Abstract.
We explored spatial variation in the prevalence of established molecular markers of antimalarial resistance across a geographically diverse, highland region of western Uganda. We identified Plasmodium falciparum CQ resistance transporter 76T mutations in all pools, but there was no evidence of spatial differences across village-based strata defined by either altitude or river valley. In contrast, we identified a significant inverse association between altitude and the prevalence of Plasmodium falciparum multidrug resistance 1 mutations with the largest proportion of Y184F mutations observed in the low-elevation, high-transmission villages. These results demonstrate the substantial heterogeneity in resistance markers observed across geographic settings, even at relatively small scales, but highlight the complex nature of these ecological relationships.
The emergence and spread of drug resistance are major threats to global public health with the most severe consequences residing in sub-Saharan Africa, where the burden of Plasmodium falciparum malaria is greatest.1,2 Although artemisinin combination therapies (ACTs) remain effective, ongoing surveillance is required to identify and contain the emergence of resistant parasites.3 The level of drug resistance in a given area can be inferred by the study of known molecular markers, which may provide an early warning of decreasing efficacy.4
In Uganda, like most of sub-Saharan Africa, chloroquine (CQ) was the mainstay of treatment for uncomplicated malaria. High rates of treatment failure, accompanied by the near universal prevalence of established CQ resistance mutations led to the adoption of CQ plus sulfadoxine–pyrimethamine (SP) as first-line therapy in June 2000.5 The efficacy of CQ–SP combination therapy was short-lived, resulting in a change to artemether–lumefantrine (AL) in June 2004, although the policy launch did not take place for another 2 years.6
With the withdrawal of CQ from routine use, reduction in the prevalence of the P. falciparum CQ resistance transporter (pfcrt) 76T mutation has been reported in Uganda, consistent with the trends observed in landmark studies from Malawi.7,8 Re-emergence of CQ-sensitive alleles depends greatly on the duration of time following the removal of selection pressure and local factors, such as transmission intensity.9 Therefore, we sought to examine spatial variation in the prevalence of established molecular markers of antimalarial resistance across a geographically diverse, highland region of western Uganda.
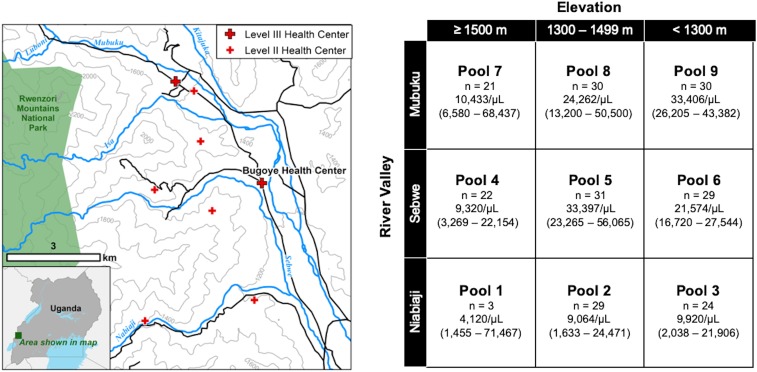
The study area, located in Bugoye subcounty of the Kasese District, experiences year-round malaria transmission marked by semiannual transmission peaks typically following the end of the rainy seasons. The geography of the subcounty is highly varied. The westernmost villages, which border the Rwenzori Mountains National Park, are characterized by steep hillsides with elevations up to 2,000 m, whereas villages located to the east are defined by low-lying, level terrain (Figure 1A). As we have previously demonstrated, this altitudinal gradient results in heterogenous malaria transmission intensity over a relatively small geographic area.10 The subcounty is also defined by three river valleys that flow from west to east converging in the low-lying areas near the health center. Artemether–lumefantrine was introduced to the area in 2008, but the supply available at public health facilities did not stabilize until 2011.
Figure 1.
(A) Topographic map of study area showing the rapid change in altitude and major river valleys. Contour lines demarcate elevation changes of 200 m. (B) Table of sample pools stratified by elevation and river valley, which can be roughly overlaid on map to left, showing number of samples and median and interquartile range of parasitemia. This figure appears in color at www.ajtmh.org.
We collected malaria rapid diagnostic test (RDT) samples from the Bugoye Level III Health Center from May to November 2015 as part of a prospective cohort study of individuals presenting with undifferentiated fever.11 DNA was extracted from each RDT sample using the Chelex method, as previously described.10 Extracted P. falciparum DNA from 219 RDT-positive samples was pooled in an isovolumetric manner, stratified by the river valley and elevation of the individual’s village of residence. The nine resulting pools (Figure 1B) then underwent PCR amplification targeting specific regions associated with putative drug resistance, including 1) pfcrt, 2) Kelch protein K13 (Kelch), and 3) P. falciparum multidrug resistance (pfmdr1) genes. The PCR conditions and the analyzed genomic regions are summarized in Supplemental Table 1. Amplicons were prepared for sequencing using the KAPA Hyper Prep Kit (Roche Sequencing, Pleasanton, CA) and were sequenced on MiSeq 300-base pair paired-end chemistry with a Phi-X spike to increase diversity (Illumina, San Diego, CA). The raw data can be downloaded from sequence read archive (PRJNA555552). All sequence analysis scripts needed to recreate haplotypes are available from GitHub (nickbrazeau/RDTSM_DrugRes_ManuscriptAnalyses).
Sequences were demultiplexed and trimmed using the extractor module in SeekDeep (Version 2.6.0) with the paired-end feature allowing for one barcode error and shortened barcodes.12 Default options were used for the cluster module, which performs initial clustering and removes chimeric haplotypes. Final haplotypes were determined with process cluster for haplotypes with a minimum frequency of 5%. Sample pools were processed with a single PCR, which warranted the higher, more conservative haplotype frequency cutoff. The resulting haplotypes were then evaluated for putative drug resistance mutations and amino acid changes using custom R scripts (R Project, Version 3.5.1, www.r-project.org).
We evaluated the effect of altitude and river valley on the frequency of the putative drug resistance phenotypes (i.e., 74I/75E/76T and N86/184F for pfcrt and pfmdr1-early, respectively) using simple linear regression weighted by the number of samples in each pool. Elevation and location were modeled as disjoint indicator variables based on the categories described in Figure 1. Models were fit a priori, and covariates were pruned using backward selection with an alpha value of 0.05.
Ethical approval of the study was provided by the institutional review boards of the Mbarara University of Science and Technology, the University of North Carolina at Chapel Hill, and the Uganda National Council for Science and Technology. Written informed consent was obtained from all the adult study participants and the caregivers of children.
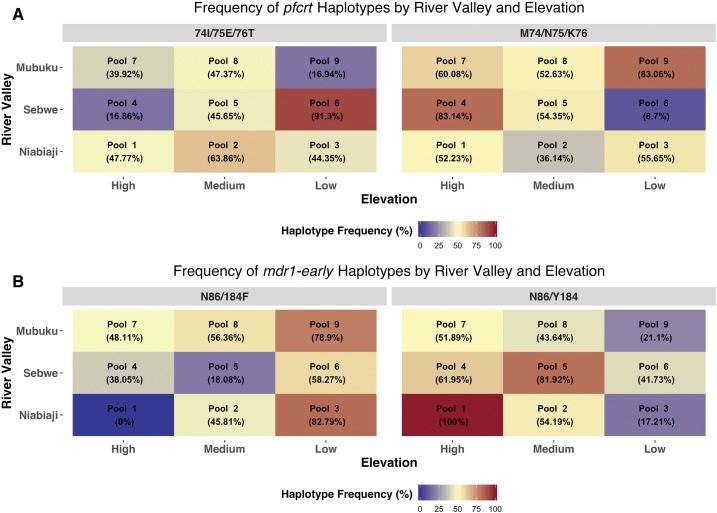
Sequencing of pfcrt showed the presence of resistant 76T haplotypes in all pools. The fraction of mutant haplotypes ranged from 16.8% to 91.3%, although six of nine of pools had fractions between 40% and 65% (Figure 2). Overall, there was no trend between elevation or river valley and the frequency of pfcrt mutations.
Figure 2.
Frequency of wild-type and nonsynonymous putative drug-resistant haplotypes by river valley (y axis) and elevation category (x axis) with respect to targeted amplicon. This figure appears in color at www.ajtmh.org.
In the early pfmdr1 PCR fragment, the Y184F mutation, which is associated with amodiaquine exposure, was identified in all pools except for the one high-elevation pool that comprised only three clinical samples (Pool 1). In contrast to the pfcrt analysis, there was evidence for a trend between the fraction of pfmdr1 mutations and elevation. The largest fraction of Y184 mutations was observed in the three low-elevation pools (pools 3, 6, and 9), whereas the smallest fraction was found in two of the three highest elevation pools (pools 1 and 7). The association was robust in the linear regression model with the fraction of N86/184F alleles being lower in both the medium (β = ‒33.04%, ‒65.59% to ‒0.49%, P = 0.05) and high (β = ‒29.85, ‒70.04% to 10.34%, P = 0.11) elevation pools compared with the low-elevation pools. No trends were seen in regard to the river valley of residence.
No mutations were observed in the pfmdr1 N86Y polymorphism, which is more consistently associated with the degree of lumefantrine susceptibility than the Y184F polymorphism. Similarly, no resistance mutations were identified in the Kelch propeller domain in any of the nine pools (Supplemental Table 2).
Overall, the results of our study provide further insight into the microepidemiological patterns of antimalarial drug resistance. Most notably, we observed a significant, inverse correlation between altitude and the frequency of pfmdr1 Y184 mutations. The ecological factors underlying this finding are, however, complex. For example, although elevation can be a surrogate measure of transmission intensity, it may also be a proxy for access to care, with the higher elevation villages to the west of the subcounty being farther from public health facilities. Residents of these lower transmission villages are likely to have less acquired immunity, be exposed to increasing clonal infections, and develop symptoms that will prompt care-seeking and treatment—factors that some models have suggested may contribute to the rapid spread of resistance in lower transmission settings.13 Yet without access to public facilities, residents are likely to seek care in the private sector where first-line ACTs such as AL are less available.14 Unfortunately, our study is not designed to disentangle these factors, and further investigation will be required. At a minimum, these results suggest that there are not clear associations between altitude of residence and the prevalence of drug resistance polymorphisms.
Our finding that approximately half of alleles in each pool were wild-type pfcrt is consistent with the well-established phenomenon of wild-type re-emergence following the withdrawal of CQ as first-line therapy. We do not have historical data from the site to assess the rate of wild-type pfcrt re-emergence, but our estimates are similar to those from contemporaneous studies from other areas of Uganda.15 The lack of an association between the frequency of pfcrt mutations and elevation may be due to the fact that alleles associated with drug resistance saturated the country at the time of the antimalarial policy change.
Our study has a number of strengths, including the unique geographic setting and use of a validated, high-throughput pooling technique. To our knowledge, this is the first study to examine differences in the frequency of putative drug resistance mutations over such a small, yet geographically diverse area. Our study also has limitations. First, our samples were collected from a single clinical site. This approach restricted the number of samples from the more distant, higher elevation villages. This sampling limitation also could have created a selection bias with patients from distant villages presenting with more severe disease. Second, the pools were not performed in replicate, which could impact the accuracy of our frequency estimations. We attempted to account for this limitation by using a very conservative minimum cutoff of 5% to determine the final haplotypes.
In conclusion, by pooling samples from a geographically diverse area, we were able to identify an association between altitude, which often serves as a surrogate marker of transmission intensity, and the frequency of the Y184F mutation, but this association was not seen across multiple drug resistance–conferring alleles, suggesting that the relationship involves factors other than transmission intensity.
Supplemental tables
Acknowledgments:
We wish to thank the clinical staff and patients of the Bugoye Health Center for their continued support. We acknowledge the cartography support provided by Corianna Keeler from the UNC Department of Geography.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.Talisuna AO, et al. 2012. Mitigating the threat of artemisinin resistance in Africa: improvement of drug-resistance surveillance and response systems. Lancet Infect Dis 12: 888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Packard RM, 2014. The origins of antimalarial-drug resistance. N Engl J Med 371: 397–399. [DOI] [PubMed] [Google Scholar]
- 3.WHO , 2011. Global Plan for Artemisinin Resistance Containment (GPARC). Geneva, Switzerland: World Health Organization. [Google Scholar]
- 4.Plowe CV, 2009. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg 103 (Suppl 1): S11–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamya MR, Bakyaita NN, Talisuna AO, Were WM, Staedke SG, 2002. Increasing antimalarial drug resistance in Uganda and revision of the national drug policy. Trop Med Int Health 7: 1031–1041. [DOI] [PubMed] [Google Scholar]
- 6.Nanyunja M, Nabyonga Orem J, Kato F, Kaggwa M, Katureebe C, Saweka J, 2011. Malaria treatment policy change and implementation: the case of Uganda. Malar Res Treat 2011: 683167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV, 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 187: 1870–1875. [DOI] [PubMed] [Google Scholar]
- 8.Mbogo GW, et al. 2014. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg 91: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talisuna AO, Erhart A, Samarasinghe S, Van Overmeir C, Speybroeck N, D’Alessandro U, 2006. Malaria transmission intensity and the rate of spread of chloroquine resistant Plasmodium falciparum: why have theoretical models generated conflicting results? Infect Genet Evol 6: 241–248. [DOI] [PubMed] [Google Scholar]
- 10.Boyce RM, et al. 2018. Reuse of malaria rapid diagnostic tests for amplicon deep sequencing to estimate Plasmodium falciparum transmission intensity in western Uganda. Scientific Rep 8: 10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyce R, Reyes R, Matte M, Ntaro M, Mulogo E, Siedner MJ, 2017. Use of a dual-antigen rapid diagnostic test to screen children for severe Plasmodium falciparum malaria in a high-transmission, resource-limited setting. Clin Infect Dis 65: 1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA, 2018. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 46: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pongtavornpinyo W, Yeung S, Hastings IM, Dondorp AM, Day NP, White NJ, 2008. Spread of anti-malarial drug resistance: mathematical model with implications for ACT drug policies. Malar J 7: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang LT, Bwambale R, Keeler C, Reyes R, Muhindo R, Matte M, Ntaro M, Mulogo E, Sundararajan R, Boyce RM, 2018. Private sector drug shops frequently dispense parenteral anti-malarials in a rural region of western Uganda. Malar J 17: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumwebaze P, et al. 2017. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 215: 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.