Abstract
Genomic analysis of cancer offers the hope of identifying new treatments or aiding in the selection of existing treatments. Rare leukemias pose additional challenges in this regard as samples may be hard to acquire and when found the underlying pathway may not be attractive to drug development since so few individuals are affected. In this case, it can be useful to identify common mutational overlap among subsets of rare leukemias to increase the number of individuals that may benefit from a targeted therapy. This chapter examines the current mutational landscape of LGL leukemia with a focus on STAT3 mutations, the most common mutation in LGL leukemia to date. We examined the linkage between these mutations and autoimmune symptoms and disorders, in cases of obvious and suspected LGL leukemia. We then summarized and compared mutations in a set of other rare leukemias that also have JAK/STAT signaling pathway activation brought about by genomic changes. These include T-cell acute lymphoblastic leukemia (T-ALL), T-cell prolymphocytic leukemia (T-PLL), cutaneous T-cell lymphoma (CTCL), select peripheral T-cell lymphoma (PTCL), and adult T-cell leukemia/lymphoma (ATLL). Though STAT3 activation is common in these leukemias, the way in which it is achieved, such as the activating cytokine pathway and/or the co-mutational background, is quite diverse.
Keywords: LGL, JAK/STAT, STAT3, T-ALL, T-PLL, CTCL, PTCL, ATLL
1. Large granular lymphocyte (LGL) leukemia.
Large granular lymphocyte (LGL) leukemia is a chronic disorder that typically results from clonal expansion of CD3+CD8+ cytotoxic T-lymphocytes (CTL) or, less frequently, CD3-natural killer (NK) cells that are localized to blood, spleen, marrow, and liver (1–5). LGL leukemia cells are constitutively activated cytotoxic cells characterized by profoundly dysregulated apoptosis, resulting in leukemic LGL survival (6,7). Clinical features include symptomatic neutropenia and anemia (8). Fatigue independent of anemia is becoming increasingly recognized as a worrisome symptom. A prominent manifestation is autoimmune disease; in particular, rheumatoid arthritis (RA) occurs in 20-30% of patients. LGL leukemia typically follows an indolent course, although studies report that 30 to 80% of patients will become symptomatic and need treatment. Retrospective studies of standard of care immunosuppressive regimens such as methotrexate, cyclophosphamide, or cyclosporine have demonstrated response rates of about 50% for each agent (5). However, these treatments are not curative (9).
1.1. STAT family members and function.
Signal transducer and activator of transcription (STAT) is a family of seven members: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6. The STATs have similar domain structure with STAT3 being depicted in Figure 1. STATs are cytoplasmic transcription factors that are activated by tyrosine phosphorylation and mediate cellular responses downstream of cytokine signaling. STAT3 regulates cell growth, differentiation and survival and is involved in cellular responses through cytokines including interferons, interleukin (IL)-6 family and the common gamma chain family ILs, and growth factors (10). Cytokine binding to receptors causes phosphorylation of Janus kinases (JAK), which in turn phosphorylate the intracellular tail of the receptor and create a docking site for STAT3. Once bound to the receptor, STAT3 becomes phosphorylated, dimerizes and translocates to the nucleus to bind directly to DNA, resulting in activation of multiple cellular pathways (11). Both activating and dominant negative germline mutations in STAT3 cause a number of immune disorders (12), underlining its importance in multiple immune cell types.
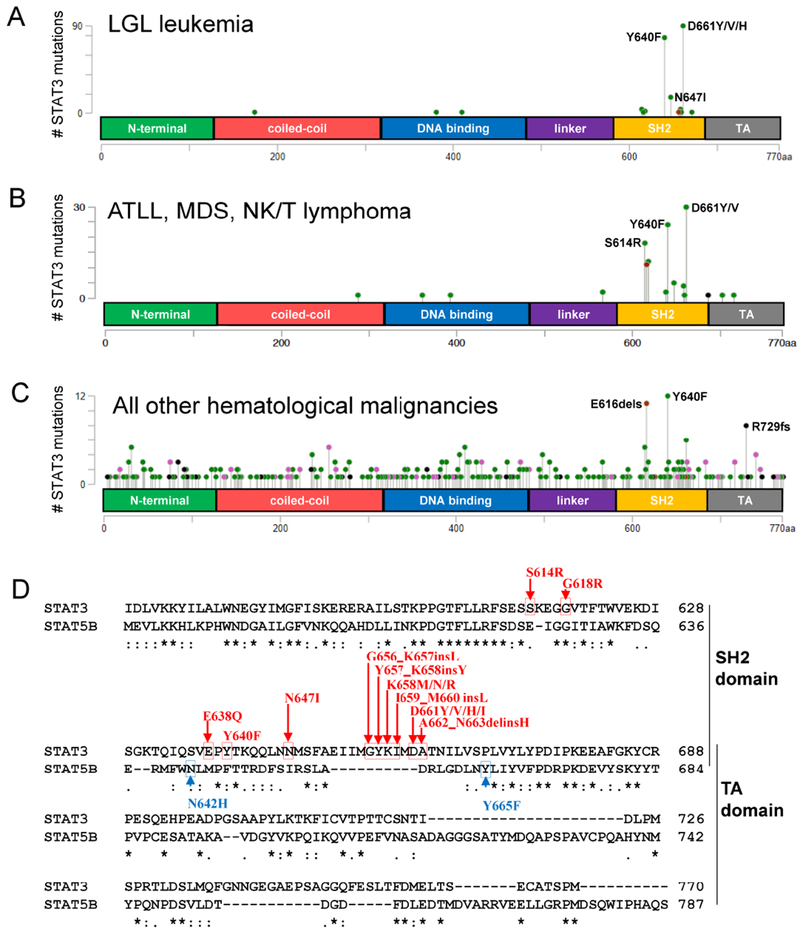
Figure 1. Somatic mutation frequencies of Homo sapiens STAT3 in hematological malignancies and comparison of STAT3 and STAT5B amino acid sequences.
STAT3 mutations specific to hematopoietic and lymphoid tissue were downloaded from COSMIC and cBio portal (STAT3 accession # NM_139276) and shown as lollipop plots. Domains are N-terminal, coiled-coil, DNA binding, linker, Src homology 2 (SH2), and transactivation (TA) (A-D). STAT3 mutation frequency is listed for (A) LGL leukemia (B) ATL, MDS, and NK/T lymphoma, and (C) all other hematological malignancies. Green, black, red, and pink circles indicate missense, truncation, in-frame, or other mutations, respectively. (D) STAT3 (alpha isoform) and STAT5B Homo sapiens amino acid sequences were retrieved from the NCBI Protein Database (accession # NP_644805 for STAT3, NP_036580 for STAT5B). The sequences were aligned using Clustal Omega, The C-terminal sequence including the SH2 and TA domains are shown. Asterisks indicate a fully-conserved amino acid at a position, while colons and periods indicate conserved and semi-conserved substitutions, respectively. STAT3 and STAT5B somatic mutations are annotated in red and blue, respectively. In two cases (Y640F and N647I) the STAT3 mutation creates the innate STAT5B amino acid.
1.2. STAT3 mutations in LGL leukemia and other hematological malignancies.
Koskela et al. showed a remarkable 40% prevalence of somatic STAT3 mutations in LGL leukemia (13). Since then, STAT3 mutations have been reported in 27-72% of LGL leukemia patient cohorts (14–16). Initially, all STAT3 mutations were found in the Src homology 2 (SH2) domain. Mutational hotspots include Y640 and D661, which account for over 70% of the mutations (Figure 1 A,D). Mutations in other regions including the DNA binding domain and coiled-coil domain have since been reported (17). Most mutations increase the activity of STAT3 in transcriptional reporter assays. STAT3 mutations have been discovered in other hematological malignancies including NK/T lymphoma, aggressive B-cell lymphoma, anaplastic large cell lymphoma, diffuse B cell lymphoma and peripheral T-cell lymphomas (18–21). Some of these disorders exhibit similar mutational hotspots to LGL leukemia as opposed to other malignancies (Figure 1B and 1C, respectively), suggesting shared functional relevance in lymphocyte biology. Interestingly, two STAT3 mutations (Y640F and N647I) generate the innate STAT5B amino acid at that same position, while many SH2 domain hotspot mutations (including D661) are located within a sequence that is absent in STAT5B (Figure 1D). The frequency and role of STAT5B mutations in LGL leukemia is described below. STAT3 mutation introduced in the marrow was not sufficient to initiate LGL leukemia in a mouse model (22), but perhaps a peripheral mutation model would be more appropriate. Work done in ALK- anaplastic large cell lymphoma showed that cytokine receptor signaling is still required for cells to survive even in the presence of JAK1/STAT3 mutations (18). Taken together, it appears that STAT3 mutations are not a fully transformative event.
1.3. STAT5B mutations in LGL leukemia.
While not as prevalent as mutations in STAT3, 2% of a cohort of 211 LGLL patients exhibited somatic STAT5B mutations. These mutations, N642H and Y665F (Figure 1D), increase the transcriptional activity of STAT5B by enhancing and prolonging STAT5B phosphorylation (23). The two N642H mutation patients, unlike the others in this study, had an aggressive disease course. N642H mutations were also found in aggressive lymphomas of NK and γδ T-cell origin. N642H appears to be a direct driver mutation in lymphoid malignancies that induces malignant transformation and expansion of CD8+ T cells (24). In the CD4+ T-cell subtype of LGL leukemia, four out of eight patients harbored these STAT5B mutations but displayed an indolent course (25).
1.4. Clinical associations with STAT3 Mutations in LGL leukemia
A prospective clinical trial demonstrated a complete response to treatment in all patients with Y640F mutations who completed methotrexate therapy. Only half of the patients with other STAT3 mutations and one quarter without measurable STAT3 mutation responded (9). Other studies suggest this sensitivity comes at a cost of increased symptoms associated with LGL leukemia. An analysis of mutation within immunophenotypes concluded that the CD8+/CD16+/CD56− LGL subtype is positively correlated with neutropenia and STAT3 mutational status, presumably through the increased Fas ligand present in mutant cells (26). Nearly 20% of LGL leukemia patients also suffer from RA (27). LGLL patients with STAT3 mutations more often presented with concomitant RA than their non-mutant counterparts (13,28,29). Additionally, the number of STAT3 mutations within the same patient correlates positively with RA in LGL leukemia (29). Felty syndrome is characterized by three symptoms in common with LGL leukemia patients: neutropenia, splenomegaly, and RA. Interestingly, 43% of sampled patients exhibit STAT3 mutations (30). Pure red cell aplasia (PRCA) is another blood disorder that can co-occur with LGL leukemia. It is characterized by anemia with a decreased erythroid production typically stemming from a T-cell mediated or autoantibody-dependent immune mechanism. In an Asian cohort of LGL leukemia patients Kawakami et al. found STAT3 mutations occurring in 77% of LGL leukemia associated PRCA patients which may indicate a direct involvement of mutated clones in disruption of red cell production (31).
1.5. STAT3 mutations in other disorders that may implicate LGL involvement
Kawakami et al. also found STAT3 mutations in three PRCA patients without LGL leukemia but did not find them in samples from (AA), myelodysplastic syndrome (MDS), or paroxysmal nocturnal hemoglobinuria (PNH) (31). An earlier study based on the same methods in an American cohort found that 2.5% of MDS and 7% of AA samples harbored STAT3 mutated clones (32). These mutations correlated with a better response to immunosuppressives in the AA patients and reduced marrow cellularity in those with MDS. Additionally, STAT3 sequencing of a large cohort of patients with leukemias, unexplained cytopenias, and other hematologic malignancies identified several previously unknown LGL leukemia cases with somatic STAT3 SH2 domain mutations that were originally misdiagnosed as MDS (33). Intriguingly, STAT3 mutations have been found in the intraepithelial lymphocyte cell lines of celiac disease patients (34). Celiac patients that develop LGL leukemia lose treatment sensitivity to gluten-free diet, which can be restored by treating the LGL leukemia (35). Given the sensitivity of Y640F and other STAT3 mutations to methotrexate, it is possible the frequent use of immunosuppressives in many disorders may be masking the presence of clonal LGL.
1.6. Beyond STAT3 mutations in LGL leukemia.
Even in the absence of STAT3 mutation, all LGL leukemia patients show constitutively active STAT signaling (36). Subsequent studies provide a mechanistic basis for these findings by identifying mutations in STAT regulators such as protein tyrosine phosphatases and upstream T-cell receptor signaling (17). However, activated STAT signaling remains largely unexplained in the ~60% of LGL leukemia patients without STAT3 mutations. There are a few possible explanations: 1) Sequencing has not been exhaustive. The largest published study details 19 exomes (37). As this was done across multiple LGL leukemia subtypes it is unlikely that this was sufficiently powered and thus commonly mutated genes may still remain to be identified. In addition, many screens for STAT3 mutations focused solely on the SH2 domain. 2) Remaining mutations may be common only at the pathway level, requiring careful review of existing LGL pathway literature (6,7,38,39) and those of other rare or common leukemias, to develop context. 3) Mutations may be present in regulatory regions. This requires whole genome sequencing combined with identification of regulatory regions in the LGL and normal cellular counterpart. 4) Mutation is not required. STAT activation is a hallmark of autoimmune disorders even without mutation. STAT3 mutation in LGL leukemia may be a later event to drive clonal dominance in a polyclonal disease (40), as opposed to being leukemogenic. It is hypothesized that an antigenic driven response is the initial activation event in LGL leukemia; the identity of this antigen is not yet known.
2. T-cell acute lymphoblastic leukemia (T-ALL)
T-ALL is an aggressive and immature lymphoid tumor that manifests both as a pediatric and adult malignancy. It is derived from T-cell precursors unlike most of the other T-cell malignancies (e.g. T-LGLL, ATLL and AITL) that derive from mature T cells. It results from accumulating genetic alterations that coordinately disrupt key genes and pathways responsible for the normal cell growth, survival and differentiation during thymocyte development (41).
2.1. Molecular sub-groups of T-ALL
Based on gene expression profiles and immunophenotypes that reflect thymocyte developmental stages, T-ALL is clinically divided into distinct molecular sub-groups (41). These are: a) early T-lineage progenitor (ETP) leukemia with developmental arrest at early stages of T-cell differentiation. They lack expression of CD4 and CD8 and share similarities with hematopoietic stem cells and myeloid progenitor cells in their global transcriptional profile. It is a high-risk subtype of T-ALL. Genetically, ETP has a lower prevalence of NOTCH1 mutations or CDKN2A deletions and is instead associated with; mutations that activate signaling factors (NRAS, FLT3) or disrupt function of key developmental transcription factors (RUNX1, GATA3, ETV6) and epigenetic regulators (DNMT3A and EZH2) (41). Aberrant activation of the JAK/STAT signaling pathway has also been seen (42). b) Early cortical, which corresponds to the early stages of cortical thymocyte maturation and is CD1a+CD4+CD8+. These have the highest prevalence of NOTCH1 mutations and deletions of the CDKN2A locus with frequent activation of TLX1 and TLX3. c) Late cortical thymocyte (CD4+CD8+CD3+) that typically show activation of the TAL1 oncogene (43). Mutations are usually non-random and appear in specific combinations. Different sub-types of T-ALL (based on expression of specific transcription factors) show cooperation of frequent oncogenic events. For example, mutations in IL7R/JAK are positively associated with mutations/deletions of PRC2 complex (EZH2, SUZ12, EED) and WT1 mutations are most prevalent in TLX3 expressing T-ALL (44). Similarly, cooperation exists between TAL1 expression and PTEN inactivation during T-cell transformation (45). Therefore, initiating lesions leading to ectopic transcription factor expression in T-ALL sensitize the cells to alterations of specific pathways.
2.2. Mutational Spectrum of Adult T-ALL
The most predominant oncogenic event in adult T-ALL is aberrant activation of NOTCH1 signaling through NOTCH1 activating mutations, rearrangements, or loss of its negative regulators. High levels of NOTCH1 in hematopoietic progenitors promotes T-cell transformation. Negative regulator of NOTCH1, FBXW7, is also commonly mutated (44). Second in prevalence is 9p21 deletion or altered methylation status affecting cell cycle regulators CDKN2A and CDKN2B (41). Dysregulation of the cell cycle and growth pathways is common with genes like PTEN, NRAS, and PI3KCA also frequently mutated. T-ALL sub-groups demonstrate an ectopic expression of T-cell specific transcription factors that could function as oncogenes (e.g. TAL1, TLX1, TLX3, HOXA, LMO2), of which the most frequent aberrantly expressed is TAL1 due to chromosomal rearrangements, super enhancer mutations and expression changes (44). Additional transcription factors mutated in at least 5% of adult T-ALL cases include MYB, ETV6, WT1, RUNX1, BCL11B, CALM-AF10, MLL-ENL, CNOT3 and SET-NUP214. Epigenetic factors such as PHF6, DNMT3A, EZH2, KDM6A/UTX, EED, and SUZ12 are also commonly mutated (44).
T-ALL demonstrates dysregulated IL7R-mediated JAK-STAT signaling. Aberrant JAK signaling was initially linked with T-ALL in the context of t(9;12)(p24;p13), a rare rearrangement encoding a constitutively active kinase fusion oncoprotein, ETV6–JAK2 (41). Later, activating mutations in IL7R, JAK1, JAK3, and/or STAT5 were observed in both pediatric and adult T-ALL cases (44). Other mechanisms of activation of IL7R signaling are also known, such as loss-of-function mutations in DNM2 that result in increased presentation of IL7R on the cell surface of thymocytes (46). Similarly, a rare translocation event involving ZEB2 increases IL7R expression and STAT5 activation, promoting T-ALL cell survival in a mouse model (47). Inactivation of PTPN2 (seen in 6% of T-ALL cases) also results in enhanced JAK1 signaling and transformation (48).
3. T-cell prolymphocytic leukemia (T-PLL)
T-PLL is a rare, mostly aggressive neoplasm of mature, predominantly CD4+, T-lymphocytes. The vast majority show cytogenetic abnormalities and complex karyotypes implicating a marked genome instability (49). The hallmark of T-PLL is the rearrangement of chromosome 14 (~80% of patients harbor these changes) involving genes of the T-cell receptor (TCR) complex, leading to overexpression of the proto-oncogene TCL1 (50). Approximately 20% (51) of patients harbor a t(X;14) translocation activating the TCL1A paralog MTCP1 (located on Xq28) and both genes promote malignant transformation of T-lymphocytes in transgenic mice (52,53). Abnormalities of chromosome 8 occur in two-thirds of patients and include trisomy of 8q (MYC amplification) or rearrangements with 8p12. Deletions in 12p13 leading to haplo-insufficiency of the CDKN1B gene may also play a role in the pathogenesis of T-PLL (54).
3.1. Mutational landscape
T-PLL displays recurrent mutations of genes in diverse pathways such as DNA repair (e.g. CHEK2), proteasomal degradation (e.g. FBXW10) and mutational activation of the IL2RG-JAK1-JAK3-STAT5B axis (49,54). Mutations are also observed in DNA damage responsive genes ATM (73%) and TP53 (14%) (55). A subset of 23 T-PLL cases exhibited recurrent mutations in epigenetic regulators i.e. EZH2 (13%), TET2 (17%), and BCOR (9%) (56). Novel recurrent mutations were also identified in chromatin regulatory genes KDM6A and KDM6B, which emphasize the importance of epigenetic dysregulation in the pathogenesis of T-PLL (54).
The majority of T-PLL patients (71%) have recurrent mutations in JAK-STAT pathway genes including JAK3 (29%), STAT5B (7%), JAK1 (6%) and IL2RG (2%) (50). The IL2RG-JAK1-JAK3-STAT5B mutations lead to increased transcriptional activation of STAT5, constitutive STAT5 hyper-phosphorylation and oncogenic transformation (54). STAT5 inhibition impairs cell proliferation and reduces viability of JAK-mutated cell lines and primary T-PLL samples with JAK-STAT pathway mutations (49). Expression based studies also confirm JAK-STAT pathway activation in T-PLL. Gene expression profiling studies indicate significant overexpression of several STAT5B target genes as well as five-fold downregulation of DUSP4 (50). DUSP4 deficiency induces STAT5 hyperactivation, enhances IL-2 signaling and promotes CD4+ T-cell proliferation in mice (57,58), which suggests an alternate mechanism to activate JAK-STAT.
4. Cutaneous T cell lymphoma (CTCL)
CTCL is a malignancy of skin-homing T-lymphocytes. While nodal non-Hodgkin lymphoma is usually of B-cell origin, 65% of primary cutaneous lymphomas are characterized by the proliferation of mature CD4+ T-helper cells (59). Mycosis fungoides (MF) and its leukemic variant Sézary syndrome (SS) represent the most common types (approximately two-thirds) of CTCL (60). CTCL displays constitutive activation of the TCR pathway.
4.1. Oncogenic drivers in CTCL
CTCL exhibits recurrent deletions of 10q and 17p; amplifications of 8q (containing MYC) and 17q and frequent deletions of TP53 and CDKN2A (61,62). However, target genes underlying many of the recurrent copy number variations (CNVs) are not well known. Mutated genes associated with CTCL include CDKN2A, CDKN2B, DLEU2, KDM6A, TP53, TP63, and VAV1 (63,64). A genomic landscape study of 40 cases of CTCL identified frequent deletions and damaging mutations in chromatin-modifying genes (ARID1A (62.5%), CTCF (12.5%) and DNMT3A (42.5%)) (61). A recent study identified recurrent mutations in CSNK1A1 (NF-kB signaling), PTPRN2 and RLTPR (T-cell receptor signaling) and RARA (T-cell differentiation)(64).
Deregulation of STAT signaling has been implicated in the pathogenesis of CTCL. STAT5 and STAT3 are constitutively activated in early and late stages of CTCL, respectively. Activation of STAT5 is believed to occur through IL-2, IL-7 and IL-15 signaling via JAK1 and JAK3 kinases. STAT5 activation increases expression of the oncogenic miR-155, which targets and decreases STAT4 expression. Thus, STAT4 expression (required for Th1 differentiation) is lost at later stages of the disease, resulting in a concomitant switch from Th1 (T helper) to Th2 malignant phenotype (65). STAT6 is often upregulated during this switch. In contrast, development of IL-21 autocrine signaling activates STAT3 in the advanced stages of the disease (65). In advanced stages, activation of STAT3 and STAT5 may become completely cytokine-independent and be driven via constitutively active JAK1 and JAK3 kinases (65).
4.1.1. Mycosis fungoides (MF):
The most recurrent oncogenic drivers in MF are large structural genomic alterations that create a complex and heterogeneous landscape of inter- and intra-chromosomal rearrangements (gain of 7q36, 7q21-7q22; loss of 5q13, 9p21) (63). Recurrently rearranged genes are ARHGAP26, ATXN1, CLEC16A, ELF1, EYS, RBPJ, RPS6KA3, SLC24A2, and SSH2 (66), which like all rearrangements seem to have no unifying theme of pathway involvement. However, translocation events in MF lead to deletion of tumor suppressors involved in commonly deregulated pathways in MF patients (ARID1A, CDKN2A/B, PTPRC, and STK11). These recurrent deletions affect cell cycle control, chromatin regulation and the PI3K pathways. Recurrent deletion of JAK-STAT signaling inhibitors i.e. HNRNPK and SOCS1, leads to an up-regulation of the JAK-STAT pathway (66).
4.1.2. Sézary syndrome (SS):
The pattern of chromosomal alterations observed in SS differs markedly from MF, as evidenced by the CNVs listed below. The genomic landscape of SS reveals frequent disruption of epigenetic modifiers, cell cycle regulators and subversion of signaling modules involved in TCR signaling. Chemokine and cytokine signaling pathways that are critical for T-cell activation, survival and differentiation are also mutated with recurrent gain-of-function mutations targeting PLCG1 being observed (9%)(67). 19% of MF and SS patients had PLCG1 mutations in one cohort (68). Recurrent loss-of-function alterations targeting the epigenetic machinery include the ARID/SMARC family chromatin remodeling complex, histone methyltransferases (MLLs, SETD1A/B) and demethylase (KDM6B), DNA methyltransferase family members and TET1/2/3 genes. These results emphasize the substantial contribution of epigenetic deregulation in the pathogenesis of SS. CNVs that lead to loss of genes include ARID1A, DNMT3A, A20/TNFAIP3, CAAP, CDKN2A-CDKN2B, PTEN, FAS, ZEB1, NFKB2, ATM, USP28, RB1, NCOR1, TP53 and E2A (62). Recurrent CNVs that lead to gain of genes are TOX, MYC, PRKCQ and STAT5B. Some of the most frequently mutated (somatic) genes in SS are ATM, CARD11, DNMT3A, KMT2C, MLL2, JAK3, TET1, TET2, SOCS7, STAT3, STAT5A, STAT5B and POT1 (62). Fusion transcripts and miRNA expression changes are also common events in SS. Several miRNAs are overexpressed in CTCL, such as miRNA155, which promotes inflammation and cancer development through STAT signaling (69,70). On the other hand, down-regulated miRNAs (e.g. miR342), may play a role in the pathogenesis of SS by inhibiting apoptosis (71).
Like MF, SS demonstrates aberrant JAK-STAT pathway activation primarily through direct activation as opposed to loss of inhibitors in MF. Somatic gain-of-function mutations affecting JAK1, JAK3, STAT3 and STAT5B (altogether 11%) (67) have been reported and elevated STAT4 expression seems to be a reliable diagnostic marker in SS (65).
5. Peripheral T-cell lymphoma (PTCL)
PTCL are a class of T-cell non-Hodgkin lymphomas (NHLs), sub-grouped into: nodal angio-immunoblastic T-cell lymphoma (AITL); anaplastic lymphoma kinase (ALK) positive anaplastic large-cell lymphoma (ALCL) (nodal); ALK− anaplastic large-cell lymphoma (ALK− ALCL) lacking ALK expression with distinctive molecular features (now classified as a separate entity); and adult T-cell lymphoma (ATL) associated with human T-lymphotropic virus type I (HTLV-1) (leukemic group). In general, chromatin modifiers and molecular players in the CD28/T-cell receptor signaling pathways are frequently mutated in PTCLs.
5.1. Angioimmunoblastic T-cell lymphoma (AITL)
is derived from malignant transformation of T-follicular helper (Tfh) cells and is the second most commonly diagnosed PTCL in the Western countries (more common in Europe). Patients with AITL are usually very immunosuppressed and often develop EBV viremia that can lead to highly aggressive forms of diffuse large B-cell lymphoma. The rudimentary immune-phenotype of a Tfh is CD3+, CD4+, and CD10+. CD30 expression is seen in 20% of AITL cases and has recently become therapeutically relevant (72).
5.1.1. Molecular drivers in AITL
AITL is characterized by high frequency co-occurring mutations in epigenetic modifiers, like TET2, IDH2, and DNMT3A (73), in contrast to myeloid disorders in which IDH2 and TET2 mutations are thought to be mutually exclusive. RHOA, a guanosine triphosphatase involved in cytoskeleton reorganization, is mutated in about 60% of AITL cases. RHOA G17V mutations are commonly seen with TET2 mutations, suggesting a multi-hit process in AITL pathogenesis (72). Large scale sequencing studies associated RHOA mutational status and other recurrent mutations in AITL (74). Therefore, three potentially distinct AITL lymphomagenic pathways have been described; a) the classical pathway with RHOA G17V mutation, associated with mutations in TET2, DNMT3A, IDH2 and CD28. b) the alternative pathway with VAV1 mutations or potentially unidentified mutations in the Rho family of GTPases or their regulatory proteins c) unknown mutations in pathways regulating Tfh differentiation (74). AITL with wild-type RHOA, shows recurrent mutations in TET2 (60.7%), CD28 (18.6%), DNTM3A (17.9%), PLCG1 (14.0%), IDH2 (13.8%), VAV1 (11.6%), FYN (7.8%) and STAT3 (7.0%) (74). Recently, gain-of-function mutations in the T-cell receptor have been described (75). In addition, a few cases of AITL exhibit karyotypic abnormalities including trisomy of chromosomes 3 and 5 (72).
5.2. ALK+ and ALK− anaplastic large-cell lymphoma (ALK+ ALCL)
ALK+ ALCL is more common in North America and shows strong and uniform expression of CD30, a member of the TNFR superfamily. It is characterized by recurrent chromosomal translocations involving the anaplastic lymphoma kinase (ALK) gene. Of note, ALK activation is considered necessary and sufficient for ALCL tumorigenesis and its inhibition is key for the therapeutic treatment of ALK+ ALCL. ALK− ALCL is immuno-phenotypically similar to ALK+ ALCL, but lacks ALK expression. ALK− ALCL is slightly more prevalent in Europe (76).
5.2.1. Genomic landscape of ALK+ ALCL
Approximately 80% of the ALK+ ALCL cases show a cytogenetic translocation t(2;5), fusing the ALK gene (2p23) to the NPM gene (5q35). This results in overexpression and constitutive activation of a chimeric ALK fusion protein (77). Other translocations leading to constitutive activation of ALK tyrosine kinase are TPM3-ALK (13% of cases), ATIC-ALK (1%), TFG-ALK (1%), and <1% of ALK fusions are with CLTC, MSN, TPM4, MYH9, RNF213 and TRAF. Constitutively active ALK fusion proteins promote tumorigenesis through the activation of multiple signal transduction pathways such as PLC-γ, PI3K/AKT, STAT3, STAT5, mTOR and MEK/ERK. Comparative genomic hybridization identified secondary chromosomal imbalances in 58% of ALK+ ALCL, including gains of 6q, 7p, 17p, and 17q24 and losses of 4q, 13q21, and 11q14 (77). Gene expression profiling studies of ALK+ ALCL show overexpression of genes (compared to ALK− ALCL) such as BCL6, PTPN12, CEBPB, and SERPINA1 (78).
Crosstalk between the JAK-STAT pathway and NPM-ALK fusion proteins occurs in ALK+ ALCL. Constitutive activation of STAT3 is consistently detected and significantly contributes to the pathogenesis. The NPM-ALK fusion protein activates STAT3 by direct phosphorylation (79). JAK3, which also binds to NPM-ALK, is highly activated in the primary tumors and contributes to the activation of STAT3 activation (80). Other members of the JAK/STAT signaling family, such as JAK2 and STAT5, exert similar mechanistic interactions and pathogenic effects (80). NPM-ALK constitutively activates STAT5, which is essential to lymphomagenesis, but STAT5 activation is not restricted to NPM-ALK fusion cases alone (81).
5.2.2. Genomic landscape of ALK− ALCL
Commonly mutated or deleted genes in ALK-ALCL are PRDM1/BLIMP1, TP53, and CSMD2, with the loss of TP53 and PRDM1/BLIMP1 having a less favorable outcome (19). Chromosomal fusion events are common, like IRF4-DUSP-22 (30% of cases) and TP63-TBL1RXR1 (8%) (77). DUSP22 is a phosphatase involved in suppression of IL-6-induced STAT3 activation and TCR signaling down-regulation in reactive T cells (76). The other gene fusions in ALK− ALCL involve ROS1 and TYK2 kinases (NKB2-ROS1, NCOR2-ROS1, NFKB2-TYK2 and PABPC4-TYK2), which activate STAT3 or JAK1 (77). ERBB4 overexpression (24% of the cases) is driven by reactivation of the long tandem repeats from endogenous retroviruses located in ERBB4 introns (82). Chromosomal imbalances are observed in 65% of cases with gains of 1q, 6p21, and 7p and losses of 6q21 (PRDM1), 13q, and 17p13 (TP53) (83).
JAK1 and STAT3 are the most recurrently mutated genes in ALK− ALCL (18%) and lead to STAT3 activation (76). Constitutive STAT3 phosphorylation is seen in nearly half (47%) of ALK− ALCL (84) including JAK1/STAT3 non-mutated cases, indicating alternative mechanisms of pathway activation (76).
6. Adult T-Cell Leukemia/Lymphoma (ATLL)
ATLL is a rare and aggressive T-cell malignancy of CD4+CD25+ cells resulting from infection with human T-lymphotropic virus type-1 (HTLV-1). ATLL is more common in Asia. The HTLV-1 proteins Tax and HBZ are the two main viral components contributing to malignancy (85). Tax is a transcriptional activator of viral and host genes that dysregulates cell cycle control and DNA repair genes, which ultimately results in genome instability and transformation (86). Tax is not expressed in all ATLL cases, most likely because it is immunogenic (87), whereas HBZ is not immunogenic and is expressed in all cases of ATLL (88). HBZ is sufficient to induce T cell lymphomas in transgenic mice (89).
6.1. Genomic landscape of ATLL
The alterations in ATLL overlap significantly with the HTLV-1 Tax interactome and are enriched for TCR and NF-κB signaling (gain of function or focal amplifications), T-cell trafficking (CCR4 or CCR7 mutated in 40% of cases), immune-surveillance (HLA-A, HLA-B and B2M) and other T-cell-related pathways (86,90). More than half of the cases harbor activating PLCG1 and/or PRKCB mutations leading to PKCβ activation (86). There is a predominance of activating mutations in CARD11, VAV1, IRF4, FYN, CCR4 and CCR7 as well as gene fusions (CTLA4-CD28 and ICOS-CD28). Frequent intragenic deletions involve IKZF2, CARD11 and TP73 (90). Frequent loss-of-function mutations and deletions affect negative regulators of TCR-NF-κB signaling.
Recurrent gene mutations and CNVs are seen in other signal-transducing molecules that are related to activation of TCR-NF-κB signaling such as JAK-STAT (JAK3, STAT3 and PTPN1), NOTCH (NOTCH1, ATXN1 and ZFP36L2), PI3K-AKT (PIK3CD and INPP4B) and casein kinase 2 (CSNK2A1 and CSNK2B) (90). Among other commonly affected genes are TP53, CDKN2A and POT1 (DNA repair and telomere maintenance); TET2, DNMT3A and IDH2 (epigenetic regulation); EP300, SETD2 and KDM6A (histone modification) and ARID2 (chromatin remodeling) (90). Potentially mediated by these epigenetic factors, ATLL genomes (40%) are characterized by extensive CpG island DNA hyper-methylation, known as the CpG island methylator phenotype (CIMP), which is associated with transcriptional silencing and aggressive phenotypes (86). Hyper-methylated and silenced genes are highly enriched for C2H2 zinc-finger genes.
There is also an ATLL-specific “epigenetic code” through PRC2-mediated H3K27me3, which is frequently reprogrammed at several genes in leukemic cells (87). Abnormal downregulation of genes is observed at early stages of disease and is explained by H3K27me3 accumulation. These genes are enriched in biological processes like transcriptional regulation and histone modification. Specifically, upon H3K27me3 gain, KDM6B is downregulated (87). Additionally, one of the key characteristics of ATLL is the global downregulation of microRNA (87).
STAT3 and STAT5 are constitutively activated in primary ATLL cells (91,92), mediated by the constitutive phosphorylation of JAK proteins. Increased expression of IL-2Rα and IL-15Rα are common features. The activation of these cytokine pathways and subsequent downstream JAK-STAT signaling are thought to play an important role in transformation by HTLV-1 (93–95).
7. DISCUSSION
The rare leukemias discussed here were selected to demonstrate the varying genomic alterations that can ultimately lead to hyperactivation of the JAK/STAT pathway (Fig. 2). LGL leukemia stands alone in that specific point mutations in STAT3 are highly suggestive of LGL leukemia. A diagnosis should be ruled out if they are found in sequencing results from cytopenic or myelodysplastic patients. They also implicate LGL leukemia or LGL cells in autoimmune disorders. The mutations indicate a clonal population, but without a substantial increase in LGL (<0.5 x 109/L), a diagnosis is unlikely to be made. We speculate this could indicate a past LGL expansion that has contracted or potential for future expansions that should be monitored. Constitutive activation of the JAK-STAT pathway is a unifying central mechanism for most T-lineage rare leukemias. AITL is an exception that is centrally driven by RHOA and mutant epigenetic modifiers. For leukemias where constitutive STAT activation is known, the mode of activation of the JAK-STAT signaling pathway seems to differ. Direct JAK or STAT mutation is seen, as well as the loss of negative regulators such SOCS, HNPRK and PTPs. Additionally, the cytokine pathway upstream of STATs is specific to each leukemia and is derived from the signaling dependencies of the normal cell counterpart. For example, it is mostly IL-15 mediated for T-LGLL (mostly CD8+ cells), IL-2, IL-7, IL-15 or IL-21 for CTCL (CD4+CD8−), IL7R for T-ALL (CD4−CD8−/CD4+CD8+), IL2RG for T-PLL (mostly CD4+) and ALK fusion driven activation of STAT for ALK+ ALCL (CD30+). This makes STAT proteins an attractive drug target due to their common role in lymphocytes. Multi-hit therapies for the STAT pathway will likely require specific combinations for each leukemia with one common therapy targeting STAT proteins and other therapies unique to the cytokine signaling specific to each leukemia.
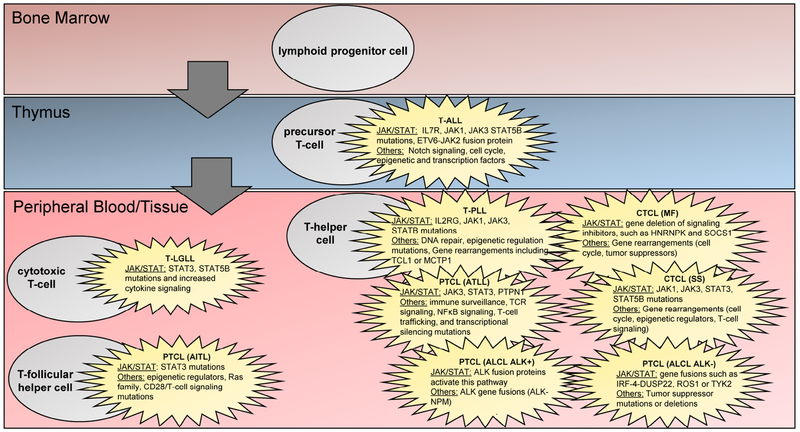
Figure 2. Rare leukemia cell lineage highlighting genomic differences from normal cell counterparts.
A simplified T-cell hematopoietic lineage is shown, along with annotations indicating which cells represent the normal counterpart of a particular leukemia. Within each cancer cell, genomic aberrations that are known to cause increased JAK/STAT signaling along with other known mutations or gene rearrangements are listed.
Each rare leukemia exhibits a recurrent driver gene/mechanism in the pathogenesis of the malignancy (Fig. 2). The most predominant oncogenic event in the pathogenesis of LGL leukemia is the constitutive activation of STAT3, aberrant NOTCH1 activation is found in T-ALL, and the over-expression of proto-oncogene TCL1 is found in T-PLL. CTCL has large chromosomal events leading to deletion of tumor suppressors like CDKN2A. AITL has mutations in RHOA and epigenetic modifiers. ALK gene fusions in ALK+ ALCL and other mechanisms in ALK−ALCL lead to STAT3 activation. The loss of negative regulators of TCR-NFκB signaling and alterations in T-cell trafficking is important in ATLL. Epigenetic modifiers play a prominent role in T-PLL, ETP subtype of T-ALL, CTCL, and PTCLs like AITL and ATLL. Targeting each of these pathways in addition to JAK/STAT may represent a strategy more effective than monotherapy, particularly in regard to eventual resistance
Practice Points.
There are a number of clinical associations with STAT mutation in LGL leukemia. STAT3 Y640F mutations have been shown to predict response to methotrexate in a small series of patients. This observation needs validation as it has implications for selecting initial best therapy for patients with LGL leukemia. STAT5B mutations in CD8+ TLGL may indicate an aggressive clinical course. The presence of STAT mutations in other disorders may implicate LGL in pathogenesis in perhaps sub-clinical or forme fruste involvement of LGL leukemia.
Rare leukemias tend to share mutations in those pathways that maintain importance across the cell types of origin. Prospective trials that target shared mutations may benefit from combining these rare subtypes together.
Research agenda.
There is an urgency to sequence as many rare leukemia patient samples as possible to identify molecular subtypes across leukemias.
Future sequencing must extend into whole genome sequencing and epigenetics to explain gaps between observed frequency of mutation and constitutive activity of pathways such as JAK/STAT.
Acknowledgements
LGL leukemia research in the Loughran lab is supported by the National Cancer Institute of the National Institutes of Health under award number R01CA178393, the Bess Family Charitable Fund, the LGL Leukemia Foundation, and a generous anonymous donor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Thomas P. Loughran, Jr. is on the Scientific Advisory Board and has stock options for both Keystone Nano and Bioniz Therapeutics. There are no conflicts of interest with the work presented in this manuscript.
Bibliography
- 1.Sanikommu SR, Clemente MJ, Chomczynski P, Afable MG, Jerez A, Thota S, et al. Clinical features and treatment outcomes in large granular lymphocytic leukemia (LGLL). Leuk Lymphoma. 2018;59(2):416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melenhorst JJ, Eniafe R, Follmann D, Molldrem J, Kirby M, El Ouriaghli F, et al. T-cell large granular lymphocyte leukemia is characterized by massive TCRBV-restricted clonal CD8 expansion and a generalized overexpression of the effector cell marker CD57. Hematol J. 2003;4(1):18–25. [DOI] [PubMed] [Google Scholar]
- 3.Loughran TP. Clonal diseases of large granular lymphocytes. Blood. 1993. July 1;82(1): 1–14. [PubMed] [Google Scholar]
- 4.Lamy T, Moignet A, Loughran TP. LGL leukemia: from pathogenesis to treatment. Blood. 2017. March 2;129(9):1082–1094. [DOI] [PubMed] [Google Scholar]
- 5.Lamy T, Loughran TP. How I treat LGL leukemia. Blood. 2011. March 10;117(10):2764–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinway SN, LeBlanc F, Loughran TP. The pathogenesis and treatment of large granular lymphocyte leukemia. Blood Rev. 2014. May;28(3):87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leblanc F, Zhang D, Liu X, Loughran TP. Large granular lymphocyte leukemia: from dysregulated pathways to therapeutic targets. Future Oncol. 2012. July;8(7):787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loughran TP, Kadin ME, Starkebaum G, Abkowitz JL, Clark EA, Disteche C, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985. February;102(2):169–175. [DOI] [PubMed] [Google Scholar]
- 9.Loughran TP, Zickl L, Olson TL, Wang V, Zhang D, Rajala HLM, et al. Immunosuppressive therapy of LGL leukemia: prospective multicenter phase II study by the Eastern Cooperative Oncology Group (E5998). Leukemia. 2015. April;29(4):886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siveen KS, Sikka S, Surana R, Dai X, Zhang J, Kumar AP, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014. April;1845(2):136–154. [DOI] [PubMed] [Google Scholar]
- 11.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013. January 10;368(2):161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014. August;46(8):812–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koskela HLM, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012. May 17;366(20):1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M, He R, Feldman AL, Viswanatha DS, Jevremovic D, Chen D, et al. STAT3 mutation and its clinical and histopathologic correlation in T-cell large granular lymphocytic leukemia. Hum Pathol. 2018;73:74–81. [DOI] [PubMed] [Google Scholar]
- 15.Fasan A, Kern W, Grossmann V, Haferlach C, Haferlach T, Schnittger S. STAT3 mutations are highly specific for large granular lymphocytic leukemia. Leukemia. 2013. July;27(7): 1598–1600. [DOI] [PubMed] [Google Scholar]
- 16.Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012. October 11;120(15):3048–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson EI, Rajala HLM, Eldfors S, Ellonen P, Olson T, Jerez A, et al. Novel somatic mutations in large granular lymphocytic leukemia affecting the STAT-pathway and T-cell activation. Blood Cancer J. 2013. December 6;3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Zhang Y, Petrus MN, Xiao W, Nicolae A, Raffeld M, et al. Cytokine receptor signaling is required for the survival of ALK-anaplastic large cell lymphoma, even in the presence of JAK1/STAT3 mutations. Proc Natl Acad Sci USA. 2017. April 11;114(15):3975–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015. April 13;27(4):516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohgami RS, Ma L, Merker JD, Martinez B, Zehnder JL, Arber DA. STAT3 mutations are frequent in CD30+ T-cell lymphomas and T-cell large granular lymphocytic leukemia. Leukemia. 2013. November;27(11):2244–2247. [DOI] [PubMed] [Google Scholar]
- 21.Ohgami RS, Ma L, Monabati A, Zehnder JL, Arber DA. STAT3 mutations are present in aggressive B-cell lymphomas including a subset of diffuse large B-cell lymphomas with CD30 expression. Haematologica. 2014. July;99(7):e105–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta A, Yan D, Hutchison RE, Mohi G. STAT3 mutations are not sufficient to induce large granular lymphocytic leukaemia in mice. Br J Haematol. 2018;180(6):911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajala HLM, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013. May 30;121(22):4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham HTT, Maurer B, Prchal-Murphy M, Grausenburger R, Grundschober E, Javaheri T, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest. 2018. January 2;128(1):387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson EI, Tanahashi T, Sekiguchi N, Gasparini VR, Bortoluzzi S, Kawakami T, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016. November 17;128(20):2465–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teramo A, Barila G, Calabretto G, Ercolin C, Lamy T, Moignet A, et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget. 2017. September 22;8(37):61876–61889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gazitt T, Loughran TP. Chronic neutropenia in LGL leukemia and rheumatoid arthritis. Hematology Am Soc Hematol Educ Program. 2017. December 8;2017(1): 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valori M, Jansson L, Kiviharju A, Ellonen P, Rajala H, Awad SA, et al. A novel class of somatic mutations in blood detected preferentially in CD8+ cells. Clin Immunol. 2017;175:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajala HLM, Olson T, Clemente MJ, Lagstrom S, Ellonen P, Lundan T, et al. The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia. Haematologica. 2015. January;100(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savola P, Bruck O, Olson T, Kelkka T, Kauppi MJ, Kovanen PE, et al. Somatic STAT3 mutations in Felty syndrome: an implication for a common pathogenesis with large granular lymphocyte leukemia. Haematologica. 2018;103(2):304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami T, Sekiguchi N, Kobayashi J, Imi T, Matsuda K, Yamane T, et al. Frequent STAT3 mutations in CD8+ T cells from patients with pure red cell aplasia. Blood Adv. 2018. October 23 ;2(20): 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jerez A, Clemente MJ, Makishima H, Rajala H, Gomez-Segui I, Olson T, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013. October 3;122(14):2453–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan EA, Lee MN, DeAngelo DJ, Steensma DP, Stone RM, Kuo FC, et al. Systematic STAT3 sequencing in patients with unexplained cytopenias identifies unsuspected large granular lymphocytic leukemia. Blood Adv. 2017. September 26;1(21):1786–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ettersperger J, Montcuquet N, Malamut G, Guegan N, Lopez-Lastra S, Gayraud S, et al. Interleukin-15-Dependent T-Cell-like Innate Intraepithelial Lymphocytes Develop in the Intestine and Transform into Lymphomas in Celiac Disease. Immunity. 2016. September 20;45(3):610–625. [DOI] [PubMed] [Google Scholar]
- 35.Malamut G, Meresse B, Verkarre V, Kaltenbach S, Montcuquet N, Duong Van Huyen J-P, et al. Large granular lymphocytic leukemia: a treatable form of refractory celiac disease. Gastroenterology. 2012. December;143(6):1470–1472.e2. [DOI] [PubMed] [Google Scholar]
- 36.Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001. February;107(3):351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppe A, Andersson EI, Binatti A, Gasparini VR, Bortoluzzi S, Clemente M, et al. Genomic landscape characterization of large granular lymphocyte leukemia with a systems genetics approach. Leukemia. 2017. February 7;31(5):1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah MV, Zhang R, Loughran TP. Never say die: survival signaling in large granular lymphocyte leukemia. Clin Lymphoma Myeloma. 2009;9 Suppl 3:S244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Shah MV, Yang J, Nyland SB, Liu X, Yun JK, et al. Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci USA. 2008. October 21;105(42):16308–16313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerr CM, Clemente MJ, Chomczynski PW, Przychodzen B, Nagata Y, Adema V, et al. Subclonal STAT3 mutations solidify clonal dominance. Blood Adv. 2019. March 26;3(6):917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016. July 25;16(8):494–507. [DOI] [PubMed] [Google Scholar]
- 42.Maude SL, Dolai S, Delgado-Martin C, Vincent T, Robbins A, Selvanathan A, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015. March 12;125(11): 1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002. February;1(1):75–87. [DOI] [PubMed] [Google Scholar]
- 44.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017. March 2;129(9):1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendes RD, Sarmento LM, Cante-Barrett K, Zuurbier L, Buijs-Gladdines JGCAM, Povoa V, et al. PTEN microdeletions in T-cell acute lymphoblastic leukemia are caused by illegitimate RAG-mediated recombination events. Blood. 2014. July 24;124(4):567–578. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay CS, Brown FC, Collett M, Saw J, Chiu SK, Sonderegger SE, et al. Loss-of-function mutations of Dynamin 2 promote T-ALL by enhancing IL-7 signalling. Leukemia. 2016. April 27;30(10): 1993–2001. [DOI] [PubMed] [Google Scholar]
- 47.Goossens S, Radaelli E, Blanchet O, Durinck K, Van der Meulen J, Peirs S, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015. January 7;6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleppe M, Soulier J, Asnafi V, Mentens N, Hornakova T, Knoops L, et al. PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood. 2011. June 30;117(26):7090–7098. [DOI] [PubMed] [Google Scholar]
- 49.Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014. August 28;124(9):1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson EI, Putzer S, Yadav B, Dufva O, Khan S, He L, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32(3):774–787. [DOI] [PubMed] [Google Scholar]
- 51.Stern MH, Soulier J, Rosenzwajg M, Nakahara K, Canki-Klain N, Aurias A, et al. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene. 1993. September;8(9):2475–2483. [PubMed] [Google Scholar]
- 52.Gritti C, Dastot H, Soulier J, Janin A, Daniel MT, Madani A, et al. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood. 1998. July 15;92(2):368–373. [PubMed] [Google Scholar]
- 53.Virgilio L, Lazzeri C, Bichi R, Nibu K, Narducci MG, Russo G, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA. 1998. March 31;95(7):3885–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laribi K, Lemaire P, Sandrini J, Baugier de Materre A. Advances in the understanding and management of T-cell prolymphocytic leukemia. Oncotarget. 2017. November 28;8(61): 104664–104686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stengel A, Kern W, Zenger M, Perglerova K, Schnittger S, Haferlach T, et al. Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Genes Chromosomes Cancer. 2016. January;55(1):82–94. [DOI] [PubMed] [Google Scholar]
- 56.Lopez C, Bergmann AK, Paul U, Murga Penas EM, Nagel I, Betts MJ, et al. Genes encoding members of the JAK-STAT pathway or epigenetic regulators are recurrently mutated in T-cell prolymphocytic leukaemia. Br J Haematol. 2016. April;173(2):265–273. [DOI] [PubMed] [Google Scholar]
- 57.Hsiao W-Y, Lin Y-C, Liao F-H, Chan Y-C, Huang C-Y. Dual-Specificity Phosphatase 4 Regulates STAT5 Protein Stability and Helper T Cell Polarization. PLoS One. 2015. December 28;10(12):e0145880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang C-Y, Lin Y-C, Hsiao W-Y, Liao F-H, Huang P-Y, Tan T-H. DUSP4 deficiency enhances CD25 expression and CD4+ T-cell proliferation without impeding T-cell development. Eur J Immunol. 2012. February;42(2):476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sokolowska-Wojdylo M, Olek-Hrab K, Ruckemann-Dziurdzmska K. Primary cutaneous lymphomas: diagnosis and treatment. Postepy Dermatol Alergol. 2015. October 29;32(5):368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005. May 15;105(10):3768–3785. [DOI] [PubMed] [Google Scholar]
- 61.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015. September;47(9):1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cristofoletti C, Narducci MG, Russo G. Sezary Syndrome, recent biomarkers and new drugs. Chin Clin Oncol. 2019. February;8(1):2. [DOI] [PubMed] [Google Scholar]
- 63.van Doorn R, van Kester MS, Dijkman R, Vermeer MH, Mulder AA, Szuhai K, et al. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood. 2009. January 1;113(1):127–136. [DOI] [PubMed] [Google Scholar]
- 64.Park J, Yang J, Wenzel AT, Ramachandran A, Lee WJ, Daniels JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood. 2017. September 21;130(12):1430–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Netchiporouk E, Litvinov IV, Moreau L, Gilbert M, Sasseville D, Duvic M. Deregulation in STAT signaling is important for cutaneous T-cell lymphoma (CTCL) pathogenesis and cancer progression. Cell Cycle. 2014;13(21):3331–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bastidas Torres AN, Cats D, Mei H, Szuhai K, Willemze R, Vermeer MH, et al. Genomic analysis reveals recurrent deletion of JAK-STAT signaling inhibitors HNRNPK and SOCS1 in mycosis fungoides. Genes Chromosomes Cancer. 2018. October 25;57(12):653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiel MJ, Sahasrabuddhe AA, Rolland DCM, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015. September 29;6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaque JP, Gomez-Lopez G, Monsalvez V, Varela I, Martinez N, Perez C, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014. March 27;123(13):2034–2043. [DOI] [PubMed] [Google Scholar]
- 69.Kopp KL, Ralfkiaer U, Gjerdrum LMR, Helvad R, Pedersen IH, Litman T, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013. June 15;12(12):1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood. 2011. November 24; 118(22):5891–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood. 2010. August 19;116(7): 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lunning MA, Vose JM. Angioimmunoblastic T-cell lymphoma: the many-faced lymphoma. Blood. 2017. March 2;129(9):1095–1102. [DOI] [PubMed] [Google Scholar]
- 73.Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014. February 27;123(9):1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willemsen M, Abdul Hamid M, Winkens B, Zur Hausen A. Mutational heterogeneity of angioimmunoblastic T-cell lymphoma indicates distinct lymphomagenic pathways. Blood Cancer J. 2018. January 17;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vallois D, Dobay MPD, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016. September 15;128(11):1490–1502. [DOI] [PubMed] [Google Scholar]
- 76.Mereu E, Pellegrino E, Scarfo I, Inghirami G, Piva R. The heterogeneous landscape of ALK negative ALCL. Oncotarget. 2017. March 14;8(11):18525–18536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montes-Mojarro IA, Steinhilber J, Bonzheim I, Quintanilla-Martinez L, Fend F. The pathological spectrum of systemic anaplastic large cell lymphoma (ALCL). Cancers (Basel). 2018. April 4;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamant L, Giuriato S. Gene-expression profiling of systemic anaplastic large-cell lymphoma reveals differences based on ALK status and two distinct morphologic ALKS subtypes. 2007;109(5):10. [DOI] [PubMed] [Google Scholar]
- 79.Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002. February 7;21(7):1038–1047. [DOI] [PubMed] [Google Scholar]
- 80.Amin HM, Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007. October 1;110(7):2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nieborowska-Skorska M, Slupianek A, Xue L, Zhang Q, Raghunath PN, Hoser G, et al. Role of signal transducer and activator of transcription 5 in nucleophosmin/ anaplastic lymphoma kinase-mediated malignant transformation of lymphoid cells. Cancer Res. 2001. September 1;61(17):6517–6523. [PubMed] [Google Scholar]
- 82.Scarfo I, Pellegrino E, Mereu E, Kwee I, Agnelli L, Bergaggio E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood. 2016. January 14;127(2):221–232. [DOI] [PubMed] [Google Scholar]
- 83.Zeng Y, Feldman AL. Genetics of anaplastic large cell lymphoma. Leuk Lymphoma. 2016;57(1):21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khoury JD, Medeiros LJ, Rassidakis GZ, Yared MA, Tsioli P, Leventaki V, et al. Differential Expression and Clinical Significance of Tyrosine-phosphorylated STAT3 in ALK•/•• and ALK•/••• Anaplastic Large Cell Lymphoma. :9. [PubMed] [Google Scholar]
- 85.Matsuoka M, Jeang KT. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene. 2011. March 24;30(12):1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vicente C, Cools J. The genomic landscape of adult T cell leukemia/lymphoma. Nat Genet. 2015. November;47(11): 1226–1227. [DOI] [PubMed] [Google Scholar]
- 87.Yamagishi M, Fujikawa D, Watanabe T, Uchimaru K. HTLV-1-Mediated Epigenetic Pathway to Adult T-Cell Leukemia-Lymphoma. Front Microbiol. 2018. July 24;9:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006. January 17;103(3):720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Satou Y, Yasunaga J-I, Zhao T, Yoshida M, Miyazato P, Takai K, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011. February 10;7(2):e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J-I, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015. November;47(11): 1304–1315. [DOI] [PubMed] [Google Scholar]
- 91.Bellon M, Lu L, Nicot C. Constitutive activation of Pim1 kinase is a therapeutic target for adult T-cell leukemia. Blood. 2016. May 19;127(20):2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takemoto S, Mulloy JC, Cereseto A, Migone TS, Patel BK, Matsuoka M, et al. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci USA. 1997. December 9;94(25):13897–13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mariner JM, Lantz V, Waldmann TA, Azimi N. Human T cell lymphotropic virus type I Tax activates IL-15R alpha gene expression through an NF-kappa B site. J Immunol. 2001. February 15;166(4) :2602–2609. [DOI] [PubMed] [Google Scholar]
- 94.Tendler CL, Greenberg SJ, Blattner WA, Manns A, Murphy E, Fleisher T, et al. Transactivation of interleukin 2 and its receptor induces immune activation in human T-cell lymphotropic virus type I-associated myelopathy: pathogenic implications and a rationale for immunotherapy. Proc Natl Acad Sci USA. 1990. July;87(13):5218–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang TT, Yang J, Zhang Y, Zhang M, Dubois S, Conlon KC, et al. IL-2 and IL-15 blockade by BNZ-1, an inhibitor of selective γ-chain cytokines, decreases leukemic T-cell viability. Leukemia. 2019. May;33(5):1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]