SUMMARY
The Golgi complex plays a central role in the intracellular sorting of proteins. Transport through the Golgi in the anterograde direction has been explained by cisternal maturation, while transport in the retrograde direction is attributed to vesicles formed by the coat protein I (COPI) complex. A more detailed understanding of how COPI acts in Golgi transport is being achieved in recent years, due in large part to a COPI reconstitution system. Through this approach, the mechanistic complexities of COPI vesicle formation are being elucidated. This approach has also uncovered a new mode of anterograde transport through the Golgi, which involves COPI tubules connecting the Golgi cisternae. We describe in this chapter the reconstitution of COPI vesicle and tubule formation from Golgi membrane.
Keywords: COPI, ARF, Golgi complex, Vesicle formation, Tubule formation
1. INTRODUCTION
Membrane traffic in the cell occurs in two general directions, exocytic (outward bound) and endocytic (inward bound). In the exocytic direction, proteins that are initially synthesized at the endoplasmic reticulum (ER) transit through the Golgi (in the anterograde direction) for sorting to other parts of the cell. In the endocytic direction, proteins at the plasma membrane also pass through the Golgi (in the retrograde direction) before reaching the ER. Thus, because the Golgi acts at the crossroad of the two general directions of transport, considerable interest exists in understanding how bidirectional transport through the Golgi complex is accomplished [1,2].
For many years, anterograde Golgi transport has been explained by the movement of the Golgi cisternae, known as cisternal maturation, while retrograde Golgi transport has been thought to be mediated by vesicles formed by the COPI complex [1,2]. Early studies identified coatomer, a multimeric complex, as the core components of the COPI complex [3,4]. The small GTPase ARF1 was then identified to regulate coatomer by dictating its distribution between the functional (on membrane) and non-functional (cytosolic) pools [5]. Moreover, key upstream regulators of ARF1 were also identified and/or characterized. These include a guanine nucleotide exchange factor (GEF) activity that catalyzes the activation of ARF1 [6,7], and a GTPase-activating protein (GAP) that catalyzes ARF1 deactivation [8].
Subsequently, our group has been making major contributions to the further understanding of COPI transport [9]. We initially elucidated a more complex role for the GAP that catalyzes ARF1 deactivation in COPI transport, known as ARFGAP1. Whereas early studies predicted that ARFGAP1 acts in the uncoating of COPI vesicles [10], we found that it has a novel function in promoting COPI vesicle formation, with mechanistic elucidation suggesting that ARFGAP1 acts as another component of the COPI complex [11,12]. We then identified BARS (Brefeldin-A ADP-Ribosylated Substrate) to act at the fission stage of COPI vesicle formation [13,14]. Further elucidating how vesicle fission occurs, we identified a key lipid, phosphatidic acid (PA), which promotes the ability of BARS to bend membranes in achieving vesicle fission [15]. Notably, rather than acting merely to recruit BARS to membrane, we found that PA participates actively with BARS to bend membranes [15].
We then uncovered an even more complex role for the COPI complex by finding that it also generates tubules that connect the Golgi stacks [16]. Mechanistically, this involves COPI coupling with distinct lipid enzymatic activities in dictating whether Golgi membranes form vesicles or tubules [16]. More recently, we have defined that transport through COPI tubules complements cisternal maturation in explaining how anterograde Golgi transport occurs [17]. A notable mechanistic detail is that the small GTPase Cdc42 plays a pivotal role in coordinating bidirectional Golgi transport by targeting the two major functions of COPI – carrier formation and cargo sorting [17].
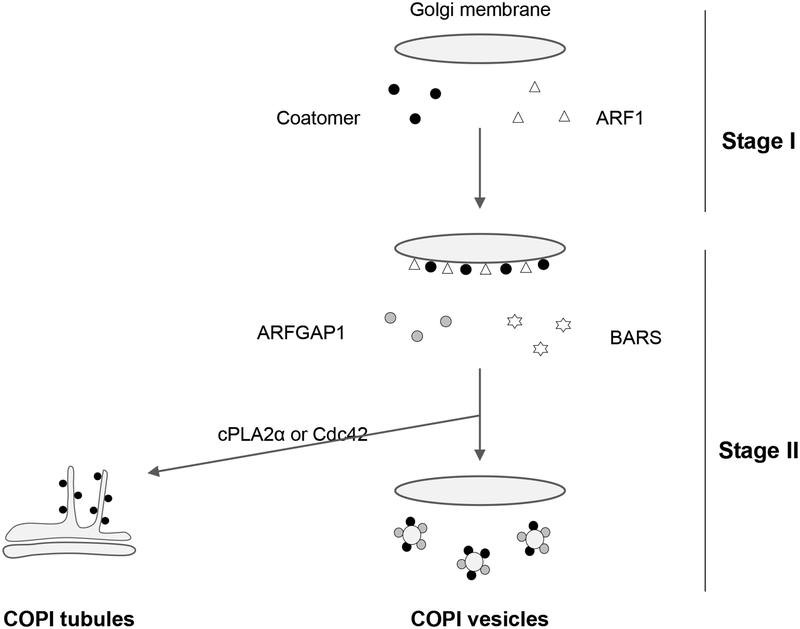
These achievements have been due, in large part, to a reconstitution system that allows novel factors to be identified, as well as elucidating how they act. The reconstitution system is divided into two stages (summarized in Figure 1). In the first stage, purified ARF1 and coatomer are incubated with Golgi membrane to achieve the recruitment of coatomer onto membrane. In the second stage, ARFGAP1 and BARS are added, which then result in COPI vesicle formation. To achieve COPI tubule formation, either cytoplasmic phospholipase type 2 alpha (cPLA2α) or Cdc42 is added additionally in the second stage, which results in vesicle formation being diverted to tubule formation. We describe below the technical details of these reconstitutions.
Figure 1.
Schematic of the two-stage incubation system to reconstitute COPI vesicles and tubules. In the first stage, Golgi membrane is incubated with purified ARF1 and coatomer. In the second stage, the incubated Golgi membrane is further incubated with purified ARFGAP1 and BARS to form COPI vesicles. For COPI tubule formation, the second-stage incubation involves the further addition of either purified cPLA2α or Cdc42.
2. MATERIALS
2.1. Recombinant proteins produced from bacteria
50 μM sodium myristate: dissolve 12.52 mg/liter culture media.
1 M isopropyl β-D-1-thiogalactopyranoside (IPTG): dissolve 238 g/ml distilled water.
1 M DL-Dithiothreitol (DTT): dissolve 154 mg/ml distilled water.
Protease inhibitor cocktail.
ARF1 lysis buffer: 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mg/ml lysozyme and protease inhibitor.
Columns: HiTrap Q HP, HiPrep 26/60 Sephacryl S-100, HiTrap phenylsepharose (GE Healthcare Life Sciences) and Ni-NTA resin (Clontech).
HiTrap Q HP running buffer: 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT and 10% glycerol.
HiPrep 26/60 Sephacryl S-100 running buffer: 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2 and 1 mM DTT.
High salt buffer: 20 mM Tris pH 8.0, 5 M NaCl, 1 mM MgCl2 and 1 mM DTT.
HiTrap phenylsepharose running buffer: 20 mM Tris pH 8.0, 3 M NaCl, 1 mM MgCl2 and 1 mM DTT.
Reaction buffer: 25 mM HEPES pH 7.2, 50 mM KCl, 2.5 mM Mg(OAc)2 and 1 mM DTT.
BARS lysis buffer: 20 mM Tris pH 8.0, 100 mM NaCl, 1 mg/ml lysozyme, 1% Triton X-100 and protease inhibitor.
BARS washing buffer: 50 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 20 mM imidazole.
BARS elution buffer: 50 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 250 mM imidazole.
2.2. Recombinant proteins produced from insect cells
BestBac 2.0 Baculovirus cotransfection kit (Expression systems).
ESF 921 insect cell culture medium, protein-free (Expression systems).
Bac-to-Bac Baculovirus Expression system (Invitrogen).
ARFGAP1 lysis buffer: 20 mM Tris pH 7.5, 100 mM NaCl, 1% Triton X-100, 1% CHAPS and protease inhibitor.
ARFGAP1 washing buffer: 50 mM Tris pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.1% CHAPS and 20 mM imidazole.
ARFGAP1 elution buffer: 50 mM Tris pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.1% CHAPS and 250 mM imidazole.
Hypotonic buffer: 20 mM sodium borate pH 10.2, 5 mM MgCl2 and protease inhibitor.
TBS-M: 50 mM Tris pH 7.2, 150 mM NaCl, 5 mM MgCl2 and protease inhibitor.
Cdc42 washing buffer: 50 mM Tris pH 7.2, 500 mM NaCl, 5 mM MgCl2, 0.1% CHAPS and 20 mM imidazole.
Cdc42 elution buffer: 50 mM Tris pH 7.2, 500 mM NaCl, 5 mM MgCl2, 0.1% CHAPS and 250 mM imidazole.
2.3. Purification of coatomer from rat liver
Rat liver (freshly prepared in the lab).
Polytron homogenizer (7 mm generator diameter, Kinematica AG).
2 mM Ethylenediaminetetraacetic acid (EDTA): dissolve 0.58 mg/ml distilled water.
Homogenize buffer: 25 mM Tris pH 8.0, 500 mM KCl, 250 mM sucrose, 2 mM EDTA, 1 mM DTT and protease inhibitor.
Saturated ammonium sulfate.
Dialysis buffer: 25 mM Tris pH 7.5, 200 mM KCl, 1 mM DTT and protease inhibitor.
0.45 μm syringe filter.
Columns: DEAE-Sepharose FF, HiTrap Q HP and Resource Q (GE Healthcare Life Sciences).
Running buffer: 25 mM Tris pH 7.5, 200 mM KCl, 1 mM DTT and 10% glycerol.
Anti-βCOP antibody (M3A5).
Dilution buffer: 25 mM Tris pH 7.5, 1 mM DTT and 10% glycerol.
2.4. Golgi membrane preparation
Cell culture media: RPMI1640 supplemented with 10% FBS, 2 mM L-glutamine, 20 mM HEPES, and 20 μg/ml Gentamycin.
ST buffer: 856 mg of sucrose/ml 10 mM Tris-HCl, pH 7.4.
Ball-bearing homogenizer and 25 μm clearance ball [18].
Sucrose gradient buffer: 62% (805 mg sucrose/ml 10 mM Tris, pH 7.4), 35% (403 mg sucrose/ml with 10 mM Tris, pH 7.4) and 29% (325 mg sucrose/ml 10 mM Tris, pH 7.4).
Refractometer.
30 ml syringe, 10 ml syringe.
50 ml ultracentrifuge tubes for SW-28 rotor (Beckman).
2.5. In vitro reconstitution
Low binding microcentrifuge tubes and tips.
Traffic buffer: 25 mM HEPES pH 7.2, 50 mM KCl, 2.5 mM Mg(OAc)2, 1 mg/ml soybean trypsin inhibitor, 1 mg/ml BSA and 200 mM sucrose.
3 M KCl solution: dissolve 224 mg KCl/ml traffic buffer.
BSA solution: dissolve 0.1 g BSA in 10 ml of traffic buffer.
Cushion buffer: 25 mM HEPES pH 7.2, 50 mM KCl, 2.5 mM Mg(OAc)2 and 15% sucrose.
2.6. Electron microscopy
Formvar carbon coated grids.
2% paraformaldehyde (PFA) in PBS (from 16% stock solution).
Uranyl acetate staining solution: carefully mix 1 ml of 4% uranyl acetate and 9 ml of 2% methyl cellulose.
JEOL 1200EX transmission electron microscope.
3. METHODS
3.1. Generating recombinant proteins using bacterial expression
3.1.1. Purification of myristoylated-ARF1
Culture BL21 cells containing Arf1/pET3 and N-myristoyltransferase/pBB131 plasmids in 1 liter LB media supplemented with 100 μg/ml ampicillin and 25 μg/ml kanamycin at 37°C.
Add 50 μM sodium myristate when O.D. is 0.6, and further culture the cells for 30 min.
Induce the expression using 0.1 mM of IPTG at room temperature, overnight.
Harvest the cells using centrifugation at 6,000g, 4°C for 10 min.
Lyse the cells using 50 ml of lysis buffer for 30 min, followed by sonication.
Centrifuge cell lysate at 100,000g, 4°C for 1 hour.
Load the supernatant to HiTrap Q HP column, which is equilibrated with running buffer at 1 ml/min.
Pool the fractions containing ARF1 determined by 15% SDS-PAGE.
Concentrate purified ARF1 using Amicon Ultra-15 (MWCO: 10,000) at 2,000g until 10 ml of volume.
Load the sample to HiPrep 26/60 Sephacryl S-100 column, which is equilibrated with running buffer, and further develop the column using running buffer at 2 ml/min.
Pool the fractions containing ARF1 determined by 15% SDS-PAGE.
Adjust salt concentration to 3M using high salt buffer (see Note 1).
Load the sample to HiTrap phenylsepharose, which is equilibrated with running buffer at 0.5 ml/min.
Wash the column using 20 ml of running buffer.
Elute myristoylated-ARF1 by decreasing salt concentration to 100 mM in 15 ml.
Pool fractions containing myristoylated-ARF1, as determined by 15% SDS-PAGE.
Dialyze against reaction buffer at 4°C, overnight.
Measure protein concentration using Bradford assay and stored at −80°C.
3.1.2. Purification of BARS
Culture BL21 cells containing BARS/pET-15b plasmid in 1 liter LB media supplemented with 100 μg/ml ampicillin at 37°C.
When O.D. is 0.6, induce the expression using 0.1 mM IPTG for 3 hours.
Harvest the cells using centrifugation at 6,000g, 4°C for 10 min.
Lyse the cells using 20 ml of lysis buffer at 4°C for 30 min, followed by sonication.
Centrifuge cell lysate at 30,000g at 4°C for 30 min.
Load the supernatant to 2 ml of Ni-NTA column.
Wash the column with 100 ml of washing buffer.
Elute recombinant BARS using elution buffer.
Pool fractions containing BARS, as determined by 10% SDS-PAGE.
Dialyze against reaction buffer at 4°C, overnight.
Measure protein concentration using Bradford assay and stored at −80°C.
3.2. Generating recombinant proteins using baculovirus expression
3.2.1. Purification of ARFGAP1 and cPLA2α
Use ArfGAP1 and cPLA2α in pVL1392 plasmids to prepare baculovirus using BestBac 2.0 Baculovirus co-transfection kit.
Infect 1:50 diluted P2 virus to 4.2 × 106 Sf9 cells/ml (2 liters) at 27°C.
Harvest the cells before viability drops below 80%.
Centrifuge at 500g for 20 min.
Lyse the cells using 100 ml of lysis buffer at 4°C for 1 hour.
Centrifuge cell lysate at 6,000g for 20 min.
Load the supernatant to 2 ml of Ni-NTA column.
Wash the column using washing buffer.
Elute recombinant proteins using elution buffer.
Pool fractions containing recombinant proteins determined by 15% SDS-PAGE.
Dialyze against reaction buffer at 4°C, overnight.
Measure protein concentration using Bradford assay, and stored at −80°C.
3.2.2. Purification of prenylated Cdc42
Use cdc42 in pFASTBAC THb plasmid to prepare Baculovirus infected insect cells (BIICS) using Bac-to-Bac Baculovirus Expression system.
Infect 1 ml of BIICS to 1.5 × 106 Sf9 cells/ml (2 liters) at 27°C.
Harvest the cells before viability drops below 80%.
Centrifuge at 500g for 20 min.
Lyse the cells using 100 ml of hypotonic buffer, followed by using dounce homogenizer (20 times).
Centrifuge cell lysate at 150,000g for 30 min.
Resuspend the pellet using 100 ml of TBS-M.
Centrifuge at 150,000g for 30 min.
Solubilize prenylated Cdc42 in the pellets using 50 ml of TBS-M containing 1% Triton X-100 at 4°C for 1 hour.
Centrifuge at 9,000g for 20 min.
Load the supernatant to 2 ml of Ni-NTA column.
Wash the column using washing buffer.
Elute prenylated Cdc42 using elution buffer.
Pool fractions containing prenylated Cdc42 determined by 15% SDS-PAGE.
Dialyze against reaction buffer at 4°C, overnight.
Measure protein concentration using Bradford assay, and stored at −80°C until use.
3.3. Purification of coatomer from tissue
Homogenize 100 g of fresh rat liver in 30 ml of homogenize buffer using polytron homogenizer (see Note 2).
Centrifuge at 10,000g for 30 min.
Take the supernatant for second centrifugation at 41,000g for 1 hour.
Dilute the supernatant with homogenize buffer to give 7 mg/ml.
Add saturated ammonium sulfate to the homogenate to give 35% ammonium sulfate concentration. Mix at 4°C for 20 min.
Centrifuge at 7,500g for 10 min.
Resuspend the pellet using 40 ml of dialysis buffer.
Dialyze against dialysis buffer at 4°C, overnight.
Centrifuge at 10,000g for 10 min.
Take the supernatant for second centrifugation at 41,000g for 45 min.
Filtrate the supernatant through 0.45 μm syringe filter.
Load the sample to DEAE-Sepharose FF, which is equilibrated with running buffer at 1 ml/min.
Elute the proteins up to 1M KCl by a linear gradient in 600 ml.
Pool the fractions containing coatomer determined by western blotting using anti-βCOP antibody.
Dilute the fractions with dilution buffer to give 200 mM KCl.
Load the mixture to HiTrap Q HP column, which is equilibrated with running buffer.
Elute coatomer using running buffer containing 500 mM KCl.
Pool the fractions containing the peak of protein.
Mix the sample with dilution buffer to give 200 mM KCl.
Load the sample to Resource Q column at 1 ml/min.
Elute coatomer using KCl gradient up to 1M in 60 ml.
Pool the fractions containing coatomer, determined by western blotting using anti-βCOP antibody.
Dialyze against reaction buffer at 4°C, overnight.
Measure protein concentration using Bradford assay, and stored at −80°C until use.
3.4. Golgi membrane preparation
Culture CHO cells in 4 roller bottles (250 ml media/bottle) until confluent.
Collect the cells using trypsin treatment, followed by centrifugation at 500g for 10 min.
Resuspend the pellet using cold ST buffer (10 ml ST buffer/ml pellet).
Centrifuge at 500g for 10 min.
Resuspend the pellet using 4 times volume of ST buffer.
Homogenize the cells using ball-bearing homogenizer with 22 μm ball for 10–12 passes (see Note 3).
Centrifuge the sample at 2,000g for 10 min.
Mix 12 ml of supernatant with 11 ml of 62% sucrose buffer and 230 μl of 100 mM EDTA, pH 7.4 (see Note 4).
Prepare sucrose gradient. Place 9 ml of 29% sucrose buffer and 15 ml of 35% sucrose buffer from the bottom of ultracentrifuge tubes using 30 ml syringe with blunt tip needle.
Gently load 12 ml of sample mixture at the bottom of sucrose gradient using 30 ml syringe with blunt tip needle.
Centrifuge at 110,000g for 2.5 hours.
Collect Golgi membrane at the 29% / 35% sucrose interface using 10 ml syringe with 18G needle gauge.
Measure protein concentration and then store in a liquid N2 tank.
3.5. COPI reconstitution system
-
1
Resuspend Golgi membrane (250 μg/200 μl) in low binding microcentrifuge tube using 1 ml of Traffic buffer, followed by centrifugation at 15,000g for 30 min.
-
2
Take out supernatant.
-
3
Wash Golgi membrane using 1 ml of 3 M KCl on ice/water bath for 5 min.
-
4
Centrifuge at 15,000g for 30 min.
-
5
Take out supernatant.
Stage I
-
6
Resuspend the pellets with 100 μl of traffic buffer and then add 50 μl of BSA solution.
-
7
Incubate the mixture at 37°C for 20 min.
-
8
Add ARF1 (6 μg/ml), coatomer (6 μg/ml), and GTP (2 mM).
-
9
Make up to 500 μl with traffic buffer.
-
10
Incubate the mixture on 37°C water bath for 15 min.
-
11
Stop reaction on ice/water bath for 5 min.
-
12
Place 10 μl of cushion buffer at the bottom of the tube (see Note 5).
-
13
Centrifuge at 15,000g for 20 min.
-
14
Take out supernatant.
Stage II
-
15
Gently resuspend the pellet using traffic buffer (see Note 6).
-
16
Add ARFGAP1 (6 μg/ml) and BARS (3 μg/ml) to generate COPI vesicles, or additionally with Cdc42 (6 μg/ml) and/or cPLA2α (3 μg/ml) to generate COPI tubules.
-
17
Make up to 50 μl with traffic buffer, followed by incubation at 37°C water bath for 20 min.
-
18
Stop reaction on ice/water bath for 5 min.
-
19
Place 5 μl of cushion buffer at the bottom of the tube.
-
20
Centrifuge at 15,000g for 10 min.
-
21
Gently collect the supernatant and pellet (see Note 7 and 8).
-
22
Determine COPI carrier formation by western blotting using anti-βCOP antibody.
3.6. Electron microscopy (EM)
EM analysis is performed to determine reconstituted COPI transport carriers (vesicle and tubule). To examine cargo sorting into the carriers, immunogold labeling approach is performed. In this case, Golgi membrane extracted from the cells expressing VSVG-Myc or VSVG-KDELR-Myc was used.
-
1
Load the sample (step 18 in 3.5 section) on grids for 10 min.
-
2
Fix the sample using 2% PFA/PBS for 10 min.
-
3
Incubate the grids using 1% BSA/PBS for 10 min.
-
4
Rinse the grids 7x using distilled water for 2 min.
-
5
Incubate the sample with uranyl acetate staining solution for 10 min.
-
6
Examine the reconstituted COPI transport carriers using TEM at 80 kV.
Immunogold labeling
-
7
Incubate the grids (step 3 in Section 3.6) with mouse anti-Myc antibody (9E10) for 1 hour.
-
8
Rinse the grids 3x using PBS for 5 min.
-
9
Incubate the grids using a rabbit anti-mouse antibody for 30 min.
-
10
Rinse the grids 3x using PBS for 5 min.
-
11
Incubate the grids using protein A conjugated with 10 nm gold particle for 30 min.
-
12
Rinse the grids 2x using PBS for 5 min.
-
13
Rinse the grids 7x using distilled water for 2 min.
-
14
Incubate the sample with uranyl acetate staining solution for 10 min.
-
15
Examine cargoes in the reconstituted COPI transport carriers using TEM at 80 kV.
4. NOTES
Mix the sample with 5M NaCl solution by the ratio between 2.94 of 5M NaCl and 2.06 of 0.1M NaCl.
Homogenize 10 g of tissue in each time.
Check how much cells were homogenized using Trypan Blue staining. 10% cells are usually homogenized by 12 passes.
Check mixture of sucrose concentration, which should be about 37%, using a refractometer.
15% sucrose in cushion buffer separates Golgi membrane from other components.
Vigorous pipetting may cause artificially generated vesicles.
Touching pellet might cause inconsistent results.
Supernatant contains a mixture of soluble proteins and vesicles, pellet contains Golgi membranes.
5. ACKNOWLEDGEMENTS
We thank Jian Li for discussions. This work was funded by grants from the National Institutes of Health to V.W.H. (R01GM058615), and also by the Basic Science Research Program of the National Research Foundation of Korea to S.-Y.P. (2014R1A6A3A03056673).
6. REFERENCES
- 1.Glick BS, Nakano A (2009) Membrane Traffic Within the Golgi Apparatus. Annual Review of Cell and Developmental Biology 25 (1):113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakano A, Luini A (2010) Passage through the Golgi. Current Opinion in Cell Biology 22 (4):471–478 [DOI] [PubMed] [Google Scholar]
- 3.Malhotra V, Serafini T, Orci L, Shepherd JC, Rothman JE (1989) Purification of a novel class of coated vesicles mediating biosynthetic protein transport through the Golgi stack. Cell 58 (2):329–336 [DOI] [PubMed] [Google Scholar]
- 4.Waters MG, Serafini T, Rothman JE (1991) ‘Coatomer’: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature 349 (6306):248–251 [DOI] [PubMed] [Google Scholar]
- 5.Donaldson JG, Cassel D, Kahn RA, Klausner RD (1992) ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc Natl Acad Sci USA 89 (14):6408–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson JG, Finazzi D, Klausner RD (1992) Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360 (6402):350–352 [DOI] [PubMed] [Google Scholar]
- 7.Helms JB, Rothman JE (1992) Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360 (6402):352–354 [DOI] [PubMed] [Google Scholar]
- 8.Cukierman E, Huber I, Rotman M, Cassel D (1995) The ARF1 GTPase-Activating Protein: Zinc Finger Motif and Golgi Complex Localization. Science 270 (5244):1999–2002 [DOI] [PubMed] [Google Scholar]
- 9.Hsu VW, Lee SY, Yang J-S (2009) The evolving understanding of COPI vesicle formation. Nat Rev Mol Cell Biol 10 (5):360–364 [DOI] [PubMed] [Google Scholar]
- 10.Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms JB, Rothman JE (1993) Hydrolysis of bound GTP by ARF protein triggers uncoating of Golgi-derived COP-coated vesicles. J Cell Biol 123 (6):1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SY, Yang J-S, Hong W, Premont RT, Hsu VW (2005) ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol 168 (2):281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang JS, Lee SY, Gao M, Bourgoin S, Randazzo PA, Premont RT, Hsu VW (2002) ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol 159 (1):69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J-S, Lee SY, Spano S, Gad H, Zhang L, Nie Z, Bonazzi M, Corda D, Luini A, Hsu VW (2005) A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J 24 (23):4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J-S, Zhang L, Lee SY, Gad H, Luini A, Hsu VW (2006) Key components of the fission machinery are interchangeable. Nat Cell Biol 8 (12):1376–1382 [DOI] [PubMed] [Google Scholar]
- 15.Yang J-S, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, Baldanzi G, Graziani A, Bourgoin S, Frohman MA, Luini A, Hsu VW (2008) A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol 10 (10):1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JS, Valente C, Polishchuk RS, Turacchio G, Layre E, Moody DB, Leslie CC, Gelb MH, Brown WJ, Corda D, Luini A, Hsu VW (2011) COPI acts in both vesicular and tubular transport. Nat Cell Biol 13 (8):996–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S-Y, Yang J-S, Schmider AB, Soberman RJ, Hsu VW (2015) Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature 521 (7553):529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balch WE, Dunphy WG, Braell WA, Rothman JE (1984) Reconstitution of the transport of protein between successive compartments of the golgi measured by the coupled incorporation of N-acetylglucosamine. Cell 39 (2):405–416 [DOI] [PubMed] [Google Scholar]