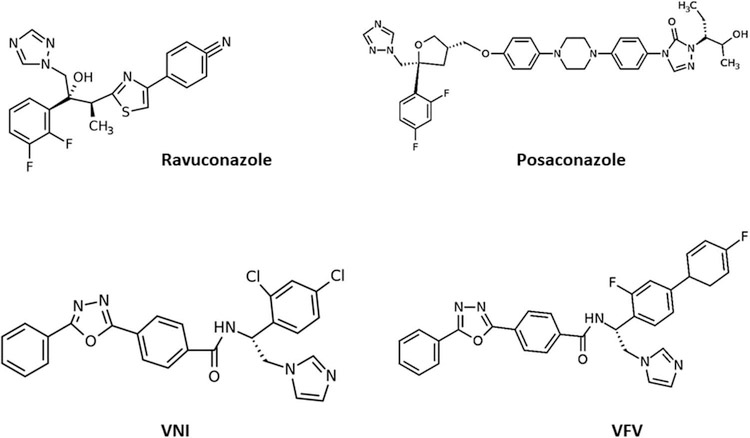
In further studies, VNI was found to be less potent as an inhibitor of Leishmania infantum 14 α-demethylase [49] and Y strain T. cruzi sterol 14 α-demethylase A [50]. The crystal structure of the T. cruzi sterol 14 α-demethylase–VNI complex revealed the molecular basis for its high potency [26] whereas the complex with the close analogue [4′- chloro-N-[(1R)-2-(1H-imidazol-1-yl)-1-phenylethyl]biphenyl4-carboxamide] [31] provided a direction for the compound redesign to broaden the spectrum of antiprotozoal activity, leading to the synthesis of VFV [(R)-N-(1-(3,4′-difluoro-[1,1′- biphenyl]-4-yl)-2-(1H-imidazol −1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl) benzamide] [39]. The additional phenyl ring filled the deepest cavity of the 14 α-demethylase active site, further increasing the strength of the interaction [50]. Having a broader spectrum of activity as a protozoan 14 α-demethylase inhibitor, VFV like VNI cured Tulahuen T. cruzi infection in mice and showed increased efficacy when compared to VNI in murine models of visceral leishmaniasis [21] and Chagas disease caused by the Y strain of T. cruzi [51]. VFV displayed better bioavailability and tissue distribution but was less metabolically stable than VNI [21]. The chemical structures of VNI, VFV, posaconazole, and ravuconazole are presented in Figure 2.
Figure 2.
Chemical structures of the new generation of azoles VNI and VFV, and old azoles ravuconazole and posaconazole that inhibit the T. cruzi sterol 14 α-demethylase. VNI [20] and VFV [21] are new promising drug candidates in pre-clinical studies, whereas ravuconazole and posaconazole entered unsuccessful clinical trials [139,11].
The human sterol 14 α-demethylase is naturally resistant to inhibition [42], which addresses the general concern that inhibitors of pathogenic sterol 14 α-demethylases might actually affect the human counterpart. All attempts to use human sterol 14 α-demethylase as a drug target have failed, and none of the known antifungal azoles or experimental inhibitors of sterol 14 α-demethylases from human pathogens inhibit the activity of the human enzyme [42]. It is believed that this natural resistance of human sterol 14 α-demethylases to inhibition is due to the higher flexibility of the substrate-binding cavity. The notion of the higher flexibility of the human sterol 14 α-demethylase active site is also supported by structural dynamic simulation experiments [52].
The Drugs for Neglected Diseases Initiative (DNDi) has discouraged the use of new azoles that target the T. cruzi sterol 14 α-demethylase including chemical libraries [17,53], because of the failure of previous azoles in clinical trials. This has the potential to delay drug discovery and development of a highly promising new generation of azoles to enter clinical trials such as VNI [20] and VFV [21]. However, rigorous PK/PD analysis is essential to translate the results of preclinical in vivo studies performed with VNI and VFV. Also, it is important to test the activity of the new azoles against T. cruzi strains that are resistant to posaconazole. Interestingly, we found that posaconazole induces resistance to the T. cruzi Tulanuen strain [37].
2.2. Sterol 24-C-methyltransferase (Tc24SMT)
A relatively unexplored and promising strategy is the inhibition of the last enzyme of the ergosterol synthesis (Tc24SMT) by azasterols. In contrast to other enzymes in the pathway, Tc24SMT is absent in humans. T. cruzi parasites treated with the 24SMT inhibitors 22,26-azasterol (AZA) and 24(R,S) 25-epiminolanosterol (EIL) showed no detectable levels of 24-alkyl sterols and strongly inhibited parasite growth [54]. Synergistic antiparasitic effects of AZA and 14 α-demethylase inhibitors were observed in vitro and in a murine model of acute Chagas disease [55]. Other azasterols were synthesized and showed to have antiproliferative and ultrastructural effects in T. cruzi and Leishmania in vitro, however further work is needed to improve the activity and selectivity of the compounds. Similarly, other 24SMT inhibitors have shown effects on epimastigote growth accompanied by severe ultrastructural alterations [56].
The suicide substrate of 24SMT, dehydrozymosterol (DHZ) strongly inhibits T. cruzi infection of myoblasts at low concentrations [Villalta, unpublished results], suggesting that DHZ is a potential chemotherapeutic agent for disrupting the sterol metabolic pathway in T. cruzi.
Inhibitors of protozoan 24SMT have the potential to be used as the next generation of drugs to treat fungal infections or neglected tropical diseases that are demonstrating resistance against current therapies.
2.3. Squalene synthase (SQS)
The SQS was originally identified as chemotherapeutic target in T. cruzi by Urbina and coworkers by evaluating the in vitro and in vivo effects of SQS inhibitors [57–60]. New isosteric analogs based on 4-phenoxyphenoxyethyl thiocyanate, which was previously described as a good non-competitive allosteric inhibitor of T. cruzi SQS [57,61], were synthesized in order to understand the effect of different atoms on the activity of this inhibitor [62]. The analogues with the selenocyanate moiety were potent inhibitors of T. cruzi proliferation, and a selenocyanate derivative with a fluorophenoxy group was also a very effective inhibitor of T. cruzi growth with very low toxicity in vitro [62]. In silico analysis showed that the presence of Se and not S increased the interaction between the compounds and SQS [62].
The x-ray crystallographic structure of T. cruzi SQS was solved using coordinates of the human SQS [62]. This enzyme catalyzes the first committed step in sterol biosynthesis leading to downstream synthesis of ergosterol. Structures of T. cruzi and human SQS bound to FSPP (substrate-like inhibitor) and TcSQS crystals could only be obtained in the presence of FSPP [63]. The determination of the structures of T. cruzi and human SQS enzymes with four classes of inhibitors that reduce T. cruzi growth in vitro provides insights into SQS inhibition. The potent inhibitor quinuclidine named E5700 binds to TcSQS and showed a synergistic effect with the 14 α-demethylase inhibitor posaconazole in inhibiting T. cruzi growth in vitro [63]. Previously it was reported that E5700 and ER-19,884 are orally active SQS inhibitors with in vivo activities and that E5700 provided better protection than ER-19,884 [60]. Although oral administration of E5700 provides full protection in terms of survival of T. cruzi infection in mice [60], no curative effects in vivo have been reported. No toxicity studies have been performed using inhibitors of the TcSQS, even though these studies suggest that the structure of squalene synthase from T. cruzi is a potentially important target for the development of novel therapeutic agents for the treatment of Chagas disease.
2.4. Farnesyl diphosphate synthase (FPPS)
The identification of FPPS as a chemotherapeutic target in T. cruzi and the mechanism of the inhibitory action of bisphosphonates against FPPS were reported [64–68]. Approaches for designing new FPPS inhibitors for T. cruzi have been reported [69,70].
The interaction of five 2-alkylaminoethyl-1,1-bisphosphonates, effective against proliferation of T. cruzi, in complex with the T. cruzi farnesyl diphosphate synthase (TcFPPS) structure was studied [71]. The structural information presented in this study provides the basis for the design of novel, more effective compounds for the treatment of Chagas disease. Although several bisphosphonate families have been shown to inhibit the TcFPPS, the lack of pharmacokinetic studies on these compounds suggests that it is still important to expand the number of compounds in the pipeline, especially for compounds of high affinity.
2.5. Oxidosqualene cyclase (OSC)
Recently using a novel biochemical assay for OSC three novel inhibitors of OSC of T. cruzi were evaluated and one found to present significant parasitological activity in vitro [72]. The identification and synthesis of inhibitors of T. cruzi OSC has been partially explored.
Among the target enzymes of the sterol pathway that participate in the synthesis of ergosterol in T. cruzi and other trypanosomatids, the 24SMT is the only enzyme with high selectivity since it is present in the parasite and not in the host. This enzyme has not yet been crystallized. The selectivity of the other enzymes can be improved during drug optimization by modifying the compound improving its affinity toward the parasite target to a higher extent than it does to the human homolog [73]. Thus, it is necessary to have structural information for both parasitic and human proteins. The first enzyme of the T. cruzi ergosterol pathway that was crystalized was the sterol 14 α-demethylase and by drug optimization, very promising inhibitors of the enzyme such as VNI and VFV were generated that cure the experimental T. cruzi infection in murine models of Chagas disease. More recently other enzymes in the ergosterol pathway in T. cruzi were crystalized such as the SQS and the FPPS. We expect that structural information about these enzymes will provide new promising inhibitors by drug optimization as well as when the 24SMT is crystallized. We also expect that the issues of toxicity, bioavailability, and PKs can be resolved rationally by minor, structure-guided optimization of the molecules as done for VNI [20] and VFV [21].
3. Other trypanosome targets
3.1. Cruzipain
Cruzipain is another substantially progressed target in Chagas disease chemotherapy. It is a lysosomal cysteine protease of T. cruzi with similarities to cathepsin L that is expressed in all life cycle stages of the parasite and is involved in intracellular replication, parasite differentiation, immune evasion, and host-parasite interactions. A vinyl sulfone irreversible inhibitor of cruzipain (K777) was advanced to preclinical development [74,75] but abandoned due to poor tolerability even at low dose in primates and dogs. K777 was found to be a reversible cruzipain inhibitor in the murine model of acute T. cruzi infection [76].
Novel scaffolds for inhibition of cruzipain were identified from high throughput screening (HTS) of GlaxoSmithKline HAT (Human African Trypanosomes) and Chagas chemical boxes. This approach identified two collections, grouping 404 non-cytotoxic compounds with high antiparasitic potency, drug-likeness, and structural diversity. A continuous enzymatic assay was adapted to a medium-throughput format and a primary screening of both collections was carried out, followed by construction and analysis of dose–response curves of the most promising hits. Using the identified compounds as a starting point, a substructure directed search against CHEMBL Database revealed plausible common scaffolds while docking experiments predicted binding poses and specific interactions between cruzipain and the novel inhibitors [77].
3.2. Cytochrome b
Chemical genomics was used to identify cytochrome b as a novel drug target for Chagas Disease [78]. It was found that the compound GNF7686 targets cytochrome b, a component of the mitochondrial electron transport chain crucial for ATP generation. GNF7686 is a potent inhibitor of T. cruzi growth in vitro. This study provides new insights into the use of chemical genetics with biochemical target validation to the discovery of additional novel drug targets and drug leads for Chagas disease.
3.3. Enzymes of the trypanothione metabolism – trypanothione reductase
The trypanothione system is central for any thiol regeneration and trypanothione reductase has been shown to be an essential enzyme in trypanosomatids. The absence of this pathway from the mammalian host and the sensitivity of trypanosomatids toward oxidative stress render the enzymes of the trypanothione metabolism attractive target molecules for the rational development of new drugs against African sleeping sickness, Chagas disease and the different forms of leishmaniasis [79].
Several existing trypanocidal drugs interact with the trypanothione system, suggesting that the trypanothione metabolism may be a good target for the development of new drugs. Three key enzymes (glutathionylspermidine synthetase, trypanothione synthetase, and trypanothione reductase) are important targets for intervention [80]. The thiol metabolism of trypanosomatids comprises the trypanothione/tryparedoxin, thioredoxin, and ovothiol systems of the parasites.
Inhibitors of trypanothione metabolism such as buthionine sulfoximine (BSO) are ideal potential candidates as drugs against T. cruzi alone, or jointly with free radical-producing drugs such as nitrofurazone (NF) and benzimidazole (BNZ) [81,82].
Trypanothione reductase catalyzes the NADPH-dependent reduction of trypanothione disulfide, but not glutathione. The enzyme is highly specific for trypanothione and has striking homology to glutathione reductase. Its crystal structure was solved alone or in complex with its substrate or inhibitors [83–87]. A number of compounds, such as nitrofurans and naphthoquinones [83,87–90], phenothiazines and related tricyclics [91–93] crystal violet [94], diphenylsulfide derivatives [95], polyamine derivatives [96–99], dibenzazepines [100], bisbenzylisoquinoline alkaloids [101],ajoene [102], acridines [103], terpyridine platinum complexes [104] Mannich bases [105] as well as some natural products [106] have been shown to inhibit T. cruzi trypanothione reductase and affect parasite growth in vitro or in vivo.
3.4. Cyclophilin
Cyclophilins (CyPs) are proteins with enzymatic peptidyl-prolyl isomerase activity (PPIase), essential for protein folding in vivo. Cyclosporin A (CsA) has a high binding affinity for CyPs and inhibits their PPIase activity. CsA has proven to have parasiticidal effects against some protozoa, including T. cruzi. Because CsA cannot be used in vivo since it has immunosuppressive properties [107], non-immunosuppressive CsA analogs originally synthesized by Novartis were used against T. cruzi. H-7–94 and F-7–62 were able to kill the three forms of the parasite, and had non-toxic effects for mammalian cells [108,109]. T. cruzi infected mice treated with H-7–94 and F-7–62 CsA analogs before and during the first 5 days post-infection survived (100%) after parasite challenge and had significant lower parasitemia than non-treated mice control (60% survival) [108].
3.5. The N-myristoylome
Protein N-myristoylation is an essential and druggable target in T. cruzi. Myristic acid accounts for ~1.5% of the total lipid content of the parasite [110], can be attached to proteins via a cysteine residue (S-myristoylation) [111] or the N-terminal glycine of specific proteins (N-myristoylation) [112,113]. This latter process is catalyzed by the enzyme N-myristoyltransferase (NMT), which utilizes myristoyl-CoA as an acyl donor and is present in all eukaryotes [113]. NMT is both a druggable and essential target in T. cruzi [114]. The specific inhibition of N-myristoylation in T. cruzi using the potent NMT inhibitor DDD85646 [115], led to a reduction of T. cruzi proliferation [114].
3.6. Carbonic anhydrases
Carbonic anhydrases are metalloenzymes with several physiological functions in all life domains and are involved in pathological processes of human and prokaryotic/eukaryotic microorganisms such as bacteria, fungi, and protozoa. This enzyme catalyzes CO2 hydration to bicarbonate and protons by nucleophilic attack of the hydroxide bound to Zn(II). In T. cruzi, an α-CA (TcCA) has a high catalytic activity for the CO2 hydration reaction and is similar kinetically to the human isoform hCA II, although it is devoid of the His64 proton shuttle which is conserved in most α-CAs. His64 is replaced by an Asn residue in TcCA [116].
For T. cruzi a large number of aromatic/heterocyclic sulfonamides were studied as TcCA inhibitors. A series of sulfonamides, some sulfamates, and thiols were tested as possible TcCA inhibitors. The tests were also done against mammalian enzymes (hCA I and hCA II) and the inhibitors showed a greater inhibition of the T. cruzi TcCA indicating a positive selectivity [116].
Anion inhibitors, and small molecules such as sulfamide, sulfamic acid, and phenylboronic/arsonic acids, interacting with zinc proteins, were also tested against TcCA. The best inhibitor was diethyldithiocarbamate but the enzyme was inhibited in the low micromolar range by iodide, cyanate, thiocyanate, hydrogen sulfide and trithiocarbonate [116].
Several research lines showed that hydroxamic acids could also act as inhibitors of the α-CA from T. cruzi and of peptidases from this pathogen, such as metallo- and cysteine peptidase [116,117]. A series of 4,5-dihydroisoxazoles incorporating hydroxamate moieties of type 34–45 were evaluated in vivo and in vitro against T. cruzi. These studies revealed that a leading compound showed excellent values for inhibition of growth for all three developmental forms of the Y strain of T. cruzi at relatively low concentrations [117].
4. Phenotypic approaches
To circumvent the challenges of target-based drug discovery, phenotypic approaches have been widely used for T. cruzi [118]. One element that is highly used in drug development for Chagas disease is HTS and in vivo imaging [119]. There are now several screening centers worldwide that do HTS for T. cruzi inhibitors. Phenotypic-based drug discovery has become the main pillar of Chagas R&D [120]. Accordingly, appropriate chemical libraries for screening, robust assays, and appropriate screening cascades have been used.
T. cruzi multiplies well inside host cells [20,21], which permits the identification of drugs that kill the parasite and those that inhibit growth and division. However, as T. cruzi evades the immune system during chronic infection, drugs that kill the parasite intracellularly are essential for eliminating Chagas. Therefore, hits need to be followed up in a parasite killing assay. One solution to the low throughput is to use an axenic (free growing) amastigote assay as the primary screen [121]. Axenic amastigotes do not occur naturally, so care must be taken in interpreting the data. Such assays also need to be designed to only identify cytocidal compounds to prevent false positives as reported [20,21]. Hits can then be confirmed in an intracellular assay.
A group of experts who advise about the screening of drugs for Chagas disease recommend that phenotypic approaches should be done with a panel of strains representing particular DTUs prevalent in patients of disease-endemic countries [122].
For all trypanosomatids, as in other areas of anti-infective drug discovery, it is also critical to measure activity against a panel of clinical isolates to be sure that activity is not laboratory-strain specific. For all cell-based assays, replication rate, starting density and rate-of-kill are key factors to correctly interpret compound potency. It is important to define these parameters as clearly as possible before interpreting data on new hits.
So far HTS has yielded several new promising inhibitors and we discuss here some representative studies. Accordingly, a high content HTS assay was described for screening chemical compounds against T. cruzi infection of myoblasts that is amenable for use with any T. cruzi strain capable of in vitro infection [123]. The assay employed a new automated image-based assay to identify new compounds against T. cruzi using the myoblastic rat cell line H9c2 as cell-cycling amastigotes hosting cells and chemical libraries [123]. From these studies, CX1 was found to inhibit T. cruzi cellular infection at 23 nM.
Ranzani et al. [124] have screened 30,000 compounds using the two-independent HTS strategies on infected cells and identified specific inhibitors that target the T. cruzi malic enzyme (TcMEs), which have trypanocidal activity against the intracellular stage of this pathogen in vitro.
A 3D quantitative structure–activity relationship (QSAR) analysis was performed using descriptors calculated from comparative molecular field analysis (CoMFA). The authors found that three hydrazones displayed highest activity against T. cruzi in vitro [125].
Over 300,000 molecules from the NIH Molecular Libraries program were screened for potential targets against T. cruzi. Development of Genome Data Base for T. cruzi and Bayesian machine learning models were used to virtually screen the compounds from the libraries [126]. Eleven compounds had EC50 <10 μM and five compounds reduced parasitemia in a murine model of Chagas disease.
A structure-based virtual screening using 3,180 FDA-approved drugs revealed that the anti-inflammatory sulfasalazine could be used as a lead compound to develop new trans-sialidase inhibitors [127]. A computer-aided identification system revealed FDA-approved drugs clofazimine, benidipine, and saquinavir as potential trypanocidal compounds at the cellular level on different parasite stages of T. cruzi in vitro [128]. These identified FDA-approved drugs have the potential to be used as new treatments for Chagas disease.
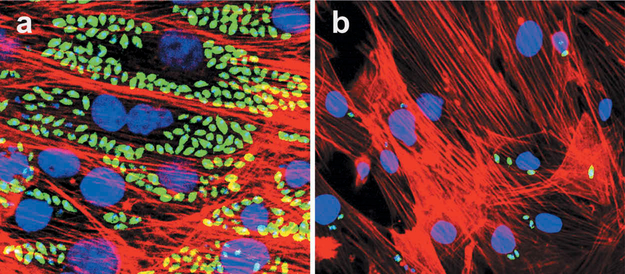
HTS of a library containing phosphodiesterases (PDE) inhibitors on infected myoblastcultures with trypomastigotes expressing GFP revealed several drugs with high activity against T. cruzi infection. One compound GKV25 strongly inhibits myoblast infection at low nM concentration (Figure 3) (Rachakonda & Villalta, unpublished results).
Figure 3. GKV25 strongly inhibit T. cruzi myoblast infection at 3.5 nM.
Cardiomyocyte monolayers were infected with GFP-expressing trypomastigotes of the Tulahuen for 24 h and then treated with different concentrations of GKV25 dissolved in DMSO. Controls were infected cells exposed to the same concentrations of DMSO. Parasite multiplication within monolayers was evaluated by fluorescence confocal microscopic observations of T. cruzi parasites inside cardiomyocytes 72 h after infection as described [20]. T. cruzi amastigotes are green, cardiomyocyte nuclei are blue, and cardiomyocyte actin filaments are red. Panel a shows control infected cells treated with DMSO whereas panel b shows the effect of GKV25 on infected myoblasts at 3 nM.
5. Challenges in drug discovery for Chagas disease
There are currently no new classes of drugs in the clinical development pipeline for Chagas disease. Drug discovery in Chagas disease must include the development of completely new classes of therapeutics, reduced host toxicity, improved administration regimens and the development of combination therapies. The advantages of combination therapies are manifold: they can increase the clinical efficacy of treatments; they can reduce side effects by allowing lower dosing of individual agents; and they can reduce the risk of resistance development. Reducing resistance is critical for safeguarding whatever new medicines do emerge from the drug discovery pipeline. Thus, significant more work is still required.
One of the key challenges of phenotypic drug discovery is how to address issues, such as potency, toxicity and pharmacokinetic problems that arise during the hit optimization process. Scaffold hopping and vector optimization become more problematic without knowledge of the molecular target. Identifying the targets of phenotypic hits should facilitate progression of these compounds and also enable more high-value target-based drug discovery in the future.
Another major challenge is defining the relevant cellular and animal models that closely mimic human clinical conditions in Chagas disease [129]. Mice detoxify drugs faster than humans and the apparent disconnect between drug efficacy observed in rodent models and the lack of success in human clinical trials is a challenge that requires a thorough pharma-cokinetics/pharmacodynamics evaluation before translating the outcomes of preclinical studies.
A major challenge for drug discovery is the diverse group of T. cruzi strains and their genetic diversity. T. cruzi presents a large and unknown number of strains and has six discrete typing units (DTUs) TcI–TcVI [130] and a proposed seventh T. cruzi branch, referred to as Tcbat, based on genetic characteristics observed across strains [131]. There is variability among strains and DTUs in terms of genetic make-up, virulence, infectivity, tissue tropism, progression of disease, drug susceptibility, and geographical distribution [132]. To overcome this challenge, we recommend that the screening of drugs should be performed using a panel of clinical isolates and representative strain members of the seven DTUs that are drug-resistant before progressing compound series too far, to be sure that the activity is not only against laboratory-strains or drug-susceptible strains of T. cruzi.
The use of pharmacogenomics [133] to identify gene variants which influence the efficacy or toxicity of currently used drugs to treat Chagas disease or new drugs entering clinical trials as clinical practice is needed for both current drugs and should be in place for future drugs. Personalized medicine or personalized treatment in Chagas disease with current drugs that present side effects, or with newly discovered drugs when they become available will help with the management of Chagas disease. In addition, there is a need to use pharmacogenomics to identify new gene targets for drug research using genomic knowledge [134] and RNA-seq to advance drug discovery in Chagas disease.
Genetically engineered parasites have significantly impacted the development of both high-throughput amenable assays to screen large chemical collections for drug discovery in vitro and in vivo screening protocols to rapidly progress hit compounds. Trypomastigotes expressing GFP or luciferase have been valuable to evaluate cellular infections for phenotypic screening or evaluation of drugs for trypanosome drug targets [20,21], and for in vivo evaluation of drug efficacy. It is recommended the use of cloned stable transgenic trypanosomes instead of non-cloned transgenic trypanosomes for accuracy in drug testing, since non-cloned transgenic trypanosomes will overgrow cloned transgenic trypanosomes and change the EC50 values of tested drugs.
More information is needed not only about the molecular mechanisms involved in the establishment of infection, but also about those involved in parasite survival and persistence to discover novel targets for intervention. Here seq RNA and CRISPR-Cas9 technologies will rapidly facilitate the identification of trypanosome targets and their validation, since very few drug targets in T. cruzi have been validated.
The screening of chemical libraries will unquestionably identify new potential targets, but they will also need to be individually curated and validated to facilitate drug discovery.
Although molecular modeling of T. cruzi proteins can be done from structural information of orthologs of other species, to develop highly selective inhibitors to target proteins it is necessary to obtain crystal structures of the specific proteins to evaluate their druggability.
A successful drug discovery campaign typically takes 10–15 years [118]. There is no doubt that identifying and developing new treatments for Chagas disease requires a large transdisciplinary team of specialists in multiple areas such as chemistry, structural biology, biology, engineering, informatics, and medicine. Ideally, several teams should be assembled and organized so that every aspect of drug development is met and, hopefully, each team should concentrate in different potential targets. Target-based drug discovery has been limited by the small number of well-validated targets, whereas the push for phenotypic-based drug discovery has become the main complementary support of Chagas R&D [129].
The dormancy of T. cruzi amastigotes in tissues [135] poses challenges since quiescent and dormant pathogens seen in the chronic phase of Chagas disease have reduced metabolism compared to actively dividing pathogens and can be less susceptible to drugs. Developing assays for these forms is essential.
6. New approaches to accelerate drug discovery for Chagas disease
Artificial intelligence (AI) has the potential power to change drug discovery for diseases including Chagas disease in the near future. Currently, investigators are employing AI to discover drugs. Machine learning and other technologies are expected to make the hunt for new pharmaceuticals quicker, cheaper and more effective [136]. The rapid growth in computer-processing power over the past two decades, the availability of large data sets and the development of advanced algorithms have driven major improvements in machine learning. An enormous figure looms over scientists searching for new drugs goes down the drain, because it includes money spent on 9 out of 10 candidate therapies that fail somewhere between phase I trials and regulatory approval [136]. Few people in the field doubt the need to be innovative. Currently, several leading pharmaceutical companies are using AI systems as machine learning, to power its search for drugs such as immuno-oncology drugs, metabolic-disease therapies, amyotrophic lateral sclerosis, cancer treatments, among others with promising results.
For instance, the function of new genes can be identified by searching public databases of T. cruzi and other trypanosomatids for essential genes in T. cruzi biology using robotics to physically test predictions in the lab. Another example is the AI approach of classifying genes according to their roles and other attributes, to look for connections between RNA-sequence variations, expression levels, molecular function and gene location.
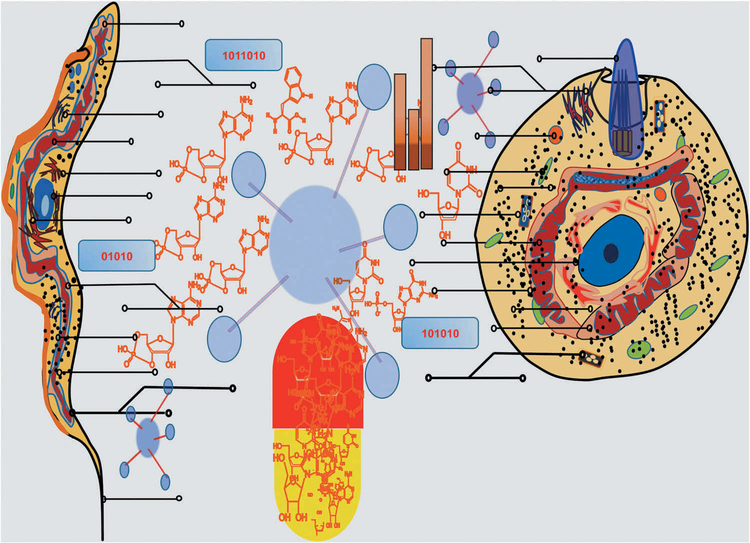
Public databases such as TriTrypDB, TDR Targets, T. cruzi RNA sequence Database (in progress), and Crystal structures Database present a great opportunity for data mining and contain a wealth of information that can be easily searched and analyzed. Furthermore, drug discovery efforts against neglected tropical diseases have increased, some pharmaceutical companies have become more engaged and several academic centers have established powerful drug discovery capabilities. Public–private partnerships such as DNDi and various foundations and governmental funding agencies have made major financial and other contributions to allow activities to proceed on the scale required for drug discovery. All these integrated resources will be beneficial for AI to accelerate drug discovery. A proposed model to advance drug discovery in Chagas disease using AI is presented in Figure 4.
Figure 4. Proposed model to advance drug discovery in Chagas disease using artificial intelligence (computer-calculated compounds targeting all molecules of T. cruzi mammalian stages).
The clinically relevant forms T. cruzi trypomastigote (left cell) and intracellular amastigote (right cell) targeted for drug discovery are depicted in the picture.
Selected verified but challenging therapeutic protein targets that would inhibit T. cruzi trypomastigotes and intracellular amastigotes are depicted by lines ending in circles. For each T. cruzi protein, pointed by lines ending in circles, AI calculates compounds targeting all selected molecules of T. cruzi, and will screen millions of compounds to predict those that bind and potentially inhibit protein function. This approach has the potential to deliver drug-like compounds that will go on to further optimization and then potential drug development. Consequently, the pill, colored in red and yellow, contains selected specific drugs that bind specifically to T. cruzi proteins, are novel targets for intervention, but do not bind to the host, are effective in vitro at low nM and in in vivo at low concentrations with no or minimal side effects in infections caused by drug-resistant T. cruzi strains and with predicted excellent PK/PD for entering clinical trials. Numbers in the interphase represent algorithms, as process or set of rules to be followed in drug calculations that target T. cruzi specific proteins by AI.
Some investigators think that the potential of AI to pinpoint previously unknown causes of disease will accelerate the trend toward treatments designed for patients with specific biological profiles. Thus, personalized medicine or precision medicine will be advanced by AI for better treatments and management of Chagas disease in the near future as new drugs become available. Furthermore, AI platforms, which data is fed from sources such as research papers, patents, clinical trials, and patient records would be important for drug discovery and management of Chagas disease. It is expected that robotic scientists using AI will generate a greater number of compounds to rapidly accelerate drug discovery in Chagas disease in the near future.
An advanced robotic colleague named Eve developed by Steve Oliver’s team at the University of Cambridge, discovered that triclosan, a common ingredient in toothpaste could potentially treat drug-resistant malaria parasites [136]. The investigators developed strains of yeast in which genes essential for growth had been replaced with their equivalents either from malaria parasites or from humans. Eve then screened thousands of compounds to find those that halted or severely slowed the growth of the strains dependent on the malaria genes but not those containing the human genes-to target the parasites while reducing the risk of toxicity. Early results were used to inform the selection of later candidates to screen. This identified triclosan as affecting malaria-parasite growth by inhibiting the DHFR enzyme - also the target of the antimalarial drug pyrimethamine. Similar approaches using AI are expected to be used for Chagas disease to advance drug discovery in the near future. Atomwise, Inc., a biotech company created in 2012, uses AI for structure-based small molecule drug discovery. In 2019, Atomwise began to collaborate with DNDi to advance drug development using AI for neglected diseases, including Chagas disease: https://www.dndi.org/2019/media-centre/press-releases/dndi-and-atomwise-collaborate-to-advance-drug-development-using-ai-for-neglected-diseases/. DNDi scientists selected three verified but challenging therapeutic protein targets that would inhibit T. cruzi. For each T. cruzi protein, Atomwise screened millions of compounds using its AI-powered screening technology to predict those that bind and potentially inhibit protein function. This research has delivered drug-like compounds that will now go on to further optimization and then potential drug development. This state-of-the-art technology is more efficient and cost-effective than standard methods. Ultimately, the next step for this partnership will be to further develop the compounds in active discovery projects and help fill the pipeline of potential treatments for patients with Chagas disease.
A recent specialized platform for innovative research exploration – ASPIRE – in preclinical drug discovery could help study unexplored biologically active chemical space through integrating automated synthetic chemistry, high-throughput biology and artificial intelligence technologies was developed [137]. ASPIRE could also help to accelerate drug discovery in Chagas disease.
7. Conclusions
T. cruzi sterol 14-α demethylase structure-based drug design has discovered a new generation of azoles, VNI and VFV, as promising small molecule candidates for Chagas disease treatment. It is important that work on these inhibitors progress to further in vitro evaluations of a panel of strains representing particular DTUs prevalent in patients of disease endemic countries and in vivo experimental infections with drug-resistant strains representative of DTUs. Furthermore, it would be important to evaluate their efficacy in clinical isolates from endemic areas. Moreover, rigorous PK/PD analysis of VNI and VFV is essential to translate the results of preclinical in vivo studies to clinical setting.
Given the fact that Tc24SMT is unique for T. cruzi with no counterpart in the human host, it is significantly important to target this critical enzyme of the sterol pathway of T. cruzi. Significant advances in developing new inhibitors that target other enzymes have been done and will be important for combinatorial therapy for the treatment of Chagas disease.
New recent promising cruzipain inhibitors as well as specific inhibitors of the enzymes of the trypanothione and tryanothione reductase that do not have host counterpart discussed here and the new hits by HTS also described here represent promising drugs for the treatment of Chagas disease. We believe that drug discovery in Chagas disease in the near future should advance by the inclusion of new therapeutics and combinational therapies with minimal or no side effects such as the promising candidates that we reviewed and discussed and the ones that rapidly would come by phenotypic screening.
Some of the trypanomatid targets discussed in this review are common for T. cruzi, T. brucei, and Leishmania parasites, so progress for one will significantly benefit the treatment of other pathogenic trypanosomatids as examples provided and discussed in this review.
The fact that CRISPR/Cas9 is in place for T. cruzi and that the strains of T. cruzi are rapidly been sequenced by RNA-seq will facilitate the rapid validation of T. cruzi targets for drug development in Chagas disease.
Given the complexity in drug discovery of Chagas disease, organized transdisciplinary collaborative efforts between basic and clinical scientists, clinicians, bioinformaticians, public health professionals, AI scientists, epidemiologists, foundations, the private and public sector would be required to advance the research enterprise for new treatments for Chagas disease.
8. Expert opinion
The enzymes that participate in the synthesis of ergosterol in T. cruzi are excellent targets for drug discovery. A set of inhibitors that target all critical enzymes of the ergosterol pathway can be used as multi-drug therapies for the treatment of Chagas disease. We believe that this strategy will produce real outcomes in the treatment of Chagas disease. Significant advances were made on the T. cruzi sterol 14 α-demethylase structure-based design of new promising inhibitors such as VNI and VFV as potential candidates for clinical trials [20,21]. However, rigorous PK/PD analysis is essential to translate the results of preclinical in vivo studies such as those with novel T. cruzi sterol 14 α-demethylase inhibitors VNI and VFV. The demonstrated drugability of the T. cruzi 14 α-demethylase is facilitating the design of new pharmacophores that do not affect the human counterpart, with high efficacy in curing T. cruzi infections in rodent experimental models and presenting excellent PKs [45]. However, Tc24SMT is the only enzyme in the biosynthesis of ergosterol in T. cruzi with no human counterpart and is a prime target for intervention. Upon crystallization of Tc24SMT and by structure-based drug design studies, it is expected that highly effective inhibitors for the treatment of Chagas disease can be designed. Ideally, the crystallization of all enzymes of the T. cruzi ergosterol pathway will facilitate to design new inhibitors for the treatment of Chagas disease.
Remarkably, phenotypic-based drug discovery has become the main pillar of Chagas disease R&D. In this regard, appropriate chemical libraries for screening, robust assays, and appropriate screening cascades have been used intensively. Significant number of new promising inhibitors that block T. cruzi infection at low concentrations have been reported and the target for some inhibitors were identified. We expect that phenotypic-based screening will also rapidly bring new promising drugs for the treatment of Chagas disease.
We believe that exploring both the ergosterol pathway and phenotypic-based drug discovery in parallel or synergistically will have a significant impact in the treatment of Chagas disease in the near future.
There are few targets that are essential for T. cruzi and a list of candidate targets need to be validated by CRISPR/Cas9 [28,138] that is now in place for T. cruzi. Furthermore, T. cruzi functional genomics to advance T. cruzi biology, infection, survival, and persistence is still in infancy. More information is needed about the molecular mechanisms involved in the establishment of infection, and also about those involved in parasite survival and persistence to discover novel targets for intervention.
We expect that RNA sequencing of all strains of T. cruzi that are classified in seven discrete typing units TcI-TcVII with emphasis in trypomastigotes and amastigotes will elucidate new targets and will facilitate the understanding of drug resistance in T. cruzi.
Although there is strong optimism and excitement to discover new effective drugs for the treatment of Chagas disease, the enterprise of drug discovery for Chagas is still challenging. One of the key challenges of phenotypic drug discovery is how to address issues, such as potency, toxicity and pharmacokinetic problems that arise during the hit optimization process. Another major challenge is defining the relevant cellular and animal models that closely mimic human clinical conditions in Chagas disease [129] and the lack of an animal model that simulates the human disease represents the greatest hindrance.
There is a need for cellular and animal models that can distinguish between compounds that are active in humans and those that are not. A major challenge for drug discovery is the diverse group of T. cruzi strains and their genetic diversity [131]. Treatment of Chagas with current or new drugs that enter or will enter clinical trials do not consider the field of pharmacogenomics [133] to identify genes which influence individual variation in the efficacy or toxicity. Screening is not performed with all T. cruzi strains of the seven discrete typing units TcI-TcVII that include drug susceptible, partial resistant and resistant strains. The absence of effective tests to monitor the course of treatment and certify the cure of patients is also a very serious problem that must be faced in parallel with efforts to develop new drugs.
Finally, we believe that AI has the potential power to change drug discovery for diseases [136] including Chagas disease. Since investigators are employing AI to discover drugs, we expect that in the near future this approach will dramatically accelerate drug discovery for Chagas disease. Interestingly, AI has identified triclosan as affecting malaria-parasite growth by inhibiting the DHFR enzyme – also the target of the antimalarial drug pyrimethamine [136] and we expect it to be used to advance drug discovery in Chagas disease. Moreover, preclinical drug discovery in Chagas disease could be accelerated by studying unexplored biologically active chemical space through integrating automated synthetic chemistry, high-throughput biology, and artificial intelligence technologies [137].
Article Highlights.
One century after Chagas disease discovery, a curative agent remains elusive. Only two drugs are available for Chagas disease treatment, benzimidazole and nifurtimox, and both drugs produce significant side effects and have low efficacy for the chronic phase of the disease.
Chagas disease has gone global due to human migration and is estimated that 8 million people are infected and more than 10,000 die every year on account of the infection. Thus, new effective, safe, and potent drugs for the treatment of Chagas diseases are urgently needed.
Various approaches including target structure-based drug design have been used to discover promising small molecule candidates for Chagas disease treatment.
Enzymes that participate in the synthesis of ergosterol in T. cruzi are excellent targets for drug discovery. A set of inhibitors that target all critical enzymes of the ergosterol pathway can be used as multi-drug therapies for the treatment of Chagas disease.
To circumvent the challenges of target-based drug discovery, phenotypic approaches have been widely used for T. cruzi. One element that is highly used in drug development for Chagas disease is HTS and in vivo imaging.
Artificial Intelligence is expected to significantly accelerate drug discovery in Chagas disease and overall other global parasitic diseases.
This box summarizes key points contained in the article.
Acknowledgments
Funding
FV was supported by National Institutes of Health grants (AI007281-28, HL007737-22, U54 MD007586-31, U54 MD007593-09, U54 MD010722-02, AI080580, and P30 AI110527-04).
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.WHO (World and Health Organization). Chagas disease - Epidemiology. 2019. Available from: http://www.who.int/chagas/epidemiology/en/
- 2.Coura PA, Jose R. Vinas. Chagas disease: a new worldwide challenge. Nature. 2010. June 24;465(7301):S6–7.. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Zhou XN. Preventing the transmission of American trypanosomiasis and its spread into non-endemic countries. Infect Dis Poverty. 2015;4:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotez PJ. The rise of neglected tropical diseases in “New Texas”. PLoS Negl Trop Dis. 2018;12(1):e0005581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conteh L, Engels T, Molyneux DH. Socioeconomic aspects of neglected tropical diseases. Lancet. 2010;375:239–47 PubMed. •Describes the geographical distribution of parasites.
- 6.Lee BY, Bacon KM, Bottazzi ME, et al. Global economic burden of Chagas disease: a computational simulation model. Lancet Infect Dis. 2013;13(4):342.PubMed-8.• Described Chagas disease burden.
- 7.Schmunis GA, Yadon ZE. Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 2010. Jul-Aug;115(1–2):14–21 [DOI] [PubMed] [Google Scholar]
- 8.Requena-Mendez A, Aldasoro E, de Lazzari E, et al. Prevalence of Chagas disease in Latin-American migrants living in Europe: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2015;9:e0003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bern C, Montgomery SP. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis. 2009;49:e52–54. [DOI] [PubMed] [Google Scholar]
- 10.Coura JR. The discovery of Chagas disease (1908–1909): great successes and certain misunderstandings and challenges. Rev Soc Bras Med Trop. 2013;46:389–90 [DOI] [PubMed] [Google Scholar]
- 11.Molina I, Gómez I Prat J, Salvador F, et al. Randomized trial of posaconazole and benznidazole for chronic Chagas’ disease. N Engl J Med. 2014;370:1899–908• Repported the failure of posaconazole in clinical trials.
- 12.Morillo CA, Waskin H, Sosa-Estani S, et al. Benznidazole and posaconazole in eliminating parasites in asymptomatic T. cruzi carriers: the STOP-CHAGAS Trial. J Am Coll Cardiol. 2017;69:939–947.• Reported the failure of posaconazole in clinical trials.
- 13.Molina I, Salvador F, Sanchez-Montalva A. The use of posaconazole against Chagas disease. Curr Opin Infect Dis. 2015. October;28(5):397–407. [DOI] [PubMed] [Google Scholar]
- 14.Urbina JA. Recent clinical trials for the etiological treatment of chronic chagas disease: advances, challenges and perspectives. J Eukaryot Microbiol. 2015. Jan-Feb;62(1):149–156. [DOI] [PubMed] [Google Scholar]
- 15.Urbina JA. Pharmacodynamics and follow-up period in the treatment of human Trypanosoma cruzi infections with posaconazole. J Am Coll Cardiol. 2017. July 11;70(2):299–300. [DOI] [PubMed] [Google Scholar]
- 16.Urbina JA. The long road towards a safe and effective treatment of chronic Chagas disease. Lancet Infect Dis. 2018. April;18(4):363–365. [DOI] [PubMed] [Google Scholar]
- 17.Chatelain E Chagas disease drug discovery: toward a new era. J Biomol Screen. 2015;20:22–35.• Interesting review on the failure of new drugs entered into the clinical trials.
- 18.Moraes CB, Giardini MA, Kim H, et al. Nitroheterocyclic compounds are more efficacious than CYP51 inhibitors against Trypanosoma cruzi: implications for Chagas disease drug discovery and development. Sci Rep. 2014. April;16(4):4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Molina JA, Poveda C, Martinez-Perez A, et al. Distribution of Trypanosoma cruzi discrete typing units in Bolivian migrants in Spain. Infect Genet Evol. 2014;21:440–442. [DOI] [PubMed] [Google Scholar]
- 20.Villalta F, Dobish MC, Nde PN, et al. VNI cures the acute and chronic experimental Chagas disease. J Infect Dis. 2013;208:504–11•• Reported first cure of experiemental Chagas disease by VNI.
- 21.Lepesheva GI, Hargrove TY, Rachakonda G, et al. VFV as a new effective CYP51 structure-derived drug candidate for Chagas disease and visceral Leishmaniasis. J Infect Dis. 2015;212:1439–1448.•• Reported the better pharmacaphore of VNI curing experimental Chagas disease.
- 22.Soeiro Mde N, de Souza EM, Da Silva CF, et al. In vitro and in vivo studies of the antiparasitic activity of sterol 14α-demethylase (CYP51) inhibitor VNI against drug-resistant strains of Trypanosoma cruzi. Antimicrob Agents Chemother. 2013. September;57(9):4151–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friggeri L, Hargrove TY, Rachakonda G, et al. Sterol 14α- demethylase structure-based optimization of drug candidates for human infections with the protozoan Trypanosomatidae. J Med Chem. 2018;61:10910–10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guedes-da-Silva FH, Batista DDGJ, Da Silva CF, et al. Successful aspects of the co-administration of sterol 14α-demethylase inhibitor VFV and benznidazole in experimental mouse models of Chagas disease caused by the drug-resistant strain of Trypanosoma cruzi. ACS Infect Dis. 2019;5:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbina JA, Lazardi K, Aguirre T, et al. Antiproliferative effects and mechanism of action of ICI 195,739, a novel bis-triazole derivative, on epimastigotes and amastigotes of Trypanosoma (Schizotrypanum) cruzi. Antimicrob Agents Chemother. 1991;35 (4):730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepesheva GI, Park H, Hargrove TY, et al. Crystal structures of Trypanosoma brucei 14 α-demethylase and implications for selective treatment of human infections. J Biol Chem. 2010;285:1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benaim G, Sanders JM, Garcia-Marchan Y, et al. Amiodarone has intrinsic anti-Trypanosoma cruzi activity and acts synergistically with posaconazole. J Med Chem. 2006;49:892–899 [DOI] [PubMed] [Google Scholar]
- 28.Soares Medeiros LC, South L, Peng D, et al. Rapid, selection-free, high-efficiency genome editing in protozoan parasites using CRISPR-Cas9 ribonucleoproteins. MBio. 2017;8(6):e01788–17.•• Reported CRISPR-Cas9 use for T. cruzi genome editing.
- 29.Lepesheva GI, Villalta F, Waterman MR. Targeting Trypanosoma cruzi sterol 14α-demethylase (CYP51). Adv Parasitol. 2011;75:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CK, Leung SSF, Guilbert C, et al. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Negl Trop Dis. 2010;4:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepesheva GI, Hargrove TY, Anderson S, et al. Structural insights into inhibition of sterol 14 alpha-demethylase in the human pathogen Trypanosoma cruzi. J Biol Chem. 2010;285:25582–25590.•• First crystallization of T. cruzi sterol 14α-demethylase.
- 32.Hoekstra WJ, Hargrove TY, Wawrzak Z, et al. Clinical candidate VT-1161′s antiparasitic effect in vitro, activity in a murine model of chagas disease, and structural characterization in complex with the target enzyme CYP51 from Trypanosoma cruzi. Antimicrob Agents Chemother. 2016;60:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lepesheva GI, Zaitseva NG, Nes WD, et al. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14alpha- demethylase. J Biol Chem. 2006;281:3577–3585. [DOI] [PubMed] [Google Scholar]
- 34.Buckner F, Bahia MT, Suryadevara PK, et al. Pharmacological characterization, structural studies, and in vivo activity of anti-Chagas disease lead compounds derived from tipifarnib. Antimicrob Agents Chemother. 2012;56:4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andriani G, Amata E, Beatty J, et al. Antitrypanosomal lead discovery: identification of a ligand-efficient inhibitor of Trypanosoma cruzi CYP51 and parasite growth. J Med Chem. 2013;56:2556–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friggeri L, Hargrove TY, Rachakonda G, et al. Structural basis for rational design of inhibitors targeting Trypanosoma cruzi sterol 14α-demethylase: two regions of the enzyme molecule potentiate its inhibition. J Med Chem. 2014;57:6704–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hargrove TY, Wawrzak Z, Alexander PW, et al. Complexes of Trypanosoma cruzi sterol 14α-demethylase (CYP51) with two pyridine-based drug candidates for Chagas disease: structural basis for pathogen selectivity. J Biol Chem. 2013;288:31602–31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hargrove TY, Wawrzak Z, Liu J, et al. Structural complex of sterol 14α-demethylase (CYP51) with 14α-methylenecyclopropyl-Δ7–24, 25-dihydrolanosterol. J Lipid Res. 2012;53:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hargrove TY, Wawrzak Z, Lamb DC, et al. Structure-functional characterization of cytochrome P450 sterol 14α-demethylase (CYP51B) from Aspergillus fumigatus and molecular basis for the development of antifungal drugs. J Biol Chem. 2015;290:23916–23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hargrove TY, Garvey EP, Hoekstra WJ, et al. Crystal structure of the new investigational drug candidate VT-1598 in complex with Aspergillus fumigatus sterol 14α-demethylase provides insights into its broad-spectrum antifungal activity. Antimicrob Agents Chemother. 2017;61:pii: e00570–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hargrove TY, Friggeri L, Wawrzak Z, et al. Structural analyses of Candida albicans sterol 14alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J Biol Chem. 2017;292:6728–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hargrove TY, Friggeri L, Wawrzak Z, et al. Human sterol 14alpha-demethylase as a target for anticancer chemotherapy: towards structure-aided drug design. J Lipid Res. 2016;57:1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lepesheva GI, Nes WD, Zhou W, et al. CYP51 from Trypanosoma brucei is obtusifoliol- specific. Biochemistry. 2004;43:10789–10799. [DOI] [PubMed] [Google Scholar]
- 44.Hargrove TY, Wawrzak Z, Liu J, et al. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14alpha-demethylase (CYP51) from Leishmania infantum. J Biol Chem. 2011;286:26838–26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lepesheva GI, Friggeri L, Waterman MR. CYP51 as drug targets for fungi and protozoan parasites: past, present and future. Parasitology. 2018;145:1820–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepesheva GI. Waterman MR Structural basis for conservation in the CYP51 family. Biochim Biophys Acta. 2011;1814:88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lepesheva GI, Ott RD, Hargrove TY, et al. Sterol 14α-demethylase as a potential target for antitrypanosomal therapy: Enzyme inhibition and parasite cell growth. Chem Biol. 2007;14:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobish MC, Villalta F, Waterman MR, et al. Organocatalytic, enantioselective synthesis of VNI: A robust therapeutic development platform for Chagas, a neglected tropical disease. Org Lett. 2012;14:6322–6325•• Novel and affordable synthesis of VNI for Chagas disease treatment.
- 49.Hargrove TY, Kim K, de Nazaré Correia Soeiro M, et al. CYP51 structures and structure-based development of novel, pathogen specific inhibitory scaffolds. Internat J Parasitol Drugs Drug Resist. 2012;2:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherkesova TS, Hargrove TY, Vanrell MC, et al. Sequence variation in CYP51A from the Y strain of Trypanosoma cruzi alters its sensitivity to inhibition. FEBS Lett. 2014;588:3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guedes-da-Silva FH, Batista DG, Da Silva CF, et al. Antitrypanosomal activity of sterol 14α-demethylase (CYP51) inhibitors VNI and VFV in the swiss mouse models of Chagas disease induced by the Trypanosoma cruzi Y strain. Antimicrob Agents Chemother. 2017;61:e02098–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu X, Nandekar P, Mustafa G, et al. Ligand tunnels in T. brucei and human CYP51: insights for parasite-specific drug design. Biochem Et Biophys Acta. 2016;1860:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sykes ML, Avery VM. 3-pyridyl inhibitors with novel activity against Trypanosoma cruzi reveal in vitro profiles can aid prediction of putative cytochrome P450 inhibition. Sci Rep. 2018;8:4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urbina JA, Vivas J, Lazardi K, et al. Antiproliferative effects of delta 24 (25)sterol methyl transferase inhibitors on Trypanosoma (Schizotrypanum) cruzi: in vitro and in vivo studies. Chemotherapy. 1996;42:294–307. [DOI] [PubMed] [Google Scholar]
- 55.Osorio-Mendez JF, Cevallos AM. Discovery and genetic validation of chemotherapeutic targets for Chagas’ disease front. Cell Infect Microbiol. 2019;8:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braga MV, Magaraci F, Lorente SO, et al. Effects of inhibitors of delta24(25)-sterol methyl transferase on the ultrastructure of epimastigotes of Trypanosoma cruzi. Microsc Microanal. 2005;11:506–515. [DOI] [PubMed] [Google Scholar]
- 57.Urbina JA, Concepcion JL, Rangel S, et al. Squalene synthase as a chemotherapeutic target in Trypanosoma cruzi and Leishmania mexicana. Mol Biochem Parasitol. 2002;125(1–2):35–45. [DOI] [PubMed] [Google Scholar]
- 58.Urbina JA, Concepcion JL, Montalvetti A, et al. Mechanism of action of 4-phenoxyphenoxyethyl thiocyanate (WC-9) against Trypanosoma cruzi, the causative agent of Chagas’ disease. Antimicrob Agents Chemother. 2003. June;47(6):2047–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Braga MV, Urbina JA, de Souza W. Effects of squalene synthase inhibitors on the growth and ultrastructure of Trypanosoma cruzi. Int J Antimicrob Agents. 2004. July;24(1):72–78. [DOI] [PubMed] [Google Scholar]
- 60.Urbina JA, Conception JL, Caldera A, et al. In vitro and in vivo activities of E5700 and ER-119884, two novel orally active squalene synthase inhibitors, against Trypanosoma cruzi. Antimicrob Agents Chemother. 2004;48:2379–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cinque GM. Structure-activity relationship of new growth inhibitors of Trypanosoma cruzi. J Med Chem. 1998;41:1540–54 [DOI] [PubMed] [Google Scholar]
- 62.Chao MN, Storey M, Li C, et al. Selenium-containing analogues of WC-9 are extremely potent inhibitors of Trypanosoma cruzi proliferation. Bioorg Med Chem. 2017;25:6435–49 [DOI] [PubMed] [Google Scholar]
- 63.Shang N, Li Q, Ko TP, et al. Squalene synthase as a target for Chagas disease therapeutics. PLoS Pathog. 2014;10(5):1004–1114.•• First crystallization of T cruzi squalene synthase.
- 64.Martin MB, Arnold W, Heath HT 3rd, et al. Nitrogen-containing bisphosphonates as carbocation transition state analogs for isoprenoid biosynthesis. Biochem Biophys Res Commun. 1999. October 5;263 (3):754–758. [DOI] [PubMed] [Google Scholar]
- 65.Martin MB, Grimley JS, Lewis JC, et al. Bisphosphonates inhibit the growth of Trypanosoma brucei, Trypanosoma cruzi, Leishmania donovani, Toxoplasma gondii, and Plasmodium falciparum: a potential route to chemotherapy. J Med Chem. 2001. March 15;44 (6):909–916. [DOI] [PubMed] [Google Scholar]
- 66.Montalvetti A, Bailey BN, Martin MB, et al. Bisphosphonates are potent inhibitors of Trypanosoma cruzi farnesyl pyrophosphate synthase. J Biol Chem. 2001. September 7;276(36):33930–33937. [DOI] [PubMed] [Google Scholar]
- 67.Garzoni LR, Caldera A, Meirelles Mde N, et al. Selective in vitro effects of the farnesyl pyrophosphate synthase inhibitor risedronate on Trypanosoma cruzi. Int J Antimicrob Agents. 2004. March;23 (3):273–285. [DOI] [PubMed] [Google Scholar]
- 68.Garzoni LR, Waghabi MC, Baptista MM, et al. Antiparasitic activity of risedronate in a murine model of acute Chagas’ disease. Int J Antimicrob Agents. 2004. March;23(3):286–290. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez JB, Falcone BN, Szajnman SH. Approaches for designing new potent inhibitors of farnesyl pyrophosphate synthase. Expert Opin Drug Discov. 2016;11(3):307–320. [DOI] [PubMed] [Google Scholar]
- 70.Ferrer-Casal M, Li C, Galizzi M, et al. New insights into molecular recognition of 1,1-bisphosphonic acids by farnesyl diphosphate synthase. Bioorg Med Chem. 2014. January 1;22(1):398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aripirala S, Szajnman SH, Jakoncic J, et al. Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzifarnesyl diphosphate synthase. J Med Chem. 2012;55(14):6445–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tani O, Akutsu Y, Ito S, et al. NMR biochemical assay for oxidosqualene cyclase: evaluation of inhibitor activities on Trypanosoma cruzi and human enzymes. J Med Chem. 2018;61:5047–5053 [DOI] [PubMed] [Google Scholar]
- 73.Kawasaki Y, Freire E. Finding a better path to drug selectivity. Drug Discov Today. 2011;16:985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choy JW, Bryant C, Calvet CM, et al. Chemical-biological characterization of a cruzain inhibitor reveals a second target and a mammalian off-target. Beilstein J Org Chem. 2013;9:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barr SC, Warner KL, Kornreic BG, et al. A cysteine protease inhibitor protects dogs from cardiac damage during infection by Trypanosoma cruzi. Antimicrob Agents Chemother. 2005;49:5160–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ndao M, Beaulieu C, Black WC, et al. Reversible cysteine protease inhibitors show promise for a Chagas disease cure. Antimicrob Agents Chemother. 2014;58(2):1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salas-Sarduy E, Landaburu LU, Karpiak J, et al. Novel scaffolds for inhibition of Cruzipain identified from high-throughput screening of anti-kinetoplastid chemical boxes. Sci Rep. 2017. September 21;7 (1):12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khare S, Roach SL, Barnes SW, et al. Utilizing chemical genomics to identify cytochrome b as a novel drug target for Chagas disease. PLoS Pathog. 2015;11(7):PubMed e1005058.• Chemical genomics strategy for Chagas drug discovery.
- 79.Krauth-Siegel RL, Meiering SK, Schmid H. The parasite specific trypanothione metabolism of Trypanosome and Leishmania. Biol Chem. 2003. April;384(4):539–549. [DOI] [PubMed] [Google Scholar]
- 80.Fairlamb AH, Cerami A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu Rev Microbiol. 1992;46:695–729 [DOI] [PubMed] [Google Scholar]
- 81.Maya JD, Cassels BK, Iturriaga-Vásquez P, et al. Mode of action of natural and synthetic drugs against Trypanosoma cruzi and their interaction with the mammalian host. Comp Biochem Physiol A Mol Integr Physiol. 2007. April;146(4):601–620. [DOI] [PubMed] [Google Scholar]
- 82.Faúndez M, López-Muñoz R, Torres G, et al. Buthionine sulfoximine has anti-Trypanosoma cruzi activity in a murine model of acute Chagas’ disease and enhances the efficacy of nifurtimox. Antimicrob Agents Chemother. 2008;52:1837–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Bond CS, Bailey S, et al. The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 A resolution. Protein Sci. 1996;5:52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bond CS, Zhang Y, Berriman M, et al. Crystal structure of Trypanosoma cruzi trypanothione reductase in complex with trypanothione, and the structure-based discovery of new natural product inhibitors. Structure. 1999;7:81–9 [DOI] [PubMed] [Google Scholar]
- 85.Krauth-Siegel RL, Sticherling C, Jost I, et al. Crystallization and preliminary crystallographic analysis of trypanothione reductase from Trypanosoma cruzi, the causative agent of Chagas’ disease. FEBS Lett. 1993;317:105–108. [DOI] [PubMed] [Google Scholar]
- 86.Lantwin CB, Schlichting I, Kabsch W, et al. The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state. Proteins. 1994;18:161–73 [DOI] [PubMed] [Google Scholar]
- 87.Saravanamuthu A, Vickers TJ, Bond CS, et al. Two interacting binding sites for quinacrine derivatives in the active site of trypanothione reductase: a template for drug design. J Biol Chem. 2004;279:29493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jockers-Scherubl MC, Schirmer RH, Krauth-Siegel RL. Trypanothione reductase from Trypanosoma cruzi. Catalytic properties of the enzyme and inhibition studies with trypanocidal compounds. Eur J Biochem. 1989;180:267–272. [DOI] [PubMed] [Google Scholar]
- 89.Ariyanayagam MR, Oza SL, Mehlert A, et al. Bis(glutathionyl)spermine and other novel trypanothione analogues in Trypanosoma cruzi. J Biol Chem. 2003;278:27612–19 [DOI] [PubMed] [Google Scholar]
- 90.Aguirre G, Cabrera E, Cerecetto H, et al. Design, synthesis and biological evaluation of new potent 5-nitrofuryl derivatives as anti-Trypanosoma cruzi agents. Studies of trypanothione binding site of trypanothione reductase as target for rational design. Eur J Med Chem. 2004;39:421–31 [DOI] [PubMed] [Google Scholar]
- 91.Benson TJ, McKie JH, Garforth J, et al. Rationally designed selective inhibitors of trypanothione reductase. Phenothiazines and related tricyclics as lead structures. Biochem J. 1992;286:9–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan C, Yin H, Garforth J, et al. Phenothiazine inhibitors of trypanothione reductase as potential antitrypanosomal and antileishmanial drugs. J Med Chem. 1998;41:148–56 [DOI] [PubMed] [Google Scholar]
- 93.Gutierrez-Correa J, Fairlamb AH, Stoppani AO. Trypanosoma cruzi trypanothione reductase is inactivated by peroxidase-generated phenothiazine cationic radicals. Free Radic Res. 2001;34:363–78 [DOI] [PubMed] [Google Scholar]
- 94.Moreno SN, Carnieri EG, Docampo R. Inhibition of Trypanosoma cruzi trypanothione reductase by crystal violet. Mol Biochem Parasitol. 1994;67:313–320. [DOI] [PubMed] [Google Scholar]
- 95.Baillet S, Buisine E, Horvath D, et al. 2-Amino diphenylsulfides as inhibitors of trypanothione reductase: modification of the side chain. Bioorg Med Chem. 1996;4:891–9 [DOI] [PubMed] [Google Scholar]
- 96.O’Sullivan MC, Dalrymple DM, Zhou Q. Inhibiting effects of spermidine derivatives on Trypanosoma cruzi trypanothione reductase. J Enzyme Inhib. 1996;11:97–114. [DOI] [PubMed] [Google Scholar]
- 97.Bonnet B, Soullez D, Davioud-Charvet E, et al. New spermine and spermidine derivatives as potent inhibitors of Trypanosoma cruzi trypanothione reductase. Bioorg Med Chem. 1997;5:1249–56 [DOI] [PubMed] [Google Scholar]
- 98.O’Sullivan MC, Zhou Q, Li Z, et al. Polyamine derivatives as inhibitors of trypanothione reductase and assessment of their trypanocidal activities. Bioorg Med Chem. 1997;5:2145–55 [DOI] [PubMed] [Google Scholar]
- 99.Li Z, Fennie MW, Ganem B, et al. Polyamines with N-(3-phenylpropyl) substituents are effective competitive inhibitors of trypanothione reductase and trypanocidal agents. Bioorg Med Chem Lett. 2001;11:251–4 [DOI] [PubMed] [Google Scholar]
- 100.Garforth J, Yin H, McKie JH, et al. Rational design of selective ligands for trypanothione reductase from Trypanosoma cruzi. Structural effects on the inhibition by dibenzazepinesbased on imipramine. J Enzyme Inhib. 1997;12:161–173. [DOI] [PubMed] [Google Scholar]
- 101.Fournet A, Inchausti A, Yaluff G, et al. Trypanocidal bisbenzylisoquinoline alkaloids are inhibitors of trypanothione reductase. J Enzyme Inhib. 1998;13:1–9. [DOI] [PubMed] [Google Scholar]
- 102.Gallwitz H, Bonse S, Martinez-Cruz A, et al. Ajoene is an inhibitor and subversive substrate of human glutathione reductase and Trypanosoma cruzi trypanothione reductase: crystallographic, kinetic, and spectroscopic studies. J Med Chem. 1999;42:364–72 [DOI] [PubMed] [Google Scholar]
- 103.Bonse S, Santelli-Rouvier C, Barbe J, et al. Inhibition of Trypanosoma cruzi trypanothione reductase by acridines: kinetic studies and structure-activity relationships. J Med Chem. 1999;42:5448–54 [DOI] [PubMed] [Google Scholar]
- 104.Bonse S, Richards JM, Ross SA, et al. (2,2′:6′,2″-Terpyridine) platinum(II) complexes are irreversible inhibitors of Trypanosoma cruzi trypanothione reductase but not of human glutathione reductase. J Med Chem. 2000;43:4812–21 [DOI] [PubMed] [Google Scholar]
- 105.Lee B, Bauer H, Melchers J, et al. Irreversible inactivation of trypanothione reductase by unsaturated Mannich bases: a divinyl ketone as key intermediate. J Med Chem. 2005;48:7400–10 [DOI] [PubMed] [Google Scholar]
- 106.Cota BB, Rosa LH, Fagundes EM, et al. A potent trypanocidal component from the fungus Lentinus strigosus inhibits trypanothione reductase and modulates PBMC proliferation. Mem Inst Oswaldo Cruz. 2008;103:263–270. [DOI] [PubMed] [Google Scholar]
- 107.McCabe RE, Remington JS, Araujo FG. In vivo and in vitro effects of cyclosporin A on Trypanosoma cruzi. Am J Trop Med Hyg. 1985;34:861–865. [DOI] [PubMed] [Google Scholar]
- 108.Bua J, Fichera LE, Fuchs AG, et al. Anti-Trypanosoma cruzi effects of cyclosporin A derivatives: Possible role of a P-glycoprotein and parasite cyclophilins. Parasitology. 2008;135:217–228. [DOI] [PubMed] [Google Scholar]
- 109.Bua J, Ruiz AM, Potenza M, et al. In vitro anti-parasitic activity of cyclosporin A analogs on Trypanosoma cruzi. Bioorg Med Chem Lett. 2004;14:4633–4637. [DOI] [PubMed] [Google Scholar]
- 110.Barreto-Bergter E, Hogge L, Steele Da CF. Lipid alterations induced by nifurtimox in Trypanosoma cruzi. Mol Biochem Parasitol. 1986;21:221–226. [DOI] [PubMed] [Google Scholar]
- 111.Armah DA, Mensa-Wilmot KS. Myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J Biol Chem. 1999;274:5931–5938. [DOI] [PubMed] [Google Scholar]
- 112.Towler DA, Eubanks SR, Towery DS, et al. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J Biol Chem. 1987;262:1030–1036 [PubMed] [Google Scholar]
- 113.Towler DA, Adams SP, Eubanks SR, et al. Purification and characterization of yeast myristoyl CoA: proteinN-myristoyl transferase. Proc Natl Acad Sci USA. 1987;84:2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roberts AJ, Torrie LS, Wyllie S, et al. Biochemical and genetic characterization of Trypanosoma cruzi N myristoyl transferase. Biochem J. 2014;459:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frearson JA, Brand S, McElroy SP, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464:728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pan P, Vermelho AB, Capaci Rodrigues G, et al. Cloning, characterization, and sulfonamide and thiol inhibition studies of an α-carbonic anhydrase from Trypanosoma cruzi, the causative agent of Chagas disease. J Med Chem. 2013;56(4):1761PubMed–71. [DOI] [PubMed] [Google Scholar]
- 117.Rodrigues GC, Feijó DF, Bozza MT, et al. Design, synthesis, and evaluation of hydroxamic acid derivatives as promising agents for the management of Chagas disease. J Med Chem. 2014;57 (2):298PubMed–308. [DOI] [PubMed] [Google Scholar]
- 118.Field MC, Horn D, Fairlamb AH, et al. Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat Rev Microbiol. 2017;15(4):217–231.•• An outstanding review highlighting challenges in Chagas drug discovery.
- 119.Lewis MD, Francisco AF, Taylor MC, et al. A new experimental model for assessing drug efficacy against Trypanosoma cruzi infection based on highly sensitive in vivo imaging. J Biomol Screen. 2015;20(1):36PubMed–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chatelain E, Loset JR. Phenotypic screening approaches for Chagas disease drug discovery. Expert Opin Drug Discov. 2018;13:141–153.•• Excellent review of cellular and animal models challenges for Chagas drug discovery.
- 121.Villalta F, Kierzenbaum F. Growth of isolated amastigotes of Trypanosoma cruzi in cell-free medium. J Protozool. 1982;29:570–576. [DOI] [PubMed] [Google Scholar]
- 122.Zingales B, Miles MA, Moraes CB, et al. Drug discovery for Chagas disease should consider Trypanosoma cruzi strain diversity. Drugs for neglected disease initiative; Chagas clinical research platform meeting. Mem Inst Oswaldo Cruz. 2014. September;109(6):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alonso-Padilla J, Cotillo I, Presa JL, et al. Automated high-content assay for compounds selectively toxic to Trypanosoma cruzi in a myoblastic cell line. PLoS Negl Trop Dis. 2015;9(1):e0003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ranzani AT, Nowicki C, Wilkinson SR, et al. Identification of specific inhibitors of Trypanosoma cruzi malic enzyme isoforms by target-based HTS. SLAS Discov. 2017;22(9):1150–1161. [DOI] [PubMed] [Google Scholar]
- 125.Costa BL, Cardoso MV, Filho GB, et al. Compound profiling and 3D-QSAR studies of hydrazone derivatives with activity against intracellular Trypanosoma cruzi. Bioorg Med Chem. 2016;24 (8):1608–1618. [DOI] [PubMed] [Google Scholar]
- 126.Ekins S, de Siqueira-Neto JL, McCall LI, et al. Machine learning models and pathway genome data base for Trypanosoma cruzi drug discovery. PLoS Negl Trop Dis. 2015;9(6):e0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lara-Ramirez E, Lopaz-Dedillo JC, Nogueda-Torres B, et al. An in vitro and in vivo evaluation of new potential trans-sialidase inhibitors of Trypanosoma cruzi predicted by a computational drug repositioning method. Eur J Med Chem. 2017;132:249–261 [DOI] [PubMed] [Google Scholar]
- 128.Bellera C, Balcazar DE, Vanrell MC, et al. Computer-guided drug repurposing: identification of trypanocidal activity of clofazimine, benidipine and saquinavir. Eur J Med Chem. 2015;93(26):338–348. [DOI] [PubMed] [Google Scholar]
- 129.Chatelain E, Konar N. Translational challenges of animal models in Chagas disease drug development: a review. Drug Des, Dev Ther 2015;19(9):4807–4823.•• Excellent review about translational challenges by presenting animal models in Chagas drug discovery.
- 130.Zingales B, Andrade SG, Briones MRS, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcIV. Mem Inst Oswaldo Cruz. 2009;104(7):1051–1054.• Description of classification of diverged groups of T cruzi.
- 131.Zingales B, Miles MA, Campbell DA, et al. The revised Trypanosoma cruzi subespecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12:240–253. [DOI] [PubMed] [Google Scholar]
- 132.Perez CJ, Lymbery AJ, Thompson RCA. Chagas disease: the challenge of polyparasitism? Trends Parasitol. 2014;30:176–182. [DOI] [PubMed] [Google Scholar]
- 133.Pang T Impact of pharmacogenomics on neglected diseases of the developing world. Am J Pharm. 2003;3(6):393–398. [DOI] [PubMed] [Google Scholar]
- 134.Sadée W. Pharmacogenomics. BMJ. 1999;319:1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sánchez-Valdéz FJ, Padilla A, Wang W, et al. Spontaneous dormancy protects Trypanosoma cruzi during extended drug exposure. Elife. 2018;7:e34039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fleming N Computer-calculated coumpounds. Researchers are deploying artificial intelligence to discover drugs. Nature. 2018;557:S55–S57.•• Excellent view of artificial intelligence for drug discovery.
- 137.Sittampalam GS, Rudnicki DD, Tagle DA, et al. Mapping biologically active chemical space to accelerate drug discovery. Nat Rev Drug Dis. 2019;18(2):83–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lander N, Chiurillo MA, Docampo R. Genome editing by CRISPR/Cas9: A game change in the genetic manipulation of protists. J Eukaryot Microbiol. 2016;63(5):679–690.•• Reported the CRISPR-Cas9 use for T.cruzi genome editing.
- 139.Torrico F, Gascon J, Ortiz L, et al. Treatment of adult chronic indeterminate Chagas disease with benznidazole and three E1224 dosing regimens: a proof-of-concept, randomised, placebo-controlled trial. Lancet Infect Dis. 2018;18(4):419–430.•• Reported the failure of ravuconazole in clinical trials.