Abstract
Objectives
To propose an operating procedure for validation of discordant trunk driver mutations.
Methods
Concordance of trunk drivers was examined by next-generation sequencing in 15 patients with two to three metastatic lung cancers and 32 paired primary and metastatic lung cancers.
Results
Tissue identity was confirmed by genotyping 17 single-nucleotide polymorphisms within the panel. All except three pairs showed concordant trunk drivers. Quality assessment conducted in three primary and metastatic pairs with discordant trunk drivers indicates metastasis from a synchronous or remote lung primary in two patients. Review of literature revealed high discordant rates of EGFR and KRAS mutations, especially when Sanger sequencing was applied to examine primary and lymph node metastatic tumors.
Conclusions
Trunk driver mutations are highly concordant in primary and metastatic tumors. Discordance of trunk drivers, once confirmed, may suggest a second primary cancer. Guidelines are recommended to establish standard operating procedures for validation of discordant trunk drivers.
Keywords: Discordance, Trunk driver mutations, Lung cancers, KRAS, EGFR, Quality assessment
Mutational profiling identifies genomic alterations for targeted therapy in metastatic non–small cell lung cancers (NSCLCs).1 The US Food and Drug Administration has approved several epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors for EGFR mutations, combined BRAF inhibitor and MEK inhibitor for BRAF p.V600E mutation, and ALK/ROS1 tyrosine kinase inhibitors for ALK and ROS1 translocations.1 Molecular testing guidelines for standard-of-care targeted therapy in patients with metastatic NSCLC have been accordingly updated by the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology.2
Driver mutations are categorized into trunk (initiating) drivers and branching drivers.3-5 Multiple trunk driver mutations cooperate to initiate the formation of a founding cancer cell. Subsequently, branching driver mutations lead to subclonal evolution of the malignancy. Trunk driver mutations, by definition, are expected to be present in each neoplastic cell of the primary and metastatic cancers. By tracking the evolution of adenocarcinomas of the lungs, most hotspot activating mutations in the EGFR, BRAF, and KRAS genes are trunk drivers Table 1.5,6 However, many reports have shown discordant EGFR and/or KRAS mutations between paired primary and metastatic lung cancer specimens, suggesting the presence of tumor heterogeneity.7-30 The discordance rates can be high and have been reported to be approximately 15% for EGFR and KRAS mutations in a meta-analysis.31
Table 1.
Common Driver Mutations of Lung Adenocarcinomas
| Gene | Hotspot |
|---|---|
| Trunk drivers | |
| BRAF | Codon 600a |
| EGFR | Exons 18-21 |
| KRAS/NRAS | Codons 12, 13, 59, 61, 117, 146 |
| MET | Exon 14 skipping mutations |
| ALK, RET, ROS1 | Translocation |
| Branching drivers | |
| PIK3CA | Codons 542, 545, 1047 |
| TP53 | Exons 5-9 |
aOther BRAF mutations involving codons 466, 469, 594, and 601 may also be trunk driver mutations of lung adenocarcinomas.
Unexpected discordance of trunk driver mutations raises concern of laboratory errors, which may occur in any step of the preanalytic, analytic, and/or postanalytic phases.32,33 Use of assays with a lower analytic sensitivity, such as Sanger sequencing, may also contribute to false detection of discordance,12,18,28 especially in a clinical diagnostics setting in which specimens with low tumor cellularity are common.34,35 Next-generation sequencing (NGS) platforms have been widely implemented in the clinical diagnostics laboratories. NGS provides mutational profiling with a higher analytic sensitivity for detection of mutations with lower variant allelic frequencies (VAFs) and a broader reportable range of mutations using a panel of genes.34 In addition, the presence of reference ranges within the NGS panel, such as germline single-nucleotide polymorphisms (SNPs), can be used to confirm tissue identity.36,37 In this retrospective study for quality assessment of clinical mutational profiling by NGS, we propose an operating procedure to confirm discordant trunk driver mutations in paired primary and metastatic lung cancer specimens and demonstrate potential clinical implications of mutational profiling of paired specimens.
Such investigations are clinically important, as concordant mutational findings would support a common clonal origin, whereas discordant trunk driver mutations, once confirmed, may suggest a secondary primary.
Materials and Methods
Materials
Between April 2013 and December 2016, the Molecular Diagnostics Laboratory at the Johns Hopkins Hospital performed NGS studies on 1,329 formalin-fixed, paraffin-embedded (FFPE) specimens with a diagnosis of lung adenocarcinoma, adenosquamous carcinoma, or non–small cell carcinoma. Patients who received prior tyrosine kinase inhibitor therapy were excluded. Paired specimens submitted from synchronous or metachronous lung nodules from the same patients were not included in this study. There were 15 patients with multiple metastatic tumors (14 with two metastatic specimens and one with three metastatic specimens) and 32 patients with primary and metastatic specimens. This included a patient with a primary tumor and two metastatic tumors Table 2 and Table 3. DNA was isolated from FFPE tissues, purified, and quantified as described previously.38 The Johns Hopkins Medicine institutional review board granted approval to this study.
Table 2.
Fifteen Patients With Two to Three Metastatic Specimens
| Case No. | Metastatic Site | Seven-Gene Profilinga |
|---|---|---|
| MM01 | Brain (Re) | KRAS p.G12D |
| Brain (Re) | KRAS p.G12D | |
| MM02 | Chest wall (Re) | KRAS p.G12A |
| Brain (Re) | KRAS p.G12A | |
| MM03 | LN, right lower paratracheal (Re) | No mutation |
| LN, supraclavicular (Bx) | No mutation | |
| MM04 | LN, right lower paratracheal (FNA) | KRAS p.G12A |
| Pleural effusion (FNA) | KRAS p.G12A | |
| MM05 | Soft tissue (Bx) | KRAS p.G12C |
| Abdominal wall (Bx) | KRAS p.G12C | |
| MM06 | Brain, frontal (Re) | KRAS p.G12C, PIK3CA p.E545K |
| Brain, occipital (Re) | KRAS p.G12C, PIK3CA p.E545K | |
| MM07 | Pleura (Re) | KRAS p.G12V, NRAS p.G12D |
| Pleural effusion (FNA) | KRAS p.G12V, NRAS p.G12D | |
| MM08 | Liver (Bx) | KRAS p.G13D |
| Pleural effusion (FNA) | KRAS p.G13D | |
| MM09 | LN, right lower paratracheal (FNA) | KARS p.A146T, BRAF p.G466V |
| LN, supraclavicular (Bx) | KARS p.A146T, BRAF p.G466V | |
| MM10 | Pleural effusion (FNA) | EGFR p.L858R |
| Pleura (Re) | EGFR p.L858R | |
| MM11 | Pleural effusion (FNA) | KRAS p.G12A, PIK3CA p.E542K |
| Pleura (Re) | KRAS p.G12A, PIK3CA p.E542K | |
| MM12 | LN, cervical (Re) | No mutation |
| Adrenal (Bx) | No mutation | |
| MM13 | LN, subcarinal (FNA) | No mutation |
| LN, right hilar (FNA) | No mutation | |
| MM14b | LN, pretracheal (FNA) | NRAS p.G12D |
| Small bowel (Re) | NRAS p.G12D | |
| MM15 | Pleura (Re) | ERBB2 p.G778_P780dup |
| Liver (Bx) | ERBB2 p.G778_P780dup | |
| Liver (Bx) | ERBB2 p.G778_P780dup |
Bx, biopsy; FNA, fine-needle aspiration; LN, lymph node; Re, resection.
a AKT1, BRAF, EGFR, ERBB2, KRAS, NRAS, and PIK3CA genes.
bCase MM14 and case PM29 in Table 3 were submitted from the same patient with a primary tumor and two metastatic tumors.
Table 3.
Thirty-Two Patients With Primary and Metastatic Specimens
| Case No.a | Specimen | Seven-Gene Profilingb |
|---|---|---|
| PM01p | Lung, left (Bx) | No mutation |
| PM01m | LN, subcarinal (FNA) | No mutation |
| PM02p | Lung (Bx) | No mutation |
| PM02m | Brain (Re) | No mutation |
| PM03p | RUL (Re) | KRAS p.A146V |
| PM03m | LN, right lower paratracheal (FNA) | No mutation |
| PM04p | RUL (Bx) | BRAF p.V600E, AKT1 p.E17K |
| PM04m | Brain (Re) | BRAF p.V600E, AKT1 p.E17K |
| PM05p | RUL (Re) | No mutation |
| PM05m | LN, right hilar (FNA) | No mutation |
| PM06p | RUL (Bx) | EGFR p.D770_N771insY |
| PM06m | Brain (Re) | EGFR p.D770_N771insY |
| PM07p | LLL (Bx) | No mutation |
| PM07m | Peritoneum (Bx) | No mutation |
| PM08p | RML/RLL (Bx) | No mutation |
| PM08m | LN, subcarinal (FNA) | No mutation |
| PM09p | RUL (FNA) | KRAS p.G12C |
| PM09m | Pleura (Re) | KRAS p.G12C |
| PM10p | LUL (Re) | KRAS p.G13D |
| PM10m | LN, right interlobar (FNA) | EGFR p.E746_A750del |
| PM11p | RUL (Bx) | No mutation |
| PM11m | Liver (Bx) | No mutation |
| PM12p | RUL (Bx) | KRAS p.G12C |
| PM12m | Pleural effusion (FNA) | KRAS p.G12C |
| PM13p | LLL (FNA) | No mutation |
| PM13m | Brain (Re) | No mutation |
| PM14p | LUL (Re) | No mutation |
| PM14m | Rib (FNA) | No mutation |
| PM15p | RUL (FNA) | No mutation |
| PM15m | Brain (Re) | No mutation |
| PM16p | RUL (Re) | EGFR p.E746_A750del |
| PM16m | LN, right lower paratracheal (FNA) | EGFR p.E746_A750del |
| PM17p | RUL (Bx) | KRAS p.G12V |
| PM17m | LN, right lower paratracheal (FNA) | KRAS p.G12V |
| PM18p | RUL (Re) | KRAS p.G12V |
| PM18m | Pleural effusion (FNA) | KRAS p.G12V |
| PM19p | Lung, left (Bx) | EGFR p.L858E |
| PM19m | Bone, acronium (Bx) | EGFR p.L858E |
| PM20p | Lung (FNA) | No mutation |
| PM20m | Brain (Re) | No mutation |
| PM21p | LLL (Bx) | No mutation |
| PM21m | LN, cervical (Bx) | No mutation |
| PM22p | Lung, left (Bx) | No mutation |
| PM22m | LN, left lower paratracheal (FNA) | No mutation |
| PM23p | RUL (Re) | KRAS p.G12A |
| PM23m | Bone, spine (Bx) | KRAS p.G12A |
| PM24p | LUL (Re) | EGFR p.E746_A750del |
| PM24m | LN, right lower paratracheal (FNA) | EGFR p.E746_A750del |
| PM25p | LUL (FNA) | KRAS p.G12C |
| PM25m | LN, right lower paratracheal (FNA) | KRAS p.G12C |
| PM26p | RML (Re) | BRAF p.V600E |
| PM26m | Brain (Re) | No mutation |
| PM27p | RUL (Re) | No mutation |
| PM27m | LN, right upper paratracheal (Bx) | No mutation |
| PM28p | LUL (Bx) | No mutation |
| PM28m | Brain (Re) | No mutation |
| PM29pc | RUL (Re) | NRAS p.G12D |
| PM29m1c | LN, pretracheal (FNA) | NRAS p.G12D |
| PM29m2c | Small bowel (Re) | NRAS p.G12D |
| PM30p | RUL (Bx) | No mutation |
| PM30m | LN, right upper paratracheal (Re) | No mutation |
| PM31p | RUL (Bx) | KRAS p.G13C |
| PM31m | Brain (Re) | KRAS p.G13C |
| PM32p | RLL (Re) | KRAS p.G12V |
| PM32m | LN, right lower paratracheal (FNA) | KRAS p.G12V |
Bx, core biopsy; FNA, fine-needle aspiration; LLL, left lower lobe; LN, lymph node; LUL, left upper lobe; Re, resection; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe.
ap, primary tumor; m, metastatic tumor.
b AKT1, BRAF, EGFR, ERBB2, KRAS, NRAS, and PIK3CA genes.
cCase PM29 and case MM14 in Table 2 were submitted from the same patient with a primary tumor and two metastatic tumors.
NGS
NGS was conducted using the AmpliSeq Cancer Hotspot Panel (v2) for targeted multigene amplification (Life Technologies), as described previously.39 Briefly, we used the Ion AmpliSeq Library Kit 2.0 for library preparation, Ion OneTouch 200 Template Kit v2 DL (or Ion Personal Genome Machine Hi-Q OT2 Kit) and Ion OneTouch 2 Instrument for emulsion polymerase chain reaction (PCR) and template preparation, and the Ion Personal Genome Machine 200 Sequencing Kit (or Ion Personal Genome Machine Hi-Q Sequencing Kit lately) with the Ion 318 Chip and Personal Genome Machine (Life Technologies) as the sequencing platform.
Mutations were identified and annotated through both Torrent Variant Caller (Life Technologies) and direct visual inspection of the binary sequence alignment/map file using the Broad Institute’s Integrative Genomics Viewer, as described previously.39,40 All specimens were initially analyzed for AKT1 (NM_005163), BRAF (NM_004333), EGFR (NM_005228), ERBB2 (NM_004448), KRAS (NM_033360), NRAS (NM_002524), and PIK3CA (NM_006218) genes for clinical reporting (seven-gene profiling). The entire 50-gene panel, including TP53 gene (NM_000546), was retrospectively examined when discordance of seven-gene profiling was observed. The performance characteristics, including analytic sensitivity (2% mutant allele) and reportable ranges of this NGS assay in lung cancer specimens, have been reported previously.39
Quality Assessment for Discordant Trunk Driver Mutations
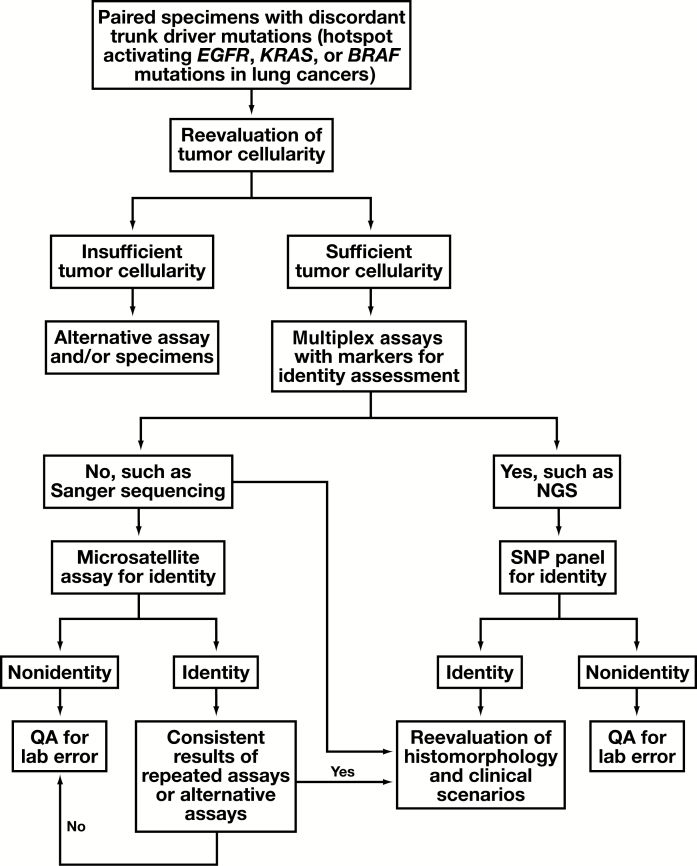
An operating procedure was proposed for clinical validation of unexpected discordant results of trunk driver mutations Figure 1. H&E-stained slides were reviewed by the molecular pathologists to reevaluate if the tumor cellularity within the designated area(s) for DNA extraction was initially overestimated for the analytic sensitivity (or limit of detection) of the assay. Tissue identity was examined using the genotypes of SNPs within the NGS panel. This was followed by review of H&E slides for histomorphology and immunohistochemistry (IHC) stains by the surgical pathologists, as well as reevaluation of clinical scenarios, such as medical history and image studies, to determine potential causes of the discordance, such as a synchronous primary cancer or a remote primary cancer.
Figure 1.
Proposed operating procedure to evaluate unexpected discordance of trunk driver mutations in lung cancers. Microsatellite assay for tissue identity may be avoided if histomorphology and/or detailed clinical history are available for reevaluation. NGS, next-generation sequencing; QA, quality assessment; SNP, single-nucleotide polymorphism.
Tissue Identity
Genotyping of 17 SNPs within the NGS panel was used to confirm tissue identity. The population minor allele frequency of these SNPs ranged from 6% to 46% according to the 1000 Genomes database Table 4. Tissue identity was also retrospectively examined by microsatellite analysis using the AmpFlSTR Identifiler kit (Applied Biosystems) as described previously.41 The assay has been validated for tissue identity and posttransplant chimerism in our laboratory. The limit of detection for the minor component is 1% to 5% of alleles.
Table 4.
Single-Nucleotide Polymorphisms With a 5% or More Minor Allele Frequency Within the Reportable Ranges of the AmpliSeq Cancer Hotspot Panel
| Gene | rs Number | cDNA Change | a.a. Change | Ref. Number | MAF, %a |
|---|---|---|---|---|---|
| APC | rs41115 | c.4479G>A | p.T1493= | NM_000038 | 33 |
| EGFR | rs1050171 | c.2361G>A | p.Q787= | NM_005228 | 43 |
| ERBB4 | rs839541 | c.421 + 58A>G | NA | NM_005235 | 36 |
| FLT3 | rs2491231 | c.1310-3T>C | NA | NM_004119 | 44 |
| HRAS | rs12628 | c.81T>C | p.H27= | NM_005343 | 30 |
| IDH1 | rs11554137 | c.315C>T | p.G105= | NM_005896 | 6 |
| KDR | rs1870377 | c.1416A>T | p.Q472H | NM_002253 | 21 |
| KDR | rs7692791 | c.798 + 54G>A | NA | NM_002253 | 46 |
| KIT | rs3822214 | c.1621A>C | p.M541L | NM_000222 | 7 |
| MET | rs35775721 | c.534C>T | p.S178= | NM_000245 | 9 |
| PDGFRA | rs2228230 | c.2472C>T | p.V824= | NM_006206 | 24 |
| PIK3CA | rs3729674 | c.352 + 40A>G | NA | NM_006218 | 27 |
| PIK3CA | rs2230461 | c.1173A>G | p.I391M | NM_006218 | 9 |
| RET | rs1800861 | c.2307T>G | p.L769= | NM_020975 | 29 |
| RET | rs1800863 | c.2712C>G | p.S904= | NM_020975 | 17 |
| SMARCB1 | rs5030613 | c.1119-41G>A | NA | NM_003073 | 15 |
| STK11 | rs2075606 | c.465-51T>C | NA | NM_000455 | 36 |
a.a. change, amino acid change; cDNA, complementary DNA; NA, not applicable, located within introns; Ref. number, reference sequence number.
aMinor allele frequency (MAF) according to the 1000 Genomes database.
Statistical Analysis
Fisher exact test or χ2 test was performed to calculate P values.
Results
Tissue Identity
Examination of 17 SNPs within the NGS panel revealed identical genotyping between all paired and trio specimens, excluding the possibility of tissue or analyte swapping during the entire processes of assays.
Multiple Metastatic Lung Cancer Specimens
Specimens were taken from two to three metastatic sites in 15 patients (Table 2). Results of the seven-gene profiling were all concordant, including three pairs with no mutation, five pairs with a KRAS mutation, one pair with an NRAS mutation, one pair with an EGFR mutation, two pairs with coexisting KRAS and PIK3CA mutations, one pair with coexisting KRAS p.G12V and NRAS p.G12D mutations, and one pair with coexisting KRAS p.A146T and BRAF p.G466V mutations. Three metastatic sites from one patient showed an ERBB2 p.G778_P780dup mutation.
Paired Primary and Metastatic Lung Cancer Specimens
Specimens were taken from the primary and metastatic tumors in 32 patients, including a patient with one primary tumor and two metastatic tumors (Table 3). Results of the seven-gene profiling were concordant in 29 pairs, including 15 pairs with no mutation, eight pairs with a KRAS mutation, four pairs with an EGFR mutation, one pair with an NRAS mutation, and one pair with coexisting BRAF p.V600E and AKT1 p.E17K mutations. Discordant results of trunk driver mutations were seen in three patients.
VAF
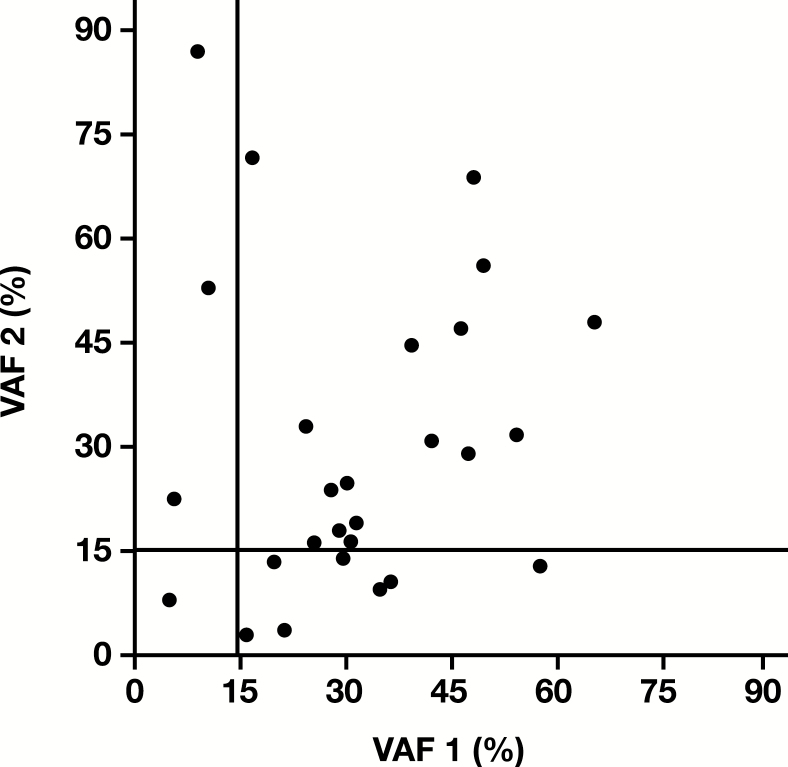
A total of 27 pairs shared the same trunk driver mutations, including activating EGFR, KRAS, NRAS, and BRAF p.V600E mutations. VAFs were less than 15% in both specimens of one pair and in one specimen of 10 (37%) of 27 pairs Figure 2. VAFs were less than 10% in both specimens of one pair and in one specimen of five (19%) of 27 pairs.
Figure 2.
Variant allele frequency (VAF) of the trunk driver mutations in the BRAF, EGFR, KRAS, and NRAS genes among the paired lung cancer specimens.
Paired Primary and Metastatic Specimens With Discordant Trunk Driver Mutations
Quality assessment was conducted to elucidate discordance observed in three pairs of primary and metastatic tumors. The entire 50-gene NGS panel, including the TP53 gene, was also analyzed. Tissue identity was confirmed by genotyping of 17 SNPs within the NGS panel. Microsatellite analysis also showed identical genotypes without sample mix-up.
In pair PM03 with the same IHC staining patterns (positive CK7 and CDX2 and negative napsin), NGS revealed a KRAS p.A146V mutation in the moderately differentiated adenocarcinoma resected from the right upper lobe after neoadjuvant therapy but not in the right paratracheal lymph node, which was taken by fine-needle aspiration 6 months before resection and contained an 11% to 30% estimated tumor cellularity Table 5. Retrospective analysis of a right paratracheal lymph node specimen, which was taken during the resection of the primary tumor and was 70% to 90% replaced by the metastatic adenocarcinoma, also did not reveal the KRAS p.A146V mutation. However, examination of two additional areas from the primary tumor showed the same KRAS mutation. No other mutations were detected within the entire 50-gene panel.
Table 5.
Discordant Trunk Driver Mutations Between Primary and Metastatic Lung Cancer Specimens
| Pairsa | Tumor %b | Trunk Driver Mutations | TP53 |
|---|---|---|---|
| PM03 | |||
| LN, 4R (FNA) | 11-30 | NMD (3/1158 reads)c | NMD |
| RUL (Re: ypT2bypN1, 6 m) | 21-40 | KRAS p.A146V (12%) | NMD |
| PM10 | |||
| LN,11R (FNA) | 61-80 | EGFR p.E746_A750del (65%) | NMD |
| LUL (Re: T1aNx, 1 m) | 41-60 | KRAS p.G13D (46%) | p.R280K (23%) |
| p.N131S (6.0%) | |||
| PM26 | |||
| RML (Re: T1aN0) | 31-50 | BRAF p.V600E (16%) | NMD |
| Brain (Re, 24 m) | 61-80 | NMD (0/1272 reads) | p.R158P (27%) |
11R, right interlobar; FNA, fine-needle aspiration; 4R, right lower paratracheal; LN, lymph node; LUL, left upper lobe; NMD, no mutation detected; Re, resection; RML, right middle lobe; RUL, right upper lobe.
aNumber of months (m) in parentheses indicates duration after the first specimens were taken.
bTumor % is estimated tumor cellularity.
cBelow the limit of detection of the next-generation sequencing assay (3/1,158 = 0.3%).
In patient PM10, the fine-needle aspiration specimen of the right interlobar lymph node revealed an EGFR mutation while the resection specimen of the left upper lobe taken 1 month later showed a KRAS mutation (Table 5). H&E slides were reviewed by a pulmonary pathologist (P.I.). The distinct morphology (poorly differentiated adenocarcinoma with signet ring cell features in the lymph node and moderately differentiated adenocarcinoma with acinar, micropapillary, and lepidic patterns in the primary tumor) and the imaging studies suggest the origin of right interlobar lymph node metastasis from a second lung primary near the right hilum, instead of the KRAS-mutated primary tumor in the left upper lobe. No specimen was taken from the lung mass near the right hilus for confirmation.
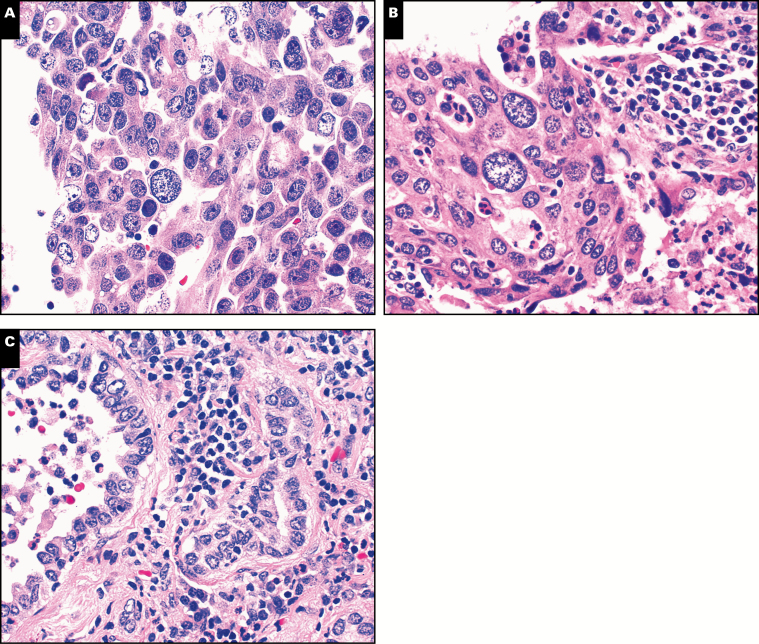
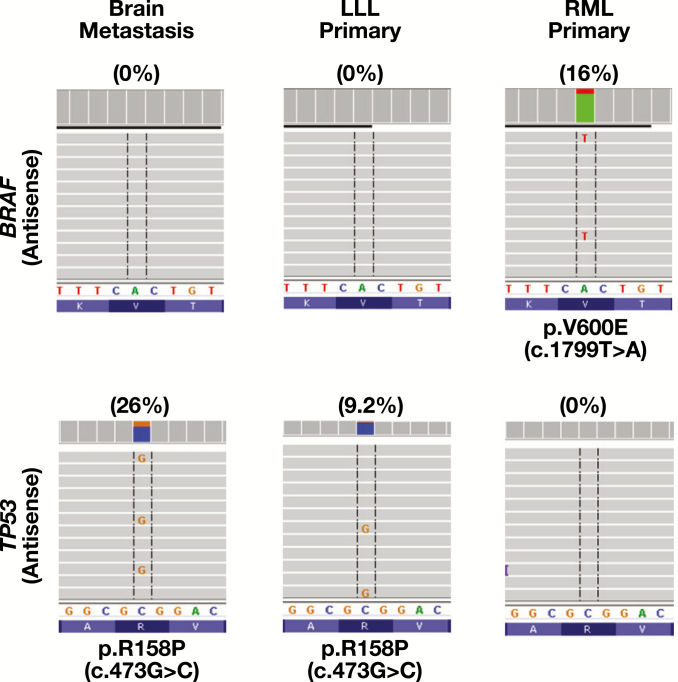
In PM26, NGS detected a BRAF p.V600E mutation in the well-differentiated adenocarcinoma involving the right middle lobe (stage T1aN0), but TP53 p.R158P mutation in the poorly differentiated TTF-1–positive brain metastasis resected 2 year later (Table 5 and Image 1). Review of the clinical history revealed a TTF-1–positive poorly differentiated adenocarcinoma (stage T2N1) involving the left lower lobe resected 7 years before brain metastasis. Retrospective NGS analysis of the remote lung cancer revealed the same TP53 p.R158P mutation but not the BRAF p.V600E mutation Figure 3, indicating brain metastasis from a remote lung primary within the left lower lobe.
Image 1.
Metachronous lung cancers with brain metastasis. Metastatic adenocarcinoma of the brain showed nuclear features and a predominantly solid growth pattern (A) similar to a remote lung cancer involving the left lower lobe (B) but different from a recent lung cancer with a predominantly acinar growth pattern involving the right middle lobe (C). (H&E, ×400)
Figure 3.
Brain metastasis from a remote lung primary. The same TP53 mutation was present in the brain metastasis and the lung primary within the left lower lobe (LLL) resected 7 years ago but not the BRAF-mutated lung primary within the right middle lobe (RML) resected 2 years ago.
Discussion
In this study for quality assessment of mutational profiling by NGS in paired primary and metastatic lung cancer specimens and multiple metastatic lung cancer specimens, trunk driver mutations are highly concordant. Trunk driver mutations in the EGFR, KRAS, or BRAF genes were concordant in 15 patients with multiple metastatic specimens but were discordant in three of 32 pairs of primary and metastatic specimens. Quality assessment according to an operating procedure indicates origin of metastasis from a synchronous lung primary in patient PM10 and from a remote lung primary resected 7 years ago instead of a recent lung primary in patient PM26. Further studies using a larger NGS panel for comprehensive multiregional analyses may be helpful to elucidate the underlying causes of discordant KRAS mutation between the lung primary and regional lymph node metastasis in patient PM03.
In a large-scale study, mutational patterns of metastatic lymph nodes were concordant with those of the primary tumors in 77 patients with an EGFR-mutated primary lung cancer and 55 patients with an EGFR wild-type primary lung cancer. Multiregional analysis of 55 EGFR-mutated lung cancers also showed identical mutational patterns within each subarea. These results indicate that heterogeneous distribution of EGFR mutations is extremely rare in lung cancers.23 However, discordant EGFR and KRAS mutations between paired primary and metastatic lung cancer specimens have been frequently reported, with a discordance rate of 14.5% and 16.7%, respectively, according to a meta-analysis.7-31 However, the discordance rates may be influenced by the prevalence of EGFR or KRAS mutations among different ethnic populations as well as the analytic sensitivity and reported ranges of the assays. When paired specimens with no mutations in both primary and metastatic tumors were removed from the denominator (ie, including only pairs with one or both specimens positive for the mutation as the denominator), the discordance rate was seen in 127 (30%) of 417 pairs for EGFR mutations from 22 studied populations Table 6 and in 67 (62%) of 108 pairs for KRAS mutations from 14 studied populations Table 7. The discordance rate of KRAS mutations was significantly higher than that for EGFR mutations in each subpopulation analysis: 40% (64/162) vs 71% (32/45) in the subgroup of metastatic lymph nodes analyzed by Sanger sequencing, 46% (39/85) vs 81% (22/27) in the subgroup of various metastatic sites analyzed by Sanger sequencing, and 0% (0/26) vs 19% (3/16) in the subgroup analyzed by NGS. These high discordance rates reported in the literature are incongruent with the fact that activating EGFR mutations are trunk driver mutations highly suitable for targeted therapy.1-3,5 Therefore, quality assessment measures are needed when potential discordance of trunk driver mutations is observed in paired primary and metastatic lung cancer specimens.
Table 6.
Discordant EGFR Mutations in Primary and Metastatic Lung Cancers Reported in the Literature
| Seriesa | Met | Assay | Met(–)/Pri(+), No. (%)b | Pri(–)/Met(+), No. (%)c | Discordant Rate, No. (%)d |
|---|---|---|---|---|---|
| Sanger: LN met | |||||
| Park et al (101)7 | LN | Sanger | 11/21 (52) | 1/11 (9.1) | 12/22 (55) |
| Schmid et al (96)8 | LN | Sanger | 3/4 (75) | 3/4 (75) | 6/7 (86) |
| Sun et al (80)9 | LN | Sanger | 2/21 (9.5) | 7/26 (27) | 7/26 (32) [2] |
| Chang et al (56)10 | LN | Sanger | 10/20 (50)b | 4/14 (29)c | 14/24 (58) |
| Han et al (22)11 | LN | Sanger | 1/7 (14) | 0/6 (0) | 1/7 (14) |
| Chen et al (49)12 | LN | Sanger | 6/20 (30) | 1/15 (6.7) | 7/21 (33) |
| Kang et al (74)13 | LN | Sanger | 6/31 (19)b | 0/25 (0)c | 6/31 (19) |
| Okada et al (14)14 | LN | PNA-Sanger | 1/3 (33) | 0/2 (0) | 1/3 (33) |
| Shimizu et al (70)15 | LN | PNA-Sanger | 10/21 (48) | 0/11 (0) | 10/21 (48) |
| Subtotal | 50/148 (34) | 16/114 (14) | 64/162 (40) | ||
| Sanger: variety met | |||||
| Matsumoto et al (8)16 | Brain | Sanger | 0/6 (0) | 0/6 (0) | 0/6 (0) |
| Kalikaki et al (25)17 | Variety | Sanger | 1/2 (50)b | 0/1 (0)c | 1/2 (50) |
| Gow et al (67)18 | Variety | Sanger | 7/16 (44)b | 18/27 (67) | 25/34 (74) |
| Munfus-McCray et al (9)19 | Variety | Sanger | 1/3 (33) | 0/2 (0) | 1/3 (33) |
| Han et al (37)20 | Variety | Sanger | 5/18 (28) | 3/16 (19) | 7/20 (35) [1] |
| Chen et al (35)12 | Variety | Sanger | 4/19 (21) | 1/16 (6.3) | 5/20 (25) |
| Subtotal | 18/64 (28) | 22/68 (32) | 39/85 (46) | ||
| Sanger: all studies | 68/212 (32) | 38/182 (21) | 103/247 (42) | ||
| Other assays | |||||
| Fang et al (219)21 | LN | TaqMan | 23/57 (40) | 0/34 (0) | 23/57 (40) |
| Luo et al (15)22 | Brain | ARMS | 0/7 (0) | 1/8 (13) | 1/8 (13) |
| Yatabe et al (127)23 | LN | Variety | 0/77 (0) | 0/77 (0) | 0/77 (0) |
| Quéré et al (18)24 | Variety | Variety | 0/2 (0) | 0/2 (0) | 0/2 (0) |
| NGS assay | |||||
| Vignot et al (15)25 | Variety | NGS | 0/1 (0) | 0/1 (0) | 0/1 (0) |
| Xie et al (35)26 | LN | NGS | 0/21 (0) | 0/21 (0) | 0/21 (0) |
| Current study (4) | Variety | NGS | 0/4 (0) | 0/4 (0) | 0/4 (0) |
| Subtotal | 0/26 (0) | 0/26 (0) | 0/26 (0) | ||
| Total | 91/381 (24) | 39/329 (12) | 127/417 (30) |
ARMS, amplification mutation refractory system; LN, lymph node metastasis; Met, metastasis; NGS, next-generation sequencing; PNA-Sanger, peptide nucleic acid mediated polymerase chain reaction (PCR) and Sanger sequencing; TaqMan, TaqMan real-time PCR.
aNumber in the parentheses indicates total pairs of primary and metastatic specimens examined. Including 77 pairs with a known EGFR mutation in the primary tumor and 50 pairs with known wild-type EGFR in the primary tumor in Yatabe et al.23 Case PM10 in the current study was not included.
bNo. (%) of mutations detected in the primary tumor but not the metastatic tumor. Discordance of uncommon EGFR mutations was not included.
cNo. (%) of mutations detected in the metastatic tumor but not the primary tumor. Discordance of uncommon EGFR mutations or p.T790M mutation in the posttreatment metastatic specimens was not included.
dNumerator: numbers of pair with discordant EGFR mutation (mutation detected only in the primary tumor or the metastatic tumor, or different EGFR mutations detected). Denominator: pairs with EGFR mutation detected in one or two of the paired specimens. Number in the bracket indicates pairs with different EGFR mutations.
Table 7.
Discordant KRAS Mutations in Primary and Metastatic Lung Cancers Reported in the Literature
| Seriesa | Met | Assay | Met(–)/Pri(+), No. (%)b | Pri(–)/Met(+), No. (%)c | Discordant Rate, No. (%)d |
|---|---|---|---|---|---|
| Sanger: LN met | |||||
| Schmid et al (96)8 | LN | Sanger | 17/28 (61) | 8/19 (42) | 25/36 (69) |
| Sun et al (80)9 | LN | Sanger | 0/1 (0) | 6/7 (86) | 6/7 (86) |
| Han et al (22)11 | LN | Sanger | 1/2 (50) | 0/1 (0) | 1/2 (50) |
| Subtotal | 18/31 (58) | 14/27 (52) | 32/45 (71) | ||
| Sanger: variety met | |||||
| Kalikaki et al (25)17 | Variety | Sanger | 3/5 (60) | 3/5 (60) | 6/8 (75) |
| Monaco et al (40)27 | Variety | Sanger | 9/11 (82) | 2/4 (50) | 9/11 (85) [2] |
| Cortot et al (21)28 | Variety | Sanger | 3/3 (100) | 4/4 (100) | 6/6 (100) [1] |
| Han et al (37)20 | Variety | Sanger | 0/1 (0) | 1/2 (50) | 1/2 (50) |
| Subtotal | 15/20 (75) | 10/15 (67) | 22/27 (81) | ||
| Sanger: all studies | 33/51 (65) | 24/42 (57) | 54/72 (75) | ||
| Other assays | |||||
| Badalian et al (11)29 | Bone | PCR/RFLP | 2/3 (67) | 2/3 (67) | 4/5 (80) |
| Munfus-McCray et al (9)19 | Variety | Pyro | 0/1 (0) | 1/2 (50) | 1/2 (50) |
| Alsdorf et al (19)30 | LN | ARMS | 0/4 (0) | 0/4 (0) | 0/4 (0) |
| Quéré et al (18)24 | Variety | Variety | 2/6 (33) | 4/8 (50) | 5/9 (56) [1] |
| NGS assay | |||||
| Vignot et al (15)25 | Variety | NGS | 0/4 (0) | 0/4 (0) | 0/4 (0) |
| Xie et al (35)26 | LN | NGS | 1/2 (50) | 1/2 (50) | 2/3 (67) |
| Current study (10) | Variety | NGS | 1/9 (11) | 0/8 (0) | 1/9 (11) |
| Subtotal | 2/15 (13) | 1/14 (7.1) | 3/16 (19) | ||
| Total | 39/80 (49) | 32/73 (44) | 67/108 (62) |
ARMS, amplification mutation refractory system; LN, lymph node metastasis; Met, metastasis; NGS, next-generation sequencing; Pyro, pyrosequencing; RFLP, restriction fragment length polymorphism.
aNumber in the parentheses indicates total pairs of primary and metastatic specimens examined. Case PM10 in the current study was not included.
bNo. (%) of mutations detected in the primary tumor but not the metastatic tumor.
cNo. (%) of mutations detected in the metastatic tumor but not the primary tumor.
dNumerator: numbers of pair with discordant KRAS mutation (mutation detected only in the primary or metastatic tumor, or different KRAS mutations detected). Denominator: pairs with KRAS mutation detected in one or two of the paired specimens. Number in the bracket indicates pair with different KRAS mutations.
In the clinical diagnostic setting, prior molecular diagnostic results should be reviewed and compared. Quality assessment should be considered in the presence of unexpected discordant findings. Trunk driver mutations should be concordant in all primary and metastatic cancer cells.3-5 Discordance of activating EGFR, KRAS, and BRAF mutations in paired lung cancer specimens taken from the same patient should raise a concern for laboratory errors. In contrast, branching driver mutation in the PIK3CA and TP53 genes or uncommon mutations in the EGFR, KRAS, or BRAF gene may be present in a subpopulation.5,33,42 While quality assessment may not be needed for discordant TP53 mutations, the same TP53 mutation seen in the brain metastasis and the remote lung primary of patient PM26 supports the common clonal origin.
Specimens with lower tumor cellularity are not uncommon in the clinical diagnostic setting.34,35 In our retrospective analysis of 1,006 lung cancers, NGS demonstrated a great analytic sensitivity, with 13% and 38% of mutations detected at a VAF at 2% to 10% and 2% to 20%, respectively.39 With an analytic sensitivity of 10% to 20% VAF depending on the context of sequences examined, Sanger sequencing might have missed 13% to 38% of mutations detected by NGS. The discordance rate reported in the literature was significantly higher when Sanger sequencing was used compared with NGS: 42% (103/247) vs 0% (0/26) for EGFR mutations and 75% (54/72) vs 19% (3/16) for KRAS mutations (Tables 6 and 7). In this study, five (or 10) of 27 pairs of primary and metastatic lung cancer specimens with concordant driver mutations might have shown discordant driver mutations if Sanger sequencing with an analytic sensitivity of 10% (or 15%) VAF had been used to detect mutations (Figure 2).
When unexpected discordant profiling is observed, H&E-stained slides should be reviewed to reevaluate if tumor cellularity within the designated area(s) selected for DNA extraction is sufficient for the analytic sensitivity of the assay. Specimens with prior neoadjuvant therapy or lymph node specimens containing minimal subcapsular or parenchymal infiltrative metastasis are particularly problematic.43,44 Therefore, mutations detected in the primary tumors may not be detected in the metastatic lymph nodes when Sanger sequencing is applied. As shown in Table 6, the same EGFR mutation was not detected in 50 (34%) of 148 paired metastatic lymph node specimens taken from patients with an EGFR-mutated primary lung cancer, while the same EGFR mutation was absent in 16 (14%) of 114 primary tumors when EGFR mutations were detected in their paired metastatic lymph nodes (P < .001). Examination of additional specimens with sufficient tumor cellularity and application of more sensitive assays may reduce the discordance rate.12,18,28,45
Unexpected discordance of trunk driver mutations in paired specimens with adequate tumor cellularity should prompt further quality assessment to identify potential laboratory errors. Laboratory errors may occur in any step of the preanalytic, analytic, and/or postanalytic phases. Mixing or swapping of analytes (tissue, DNA, PCR products, etc) or data files can lead to discordant results. When discordant profiling is detected by a simplex assay, such as Sanger sequencing, tissue identity should be examined by microsatellite analysis to exclude mixing or swapping during the process of tissue preparation or DNA extraction. Assays should be repeated to exclude laboratory errors during the sequencing process and data analysis. Alternative assays may be needed to exclude artifacts, especially when the incidence of rare mutations is relatively higher in the study cohort.17 When NGS analysis is conducted for mutational profiling, we recommend that tissue identity be simultaneously examined by a panel of SNPs.
Discordant profiling of trunk driver mutations in the EGFR, KRAS, and BRAF genes, once confirmed, suggests a different clonal origin. In paired specimens taken from separate lung nodules, it indicates two primary lung cancers as reported previously.46,47 In paired primary and metastatic specimens, it suggests tumor heterogeneity or metastasis from another primary cancer. H&E slides and IHC stains should be reviewed by surgical pathologists. Medical history and image studies should be evaluated to search for possible synchronous or remote primary cancer as shown in patients PM10 and PM26 in the current study.
Mutational profiling of lung cancers identifies mutations for targeted therapy and determines clonal origin of multiple specimens submitted from the same patient. In this retrospective quality assessment study, we showed a high concordance rate of trunk driver mutations in patients with primary and metastatic lung cancers and in patients with multiple metastatic lung cancers. While concordance of somatic mutations supports a common clonal origin, discordance of trunk driver mutations, once confirmed, may suggest a second primary cancer. Guidelines from official organizations are recommended for validation of discordant trunk drivers in both research and clinical diagnostic settings.
Acknowledgment
This work was supported by the National Institute of Health–National Cancer Institute of the United States (1UM1CA186691-01 to Michael Carducci).
References
- 1. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377:849-861. [DOI] [PubMed] [Google Scholar]
- 2. Lindeman NI, Cagle PT, Aisner DL, et al. . Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321-346. [DOI] [PubMed] [Google Scholar]
- 3. Gerlinger M, Rowan AJ, Horswell S, et al. . Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogelstein B, Papadopoulos N, Velculescu VE, et al. . Cancer genome landscapes. Science. 2013;339:1546-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. ; TRACERx Consortium Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109-2121. [DOI] [PubMed] [Google Scholar]
- 6. de Bruin EC, McGranahan N, Mitter R, et al. . Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park S, Holmes-Tisch AJ, Cho EY, et al. . Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809-815. [DOI] [PubMed] [Google Scholar]
- 8. Schmid K, Oehl N, Wrba F, et al. . EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554-4560. [DOI] [PubMed] [Google Scholar]
- 9. Sun L, Zhang Q, Luan H, et al. . Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res. 2011;30:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang YL, Wu CT, Shih JY, et al. . Comparison of p53 and epidermal growth factor receptor gene status between primary tumors and lymph node metastases in non-small cell lung cancers. Ann Surg Oncol. 2011;18:543-550. [DOI] [PubMed] [Google Scholar]
- 11. Han CB, Ma JT, Li F, et al. . EGFR and KRAS mutations and altered c-Met gene copy numbers in primary non-small cell lung cancer and associated stage N2 lymph node-metastasis. Cancer Lett. 2012;314:63-72. [DOI] [PubMed] [Google Scholar]
- 12. Chen ZY, Zhong WZ, Zhang XC, et al. . EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17:978-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang HJ, Hwangbo B, Lee JS, et al. . Comparison of epidermal growth factor receptor mutations between metastatic lymph node diagnosed by EBUS-TBNA and primary tumor in non-small cell lung cancer. PLoS One. 2016;11:e0163652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okada H, Anayama T, Kume M, et al. . Comparison of epidermal growth factor receptor mutation analysis results between surgically resected primary lung cancer and metastatic lymph nodes obtained by endobronchial ultrasound-guided transbronchial needle aspiration. Thorac Cancer. 2012;3:262-268. [DOI] [PubMed] [Google Scholar]
- 15. Shimizu K, Yukawa T, Hirami Y, et al. . Heterogeneity of the EGFR mutation status between the primary tumor and metastatic lymph node and the sensitivity to EGFR tyrosine kinase inhibitor in non-small cell lung cancer. Target Oncol. 2013;8:237-242. [DOI] [PubMed] [Google Scholar]
- 16. Matsumoto S, Takahashi K, Iwakawa R, et al. . Frequent EGFR mutations in brain metastases of lung adenocarcinoma. Int J Cancer. 2006;119:1491-1494. [DOI] [PubMed] [Google Scholar]
- 17. Kalikaki A, Koutsopoulos A, Trypaki M, et al. . Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gow CH, Chang YL, Hsu YC, et al. . Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696-702. [DOI] [PubMed] [Google Scholar]
- 19. Munfus-McCray D, Harada S, Adams C, et al. . EGFR and KRAS mutations in metastatic lung adenocarcinomas. Hum Pathol. 2011;42:1447-1453. [DOI] [PubMed] [Google Scholar]
- 20. Han HS, Eom DW, Kim JH, et al. . EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer. 2011;12:380-386. [DOI] [PubMed] [Google Scholar]
- 21. Fang Q, Yang H, Ou W, et al. . Discordance of epidermal growth factor receptor mutations between primary tumors and corresponding mediastinal nodal metastases in patients operated on for stage N2 non-small cell lung cancer. Thorac Cancer. 2012;3:313-319. [DOI] [PubMed] [Google Scholar]
- 22. Luo D, Ye X, Hu Z, et al. . EGFR mutation status and its impact on survival of Chinese non-small cell lung cancer patients with brain metastases. Tumour Biol. 2014;35:2437-2444. [DOI] [PubMed] [Google Scholar]
- 23. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972-2977. [DOI] [PubMed] [Google Scholar]
- 24. Quéré G, Descourt R, Robinet G, et al. . Mutational status of synchronous and metachronous tumor samples in patients with metastatic non-small-cell lung cancer. BMC Cancer. 2016;16:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vignot S, Frampton GM, Soria JC, et al. . Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol. 2013;31:2167-2172. [DOI] [PubMed] [Google Scholar]
- 26. Xie F, Zhang Y, Mao X, et al. . Comparison of genetic profiles among primary lung tumor, metastatic lymph nodes and circulating tumor DNA in treatment-naïve advanced non-squamous non-small cell lung cancer patients. Lung Cancer. 2018;121:54-60. [DOI] [PubMed] [Google Scholar]
- 27. Monaco SE, Nikiforova MN, Cieply K, et al. . A comparison of EGFR and KRAS status in primary lung carcinoma and matched metastases. Hum Pathol. 2010;41:94-102. [DOI] [PubMed] [Google Scholar]
- 28. Cortot AB, Italiano A, Burel-Vandenbos F, et al. . KRAS mutation status in primary nonsmall cell lung cancer and matched metastases. Cancer. 2010;116:2682-2687. [DOI] [PubMed] [Google Scholar]
- 29. Badalian G, Barbai T, Rásó E, et al. . Phenotype of bone metastases of non-small cell lung cancer: epidermal growth factor receptor expression and K-RAS mutational status. Pathol Oncol Res. 2007;13:99-104. [DOI] [PubMed] [Google Scholar]
- 30. Alsdorf WH, Clauditz TS, Hoenig T, et al. . Intratumoral heterogeneity of KRAS mutation is rare in non-small-cell lung cancer. Exp Mol Pathol. 2013;94:155-159. [DOI] [PubMed] [Google Scholar]
- 31. Wang S, Wang Z. Meta-analysis of epidermal growth factor receptor and KRAS gene status between primary and corresponding metastatic tumours of non-small cell lung cancer. Clin Oncol (R Coll Radiol). 2015;27:30-39. [DOI] [PubMed] [Google Scholar]
- 32. Pfeifer JD, Liu J. Rate of occult specimen provenance complications in routine clinical practice. Am J Clin Pathol. 2013;139:93-100. [DOI] [PubMed] [Google Scholar]
- 33. De Marchi F, Haley L, Fryer H, et al. . Clinical validation of coexisting activating mutations within EGFR, mitogen-activated protein kinase, and phosphatidylinositol 3-kinase pathways in lung cancers. Arch Pathol Lab Med. 2019;143:174-182. [DOI] [PubMed] [Google Scholar]
- 34. Frampton GM, Fichtenholtz A, Otto GA, et al. . Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen G, Yang Z, Eshleman JR, et al. . Molecular diagnostics for precision medicine in colorectal cancer: current status and future perspective. Biomed Res Int. 2016;2016:9850690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huijsmans R, Damen J, van der Linden H, et al. . Single nucleotide polymorphism profiling assay to confirm the identity of human tissues. J Mol Diagn. 2007;9:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eduardoff M, Santos C, de la Puente M, et al. . Inter-laboratory evaluation of SNP-based forensic identification by massively parallel sequencing using the Ion PGM™. Forensic Sci Int Genet. 2015;17:110-121. [DOI] [PubMed] [Google Scholar]
- 38. Zheng G, Lin MT, Lokhandwala PM, et al. . Clinical mutational profiling of bone metastases of lung and colon carcinoma and malignant melanoma using next-generation sequencing. Cancer Cytopathol. 2016;124:744-753. [DOI] [PubMed] [Google Scholar]
- 39. Illei PB, Belchis D, Tseng LH, et al. . Clinical mutational profiling of 1006 lung cancers by next generation sequencing. Oncotarget. 2017;8:96684-96696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tseng LH, Tang JL, Haley L, et al. . Microsatellite instability confounds engraftment analysis of hematopoietic stem-cell transplantation. Appl Immunohistochem Mol Morphol. 2014;22:416-420. [DOI] [PubMed] [Google Scholar]
- 42. Zhang LL, Kan M, Zhang MM, et al. . Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int J Cancer. 2017;140:103-108. [DOI] [PubMed] [Google Scholar]
- 43. Dudley J, Tseng LH, Rooper L, et al. . Challenges posed to pathologists in the detection of KRAS mutations in colorectal cancers. Arch Pathol Lab Med. 2015;139:211-218. [DOI] [PubMed] [Google Scholar]
- 44. Chen G, Dudley J, Tseng LH, et al. . Lymph node metastases of melanoma: challenges for BRAF mutation detection. Hum Pathol. 2015;46:113-119. [DOI] [PubMed] [Google Scholar]
- 45. Sherwood J, Dearden S, Ratcliffe M, et al. . Mutation status concordance between primary lesions and metastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res. 2015;34:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patel SB, Kadi W, Walts AE, et al. . Next-generation sequencing: a novel approach to distinguish multifocal primary lung adenocarcinomas from intrapulmonary metastases. J Mol Diagn. 2017;19:870-880. [DOI] [PubMed] [Google Scholar]
- 47. Roepman P, Ten Heuvel A, Scheidel KC, et al. . Added value of 50-gene panel sequencing to distinguish multiple primary lung cancers from pulmonary metastases: a systematic investigation. J Mol Diagn. 2018;20:436-445. [DOI] [PubMed] [Google Scholar]