Abstract.
National border areas are special places for the spread of Mycobacterium tuberculosis (MTB). These regions concentrate vulnerable populations and constant population movements. Understanding the dynamics of the transmission of MTB is fundamental to propose control measures and to monitor drug resistance. We conducted a population-based prospective study of tuberculosis (TB) to evaluate molecular characteristics of MTB isolates circulating in Roraima, a state on the border of Venezuela and Guyana. Eighty isolates were genotyped by IS6110-RFLP (restriction fragment length polymorphism), spoligotyping, and 24-locus mycobacterial interspersed repetitive unit-variable number of repeats tandem (MIRU-VNTR). Drug susceptibility tests were performed by using the proportion method and GeneXpert® MTB/RIF (Cepheid, Sunnyvale, CA). Isolates showing a phenotypic resistance profile were submitted to polymerase chain reaction (PCR) and sequencing. Spoligotyping showed 40 distinct patterns with a high prevalence of Latin-American and Mediterranean (LAM), Haarlem (H), and the “ill-defined” T clades. Mycobacterial interspersed repetitive unit -VNTR and IS6110-RFLP showed clustering rates of 21.3% and 30%, respectively. Drug resistance was detected in 11 (15.1%) isolates, and all were found to have primary resistance; among these, six (8.2%) isolates were streptomycin mono-resistant, four (5.4%) isoniazid mono-resistant, and one (1.3%) multidrug resistant. This is the first study on the molecular epidemiology and drug resistance profile of MTB from Roraima. Herein, we describe high diversity of genetic profiles circulating in this region that may be driven by the introduction of new strain types because of large population flow in this region. In summary, our results showed that analyses of these circulating strains can contribute to a better understanding of TB epidemiology in the northern Brazilian border and be useful to establish public health policies on TB prevention.
INTRODUCTION
The global increase in human migrations, especially in transnational border regions with high proportion of vulnerable indigenous population, favors the spread of tuberculosis (TB) between countries.1 The Roraima state, located in the northern region of Brazil, has approximately 576,568 inhabitants2 and in 2016, had an incidence of 27.6 TB cases/100,000 inhabitants and a mortality of 0.8/100,000 inhabitants.3 In this region, the indigenous segment represents 10% of the general population, which is distributed in 35 indigenous territories. The Yanomami territory is located at the border with Venezuela and the Macuxi/Wapichana territory at the border with Guyana and Venezuela.4 After the National Integration Plan was implemented two decades ago, the local population increased 39%, where 30.5% migrated from northeastern and northern regions.2
Among few studies conducted in Brazil to assess the dynamics of TB transmission in border regions, there was a higher incidence of TB in the indigenous ethnicity,5,6 significant molecular diversity in circulating isolates,7 and higher resistance rates in frontier regions.8 The rate of multidrug/rifampicin-resistant tuberculosis in Brazil is lower than that in other Latin American countries (1.5% in new cases and 8% in previously treated patients). However, an enhanced monitoring of these regions is particularly relevant, considering the resistance rates in the two countries at the northern Brazilian border are significantly higher corresponding to 2.9% in new cases and 17% in previously treated patients (Guyana), and 2.9% and 13% in Venezuela.9
The contribution of an intensified migratory flow and consequent introduction of new Mycobacterium tuberculosis (MTB) strains requires a greater investment in integrated TB control programs in the border regions, as well as the monitoring of drug resistance and recent transmission in this region. In this sense, combining classical epidemiological data with molecular data obtained from genotypic analysis can provide an additional layer of information that enables a better understanding of local TB transmission dynamics while allowing a comparison against macro-epidemiological scenario. Presently, no genotypic data are available for this specific region of Brazil, and molecular epidemiological data could help identify cryptic transmission clusters that are otherwise undetected by classical epidemiological investigation. Moreover, being a border state, the identification of clustered MTB clinical isolates associated with migrants can help identify risk factors for the dissemination of imported cross-border strains.
Under this rationale, we conducted a cross-sectional study consisting of a clinical-epidemiological description of the TB cases and molecular characterization of MTB isolates circulating in the region to obtain a high-resolution snapshot of the TB transmission in Roraima.
MATERIALS AND METHODS
Study population and data collection.
From April 2015 to September 2016, we established a surveillance network composed of four laboratories: Casa de Saúde Indígena (CASAI), Hospital Geral de Roraima, Laboratório Regional Municipal, and Laboratório Central do Estado Roraima (LACEN/RR) located in the city of Boa Vista, the capital of Roraima. The inclusion criteria were pulmonary and/or pulmonary plus extrapulmonary TB (PEPTB) cases with positive culture, and we were included only patients with age ≥ 13 years who answered a standardized questionnaire and signed a written informed consent. Patients with no culture were excluded. The variables obtained during the interview included birthplace, address, time duration in the actual city, job activities in other municipality/countries, previous history of TB treatment, last TB treatment outcome, contact with pulmonary tuberculosis (PTB), classification of the current TB case (new case or retreatment), and family relationship (city of residence, length of residence, and frequency meeting with relatives). Complementary sociodemographic data were obtained from the National Reporting System on Notifiable Diseases (SINAN), which included race/skin, education level, TB treatment outcome, vulnerable population, and comorbidities (diabetes, mental disorder, drug addiction, smoking history, and alcohol abuse). All collected data were stored and analyzed by IBM SPSS Statistics 13.0 Program (IBM Corp., Armonk, NY). The frequency and percentages were analyzed for all categorical variables. In accordance with the Brazilian guidelines, HIV testing was offered to all TB patients included in the study. This study was conducted with the approval of the Research Ethics Committee from the National Public Health School/Oswaldo Cruz Foundation (number CAE 32357114.4.00005240).
Clinical samples and susceptibility testing.
All sputum specimens collected in the participating laboratories were examined by microscopy and/or GeneXpert® MTB/RIF, processed with the sodium hydroxide method, and inoculated in the Ogawa–Kudoh culture medium. The cultures were incubated at 37°C for up to 8 weeks and checked weekly for visible colonies. Detection of acid-fast bacilli was performed in the LACEN/RR with evaluation of microscopic aspects (Ziehl–Neelsen staining, bacillus size, strand of bacilli in cords),10 colony aspect, growth time, and ρ-nitrobenzoic acid inhibition test.11 Mycobacterium tuberculosis identification was performed with the MPT64 protein detection–based immunochromatographic rapid test (SD Bioline Kit, Standard Diagnostics, Inc., Kyonggi-do, Korea).12 Drug susceptibility tests (DSTs) were performed using the proportion method in the Lowestein–Jensem medium. The concentrations tested were streptomycin (SM), 4.0 μg/mL; isoniazid (INH), 0.2 μg/mL; rifampicin (RIF), - 40 μg/mL; and ethambutol, 2.0 μg/mL.13 Isolates showing a phenotypic resistance profile to RIF and INH were classified as multidrug resistant (MDR).8
Detection of mutation in genes related to drug resistance and phylogenetic analysis.
Positive cultures with confirmed MTB complex were sent to Laboratories of Universidade Federal da Grande Dourados, Brazil, and Universidade de Lisboa, Portugal, for molecular analysis. DNA was extracted from a fresh culture to investigate chromosomal mutational events related to drug resistance. The nucleotide sequences of rpoB, katG, inhA, rrs, rpsL, and gidB genes were analyzed by PCR and sequencing.14 Rifampicin resistance was reconfirmed using the GeneXpert MTB/RIF assay.15 The genetic relationship of MTB isolates and the recent transmission was evaluated using IS6110-RFLP (restriction fragment length polymorphism),16 spoligotyping,17 and MIRU-VNTR typing 24 loci.18 Restriction fragment length polymorphism patterns were analyzed using the IS6110-RFLP database (RIVM–Bionumerics; Applied Maths, Sint-Martens-Latem, Belgium), and a genotypic cluster was defined as a group of two or more isolates from different patients whose RFLP patterns were identical with respect to the number and size of bands. Spoligo patterns were further analyzed using SITVITWEB/SpolDB4 database19,20 and SPOTCLUST.21 Twenty-four loci MIRU-VNTR typing was performed as previously described22 and were considered cluster strains with 100% of similarity. The patterns were analyzed using the algorithms available within the BioNumerics software v.7.1 (Applied Maths, Sint-Martens-Latem, Belgium). Percentage similarity between fingerprints was scored by the Dice coefficient. The unweighted pair group method, with arithmetic mean, with a 0.5% limit tolerance, was used to construct a dendrogram.23
RESULTS
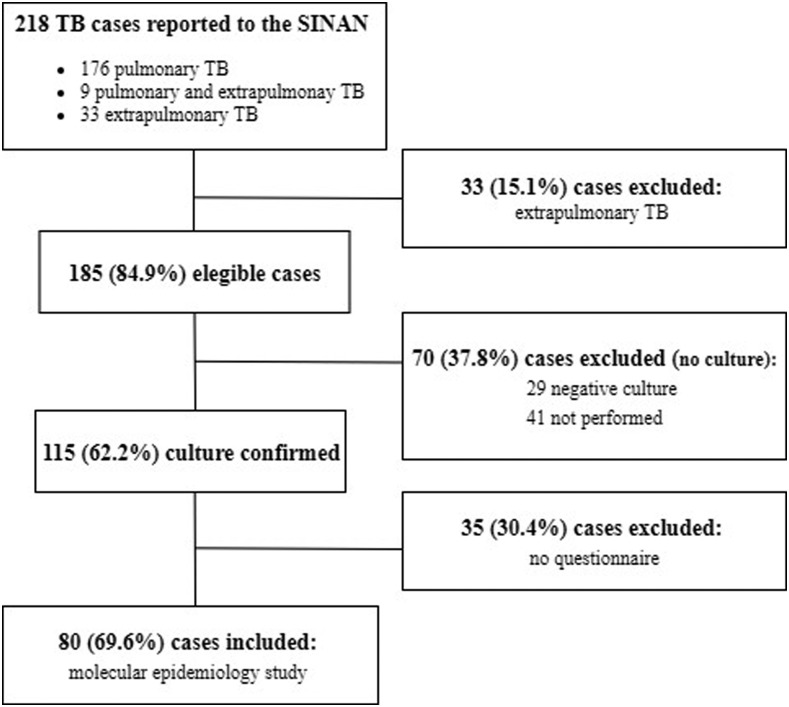
In the study period, 218 TB patients were notified in the SINAN, of which 176 patients presented PTB, nine PEPTB, and 33 extrapulmonary TB. Among the 185 PTB and PEPTB cases, 115 (62.2%) were culture confirmed, and among them 80 (69.6%) were included in the epidemiological and molecular study (Figure 1).
Figure 1.
Flowchart of recruitment of patients with tuberculosis. SINAN = Sistema de Informação de Agravos de Notificação (National Notifiable Diseases Information System).
Among the patients included in the study (n = 80), the majority were male (62.5%), nonwhite (81%), and age ranged between 21 and 60 years (76.3%). Most patients had an educational level below 8 years (32.3%), and a low proportion was registered in the social protection government programs, as Bolsa Família Program (17.7%). Patients had a history of smoking (24.6%), illicit drug use (20.8%), and alcoholism (20%). Most of these cases were reported by hospitals (60%) and among indigenous, 41.7% (n = 5/12) were reported by CASAI (Table 1).
Table 1.
Sociodemographic and clinical characteristics of TB patients from the state of Roraima, Brazil
| Characteristics | TB patients, n (%) |
|---|---|
| Gender | |
| Male | 50/80 (62.5) |
| Female | 30/80 (37.5) |
| Age | |
| Mean/SD | 40.1/16.559 |
| Median | 37.5 |
| Age range (years) | |
| > 14–20 | 8/80 (10) |
| > 20–40 | 37/80 (46.3) |
| > 40–60 | 24/80 (30) |
| > 60 | 11/80 (13.7) |
| Race | |
| White | 3/79 (3.8) |
| Nonwhite | 64/79 (81) |
| Indigenous | 12/79 (15.2) |
| Schooling level | |
| Illiterate | 11/65 (16.9) |
| ≤ 4 years | 15/65 (23.1) |
| > 4–8 years | 18/65 (27.7) |
| > 8 years | 21/65 (32.3) |
| Government social program | 11/62 (17.7) |
| Vulnerable population: | |
| 1. Inmates | 7/80 (8.7) |
| 2. Homeless | 3/80 (3.7) |
| 3. Immigrants | 2/80 (2.5) |
| Tb clinical presentation | |
| Pulmonary | 77/80 (96.3) |
| Pulmonary and extrapulmonary | 3/80 (3.7) |
| Commorbidities | |
| 1. Smoking history | 18/73 (24.6) |
| 2. Drug addiction | 16/77 (20.8) |
| 3. Alcoholism | 16/80 (20) |
| 3. Diabetes | 10/80 (12.5) |
| 4. Mental disorder | 1/80 (1.3) |
| 5. Aids | 8/72 (11.1) |
| Health care | |
| Primary health units | 27/80 (33.8) |
| Hospital | 48/80 (60) |
| Casa de Saúde Indígena | 5/80 (6.2) |
| Treatment outcome | |
| Favorable | 57/77 (74) |
| Unfavorable* | 20/77 (26) |
TB = tuberculosis.
* Death by other causes (n = 9); failure (n = 2); lost of follow-up (n = 10); tbdr (n = 6); changed treatment (n = 1).
GeneXpert MTB/RIF showed that 1/55 (1.8%) strain was resistant to RIF. Conventional DST detected resistance to one or more drugs in 11/73 (15.1%) strains: six resistant to SM, four resistant to INH, and one MDR-TB. Sequencing analysis of rrs, rpsL, and gidB (SM resistance) showed that four strains presented a point mutation in the gidB (C413T/A138V), whereas rrs and rpsL genes showed a wild type sequence (WT) in all isolates analyzed. Finally, no mutations in the aforementioned genes were found in two SM-resistant isolates. Analysis of katG and inhA promoter region (INH resistance) genes showed that three isolates carried a point mutation in the promoter region of inhA (C-15T) gene, although in one isolate carried a point mutation in the katG gene (G994C/S315T). In addition, the MDR-TB strain presented point mutations in the katG (G994C/S315T) and rpoB gene (C1332T/H445Y), causing INH and RIF resistance, respectively.
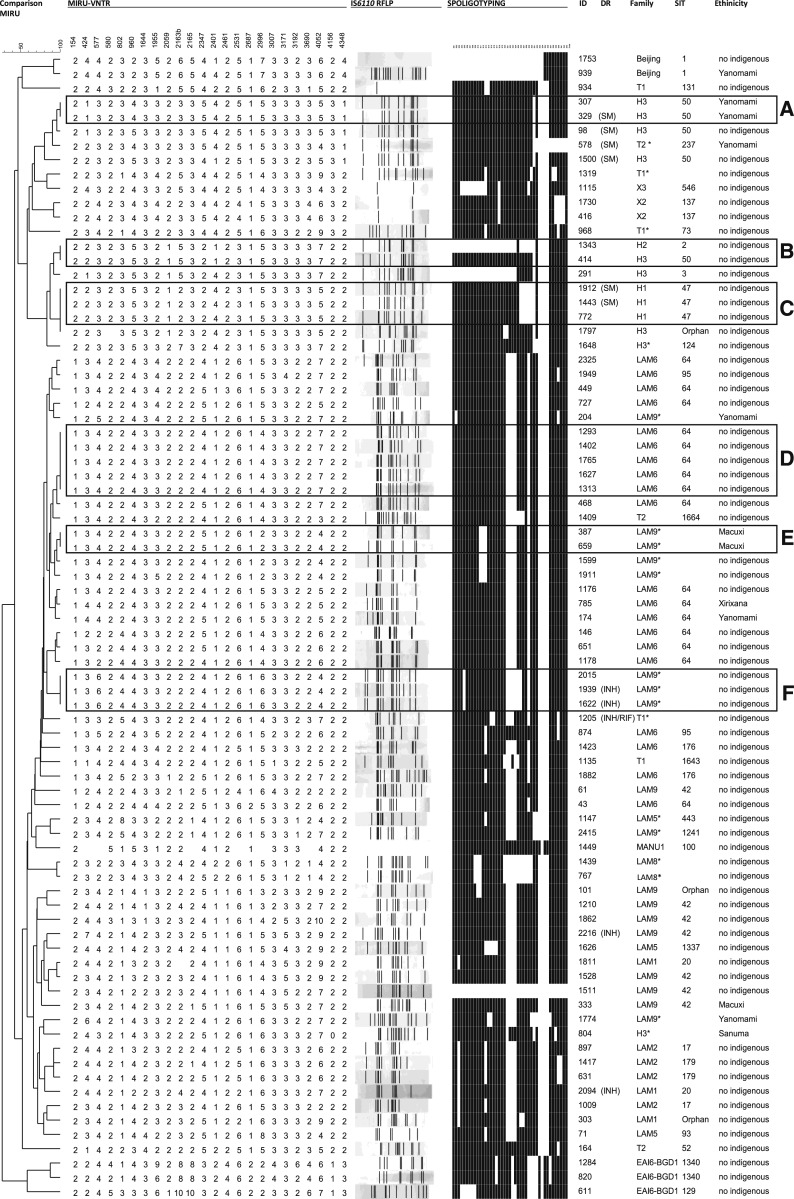
Next, to evaluate the genetic diversity and clonality underpinning local TB transmission dynamics in the region, all 80 MTB isolates included in the study were typed by IS6110-RFLP, spoligotyping, and 24-loci MIRU-VNTR (Figure 2). IS6110-RFLP analysis showed 67 distinct profiles, of which 24 isolates were grouped into 11 different genetic clusters composed of two isolates (n = 10 clusters) and four isolates (n = 1 cluster), yielding an estimated recent transmission rate of 30% (24/80). Spoligotyping showed a high genetic diversity with 40 distinct patterns and a higher frequency of SIT 64/LAM 6 (n = 16, 20.0%), SIT42/LAM 9 (n = 7, 8.75%), and SIT50/H3 (n = 5, 6.25%). Noteworthy, uncommon profiles in Brazil were also detected: SIT1/Beijing (n = 2, 2.5%) and SIT100/MANU 1 (n = 1, 1.25%). The EAI6-BGD1 family was represented by three isolates (3.75%) with sublineages SIT 129 (n = 1) and SIT 1,340 (n = 2). In addition were found three non-SIT (orphan patterns) and nine unknown profiles.
Figure 2.
Dendrogram based on MIRU-VNTR (24 loci) and presentation of the IS6110-RFLP (restriction fragment length polymorphism) and spoligotyping profiles of Mycobacterium tuberculosis strains obtained from Roraima, Brazil.
The genotypic analysis by MIRU-VNTR (24 loci) showed 69 distinct profiles, including 17 isolates (21.3%) grouped into six clusters, of which three contained INH- or SM-resistant isolates. The largest cluster (Cluster D/n = 5) was composed only of nonindigenous patients and non–drug-resistant isolates. Two clusters (Clusters C and F) were composed of three isolates exclusively from nonindigenous patients presenting SM- and INH-resistant isolates. Cluster B presented two isolates with different IS6110-RFLP and spoligotyping profiles, which may occur in case of mixed bacterial populations. Two clusters grouped isolates from indigenous patients of the Yanomami (n = 2) and Macuxi (n = 2) ethnicity (Cluster A and E), where one of these isolates presented SM resistance (Figure 2).
Comparing clustered MTB isolates with those nonclustered isolates, no statistically significant difference was observed with the following variables: gender, ethnicity, scholar level, length residence in Boa Vista, previous contact with TB, HIV seropositivity, and unfavorable TB treatment (Supplemental Table S1). Cluster D was the only one where evidence of possible epidemiologic links was found in two patients living on the same street. In the remaining clusters, no epidemiological correlations were found. An interactive version of the global phylogeny, with strains labeled by genotype, city of origin, and year of isolation, is available at https://microreact.org/project/rJmSTBEMV.
DISCUSSION
In this study, the genotypic diversity and drug resistance of MTB isolated from a Brazilian state (Roraima) were characterized, comparing demographic, clinical, and epidemiologic characteristics of the patients. To investigate the transmission between patients, the RFLP technique IS6110 and MIRU-VNTR analysis were performed. Our results showed that most of the MTB isolates had unique profiles, with a recent transmission rate lower than those described in other studies carried out in Brazil.7,24,25 This might be explained by several factors, including period of sample collection and constant migration flow (legal and illegal crossing of the border).
The capital Boa Vista gathered most of the cases. Since 2000, the region has been experiencing an increasing accessibility, leading to a high population growth, industrial, and commercial development.26 People from Venezuela and Guyana cross the borders to Brazil because of a lack of universal health care and/or economic and political crisis in their countries.27 In 2017, the incidence of TB in the state increased to 8.4 TB cases/100,000 inhabitants compared with that in 2016. Among the TB cases notified, 90% were from residents in Roraima, 3.2% from Amazonas, and 6.9% from Guyana and Venezuela.28
The exact transmission source between individuals could not be established in the clusters with similar genotypes; however, transmission insights can be addressed, given that most of our samples are from the same city. Even without explicitly reconstructing transmission events, some conclusions can be drawn because the combination of spatial and genotyping data suggested ongoing neighborhood transmission. In addition, patient history showed that 59% of the individuals grouped into clusters had a history of close contact with a family member or a friend with TB.
Spoligotyping analysis showed that the LAM (62.5%), H (16.25%), and the “ill-defined” T (10%) lineages presented the highest frequency in this study, similar to the results described in South American countries, including Venezuela.29–32 These findings are also similar to reports of other Brazilian states, describing the predominance of the LAM (46%), T family (18.6%), and H family (12.2%), in Minas Gerais, Belém, and Paraná, a Brazilian state bordering Paraguay and Argentina.7,24,33,34 An interesting finding was the identification of untypical spoligotype profiles in Brazil (EAI, Beijing, and MANU1), which are more common in the Far and Middle East, Asia, Oceania, and India.19,35
Regarding the indigenous population, the most frequent patterns (SIT50/H3, SIT64/LAM6, SIT42/LAM9, and LAM9*) were also identified in the nonindigenous patients. Surprisingly, unlike the other common spoligotypes found in indigenous communities, the Beijing strains were identified in our study in two patients (indigenous and nonindigenous), living in the same neighborhood. These findings support the introduction of new lineages in the indigenous population of this region, which may be a result of social activities because indigenous populations often go to urban areas.36
In contrast to previous studies,8,24 we have found a lower frequency of resistance cases, which could be related to the high rate of favorable treatment outcomes (80.6%) observed in the Roraima state over a period of 13 years.37 Nucleotide sequences of rpoB, katG, inhA, rrs, rpsL, and gidB genes were analyzed and were compared with sequences from a susceptible MTB reference strain. All mutations identified in the isolates were described in previous studies.38–41 No mutations were found in the SM-resistant related genes in two isolates, indicating that other mechanisms were responsible for the resistance of this isolates.
Our findings highlight a great diversity of genetic profiles, with the identification of main clades (LAM, H, and T) circulating in this region. Although the lower recent transmission rate and decreased number of resistant isolates were identified in the study, the dissemination of these strains always remains a public health problem. Implementation of prevention and infection control measures is extremely important especially in a region with large population flow.
Supplemental table
Acknowledgments:
We would like to thank the Programa Estadual de Controle de Tuberculose and Laboratório Central de Roraima (LACEN/RR).
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Rendon A, Centis R, Zellweger JP, Solovic I, Torres-Duque C, Cordeiro CR, de Queiroz Mello F, Manissero D, Sotgiu G, 2018. Migration, TB control and elimination: whom to screen and treat. Pulmonology 24: 99–105. [DOI] [PubMed] [Google Scholar]
- 2.IBGE , 2018. Censo Demográfico 2018: Características da População. Available at: https://cidades.ibge.gov.br/brasil/rr/panorama. Accessed December 19, 2018. [Google Scholar]
- 3.Saúde Md, 2017. Indicadores prioritários para o monitoramento do Plano Nacional pelo Fim da Tuberculose como Problema de Saúde Pública no Brazil. Boletim Epidemiológico 48: 1–11. [Google Scholar]
- 4.CIR , 2018. Atuação do Conselho Indígena de Roraima. Available at: http://www.cir.org.br/site/?page_id=158. Accessed December 10, 2018. [Google Scholar]
- 5.Belo EN, Orellana JD, Levino A, Basta PC, 2013. Tuberculosis in Amazonian municipalities of the Brazil-Colombia-Peru-Venezuela border: epidemiological situation and risk factors associated with treatment default [article in Portuguese]. Rev Panam Salud Publica 34: 321–329. [PubMed] [Google Scholar]
- 6.Correia Sacchi FP, et al. 2018. Genetic clustering of tuberculosis in an indigenous community of Brazil. Am J Trop Med Hyg 98: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machado LNC, Marcondes NR, Leite CQF, Santos ACB, Pavan FR, Baldin VP, Castilho AL, Siqueira VLD, Baeza LC, Berghs H, 2014. First baseline of circulating genotypic lineages of Mycobacterium tuberculosis in patients from the Brazilian borders with Argentina and Paraguay. PLoS One 9: e107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marques M, Cunha EAT, Evangelista M, Basta PC, Marques AMC, Croda J, 2017. Antituberculosis-drug resistance in the border of Brazil with Paraguay and Bolivia [Article in Portuguese]. Rev Panam Salud Publica 41: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO , 2017. Global Tuberculosis Report 2017. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 10.Coelho AGV, Zamarioli LA, Reis CMPV, de Lima Duca BF, 2007. Avaliação do crescimento em cordas na identificação presuntiva do complexo Mycobacterium tuberculosis. J Brasileiro Pneumologia 33: 707–711. [DOI] [PubMed] [Google Scholar]
- 11.Giampaglia CMS, Martins MC, Chimara E, Oliveira RS, Vieira GBdO, Marsico AG, Mello FCQ, Fonseca LdS, Kritski A, Telles MAdS, 2007. Diferenciação entre Mycobacterium tuberculosis e outras Micobactérias com ácido ρ-nitrobenzóico utilizando o sistema MGIT960 Int J Tuberc Lung Dis 11: 803–807. [PubMed] [Google Scholar]
- 12.Arora J, Kumar G, Verma AK, Bhalla M, Sarin R, Myneedu VP, 2015. Utility of MPT64 antigen detection for rapid confirmation of Mycobacterium tuberculosis complex. J Glob Infect Dis 7: 66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ministério da Saúde, Secretaria de Vigilância em Saúde, Departamento de Vigilância Epidemiológica, Brazil , 2008. Manual Nacional de Vigilância Laboratorial da Tuberculose e Outras Micobactérias. Brasilia, Brazil: Ministério da Saúde. [Google Scholar]
- 14.Maningi NE, Daum LT, Rodriguez JD, Said HM, Peters RPH, Sekyere JO, Fischer GW, Chambers JP, Fourie PB, 2018. Multi- and extensively drug resistant Mycobacterium tuberculosis in South Africa: a molecular analysis of historical isolates. J Clin Microbiol 56: e01214–e012117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, 2010. Rapid molecular detection of tuberculosis and rifampin resistance. New Engl J Med 363: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Embden J, Cave MD, Crawford JT, Dale J, Eisenach K, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T, 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31: 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C, 2000. Variable human minisatellite‐like regions in the Mycobacterium tuberculosis genome. Mol Microbiol 36: 762–771. [DOI] [PubMed] [Google Scholar]
- 19.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N, 2012. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 12: 755–766. [DOI] [PubMed] [Google Scholar]
- 21.Vitol I, Driscoll J, Kreiswirth B, Kurepina N, Bennett KP, 2006. Identifying Mycobacterium tuberculosis complex strain families using spoligotypes. Infect Genet Evol 6: 491–504. [DOI] [PubMed] [Google Scholar]
- 22.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peres RL, et al. 2018. Risk factors associated with cluster size of Mycobacterium tuberculosis (Mtb) of different RFLP lineages in Brazil. BMC Infect Dis 18: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dantas NGT, Suffys PN, da Silva Carvalho W, Gomes HM, De Almeida IN, De Assis LJ, Augusto CJ, Gomgnimbou MK, Refregier G, Sola C, 2015. Genetic diversity and molecular epidemiology of multidrug-resistant Mycobacterium tuberculosis in Minas Gerais state, Brazil. BMC Infect Dis 15: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribeiro FKC, Pan W, Bertolde A, Vinhas SA, Peres RL, Riley L, Palaci M, Maciel EL, 2015. Genotypic and spatial analysis of Mycobacterium tuberculosis transmission in a high-incidence urban setting. Clin Infect Dis 61: 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Mello SB, Silva BCN, 2005. Roraima: problemas de desenvolvimento sustentável em uma região de fronteira. Redes 10: 129–149. [Google Scholar]
- 27.Fraser B, Willer H, 2016. Venezuela: aid needed to ease health crisis. Lancet 388: 947–949. [DOI] [PubMed] [Google Scholar]
- 28.Benedetti MSG, 2018. Relatório Anual de Epidemiologia de Roraima 2017. Boa Vista, Brazil: Secretaria de Saúde do Estado de Roraíma, Governo do Estado de Roraíma. [Google Scholar]
- 29.Lagos J, Couvin D, Arata L, Tognarelli J, Aguayo C, Leiva T, Arias F, Hormazabal JC, Rastogi N, Fernandez J, 2016. Analysis of Mycobacterium tuberculosis genotypic lineage distribution in Chile and neighboring countries. PLoS One 11: e0160434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abadia E, et al. 2009. Mycobacterium tuberculosis ecology in Venezuela: epidemiologic correlates of common spoligotypes and a large clonal cluster defined by MIRU-VNTR-24. BMC Infect Dis 9: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Candia N, Lopez B, Zozio T, Carrivale M, Diaz C, Russomando G, de Romero NJ, Jara JC, Barrera L, Rastogi N, 2007. First insight into Mycobacterium tuberculosis genetic diversity in Paraguay. BMC Microbiol 7: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mokrousov I, Vyazovaya A, Narvskaya O, 2014. Mycobacterium tuberculosis Latin American-Mediterranean family and its sublineages in the light of robust evolutionary markers. J Bacteriol 196: 1833–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furlaneto IP, Conceição EC, Brito MLd, Costa ARFd, Monteiro JJB, Gonçalves NV, Gomes HM, Lima KVB, 2013. Genotipagem por spoligotyping de Mycobacterium tuberculosis obtidos de lâminas de Ziehl-Neelsen em Belém, estado do Pará, Brasil. Revista Pan-Amazônica de Saúde 4: 33–41. [Google Scholar]
- 34.Gomes HM, et al. 2012. Spoligotypes of Mycobacterium tuberculosis complex isolates from patients residents of 11 states of Brazil. Infect Genet Evol 12: 649–656. [DOI] [PubMed] [Google Scholar]
- 35.Banuls AL, Sanou A, Anh NT, Godreuil S, 2015. Mycobacterium tuberculosis: ecology and evolution of a human bacterium. J Med Microbiol 64: 1261–1269. [DOI] [PubMed] [Google Scholar]
- 36.Malacarne J, Kolte IV, Freitas LP, Orellana JDY, Souza MLP, Souza-Santos R, Basta PC, 2018. Factors associated with TB in an indigenous population in Brazil: the effect of a cash transfer program. Rev Inst Med Trop Sao Paulo 60: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues HAdN, Barden JE, da Silva Laroque LF, 2016. A geografia da tuberculose em Roraima. Hygeia 12: 38–49. [Google Scholar]
- 38.Wang Q, Lau SKP, Liu F, Zhao Y, Li HM, Li BX, Hu YL, Woo PCY, Liu CH, 2014. Molecular epidemiology and clinical characteristics of drug-resistant Mycobacterium tuberculosis in a tuberculosis referral hospital in China. PLoS One 9: e110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molodtsov V, Scharf NT, Stefan MA, Garcia GA, Murakami KS, 2017. Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis. Mol Microbiol 103: 1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lempens P, Meehan CJ, Vandelannoote K, Fissette K, de Rijk P, Van Deun A, Rigouts L, de Jong BC, 2018. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci Rep 8: 3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kandler JL, Mercante AD, Dalton TL, Ezewudo MN, Cowan LS, Burns SP, Metchock B, Cegielski P, Posey JE, 2018. Validation of novel Mycobacterium tuberculosis isoniazid resistance mutations not detectable by common molecular tests. Antimicrob Agents Chemother 62: e00974–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.