Abstract
Animals can perform complex and purposeful behaviors by executing simpler movements in flexible sequences. It is particularly challenging to analyze behavior sequences when they are highly variable, as is the case in language production, certain types of birdsong and, as in our experiments, flies grooming. High sequence variability necessitates rigorous quantification of large amounts of data to identify organizational principles and temporal structure of such behavior. To cope with large amounts of data, and minimize human effort and subjective bias, researchers often use automatic behavior recognition software. Our standard grooming assay involves coating flies in dust and videotaping them as they groom to remove it. The flies move freely and so perform the same movements in various orientations. As the dust is removed, their appearance changes. These conditions make it difficult to rely on precise body alignment and anatomical landmarks such as eyes or legs and thus present challenges to existing behavior classification software. Human observers use speed, location, and shape of the movements as the diagnostic features of particular grooming actions. We applied this intuition to design a new automatic behavior recognition system (ABRS) based on spatiotemporal features in the video data, heavily weighted for temporal dynamics and invariant to the animal’s position and orientation in the scene. We use these spatiotemporal features in two steps of supervised classification that reflect two time-scales at which the behavior is structured. As a proof of principle, we show results from quantification and analysis of a large data set of stimulus-induced fly grooming behaviors that would have been difficult to assess in a smaller dataset of human-annotated ethograms. While we developed and validated this approach to analyze fly grooming behavior, we propose that the strategy of combining alignment-invariant features and multi-timescale analysis may be generally useful for movement-based classification of behavior from video data.
Keywords: Grooming, Neuroethology, Behavior, Machine Learning
INTRODUCTION
Quantifying variable and complex animal behavior is challenging: observers must record many instances of a given behavior in order to detect changes with statistical power. For example, researchers analyzed thousands of hours of acoustic data from songbirds to detect small changes in song structure that result from learning (Ravbar et al. 2012). Large amounts of video data also present analysis challenges: manual human annotation of behavior can be slow, labor intensive, error-prone, and anthropomorphically biased.
Recently, several machine-learning algorithms have been developed to compress video data, extract relevant features, and automatically recognize various animal behaviors (Todd, Kain, and de Bivort 2017; Robie, Seagraves, et al. 2017; Mathis et al. 2018). Unfortunately, we were not able to employ these techniques to our video of fly grooming behavior because our experimental setting made it difficult to reliably detect fly body parts and orientation as they remove the dust. This limits the range of behaviors and experimental manipulations that can be studied. For example, to recognize fly antennal cleaning events from video by a machine vision algorithm, the front legs must be visible in each frame of the video with sufficient pixel resolution, the illumination should be constant enough for background subtraction, the animal’s appearance cannot change during the experiment and at the very least, the animal’s position and orientation should be easy to identify by machine (Hampel et al. 2015). Such dependencies limit the use of existing behavior recognition methods that rely on either good spatial resolution, few occlusions of body parts, easy background subtraction, or constant appearance. These problems become especially significant when it is desirable to record behavior in either more natural environments or in experimental conditions like ours, where flies are covered in dust, and as such they present a particular obstacle to research projects like our work on fly grooming. While here we describe how we address the machine vision challenge that arose from our specific experimental settings, we believe our solution should be generally applicable to a wide range of behavioral data where experimental settings are similarly unconstrained.
Our motivation to develop new behavior recognition software comes from ethological investigation of grooming in fruit flies (Drosophila melanogaster). This behavior is composed of individual grooming movements (IGMs) performed in a flexible sequence when the fly is coated in dust (Seeds et al 2014). IGMs include leg rubbing and leg sweeps directed toward different body parts to remove debris. The IGMs are organized into subroutines. For example, flies use their front legs to alternate between head sweeps and front leg rubbing. We refer to this subroutine as the anterior grooming motif. Alternation between abdominal or wing sweeps and back leg rubbing constitutes posterior grooming motifs. When we experimentally apply dust all over the fly, it executes grooming motifs in quick succession, beginning with anterior body parts and then gradually progressing towards posterior body parts. The sequence of grooming motifs is flexible (variable). Because one of our research goals is to quantify subtle changes in this variable sequence with respect to phenotypes resulting from genetically inhibiting or activation various neural circuits, we need to reliably analyze hundreds of hours of video recordings of grooming behavior for potentially subtle perturbations, which drove us to explore automatic behavior detection methods and ultimately to develop our own.
To elicit naturalistic grooming behavior, we film flies covered in dust in chambers that allow them to walk and move freely. The dust often creates patterns over the animal’s body that can make it hard to separate their whole body from the background. Furthermore, as flies remove the dust, their appearance changes: for example, when the wings are covered in dust, the back legs are obscured, but as the wings are cleaned, the back leg movements become more visible. The appearance of the background varies as well: the floor of our recording arena is mesh to allow the dust to drop through, but this makes maintaining uniform background and lighting difficult. The flies perform the same grooming movement with their limbs while their bodies are in different orientations and positions. All of these aspects of the video data confound the current behavior classification methods. We had two possible strategies for achieving accurate automatic behavior recognition: we could either design a more constrained experimental setting to ease the computer vision problem or we could rely on more sophisticated computer vision methods that allow more flexibility in the types of data to which they can be applied. Here we chose the latter strategy. We developed a method for behavior recognition from massive data sets that functions in less-constrained experimental settings such as ours and presents a major step toward naturalistic behavior analysis.
In general, behaviors can be either identified from particular configuration of body parts – spatial information (for example, a posture of a golf player may indicate what move she does next), from the dynamics of the movements – temporal information (for example, the periodic timing of the swings) or from a combination of both – spatiotemporal information (for example, the shape of the swinging movement). Our solution is to combine spatial with temporal information, putting more weight on temporal information in cases where spatial information is limited. This idea draws inspiration from biology: in peripheral vision, it is very difficult to count the number of fingers on a hand if presented far from the center of the visual field (poor spatial information) but it is relatively easy to count the number of hand motions (sufficient temporal information) because our peripheral vision is more sensitive to movement than to spatial information. As we will describe below, our strategy for automatically recognizing grooming movements is based on the principle to “gain in time what we lose in space”.
We combine spatial with temporal information to obtain spatiotemporal features of our data that do not vary as a function of animal’s position or horizontal orientation in the scene. They encode useful information about movement class even when behavior would be difficult to discern from individual movie frames. While this approach allows us to recognize certain movements without having to determine the fly’s orientation or locate individual body parts, it introduces a new problem of differentiating between behaviors that have similar temporal features but differ in spatial features (e.g. front vs. back leg rubbing). We solve this problem by carrying out the behavior classification, based on supervised machine learning, in two steps, corresponding to two time-scales: first we determine broad time-scale behavioral context (e.g. anterior vs. posterior grooming behavioral motifs); and second, we determine individual grooming movements that happen on a much faster time-scale (leg rubbing vs. body sweeps). In other words, what we lose in spatial context (the location of body parts engaged in the behavior), we regain in temporal context (broad time-scale).
RESULTS
Method summary
To foster readability for diverse audiences, we present a conceptual overview of the ABRS pipeline in the Methods summary, in this section, and the detailed technical description of the methods in MATERIALS AND METHODS. We also provide the Python code on GitHub repository (https://github.com/AutomaticBehaviorRecognitionSystem/ABRS). However, this section should be sufficient for understanding of the ABRS method and for reviewing the results.
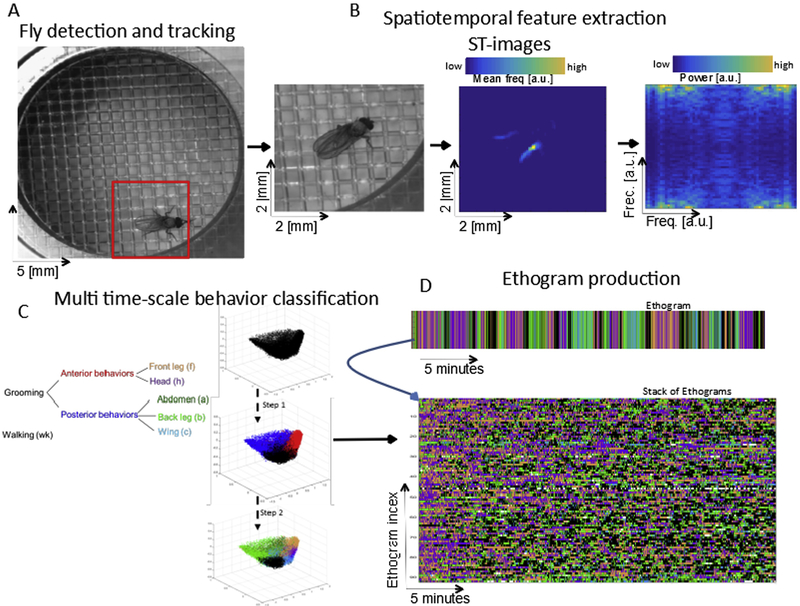
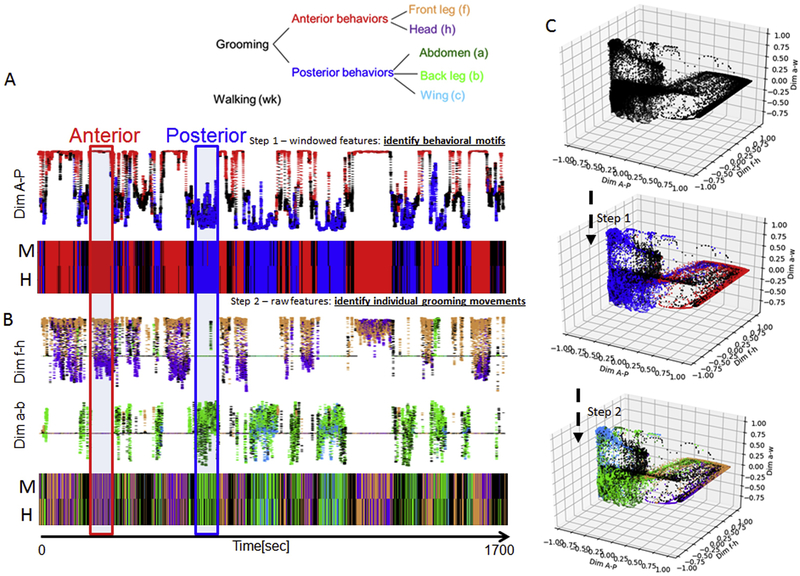
Here we describe our Automatic Behavior Recognition System (ABRS) as applied to fly grooming, demonstrate that this system can reliably and quickly recognize various behaviors without any image segmentation or detection of body parts. We show the results of behavioral analysis of 91 stimulated wild-type grooming flies, including the dynamics of behavioral sequence structure at multiple time-scales. Figure 1 presents an overview of the steps in the ABRS workflow: fly detection and tracking (Fig. 1A), extraction of spatiotemporal features (Fig. 1B), and dimensionality reduction and behavioral classification (Fig. 1C). Subsequent figures explain how each step is accomplished.
Figure 1. Overview of steps in grooming behavior analysis.
A: A single fly in the recording arena is detected by its movement and the area around the fly is cropped out. B: Space-time images (ST-images) are produced (middle panel) from consecutive frames (left panel) and are made orientation-translation invariant (right panel) by Radon Transformation (Spectra of RT). C: Dimensionality reduction is accomplished by unsupervised learning with singular value decomposition (SVD) and classification of behavior is carried out in two steps, corresponding to two different time-scales. D: Large numbers of behavioral records (ethograms) can be produced by this method efficiently.
Fly detection and tracking
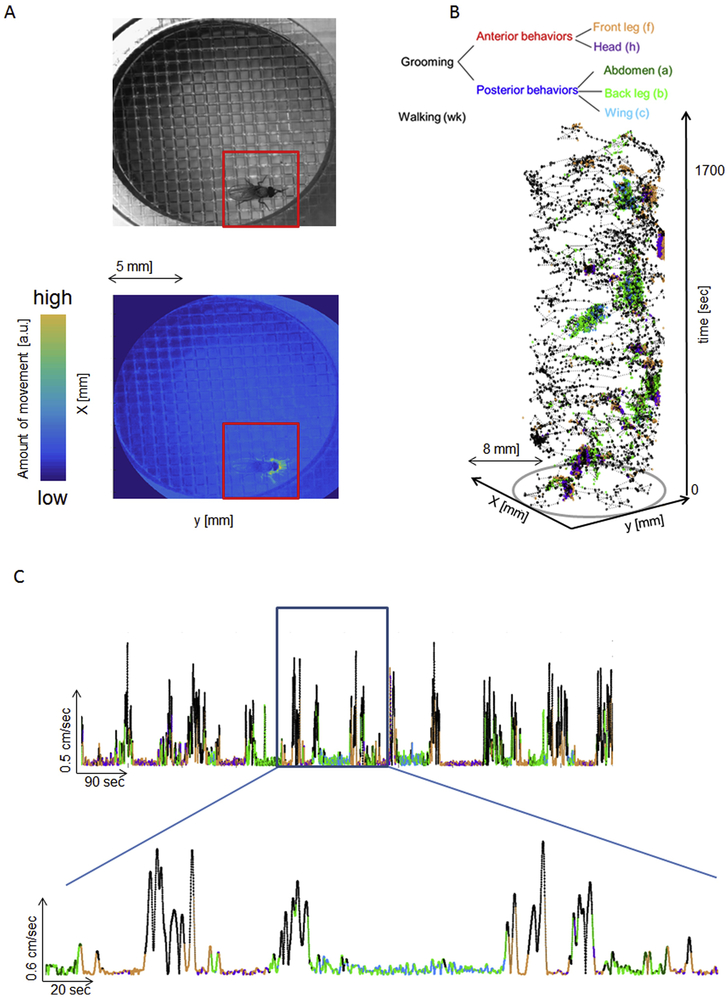
We record video of a fly freely moving in a flat arena large enough to facilitate natural (non-flying) behavior, which consists of grooming and walking bouts (radius = 0.5 cm, height = 3 mm, 1024×1024 pixels at 30 Hz). Since individual flies occupy only a small portion of the space in each frame, the first step of the video-processing pipeline is to remove uninformative pixels. We locate the fly by detecting movement in a 17-frame time window (~500 msec) and crop to a region of interest (Fig. 2A, 5×5 mm). The time window is wide enough to distinguish the animal’s movements from noise in light fluctuations but still narrow enough to detect changes in its location. We accurately track flies for the entire 30 min (1700 sec) video; when the resulting fly trajectory is annotated with human-provided behavior labels, we see that the fly remains relatively stationary while grooming (Fig. 2B and C).
Figure 2. Fly detection and tracking.
A: The animal’s position in the arena is determined by identifying pixels that show activity, as reflected in the change in light intensity in a 17-frame (500ms) time window. The red box shows the region of interest around the fly selected for subsequent processing steps. B: The plot of position over time shows that the fly can be reliably tracked throughout the entire recording session (1700 sec/28 min). Color-coding the positions in time using human annotations of grooming movements suggests that a fly remains stationary for several seconds while performing each movement. C: Animal’s body velocity over 28 minutes of the movie. The framed area is enlarged below. Color-coding is done by same human annotation as in B.
Spatiotemporal feature extraction
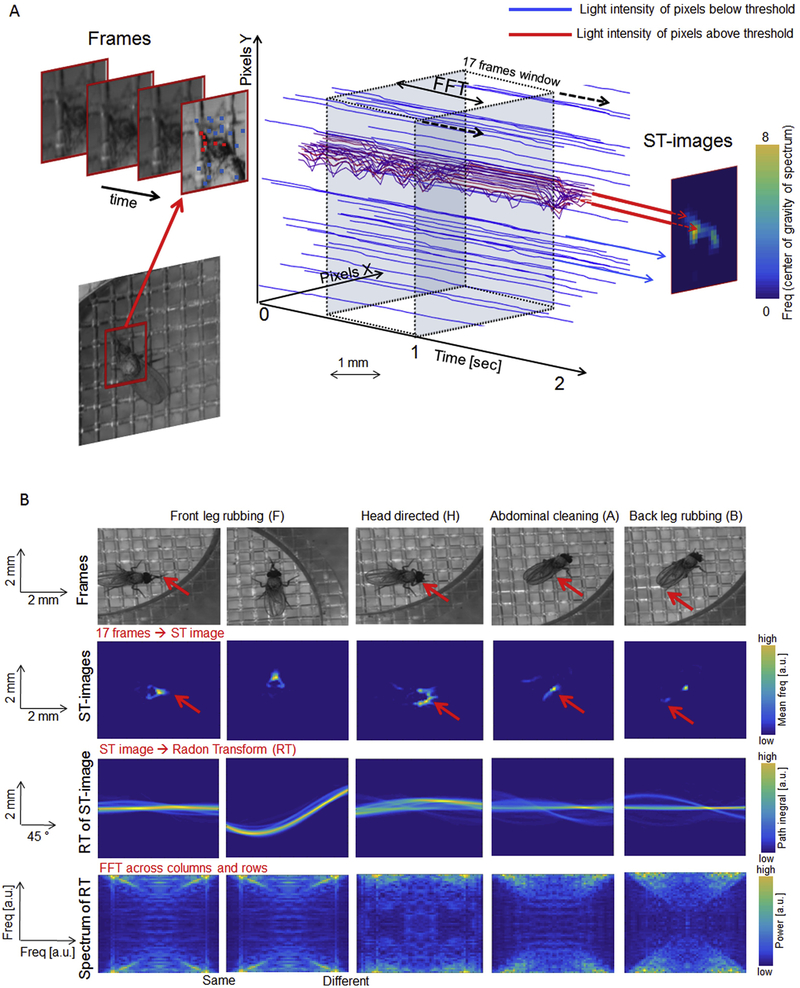
Grooming movement signatures can be identified by spatiotemporal features: adjacent pixels in which light intensity changes with time in a periodic manner. For example, as the front legs move back and forth during leg rubbing, the light intensity in the affected pixels changes periodically. We compute the frequency of the light intensity fluctuations in those pixels by applying Fourier Transformation to light intensity time-traces over a 17-frame, 500 msec sliding window. We refer to this combination of spatial information (position of pixels) with temporal changes of light intensity in each pixel as spatiotemporal images (ST-images; Fig. 3A). An ST-image thus represents the “shape of movement” at a given time in the video. Fig. 3B, top row, shows still images of four different grooming movements (front leg rubbing, head cleaning, abdominal cleaning and back leg rubbing) and the corresponding ST-images (second row). (See Video 1 in Supplemental Data for more examples of grooming movements.) The fly can perform the same type of grooming movement anywhere in the arena, resulting in the shape of the movement located and oriented differently in the ST-images (the first two ST-images in Fig. 3B). But the spatiotemporal features should be invariant to position or orientation of the fly in the arena. We accomplished this by Radon Transformation (RT) of all ST-images (M. van Ginkel, 2004) Briefly, the RT computes integrals along lines placed at all angles and displacements across the image. The computed values of these line integrals are then plotted according to the angles (x-axis) and displacements (y-axis) of each line. The results obtained by this operation are illustrated in Fig. 3B, where different instances of front leg rubbing behavior in different positions and orientations produce similar triangular shapes in the ST-images (first two ST-images), while head cleaning, abdominal cleaning, and back leg rubbing produce ST-images with distinct shapes. After applying the RT, however, the same shapes in the ST-images result in the same Spectra of RT, regardless of their position or orientation in the original ST-image (first two images in Fig. 3B), effectively bringing the signatures of the same types of movements into a common frame of reference.
Figure 3. Spatiotemporal feature extraction.
A: The image in each frame is cropped to an area of 25mm2 (80×80 pixels) around the fly’s position and the light intensity is recorded in each pixel across a sliding 17-frame time-window (~0.5 sec) to produce 6400 time-traces of light intensity. Each time-trace is decomposed by Fourier Transform and the center of gravity of each spectrum is computed to generate a movement map called a Spatiotemporal image (ST-image). Time-traces corresponding to pixels where light intensity changes periodically tend to have higher centers of gravity of their spectra - these are indicated by red traces and yellow pixels with high values in the resulting ST-image on the right. The time-traces where light intensity does not change above the threshold (blue traces) are discarded and their corresponding value in the ST-image is set to zero.
B: Examples of grooming behaviors (central frame of sliding window shown for each) and the corresponding ST-images. The first two ST-images are produced from the same behavior (front leg rubbing) but the flies differ in the position and orientation. The other three ST-images are produced from different grooming behaviors. Identification of a grooming movement does not depend on the position or orientation of the fly because ST-images are transformed by Radon Transformation (third row of images) and a second Fast Fourier Transformation to produce 2 D power spectra. Two examples of front-leg rubbing show the same 2 D spectra, while the spectrum from head cleaning is different.
Dimensionality reduction using unsupervised learning
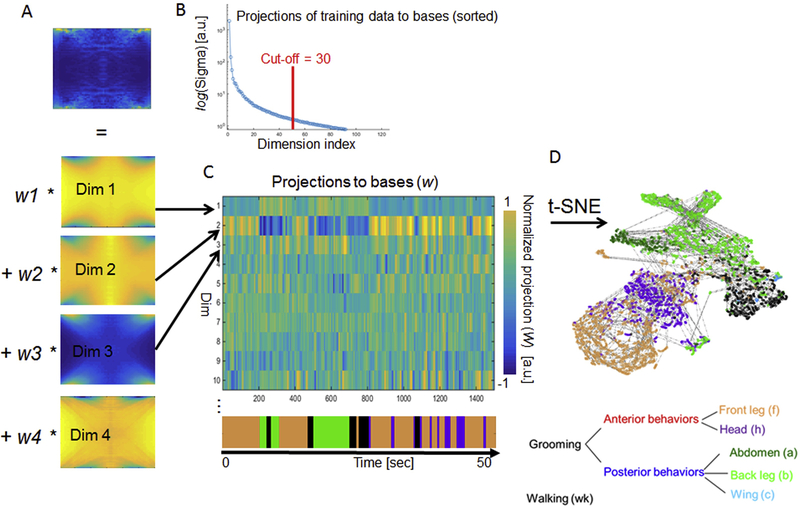
In the previous section, we described how the original video of grooming flies is converted into ST-images and further processed by Radon Transformation so that they can be compared in orientation and translation invariant manner. The next step is to determine which features best separate these images into discrete behavioral categories. The images shown in Fig. 3B have 80 × 80 pixels and 6400 dimensions, but many of these dimensions may be redundant or may represent noise in the data. We reduce the number of dimensions to only the most informative ones (or their linear combinations) using a representative training data set that contains instances of all relevant movements. We then apply Singular Value Decomposition (SVD), to the training data, compressing it to the 30 most informative dimensions, as shown in Fig. 4 A-D. These 30 dimensions are equivalent to the eigenvectors of the squared data matrix, and thus similar to the principal components. They form the bases of the low-dimensional space in which data can be more efficiently classified. A significant advantage of the ABRS method is that the low-dimensional representation only needs to be learned once – the 30 bases obtained by SVD can be stored and reused with new data. The new data can be projected to the 30 bases to obtain the 30 spatiotemporal features. These 30 bases therefore act as filters through which all new data can be passed for the purpose of compressing it into 30 spatiotemporal features used for behavior classification. Fig. 4C shows an example of how these spatiotemporal features relate to distinct behaviors observed in a short segment of a movie. Note, for example, that the first feature tends to be high during abdominal grooming behaviors while the second feature to correlates with anterior grooming. The changes in these features correspond well to the changes in grooming movements identified by human observers, suggesting that they reliably describe recognizable aspects of the behavior (Fig. 4C).
Figure 4. Dimensionality reduction with unsupervised learning.
A: A training set of these spectra (68,688 images) is decomposed by Singular Value Decomposition (SVD) and the first most informative 30 bases are selected according to singular value Sigma (B) - unsupervised learning will be carried out in the space of these bases. These bases are sufficient to reconstitute the diagnostic features of each input image with appropriate weighting factors (w1-4 shown). This reduces the original 6400 dimensions (number of pixels in a Spectrum of RT) to 30 dimensions we refer to as spatio-temporal features. C: To demonstrate that even the first 10 spatiotemporal features are sufficient to discriminate among grooming movements, we aligned them to manually scored grooming behavior for 50 seconds of video; note that different combinations of spatiotemporal features correlate with different grooming behaviors and the combination of features shifts when the behaviors do. D: An alternative way to visualize how these spatiotemporal features explain the behavioral variance is shown in the t-Distributed Stochastic Neighbor Embedding (t-SNE) map that projects 30 dimensions into two-dimensional space, preserving distance between neighboring data points and showing temporal connections between the data points as dashed lined. For example, the data points that correspond to frames a human observer labeled front leg grooming (orange) are clustered in the t-SNE map.
To illustrate how these features separate distinct behaviors, we used t-Distributed Stochastic Neighbor Embedding (t-SNE) (Maaten, 2008) to display the distribution of our 30 dimensional data onto two axes (Fig. 4D; note that we are only using this technique for visualization; classification is performed using 30 dimensions). The different behaviors identified by human observers are color-coded and clearly fall in distinct areas of t-SNE projection space, suggesting that our features do indeed separate grooming behaviors sufficiently for meaningful classification.
Behavior can be classified from spatiotemporal features
Both spatial and temporal features of video data help categorize the fly’s grooming behaviors. We have described how raw movie data is reduced to 30 spatiotemporal features, and the main advantage of these features is that they are invariant to the precise orientation or location of the animal in the arena, which makes them useful for challenging experimental settings like ours. This advantage unfortunately creates a new problem: movements with similar shapes in the ST-images are difficult to separate. For example, front leg rubbing and back leg rubbing are similar movements and so produce similar ST-images. What makes front and back leg rubbing distinct to human observers is where the movements occur in relation to the fly’s body: we observe that front leg rubbing occurs during the anterior grooming motif, while back leg rubbing is nested in the posterior motif. To overcome the difficulty, we designed a two-step classification method, described below, which was inspired by the way humans recognize these behaviors and by how flies organize grooming in nested time-scales.
In the first step of the classification, we determine whether the fly is engaged in anterior or posterior grooming, walking, or standing. These motifs provide the context for grooming behavior and occur over longer time scales (seconds). To do this, we perform a supervised technique, Linear Discrimination Analysis (LDA) (Welling, 2005), which can be used to find a direction that best separates data according to human labels. The LDA was applied to the 30 spatiotemporal features evaluated over a broader time-window (90 frames, 3 seconds) that robustly partitions behavior into anterior or posterior grooming motifs, walking or standing (Fig. 5A, C). Two examples of behavioral motifs, one anterior, one posterior, are indicated by red and blue frames respectively in Fig. 5A, B. The broad, 3-second, time-window smooths the features such that individual grooming movements (e. g. front leg rubbing) are no longer discernible, while the broad time-scale behavioral motifs survive the smoothing. We have already shown that our 30 spatiotemporal features are sufficient to separate these broad time-scale motifs from each other (Fig. 4C), so it is not surprising that a simple supervised machine learning method such as LDA can be used to efficiently perform the separation. LDA is an algorithm that can find the direction, in a multi-dimensional space, that maximizes the distances between the data points belonging to different categories (different behavioral motifs, in our case), while minimizing the variance within a category (Welling, 2005). The resulting dimension therefore best separates the data belonging to different categories. In our case the output of the first step is expressed in terms of probabilities of behaviors (anterior, posterior and walking). Subtracting probability of anterior motif from probability of posterior motif gives us the time-series (a dimension) that best separates those two behavioral motifs. We named this dimension “Dim A-P” (the y-axis in Fig. 5A). This first step of the classification therefore provides the behavioral context, dividing various behaviors into anterior or posterior motifs.
Figure 5. Classification of grooming movements using two time-scales.
A: Longer Timescale (Step 1): The 30 spatiotemporal features, smoothed over a 3 second window, are sufficient to reliably separate broad behavioral motifs (anterior grooming - red, posterior grooming – blue, and walking - black). The motifs are separated by Linear Discrimination Analysis (LDA) trained by human labels. The y-axis is the LDA-obtained dimension (Dim A-P) which best separates anterior behaviors from posterior. The colors are human labels. Dynamic thresholds are then used to produce ethograms of the behavioral motifs. An ethogram thus produced by the automatic method (M) is compared to that labeled by humans to illustrate agreement (H).
B: Shorter Timescale (Step 2): Using the 30 spatiotemporal features and no smoothing (time window of one frame or 33msec) enables the three behavioral categories, identified in A, to be further subdivided into 5 classes that correspond to the individual grooming movements (IGMs) as recognized by human observers. As in A, the LDA is used sequentially to separate pairs of IGMs from each other, using human labels. The LDA-obtained dimensions that best separate IGMs (Dim f-h and Dim a-b) are shown (see color legend above). The classification was performed separately for anterior behaviors (Dim f-h) from posterior behaviors (Dim a-b). Human (H) and machine-produced (M) ethograms obtained by this method are shown for comparison.
C: For both time-scales, the classification of behaviors is done by LDA. This is illustrated schematically in three dimensions obtained by LDA, with the color code corresponding to the human labeled behaviors at each step (see color legend above). First (Step 1), all of the data is clustered into the three broad behavioral motifs. Then (Step 2) the data is further divided into six classes corresponding to the IGMs.
In the second step, we apply another iteration of the LDA (again using human labels) within each motif separately to identify individual grooming movements (IGMs): front leg rubbing and head cleaning subdivide the anterior motif, while abdomen, wing sweeps, and back leg rubbing happen within the posterior one (Fig. 5B, C). In this step of classification, we feed the LDA model with raw features (no smoothing) and the outputs of the first step LDA model (probabilities of behavioral motifs). The outputs of the second step are the probabilities of IGMs. Again, as in the first step we separate IGMs from each other by computing the differences between their probabilities, obtaining time-series that best separate the IGMs from each other (in Fig. 5B “Dim f-h” separates front leg rubbing from head cleaning, for example). We can now solve the problem of distinguishing between grooming behaviors with similar movement shapes in the ST-images (e.g. back leg rubbing vs. front leg rubbing) because we have already determined the temporal context in which they occur. In other words, while some different behaviors may result in similar ST-images on the short time-scale, they are distinguishable on a broad time-scale.
Final production of ethograms involves separating the data according to outputs of the first and the second step (Dim A-P, Dim f-h, etc) by stationary or dynamic thresholds. (Setting these thresholds involves only three user-set parameters – the only manual step in the pipeline.) While the usage of LDA effectively separates behavioral classes from each other, it also generalizes well within a class. For example, a fly performing head sweeps on the vertical side of the arena appears different from a fly performing head sweeps while standing on the floor of the arena, yet in both cases the data from this behavior will fall on the same side of the LDA-determined threshold and both cases will be classified as head cleaning behavior. An example of the final form of an ethogram is shown in Fig. 5B, aligned with a human-generated ethogram for comparison.
As with dimensionality reduction step, described in the previous section, the LDA training with human labels needs only be done once to produce the LDA models. New data can then be fit into the LDA models to obtain behavioral probabilities. This saves time and provides consistency between subsequent data-sets. For example, once the 30 spatiotemporal features are computed, it takes only about 60 seconds to complete the behavior recognition and ethogram production of the dataset containing 91 27.8-minute movies. This processing speed makes it possible to consider real-time behavior recognition and closed-loop experiments in future. (In the ABRS GitHub repository we include a Python script that can be used to classify grooming behaviors in real time. Although the results are preliminary, we anticipate that the real-time output can be improved with better LDA training or with application of neural networks.)
Validation
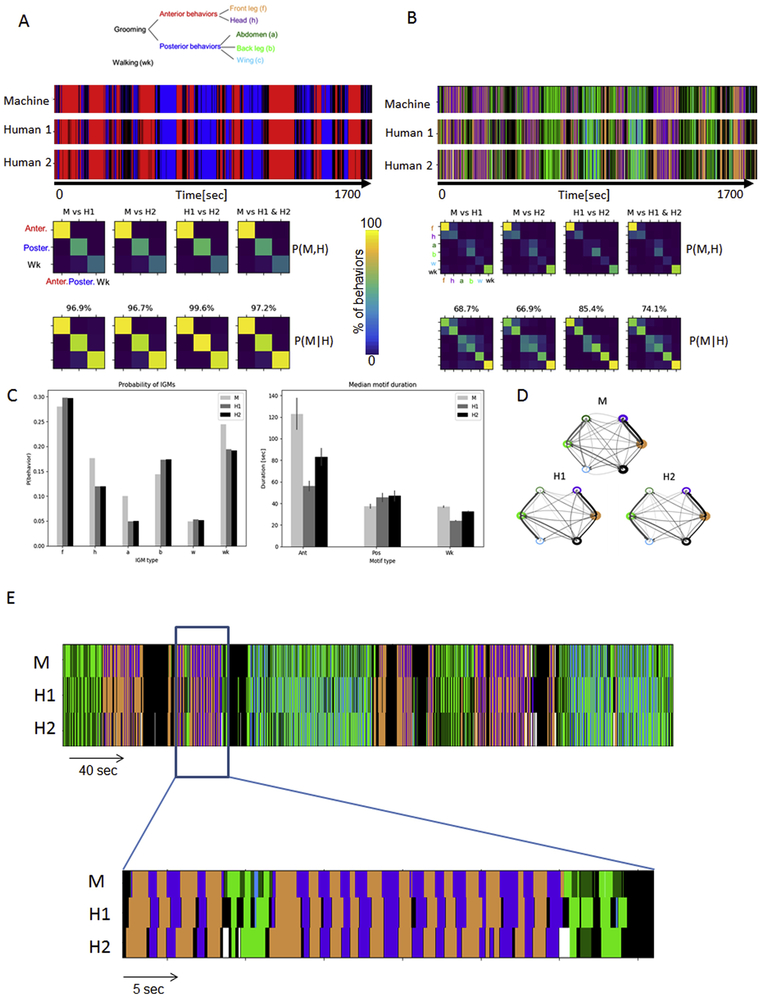
We validated the automatically produced ethograms by comparing them to human-annotated behavioral records. Encouragingly, the agreement between our automatic method and individual humans was not significantly different from the agreement between individual human observers (Fig. 6 A and B). At the level of grooming motifs (Anterior, Posterior, and Walking), the agreement between automatic behavior classification and the consensus between two humans was 97.2% (Fig. 6A) while human agreement is 99.6%. At the level of individual grooming movements (IGMs) the agreement between machine and human consensus was 74.1% while between the human observers the agreement was 85.4% (Fig. 6B), with most of the disagreements originating from identification of precise time of behavioral onsets and offsets (Fig. 6E). ABRS and humans agree on the general pattern of alternations between anterior IGMs (front leg rubbing (f) and head cleaning (h) but disagree on specific IGMs precise initiations, terminations and durations.
Figure 6. Validation of automatic behavior recognition system (ABRS).
A: Ethograms of grooming motifs (Anterior, red; Posterior, blue; and Walking, black) obtained by machine (M) and human observers (H1, H2) show good agreement, which is quantified below in confusion matrices (rows – machine; columns – humans). Diagonal terms show agreement and off-diagonal terms show disagreement; comparisons between machine (M) and humans (H1 and H2), between two human observers, and between machine and the consensus between humans all show greater than 90% agreement.
B: The accuracy of detecting individual grooming movements (IGMs) was also analyzed by comparison of human and machine ethograms using confusion matrices and shows 60-90% agreement.
C: The ABRS performs similarly to human observers when aggregate characteristics of grooming behavior are considered. The percentage of time spent doing each individual grooming movement (f, h, a, b, w) and walking (wk) are very similar, as assessed by comparing automatic (light grey) and manual (gray and black bars) annotation (left). The median behavioral motif durations are also very similar between all three methods of annotation.
D: Syntax structure of grooming behavior obtained by human and machine. Thickness of edges shows transition probabilities, while thickness of nodes and shade of edges indicates probability of behavior. As an example, front leg <--> head transition probabilities: for machine PM(h∣f)= 0.92 and PM(f∣h)= 0.98; while for humans PH1(h∣f)= 0.92, PH1(f∣h)= 0.73 and PH2(h∣f)= 0.97, PH2(f∣h)= 0.80.
E: Most disagreements between Machine and Humans are in determining the precise beginnings and ends of IGMs. The general pattern of alternations between f and h is similar but the exact start and end points differ.
Because we are particularly concerned with quantifying behavioral characteristics, such as the total amount of time spent in a particular grooming behavior or duration of behavioral motifs, we also compared these aggregate properties. The results obtained from the automated method (M) and human-annotated ethograms (H1, H2) show similar total amount of time spent in a behavior (Fig. 6C, left). The durations of behavioral motifs are also similar (Fig. 6C, right). For each type of behavioral motif (anterior, posterior and walking) the difference in median durations of the motifs between automated method (M) and human observers (H1, H2) are not significant (p-values > 0.1 between all pairwise comparisons; two-sample t-test).
Another behavioral characteristic that is especially relevant for sequential behavior such as grooming movements is syntax, defined as the transition probabilities between behaviors. In Fig. 6D we thus compare the syntax obtained from human-labeled (H) and machine labeled (M) ethograms. While the overall syntactic structure appears similar, there noticeably are more transitions between abdominal grooming (a) and wing grooming (w) in the automatically produced ethograms, likely due to the short transition through abdominal grooming movement, on the way from back-leg grooming to the wing grooming behavior.
We thus conclude that performance of our automated method for behavior recognition is comparable to human observers in recognition of broad time-scale behavioral motifs, IGMs, total time spent in a behavior, durations of behavioral motifs and the syntax.
Behavior analysis using large datasets
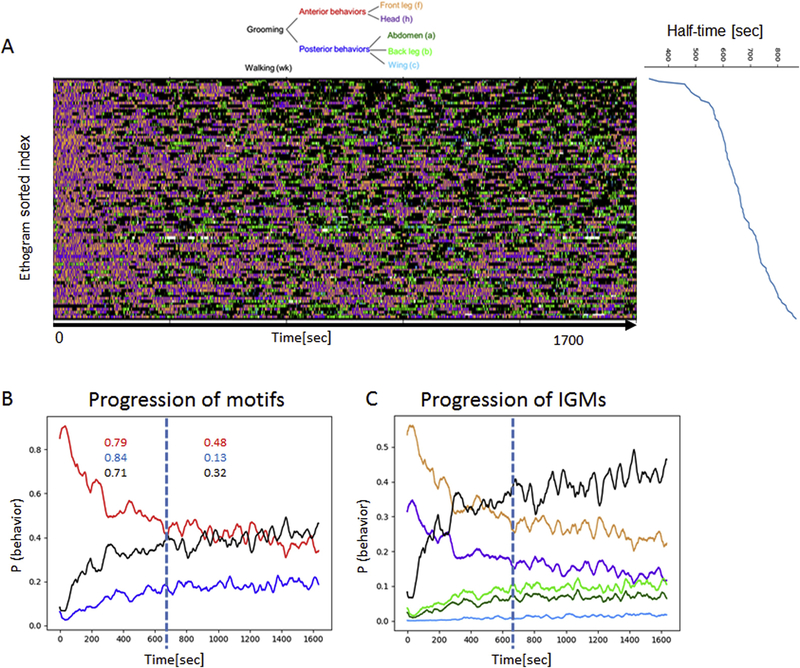
With the performance of ABRS validated, we can use this method to quickly and reliably annotate massive amounts of video data, resulting in ethograms representing behavior in large populations of animals. Analysis of ethograms produced from 91 videos of stimulated wildtype flies (each video is 27.8 minutes long) (Fig. 7A) confirm the anterior-to-posterior progression of grooming movements that we had previously observed and quantified manually (Seeds et al., 2014), but with a much greater sample size (Fig. 7B and C). We can now quantify significant changes in the frequency of grooming behaviors and walking over time as the flies remove the dust. For each ethogram we compute the time when the cumulative amount of anterior behavior reaches half of the total amount of anterior behavior. We refer to this point in time as “half-time”. The average half-time across all the flies equals 663 sec (SD = 109 sec, n=91) after the time when flies are stimulated by the irritant. The ethograms in Fig. 7A are sorted according to the half-time. Prior to half-time, the probabilities of broad time-scale behavioral motifs change sharply (r-squared = 0.79, 0.84 and 0.71 for anterior motifs, posterior motifs and walking respectively; p-values << 0.001). Specifically, anterior grooming movements decrease sharply within the first 663 sec post-stimulation, while posterior behaviors increase. After the 663 sec these grooming behavioral dynamics become more stable (r-squared < 0.38, <0.13 and 0.32) although residual changes in probabilities remain significant due to continuing increase of walking behavior, and the animals switch between anterior and posterior motifs with approximately constantly higher probability of anterior behavior. Similar dynamics are also reflected in terms of IGMs (Fig. 7C). This result suggests that the stimulus-related drive for posterior and anterior behaviors stabilizes after a period of time from the initial stimulation and provides a new signal upon which to align the ethograms of individual flies.
Figure 7. Analysis of sequences of grooming behavior in flies stimulated by dust.
Automatic annotation allows detection of behavioral features not immediately obvious to human observers and trends that emerge only after analyses of large quantities of behavioral records.
A: Here, 91 automatically generated ethograms (1700 sec each) of dusted wild-type flies are analyzed to show the temporal progression of grooming behavior. The ethograms are sorted according to “half-time” (shown on the right), the time at which half of the total anterior grooming behavior is completed, for each ethogram.
B: The probability of performing each behavioral motifs changes over the course of the assay as flies remove dust from various body parts; anterior grooming motifs occur very frequently at the beginning and sharply decrease with time, while posterior motifs and walking behavior gradually increase. On average, flies complete half of the anterior behavior in 663 seconds after the stimulation, as indicated by the dashed line (SD = 129 sec). The behavior dynamics largely stabilize after the half-time point (as reflected in the R-squared values shown in B-left panel, for pre- and post-half-time) although they are still statistically significant (p-value < 0.02 for all three motifs).
C: These trends are reflected in IGMs as well.
Quantification of intra-motif behavioral dynamics
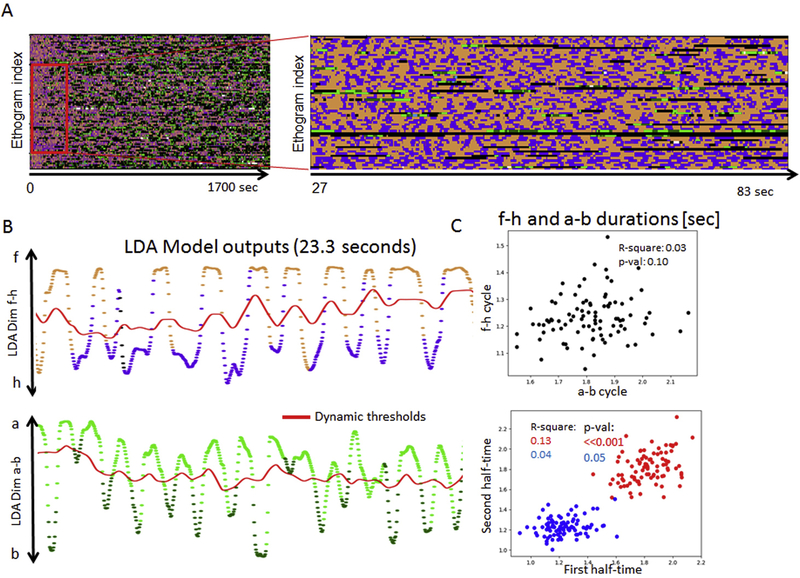
Automatic analysis and recognition of behavior in dust-stimulated flies also enables us to quantify short time-scale behavioral dynamics that would be difficult to observe in smaller data sets. In Fig. 8 we show strong periodicity observed in alternations between leg cleaning (f, b) and body-directed movements (h, a, w). This data set, together with LDA derived features described in the previous sections (probabilities of behaviors Dim f-h and Dim a-b), can be used to measure the durations of these alternation cycles (Fig. 8 B). Weak periodicity would be reflected in highly dispersed (high spread) of average f-h/a-b cycle lengths, while strong periodicity would result in tight distribution of cycle lengths. The top panel in Fig. 8 C shows the mean durations of f-h vs a-b cycles of the 91 ethograms. While there is no significant correlation between durations of f-h and a-b cycles of the same flies, almost all ( > 99%) average cycle lengths fall between 1.0 and 1.5 seconds for f-h and 1.5 – 2 seconds for a-b alternations. Such tight distribution of cycle lengths across a large number of animals suggests a strong periodicity of alternations between IGMs. Next we asked whether f-h/a-b cycle lengths sampled from first half-time of the ethograms are correlated to those cycle lengths sampled from the second half-time. If the alternations between leg rubbing and body-directed movements (f-h or a-b) were governed by pattern generation circuits, we would expect a correlation between their durations. While we anticipate that individual grooming movements (eg. leg rubbing) will be controlled by central pattern generator circuits, the periodic alternations between individual grooming movements performed by the same leg (f-h) suggests the possibility of inter-limb, intermediate time-scale pattern generators as well, which we are now experimentally investigating. Fig 8C, bottom panel, shows strong correlations of cycle lengths between the first and the second half-time (r-squared=0.13 and 0.04 for f-h and a-b cycle lengths respectively; p-values are <0.001 and 0.05 respectively.
Figure 8. Analysis of intra-motif dynamics in dust-stimulated flies.
A: 91 ethograms of dust-stimulated flies (same as in Fig. 7 but sorted according to f-h cycle length). Periodic alternations between IGMs (f-h and a-b) seem ubiquitous across animals (enlarged area indicated by the red frame).
B: We use LDA Model-derived probability time-series (outputs of the LDA model, expressed as probabilities of behaviors), LDA dim f-h –above, LDA dim a-b – below, to analyze the intra-motif alternations. The behavior probability values (colored dots) are colored by behavioral categories; red lines are smoothened versions of the LDA features for anterior and posterior behaviors respectively. The smoothened features are used as the thresholds for the classification. Examples of anterior motif (f-h cycles) and posterior motif (a-b cycles) are shown. The data are colored by behavior (same legend as in previous figures).
C: The intra-motif cycle length is highly conserved across the 91 stimulated flies. Ethograms are sorted according to f-h cycle length (first 90 seconds shown in the left panel). Each data-point represents one ethogram. Top: there is no significant correlation between average f-h and a-b cycle durations. Bottom: mean f-h cycle durations (red) and mean a-b cycle durations (blue) from the first half-time are strongly correlated with the f-h/a-b cycle durations from the second half-time (p<0.001). Each data-point shows f-h cycle (red) and a-b cycle (blue) lengths, sampled from first vs second half-time of the ethograms.
MATERIALS AND METHODS
Animals and Assay
These analyses used the Canton S wild-type strain of Drosophila melanogaster. Flies were reared at room temperature (21°C, ~50% humidity) with day/night cycle of 16:8hrs and assayed at matched circadian time. Males of 3-8 days old were selected and transferred without anesthesia. The dusting assay was performed according to the detailed methods described in (Seeds et al. 2014) using Reactive Yellow 86 powder (Sigma). Four flies were videotaped simultaneously, in four separate arenas (each 15mm in diameter and 5 mm in height) where they can walk and groom but not fly. The chamber walls and ceiling were coated with insect-slip and Sigmacoat to encourage flies to remain on the floor where the camera focus was optimal.
The flies were videotaped through a magnifying lens (camera model: TELEDYNE DALSA, CE FA-81-4M180-01-R 18007445) to produce 2048 × 2048 pixel images that include all four chambers. These videos were then processed in with custom MATLAB scripts to divide spatially into four separate movies.
Video was collected for 27.8 minutes to capture essentially all grooming movements, at a frame rate of 30Hz, which is sufficient temporal resolution to detect individual leg sweeps or rubs. The video was then divided into 1000 frame AVI clips for subsequent image processing. For each fly in the chamber the first 50 AVI clips were selected (50,000 frames in total).
Pre-processing of video data and fly tracking
The position of each fly was tracked by detecting changes in light intensity in every pixel across a sliding time-window, W = 30 frames (1 sec). The window slidies with one-frame steps across all frames, i=1,2,...I, where i is the frame index and I the total number of frames and clips were stitched together to avoid gaps. Differences in light intensity were computed by subtracting light intensity in each frame Fi from the preceding frame Fi-1 and then adding the resulting differences across the time-window W (Eq. 1).
| (Eq. 1) |
The differences Di where normalized by dividing them by total light intensity in corresponding frames Fi to compensate for uneven lighting across the frame.
Animal’s position in the arena, for each frame Fi (xi, yi) was then determined by finding the peak of Di (Eq 2). The positions xi, yi are stored in vectors x and y.
| (Eq. 2) |
Fig. 2A shows the total change in light intensity for all pixels in the time-window (Di) and Fig. 2B is presenting the tracked position of the fly across the entire recording session of 1500 sec (vectors x and y plotted across time). Fig. 2C shows the speed of the animal as measured by changing of the position.
The fly’s position was used to crop the 400 × 400 region of interest (ROI) around it, which reliably covers the entire body of the fly, regardless of where the peak of Di was discovered.
The data in the ROI were sub-sampled by averaging pixel values in 5 × 5 patches. This resulted in 80 × 80 pixel frames, Ti,where i=1,2,... I. These Ti regions were subsequently used for extraction of spatial-temporal features. The pre-processing steps described here is done fully automatically and do not require any human input. However, several parameters can be modified by the user in order to extend this method to other animal models. For example, the sub-sampling can be reduced (increasing the spatial resolution) or the threshold for minimal movement can be changed to discard the frames where nothing is happening (by default there is no minimal movement threshold).
Feature extraction
Spatial-temporal features were extracted from a stack of 17 consecutive frame, 80 × 80 pixel regions (Ti), a time-window of 0.57 seconds (Fig. 3A). The time-window was sliding across the entire data-set, with the step of 1 frame. Each Ti area in the time-window was turned into a column vector tw(the length of which equals the number of pixels in Ti, P= 6400), where index w=1,2,... W, and W is the size of the time window (W=17). We stacked column vectors tw to construct a P × W matrix T (Eq. 3):
| (Eq. 3) |
The rows of matrix T range across space and the columns of T range across time. For each member of T (each pixel in the time-window) we read the light intensity value. This produced 64,000 light intensity time-traces (64,000 rows in matrix T, or one time-trace for each pixel in each region Ti). This is illustrated in Fig. 3A (middle).
Time-traces were relatively flat for those pixels where light intensity did not change much within the time-window (blue traces in Fig. 3A), whereas for pixels where light intensity changed periodically (notably when the fly was engaged in grooming behavior) the time-traces were roughly sinusoidal (red traces Fig. 3A, middle). In order to quantify the shape of each time-trace, we decomposed each of the 64,000 time-traces by Fast Fourier Transformation (FFT) to obtain Fourier transform, Fp ,for each time-series. The magnitude of each Fp (spectrum) was retained and stored in vector fp. Vectors fp were stacked as rows of P × F matrix F (P=number of pixels, F=length of spectrum fp). We obtained matrix F by running the Python Numpy function, across the fft(), across the P rows of matrix T (Eq. 4).
| (Eq. 4) |
In order to obtain a single value for each spectrum fp we computed its center of mass, cp (Eq. 5).
| (Eq. 5) |
In Eq.5 j = 1, 2, 3....F. Thus for P rows of F (6400 rows) we obtained a vector c containing centers of mass, cp, of each spectrum in F (Eq. 5b):
| (Eq. 5b) |
We assigned the values from vector c (containing the centers of mass of spectra) to each corresponding pixel p as shown in Fig. 3A (right). (While here we are using a single value (center of gravity) as a spectral feature, other temporal features can also be added to the ST-image. It is possible to design ST-images with three or more channels (colors) which correspond to different temporal features. See the ABRS GitHub page for experimentation with such multichannel ST-images.)
(Vector c was re-shaped into an 80 × 80 matrix.) Thus we produced spatial-temporal images (ST-images), IST, where each pixel is assigned the value of center of mass, cp, of the spectrum computed from its corresponding time-trace (Fig. 3A, right). The ST-image can be represented by the matrix I in Eq. 6, where x and y are coordinates of pixels.
| (Eq. 6) |
Achieving rotation/translation invariance
Fig. 3B shows individual frames (ROI) sampled from video clips where flies were engaged in various grooming behaviors (top row) and ST-images produced from data sampled from the time-window around these frames (second row). Because we include no a priori knowledge of fly’s orientation and position of its body parts, we cannot directly compare ST-images from cases where animal’s orientation and position are not the same. Note the first two frames in Fig. 3B and their corresponding ST-images. The shapes in these two images are identical except for their orientation and position (one image is a rotated and shifted version of the other). The spatiotemporal features that we extract from images with identical shapes should be the same, no matter how the shape is oriented and positioned in the image. In order to compute such rotation/translation invariant features we performed three additional operations on the ST-images. First, we transformed the ST-images (IST) by Radon Transformation (RT) to produce Radon Transforms of the ST-images, shown in Fig. 3B, third row from top. The RT transforms rotation into translation by computing intensity integrals along lines cutting through the original image ST-image (IST ) at various slopes and intercepts. The images in Fig. 3B, third row, represent Radon Transforms of the ST-images shown above them (second row), where each column is the slope of the line cutting through the original ST-image and each row is the intercept of the line. There are 180 columns, corresponding to 180 slopes (1 ° to 180 °, with 1 ° increments) and 120 rows, corresponding to 120 intercepts (so the size of Radon Transform images is 120 × 180). The value in each point in a Radon Transform image is the magnitude of the intensity integral of the cutting line. Radon Transformation is an invertible operation, so no information is lost (we can reconstruct the original ST-image by Inverse Radon Transformation).
In the next two steps we decomposed the resulting Radon Transform images (such as those shown in Fig. 3B, third row) by Fourier Transformation, first along the columns and then along the rows (Eq. 7), obtaining spectra of Radon Transform images, ISTR, which we store in matrix R (Fig. 3B, third row) (Eq. 8).
| (Eq. 7) |
| (Eq. 8) |
In order to understand what this operation does, compare the first two Radon Transforms in Fig. 3B (third row). The spread of energy in these two images is identical but its position in the image differs between the two images (see the third Radon Transform as an example of different spread). The first FT decomposition, across the columns produces spectra of the columns (image not shown). The magnitudes of these spectra are the same, no matter where, along the columns of Radon Transform images, the energy spread is positioned (the position information is now contained in the phase of the Fourier Transforms and is discarded). Notice that the spread in the two Radon Transform images is also shifted along the x-axis (representing the angles). That shift persists in the spectra resulting from the first FT (the intermediate result not shown). In order to get rid of this shift, the second FT is performed across the rows of the spectra.
To summarize, we retained the energy spread in the Radon Transform images, which contains information about the shape in the original ST-image, and discarded the phase (which contains information about translation and rotation of the shape in the original ST-image). Again, note that the first two ST-images (IST), in Fig. 3B (second row), correspond to the same behavior but differ in their orientation and translation. The final spectra of those two ST-images (ISTR), resulting from operations described above (and Eq. 7), however, are identical (Fig. 3B, last row, first two images). The sizes of the spectra of the ST-images, ISTR, are the same as the sizes of the ST-images.
These transformations complete our final pre-processing step.
Dimensionality reduction and training
We applied Single Value Decomposition (SVD) to reduce the dimensionality of the data before the classification can be carried out. The training data set consisted of 216 video clips of wildtype dusted flies, capturing various time-points from the onset of grooming behavioral sequences. We used every tenth 30 sec clip from several independent movies to obtain examples of all the individual grooming movements. The training data was pre-processed as described in previous sections and the total of 68688 spectra of ST-images (ISTR) were produced (corresponding to the total number of frames contained in the training data). The SVD was then used for dimensionality reduction as described below.
The rotation/translation invariant spectra of ST-images (ISTR), stored in matrix 6400 × 68688 R, were used as the input for SVD. The input matrix R was decomposed as shown in Eq. 9:
| (Eq. 9) |
The matrix U contains the new orthogonal bases, Bd, learned during the training phase.
Fig. 4A (left) shows examples of bases (filters) learned by applying the SVD to the training data. The contribution of each base to the training set is shown in Fig. 4B. These contributions are stored in diagonal matrix sigma Σ (Eq. 9). The first 30 bases were selected as shown in Fig. 4B. Each spectrum of ST-image (ISTR) from the training set can be composed from 30 bases weighted by values stored in matrix V* (Fig. 4A, B) (Eq. 9).
Dimensionality reduction of the new data (data not used in the training) is carried out by computing projections of ISTR onto the 30 bases stored in matrix U (the new bases learned during the training phase) (Eq. 10). The ISTR images of new data are stored as column vectors Si (where i is the frame index) and the 30 bases are stored as column vectors Bd (where d=1, 2, ... D; D=30). Then the projections are computed as shown below (Eq. 10):
| (Eq. 10) |
The I × D matrix M now contains projections of ISTR images to D bases.
| (Eq. 11) |
In Fig. 4C the projections of ~1500 consecutive images (~ 50 sec of video) to the first 10 bases are shown. These projections can be thought of as firing rates of artificial neurons responding to different features of stimuli (ISTR). The first projection responds mostly to posterior grooming motifs while the second projection responds to anterior motifs (the first two rows in Fig. 4C). We computed the 30 projections for the entire data-set of 91 movies (50,000 frames per movie or ~30 minutes) and obtained 4.9 million 30 dimensional vectors (stored as columns in matrix M). These data were used for classification, described in the next section.
There are two significant advantages of our method: first, it does not require re-training of the bases with each new dataset. Once the bases are computed during the training phase, they can be used for dimensionality reduction of new data (the same matrix U can be used) and thus the new data is always represented by the same spatiotemporal features. The second advantage is that computing the projections of new data to the bases is much faster than initial training with SVD because it can be parallelized.
The 30 bases were sufficient for representation of various grooming behaviors, as is shown by the t-SNE map in Fig. 4D. The t-SNE analysis (Maaten, 2008) allows us to visualize high dimensional data (30 dimensions in this case) in just two dimensions. (It preserves local distances between data-points.) Note the good separation between anterior and posterior grooming motifs (red and blue colors, respectively). The edges between the data points indicate temporal transitions between data-points (consecutive images). Most transitions occur within clusters representing the anterior or posterior motifs, with only a few transitions connecting the two clusters. This distribution of the data in the t-SNE map strongly suggests the hierarchical organization of grooming behavior, with most transitions occurring between individual grooming movements (IGMs) belonging the same motif, e.g. head cleaning to front leg rubbing, and few transitions between different motifs (between posterior and anterior grooming behaviors).
Two-step classification using LDA
The classification of behavior was performed by Linear Discriminant Analysis (LDA) (Welling, 2005) on the 30 spatiotemporal features, in two steps corresponding to two behavioral time-scales, using human labels of behavior as a reference (training data). The LDA is implemented in Python using sklern library (https://scikit-learn.org/stable/modules/generated/sklearn.discriminant_analysis.LinearDiscriminantAnalysis.html).
The full dataset presented in this paper included 91 movies of dusted flies, each 27.8 minutes in length, starting from the time when the flies were dusted. Behavioral labels and corresponding data from two movies (57.6 minutes in total) were used for LDA training..
The 30 translation/rotation invariant spatiotemporal features were extracted as projections to the bases trained with the training data-set. This produced 4,600,000 data-points populating the 30-dimensional space (30 dim vectors). Thus, the input data came in the form of a 30 × 4,600,000 matrix M.
The LDA classification is performed in two steps, corresponding to two different time-scales. In the first step the general behavioral context was identified (anterior and posterior grooming motifs, whole-body movements and periods with no detectable movements) then, in the second step, the LDA was again applied, separately, to anterior and posterior motifs to identify 6 behaviors: front leg rubbing (f), head cleaning (h), abdominal cleaning (a), back leg rubbing (b), wing cleaning (w) and whole-body movements (Wk). Below we describe both steps in detail. During the first step, windowed features are computed from the raw spatio-temporal features (matrix M). We convolved the first five rows of M, with the Savitzky-Golay kernel h (https://docs.scipy.org/doc/scipy-0.15.1/reference/generated/scipy.signal.savgol_filter.html). The size of the sliding Savitzky-Golay kernel is 90 (3 sec of a 30 Hz movie) and the step =1 (Eq. 12):
| (Eq. 12) |
The 30 × I matrix W now contains the 30 (smoothened) windowed features of the entire dataset. Using human-generated behavior labels (of anterior, posterior and walking behaviors) and the corresponding training data from two movies (57.6 minutes in total)) we trained the LDA to obtain a LDA-model for predicting the anterior, posterior and walking behaviors - ModelAP. This model is used to predict the behaviors from the smoothened spatio-temporal features (matrix W). Predictions are expressed as probabilities of behaviors (outputs of the modelAP). Thus in the first step the outputs of the modelAP are four time-series, corresponding to probabilities of the behavioral motifs (anterior, posterior, walking and no movement or “standing”). When we subtract the probability of anterior grooming (Ant) from the probability of posterior grooming (Pos), we obtain a time-trace that best separates Ant from Pos. We refer to it as Dim A-P. This is illustrated in Fig. 5A.
An example of Dim A-P, corresponding to one 1700-second movie is shown in Fig. 5A and 5C (the colors correspond to behaviors as labeled by humans, however, these labels were NOT used as a reference for LDA training). A stationary threshold is then used to separate anterior from posterior behaviors. In Fig 5A the human-labeled ethogram (H) and the machine-labeled ethogram (M), obtained by this procedure, are placed below the Dim A-P projections as a reference and for comparison.
Fig. 5C is showing how the same data as in 5A is projected to the 3 dimensions obtained by LDA (Dim A-P, Dim f-h and Dim a-w – the last 2 are described in the next paragraph). Note that along the Dim A-P the data fall in different parts of this space, so it can be reliably separated by stationary thresholds.
Next, we carried out the second step of the classification. In this step Anterior and Posterior motifs are further subdivided into behavioral classes that correspond to IGMs as well as walking (Wk). The second step is similar to the first step, except that raw spatiotemporal features are used (as opposed to the windowed features used in the first step). Crucially, however, the raw features are now added to the outputs of the first step (i.e. probabilities of anterior, posterior and walking behaviors) as the inputs to the second step. The outputs of the first step provide the temporal context. Thus the input training data for the second step consists of: 30 raw spatio-temporal features and 3 outputs of the first step (so 33 inputs in total). This expanded set of inputs informs the LDA about the values of the raw features as well as about the probabilities that an animal is engaged in a particular behavioral motif.
After LDA training with human labels we obtained a model for IGMs – modelComb. The outputs of modelComb are expressed as probabilities of grooming behaviors (IGMs) and walking (probability time-series). Just as we did in the first step, we can subtract the probability of one behavior from probability of another behavior to obtain the time-series that best separates the two behaviors. In Fig. 5B we show the time-series that separates the f and h behaviors – Dim f-h (top) and the time-series that separates a from b – Dim a-b.
In Fig. 5A,B examples of Anterior and Posterior behaviors are shown in red and blue frames respectively. The final behavioral classes (f,h,a,b,wk) are obtained by applying dynamic thresholds to the probability time-series (outputs of modelComb), specifically to Dim f-h, Dim a-b and Dim a-w – i.e. the time-series that best separate pairs of grooming behaviors. Stationary thresholds were applied to Dim A-P and to the probability of walking. The dynamic thresholds are obtained by smoothing data with a user-specified window (a 1-second window was used here) and the stationary thresholds, manually set by the user (set at zero here). Both, the dynamic and stationary thresholds need to be set only once and they represent the only user-specified parameters of this classifier. To improve the accuracy of classification we also added two more time-series: the speed of displacement (whole body movement) and the max light intensity change in the ST window (strength of signal). See GitHub repository code for details of the implementation.
The final classes are used to construct the “machine-labeled” ethograms (M) exemplified in Fig. 5B. Human-labeled ethogram (H) is also shown for comparison. Fig. 5C (bottom) shows where the classified behaviors (f, h, a, b, w, wk) fall in the space of the 3 LDA dimensions (Dim A-P, Dim f-h and Dim a-w) after the second step of the classification.
Alternative supervised or unsupervised machine learning methods (other than the LDA) could be used on the 30 spatio-temporal features in a similar two-step protocol. . The modularity of the ABRS process allows us to test alternatives. For example, we initially applied a GMM (Gaussian Mixture Modeling) based classifier that estimated the probability density, as well as k-means unsupervised method, which does not depend on probability density estimation. The GMM-based classifier discovered a high number of clusters in the training data, which had to be then matched to human labels to assign the behavioral identity to each of the discovered clusters (self-supervised learning). This procedure occasionally resulted in inaccurate matching of machine-discovered classes to human labels because of small temporal discrepancies, such as occur at the initial and final frame of a behavior bout. We considered time-independent centroid matching but this fails to incorporate modeling of probability density, which disproportionately affects rare behaviors. The GMM is also relatively slow, which is problematic because it requires re-training with the whole dataset every time it is run. We also tested other supervised machine learning methods, such as dense neural networks or Convolutional Neural Networks (CNNs) and obtained reasonable accuracy (see the “dense neural network” code in the GitHub repository.) For our current implementation of the ABRS, we selected LDA for its accuracy, speed, and simplicity. LDA assigns identity to each point based on its position on the axis which best separates the behavioral labels (by simultaneously minimizing intra-cluster variability and maximizing the distance between the clusters). LDA is also much more time-efficient and does not require the re-training with the whole dataset each time it is run.
Post-processing and application of heuristics
One of the advantages of machine learning approaches to behavior analysis is the unbiased identification of repeated elements and the principled clustering to show similarities and differences between these elements. One of the advantages of human annotation is the ability to generalize among similar actions and to reject illogical or impossible transitions. During the development of ABRS, we hand-coded a lot of grooming data, and we manually checked a lot of the video annotated by ABRS. This lead to some insights into the behavior organization rules that we have encoded in a final proofreading step applied after clustering - essentially data-scrubbing to remove impossible things. We find that grooming and walking do not occur simultaneously. We have not seen extremely rapid transitions between front and back leg grooming movements (anterior and posterior motifs). The speed of whole-body motion can be used as the heuristic in the first step of the classification or in the post-processing of ethograms, or both. It can also be used as an additional feature (added to the 30 spatiotemporal features). The speed (of body displacement) is calculated from the changes in position of the fly, obtained in the tracking step (see Fig. 2C and Pre-processing of video data and fly tracking).
Sometimes the back legs are completely occluded by the dusted wings while flies are engaged in the back leg rubbing behavior and those data points by themselves cannot be distinguished from periods of inactivity (when nothing is moving). We have never observed a fly engaging in back leg rubbing alone for an extended period of time (i.e. they do not rub their legs without transitioning to another grooming movement or whole-body movement.) Thus when no movement is detected for 20 sec or more we classify those data points as standing rather than back leg rubbing. Conversely, when no movement is detected for less than 1/6 of a second (5 frames at 30 Hz) we do not count such a short period as “standing”.
We have never observed a fly transitioning from anterior grooming motif to posterior grooming motif and then right back to anterior motif (or conversely) in less than two seconds. These transitions (Anterior → Posterior → Anterior and Posterior → Anterior → Posterior, lasting less than a second) were never observed even when we specifically looked for them in the entire data-set of human labels (several hours of video randomly sampled from hundreds of flies) nor can any such transitions be seen in previously published ethograms (Seeds et al. 2014). There is almost always more time spent during a motif transition itself, as the flies need to change their posture. Even though some transitions can be as fast as one second, we have never seen two such transitions performed within a one-second-window. Thus we assume that this type of a transition present in an automatically generated ethogram is an error (we compared such machine-discovered transitions with human-produced ethograms and none turned out to be real). Therefore, very rapid transitions back and forth between anterior and posterior grooming motifs are eliminated from the ethograms during post-processing (for example, if such a short period of posterior behavior is found within an anterior motif we ignore it – i.e. we replace it with the anterior behavior). To further de-noise the ethograms we also eliminate behavioral motifs (anterior and posterior motifs) that last less than 1/6 of a second (5 frames at 30 Hz). Analysis of human labels of the training data reviles that such short motifs do not occur there. We set the minimum duration for walking (wk) at 1/3 of a second.
It is key to point out that these are heuristics are based on the behaviors we have observed in wild-type flies, and that these corrections are applied in the last, optional curation step. We do not implement this step when using ABRS for behavior discovery or on datasets of mutant flies, which do indeed violate many of these assumptions in experimentally interesting ways.
Validation
To assess the quality of automatic behavior classification method we compared the final output ethograms to human-scored ethograms. We manually scored fly behavior using VCode software (ref:VCode and VData) Fig. 6A shows one machine (M) and two human (H) labeled ethograms of a 27.8-minute movie of wildtype dusted fly’s behavior. Here the identified behaviors include only anterior grooming motifs (Anterior), posterior grooming motifs (Posterior) and the whole-body movements. The total agreement between human two observers is 99.2% and the agreement between machine and the human consensus is 96.0%.
The human observers were specifically looking for the 5 categories of grooming movements that are easy to recognize from the video clips. When uncertain, the observers could play the same behaviors back and forth at low speed. Behaviors that could not be clearly identified were not labeled. The human observers did not communicate with each other and did not compare results (so their observations were independent). Most disagreements between them occurred on the precise on/offsets of the behaviors - see Fig. 6E. For consensus we used human generated ethograms and retained the intersection where both humans agreed. The confusion matrices below show the breakdown of the agreements and disagreements between M and H (two human observers, H1 and H2) for each behavioral category. The top row matrices show the absolute number of agreements on the diagonal (H = rows, M = columns). The off-diagonal terms are disagreements. The most common disagreements arise when the output of M is “Wk” (whole-body movements) and the human observation is “Posterior” (posterior grooming), for both human observers (H1 and H2). The second row of matrices shows the same data as above but normalized by the sum across columns (so that all columns add up to 1).
Fig. 6B is analogous to 6A but subdivides the Anterior and Posterior motifs into individual grooming movements (f, h, a, b and w). Not surprisingly the agreements are less consistent than for the whole grooming motifs but nevertheless good in terms of comparing agreements between individual humans (H1, H2) and the agreements between the machine (M) and human consensus (H1 + H2). Notice that the most common disagreements are those between individual grooming movements within the same motif, i.e. between f and h (anterior grooming motifs) and between a and b (posterior grooming motifs). After visually inspecting ethograms produced by both humans and machine we conclude that most of these disagreements are due to determination of the exact timing of individual grooming movements (Fig. 6E). There is often ambiguity about when exactly an individual grooming movement starts and ends, e.g. there is a short transition period between pure front-leg rubbing (f) and pure head cleaning (h) behaviors as the two do not appear to be completely discrete (see also Fig. 8B for illustration). The transition periods between semi-continuous behaviors are not given a special label; therefore they are assigned f or h, depending on the side of the classification threshold that they fall on during the LDA step.
Population analysis using automatic behavioral recognition method
We used the ABRS ethograms (Fig. 7A) to estimate the dynamics of behavioral probabilities throughout the grooming period. The probabilities of individual grooming movements typically change over time, from when the flies are stimulated by dust until later, when they stabilize. To measure these dynamics we first calculated the frequencies of behavioral motifs as well as IGMs. This was achieved by counting all the instances of the behaviors in the ethograms within a sliding time-window (window width was 1000 frames or ~3.3 sec and the step was one frame). We compared the changes in behavioral frequencies in two epochs, early and late. To find a biologically relevant time-point which would separate the early and late epochs we determined the average “half-time”, defined as the average time it takes a fly to finish half of its anterior grooming behavior. The half-times of all ethograms were then computed (the cumulative frequency was equal to total frequency of anterior grooming behavior divided by two) and we took the average of that. The dynamics in behavioral frequencies for each behavior, in both epochs, were measured by R-square (coefficient of correlation squared).
Intra-motif dynamics analysis
The f-h and a-b cycle lengths were computed by counting the number of transitions (transition frequency) between the respective IGMs in each fly ethogram (i.e. 1700 sec of behavior). The number of cycles per motif per ethogram then equals transition frequency divided by the total amount of time spent in the motif. This analysis therefore does not account for variable durations of the motifs, i.e. it is agnostic to whether cycle lengths are correlated with motif lengths. We sorted the ethograms according to the cycle lengths (for anterior and posterior motifs separately) as shown in Fig. 8 C. To find out how the cycle lengths of different motifs or different epochs are related to each other (as very similar cycle-lengths may suggest that the cycles are produced by a shared pattern generation) we computed the correlations between the anterior and posterior motif cycle lengths and the correlations between the epoch’s cycle lengths (for anterior motifs), using a correlation function and obtained coefficients and p-values for each comparison.
DISCUSSION
Exploring solutions to our problem of objectively quantifying fly grooming behaviors from video led us to develop a method that is potentially broadly applicable. The recent fusion of neuroethology and computer science to form a new field of computational ethology (Anderson and Perona, 2014; Egnor and Branson, 2016) allows a virtuous cycle: ideas from biology inform and inspire technical advances and innovation in machine learning/computer vision, while large-scale computer-generated annotation of behavior from video enable both behavior discovery and objective quantification.
Several recent methods recognize and quantify animal behavior from video (reviewed in Robie, Seagraves, et al. 2017; Todd, Kain, and de Bivort 2017)). Many of these methods have been developed using Drosophila. For example, JAABA is a supervised machine learning system that analyzes the trajectories of flies walking and interacting in large groups and identifies temporal features based on trajectories of animal body positions and orientations (obtained from ellipse fit to the whole body) which does not provide information about such behaviors as antennal grooming, for example. These features are used to generate behavioral classifiers by with human labeled behavioral data (Kabra et al., 2013). The method mainly uses spatial features to extract information about spatial relationships between different animals. It has been used to study fly aggression and several other behaviors (Hoopfer et al. 2015; Robie, Hirokawa, et al. 2017). Other methods using Drosophila are based on unsupervised learning algorithms (reviewed in Todd, Kain, and de Bivort 2017). One such method that discovers stereotyped and continuous behavioral categories from postures of spontaneously behaving flies in an arena, (Berman et al., 2014) uses spatial features and unsupervised learning techniques to identify low-dimensional representation of postures (“postural modes”), followed by time-series analysis of these “modes” and low-dimensional spatial embedding. We refer to this as the “spatial embedding method”, and it has been used to explore how flies shift between behaviors according to hierarchical rules (Berman et al., 2016). This method applies Radon transformation to align images, so that rotation and translation invariance is achieved early on in the pipeline (In contrast, the ABRS does not require such alignment, employs the Radon transform late in the pipeline, and applies the human-supervised step of label-matching last.)
For fly grooming specifically, two previous behavior quantification methods have been reported. One relies on the amount of residual dust to detect grooming defects (Barradale, 2017), and the other adapts a beam-crossing assay usually used in for analysis of circadian rhythms to detect periods when the flies move but do not walk as a marker for time spent grooming (Qiao, 2018).
More recently two powerful methods were reported for analysis of animal pose from video data using deep neural networks: Deep Lab Cut (Mathis, 2018) and LEAP (Pereira, 2019). Both methods rely on labeling of anatomical features in videos of animals and can extract poses (configurations of the anatomical features).
Unfortunately, none of the existing methods suited the recording conditions and intermediate spatiotemporal resolution required for our particular research questions. Our behavioral paradigm requires freely moving flies covered with changing amounts of dust. Whole-body movement trajectories do not reveal which grooming movements the fly is performing, some of the limb movements involved in grooming occur hidden under the body (occlusions), the same grooming behavior can occur in many different positions/locations/viewing angles, and the fly’s appearance changes with time as the dust is removed. These attributes of our assay create challenges for the existing behavior recognition systems.
To develop an automatic behavior recognition system suitable for our experimental assay, we focused instead on temporal features. When humans annotate grooming behavior, they often play videos backwards and forwards at different speeds to label grooming movements, suggesting that it is the dynamics of the movements that help them to recognize specific behaviors. With the importance of temporal features in mind, we designed ABRS to achieve this automatically. ABRS enables reliable recognition of flexible sequences of fly grooming behavior despite limited knowledge of the animal’s limb position, pose and orientation. The system is based on recognizing shapes of movements in space and time. The pipeline consists of signal processing techniques for pre-processing of video data, followed by unsupervised learning methods on two separate time-scales, and finally a supervised learning step to label behavior. The primary advances include the strategy to combine spatial and temporal features that extract movement signatures without requiring any knowledge of the spatial context or even the animal’s anatomy. As such, these features are invariant to the subject’s location, orientation, and appearance. We compensate for the lack of spatial information with additional temporal context, applying a second time-scale. Here, we applied this method to reliably and quickly recognize long sequences of fly grooming behaviors in response to dust stimulus from massive amounts of video.
The ABRS, JAABA, spatial embedding, and pose estimation methods have different strengths. An ideal behavior quantification/recognition system might incorporate them all, with adaptive switching among them depending on the nature of the video data (perhaps employing “artificial attention” (Lanillos, 2015). To facilitate this future goal for the computational ethology community, we have constructed the code for the ABRS in Python with separable modules. The code is available on GitHub (https://github.com/AutomaticBehaviorRecognitionSystem/ABRS). At present there is no GUI available but it may be developed in the future. We hope the open source code will be improved and expanded by the broader community, including the addition of functionality for closed-loop experiments.
Here we have validated our method for recognition of fly grooming behaviors, but we propose that because of its focus on dynamics and temporal features rather than image alignment or anatomical features, it could be much more generally applicable. We are now applying the ABRS to decapitated flies (which also exhibit a range of behaviors, including grooming), to genetically manipulated strains and different species, as part of ongoing screens in our lab and even further diversification of experimental settings and animal models may be required in future screens. These experiments suggest that with minimal modification of parameters and training data, the ABRS software could be used to quantify a broad range of behaviors where spatial context, anatomy and even species are not pre-defined and where weighting temporal context more heavily is advantageous. For example, it may be possible to increase the time-window for ST-images to recognize behaviors in animals that move more slowly or less repetitively than flies during grooming. To increase the granularity of behavior (e.g. “antennal cleaning” and “eye cleaning” rather than just “head cleaning”) spatial resolution can be increased by reducing the spatial subsampling. ST-images themselves can be expanded as well. We are currently testing the utility of 3-channel ST-images (three colors), where the first channel is a spectral feature (center-of-gravity), the second channel is moment-to-moment movement (difference between consecutive frames) and the third channel is the raw image. Other modifications that can be done in order to apply the ABRS to other animal/behavior models include changing the number of spatio-temporal features (dimensionality reduction) and replacing the supervised learning method (currently LDA) with neural networks. For example, in attempting to improve the accuracy of behavior recognition in decapitated flies, we are using full dimensionality of the ST-images (no dimensionality reduction at all) and we are applying convolutional neural networks (ConvNets) directly to the ST-images (see https://github.com/AutomaticBehaviorRecognitionSystem/ABRS for examples of ConvNet models).
The core of our method is the rotation/translation invariant spectra of ST-images (Fig. 3B), which represent temporal information extracted from movements agnostic to their spatial location. As such, ABRS has conceptual similarities to the bag-of-words approach, in which the invariances to various input transformations are achieved by discarding the locations of image feature vectors (“visual words”) and only their frequencies of occurrence in the image are retained (as in counting the number of same words in a document and discarding the syntax of sentence structure) (Sivic, 2009). Thus in our case we similarly discard the phase of the spectra of ST-images while retaining the power of the spectra (see Achieving rotation/translation invariance in METHODS and Fig. 3B).
Convolutional neural networks (CNNs) can also achieve invariances to input transformations such as translation (in the first layers of the network) and rotation (learned in deeper layers) (Pereira, 2018). However, how these invariances are represented is still not clear and is a subject of current research (Furukawa, 2017; Lenc, 2015) Invariances (or, more generally, equivariances) to various input transformations can be achieved by data augmentation methods (Ratner, 2017). Building invariances into feature representations is an important strategy in making computer vision work in less constrained settings, and we have developed a translation and rotation invariant feature representation that seems to be useful for unsupervised and supervised behavior analysis.
Conceptually, our biologically-inspired emphasis on using temporal features, on two separate time-scales, should serve as a useful model for design of other behavior recognition software systems, including those based on ConvNets. Multi-time-scale analysis or combinations of temporal features with spatial context, can improve behavior classification methods. The leg movements associated with human walking or swimming may look roughly similar if only temporal features are considered, but adding a spatial feature such as orientation (vertical vs. horizontal) or environmental context (water vs. land) will differentiate the two types of movements even though they initially appear similar outside of the context. This two-step, two-time-scale approach separates similar movements efficiently by providing the temporal context in which they occur.
There are some natural extensions to ABRS itself. With minimal modification, our method could be used for behavior discovery, as well as automatic recognition of human-selected behaviors as we show here. This option is available at low cost because we perform unsupervised learning before the supervised step. Here we use human-selected behaviors (labels of grooming movements) to generate ethograms at the very end in our work-flow, but we can easily apply unsupervised learning methods (clustering algorithms) to the same spatiotemporal features to discover new behaviors. In the future we can also combine supervised with unsupervised methods by using human labels to find all instances of a behavior (e.g. head cleaning) and then apply unsupervised learning (without specifying how many behavioral classes to expect) to discover variations on the behavioral themes (e.g. cleaning of different parts of the head). Therefore we suggest using ABRS in combination with unsupervised behavior classification methods to first discover new behavioral varieties, create labels of those behaviors, and then apply the supervised step of the ABRS to find those behaviors in new data. In careful observations of the behaviors that we found by applying unsupervised learning in wild-type grooming flies, we did not identify any previously unknown types of repeated movements that could be associated with grooming, but we expect that this may change when we include genetically modified flies in our analysis. New or deviant grooming movements should be relegated to distinct clusters, enabling us to recognize more behaviors important to the fly than our human biases may perceive. Currently we are only identifying behaviors that are usually mutually exclusive in normal flies (grooming movements OR walking), however if a user is interested in identifying behaviors that occur simultaneously (e.g. grooming AND walking), this could probably be accomplished by creating labels for combinations of the behaviors (this could be done manually or by applying an unsupervised learning method to identify clusters of such behavioral combinations). Finally, due to the flexibility of ABRS and less reliance on specific anatomical features, it could in the future be used with other animal models such as mice or worms.
By focusing on movements and being robust to changing subject appearances the ABRS fills an empty niche in the computational ethology ecosystem. Together with JAABA, spatial embedding, Deep Lab Cut, LEAP and other methods researchers now have an extensive toolkit to choose from. Once again, biology suggests computational solutions and computational methods advance biological knowledge by revealing new phenomena.
Supplementary Material
Highlights:
ABRS reliably and quickly recognizes fly behaviors such as grooming from video, even in sub-optimal recording conditions.
The algorithm does not require image alignments or labeling of animal's body parts.
Useful orientation/translation invariant spatio-temporal features are extracted from video.
ABRS is freely available on GitHub for usage, modification and future development.
The method is flexible in implementation and can be extended to other behaviors or animal models.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson DJ, and Perona P (2014). Toward a science of computational ethology. Neuron 84, 18–31. [DOI] [PubMed] [Google Scholar]
- Berman GJ, Bialek W, and Shaevitz JW (2016). Predictability and hierarchy in Drosophila behavior. Proceedings of the National Academy of Sciences of the United States of America 113, 11943–11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman GJ, Choi DM, Bialek W, and Shaevitz JW (2014). Mapping the stereotyped behaviour of freely moving fruit flies. J R Soc Interface 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egnor SE, and Branson K (2016). Computational Analysis of Behavior. Annual review of neuroscience. [DOI] [PubMed] [Google Scholar]
- Furukawa Hidetoshi. "Deep learning for target classification from sar imagery: Data augmentation and translation invariance." arXiv preprint arXiv:1708.07920 (2017). [Google Scholar]
- Data Augmentation and Translation Invariance Hagedorn J, Hailpern J, and Karahalios KG (2008).
- VCode and VData: Illustrating a new Framework for Supporting the Video Annotation Workflow. Extended Abstracts of AVI.
- van Ginkel Michael, Hendriks CL Luengo, and van Vliet Lucas J.. "A short introduction to the Radon and Hough transforms and how they relate to each other." Delft University of Technology; (2004). [Google Scholar]
- Hoopfer ED, Jung Y, Inagaki HK, Rubin GM, and Anderson DJ (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra M, Robie AA, Rivera-Alba M, Branson S, and Branson K (2013). JAABA: interactive machine learning for automatic annotation of animal behavior. Nature methods 10, 64–67. [DOI] [PubMed] [Google Scholar]
- Lenc Karel, and Vedaldi Andrea. "Understanding image representations by measuring their equivariance and equivalence." Proceedings of the IEEE conference on computer vision and pattern recognition 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravbar P, Lipkind D, Parra LC, and Tchernichovski O (2012). Vocal exploration is locally regulated during song learning. J Neurosci 32, 3422–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner Alexander J., et al. "Learning to compose domain-specific transformations for data augmentation." Advances in neural information processing systems. 2017. [PMC free article] [PubMed] [Google Scholar]
- Robie AA, Hirokawa J, Edwards AW, Umayam LA, Lee A, Phillips ML, Card GM, Korff W, Rubin GM, Simpson JH, et al. (2017a). Mapping the Neural Substrates of Behavior. Cell 170, 393–406 e328. [DOI] [PubMed] [Google Scholar]
- Robie AA, Seagraves KM, Egnor SE, and Branson K (2017b). Machine vision methods for analyzing social interactions. The Journal of experimental biology 220, 25–34. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Ravbar P, Chung P, Hampel S, Midgley FM Jr., Mensh BD, and Simpson JH (2014). A suppression hierarchy among competing motor programs drives sequential grooming in Drosophila. Elife 3, e02951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling Max. "Fisher linear discriminant analysis." Department of Computer Science, University of Toronto; 3.1 (2005). [Google Scholar]
- Sivic Josef, and Zisserman Andrew. "Efficient visual search of videos cast as text retrieval." IEEE transactions on pattern analysis and machine intelligence 31.4 (2009): 591–606. [DOI] [PubMed] [Google Scholar]
- Todd JG, Kain JS, and de Bivort BL (2017). Systematic exploration of unsupervised methods for mapping behavior. Phys Biol 14, 015002. [DOI] [PubMed] [Google Scholar]
- van der Maaten LJP and Hinton GE. Visualizing High-Dimensional Data Using t-SNE. Journal of Machine Learning Research 9(November):2579–2605, 2008. [Google Scholar]
- Mathis Alexander, et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nature Publishing Group, 2018. [DOI] [PubMed] [Google Scholar]
- Pereira Talmo D., et al. "Fast animal pose estimation using deep neural networks." Nature methods 16.1 (2019): 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Bing, et al. "Automated analysis of long-term grooming behavior in Drosophila using a k-nearest neighbors classifier." Elife 7 (2018): e34497 https://github.com/AutomaticBehaviorRecognitionSystem/ABRS [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.