Highlights
-
•
Three 3CLpro mutations (N25S, A252S or K260N) were identified from a FIP cat treated with the protease inhibitor (GC376) for prolonged duration.
-
•
The 3CLpros containing the mutations were generated and their susceptibility to protease inhibitors were determined in the enzyme assay.
-
•
Only N25S showed a small change in susceptibility to GC376, which may explain the absence of clinical resistance to long-term treatment in the trial.
Keywords: Feline infectious peritonitis virus, 3C-like protease, Antivirals, Genetic barrier, Feline coronavirus, Resistance
Abstract
Feline infectious peritonitis (FIP) is a highly fatal disease caused by a virulent feline coronavirus in domestic and wild cats. We have previously reported the synthesis of potent coronavirus 3C-like protease (3CLpro) inhibitors and the efficacy of a protease inhibitor, GC376, in client-owned cats with FIP. In this study, we studied the effect of the amino acid changes in 3CLpro of feline coronavirus from a feline patient who received antiviral treatment for prolonged duration. We generated recombinant 3CLpro containing the identified amino acid changes (N25S, A252S or K260 N) and determined their susceptibility to protease inhibitors in the fluorescence resonance energy transfer assay. The assay showed that N25S in 3CLpro confers a small change (up to 1.68-fold increase in the 50% inhibitory concentration) in susceptibility to GC376, but other amino acid changes do not affect susceptibility. Modelling of 3CLpro carrying the amino acid changes was conducted to probe the structural basis for these findings. The results of this study may explain the observed absence of clinical resistance to the long-term antiviral treatment in the patients.
1. Introduction
Coronaviruses are a group of diverse RNA viruses that can cause a number of diseases in animals and humans. Coronaviruses have a single-stranded, positive-sense RNA genome and belong to the family Coronaviridae, which are further classified into four genera, Alpha-, Beta-, Gamma-, and Deltacoronaviruses (King et al., 2011). Feline coronaviruses belong to the genus Alphacoronavirus and are the causative agent of enteritis in cats. Feline enteric coronavirus infections are quite common in multi-cat environments, such as shelters, rescues and catteries. Since more than a third of the affected cats chronically shed viruses in the stool and feline coronaviruses are relatively stable in the environment, it is difficult to eliminate feline coronaviruses from the affected cat populations (Addie et al., 2019; Pedersen et al., 2008). Cats infected with feline coronaviruses may show mild to severe enteritis, but they usually recover without complications. However, a small number of cats may develop feline infectious peritonitis (FIP), a highly fatal, systemic disease, following enteric coronavirus infection (Pedersen, 2009, 2014). The detailed mechanism of FIP development is still elusive, but it is believed that FIP arises in individual cats through altered viral tropism that allows viral replication in the macrophages (Barker et al., 2013; Chang et al., 2012; Licitra et al., 2013; Pedersen et al., 2009, 2012) in the presence of inadequate cellular immunity of the affected cats (Pedersen, 2014). FIP are commonly described as wet or dry forms based on the presence or absence of effusion, respectively, but these forms may switch as the disease progresses in some cats. The clinical signs of FIP are variable and depend on the affected organs and may include fever, jaundice, effusion, weight loss or neurological signs and ocular lesions (Pedersen, 2014). Although FIP is almost invariably fatal once cats develop clinical signs, there is no commercially available treatment licensed for FIP with only supportive treatment being available.
Following viral entry into host cells, coronaviruses produce viral polyproteins composed of non-structural viral proteins, which are subsequently cleaved into mature, functional viral proteins by virally-encoded 3C-like protease (3CLpro) and papain-like protease. The released viral proteins form the replicase-transcriptase complex, which is required for RNA replication and transcription of the subgenomic RNAs, thus inhibition of viral proteases blocks viral replication. We have previously reported the synthesis of protease inhibitors for feline coronavirus (Kim et al., 2012a, 2013; Kim et al., 2015), and showed the efficacy of one of the inhibitors, GC376, in cats with experimentally induced FIP (Kim et al., 2016) and naturally-occurring FIP in a field trial (Pedersen et al., 2018). We have also reported the amino acid changes (N25S, A252S and K260 N) in the 3CLpro of feline coronavirus collected from a patient who was treated with GC376 but did not show clinical resistance to multiple rounds of treatment in the field trial (Pedersen et al., 2018).
In this study, we assessed the effects of the amino acid changes in the feline coronavirus 3CLpro against GC376 in florescence resonance energy transfer (FRET) assay by generating 3CLpro carrying the amino acid changes. We also conducted modelling of 3CLpro carrying the amino acid changes and compared them with wild-type (pre-treatment) 3CLpro to probe the structural basis for these findings. The results showed that N25S, but not A252S and K260 N, confers a statistically significant but small reduction in susceptibility to GC376 with up to 1.68-fold increase in the 50% inhibitory concentration (IC50), compared to that of wild-type 3CLpro. Interestingly, none of these amino acid changes in 3CLpro decreased susceptibility to another 3CLpro inhibitor (NPI52) that shares the similar structure with GC376. The results of this study may explain the observed absence of clinical resistance to the long-term antiviral treatment in the patients in the field trial.
2. Materials and methods
2.1. Patient samples and compound
One and ½ year-old castrated male Himalayan cat (CT10) that was presented with a wet form of FIP was treated with GC376 in a field trial during 2016–2017 (Pedersen et al., 2018). He was diagnosed with FIP based on physical examination, signalment, clinical history, laboratory test results on blood and effusion prior to the enrollment in the trial (Pedersen et al., 2018). At admission, he exhibited lethargy, inappetance and abdominal effusion with enlarged colonic lymph node and had been treated with antibiotics and prednisolone prior to entry into the trial. The first round of treatment (GC376 at 10 to 30 mg/kg/injection, twice daily) lasted for 9 weeks. One month after discontinuation of the treatment, he relapsed with fever and lethargy and was re-treated (GC376 at 7.5 to 15 mg/kg/injection, twice daily) for 12 weeks. During these treatments, he remained in clinical remission. However, about 5 weeks following the discontinuation of the second treatment, he relapsed again with typical intra-abdominal lesions without neurological signs and was euthanized (Pedersen et al., 2018). The time between the first day of treatment and euthanasia is 236 days. On necropsy, tissue samples were collected and prepared for sequence analysis of feline coronavirus 3CLpro (Pedersen et al., 2018). Comparison of the amino acid sequences of feline coronavirus 3CLpro from the pre-treatment ascites sample and the postmortem tissue samples has previously revealed amino acid changes of N25S and K260 N in the kidney and N25S, A252S and K260 N in the lung and spleen (Pedersen et al., 2018) (Fig. 1 ). Synthesis of 3CLpro inhibitors GC376 and NPI52 was previously reported by our group (Kim et al., 2012a).
Fig. 1.
Comparison of 3CLpro amino acid sequences of feline coronavirus from the pre-treatment (MO, macrophages) and postmortem tissue samples (lung, kidney and spleen) from CT10. The amino acid changes in the postmortem tissue samples are indicated by asterisks. The C-terminal residues 294–302 are not included in the alignment.
2.2. Expression of feline coronavirus 3CLpro carrying the amino acid changes
The full-length sequence of feline coronavirus 3CLpro from the pre-treatment ascites sample (designated as wild type, WT) was amplified by RT-PCR and cloned into a pET28+ vector (GenScript, Piscataway, NJ). To construct recombinant 3CLpros containing the identified amino acid changes, single (N25S, A252S or K260N), double (N25S and K260N) or triple (N25S, A252S and K260N) mutations were introduced into WT 3CLpro gene using a QuickChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions. The WT and mutant clones were verified by sequence analysis, and the clones were transformed into BL21 cells (Invitrogen, Carlsbad, CA) for expression. Protein expression was induced in Luria Bertani broth with 1 mM isopropyl β-D-thiogalactopyranoside for 4–6 h at 37 °C. The 3CLpros were purified using Ni-NTA affinity columns (QIAGEN, Valencia, CA) and stored at −80 °C.
2.2.1. FRET assay
The activities of the expressed WT and mutant 3CLpros were determined in the FRET assay as described previously (Kim et al., 2012a). Each 3CLpro in 25 μl of assay buffer was mixed with 25 μl of assay buffer containing fluorogenic substrate (dabacyl-KTSAVLQ/SGFRKME-edans) in a 96-well imaging microplate. The assay buffer consists of 120 mM NaCl, 4 mM DTT, 50 mM HEPES and 30% Glycerol at pH 6.0. Following the incubation of the plate at 37 °C for 30 min, fluorescence was measured on a fluorescence microplate reader (FLx800, Biotek, Winnooski, VT). Background (substrate only) was subtracted from the fluorescence readings and the readings were compared among the 3CLpro using one-way analysis of variance (ANOVA) with Tukey’s post hoc test in GraphPad Prism version 6.07 (GraphPad Software, La Jolla, CA).
After confirmation of the activity of the recombinant 3CLpros, inhibition assays using GC376 or NPI52 were conducted as previously described (Kim et al., 2016, 2012a). Serial dilutions of GC376 or NPI52 in assay buffer were incubated with 25 μl of assay buffer containing 3CLpro at 37 °C for 30 min. The mixture was then added to 25 μl of assay buffer containing fluorogenic substrate in a 96 well plate. The plate was further incubated at 37 °C for 30 min and the fluorescence readings were measured on a florescence microplate reader. Relative fluorescence was calculated by subtracting background fluorescence from each fluorescence reading. The IC50 of each compound was calculated by non-linear regression analysis (four-parameter variable slope) using GraphPad Prism software. At least four independent experiments were conducted to determine the IC50 values, and a two-tailed student’s t-test was used for statistical comparison between the IC50 values.
2.3. Multiple sequence alignment and three-dimensional homology structure of feline coronavirus 3CLpro
To determine whether the residues at positions 25, 252 and 260 in 3CLpro are conserved, the 3CLpro amino acid sequences of 44 strains of feline coronaviruses in the GenBank were used for multiple sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (McWilliam et al., 2013). Three-dimensional structures of WT and mutant 3CLpro were built by the EasyModeller program (version 4.0) (Kuntal et al., 2010) using the crystal structures of transmissible gastroenteritis virus (TGEV) 3CLpro (PDB accession number: 4F49) and feline coronavirus (1146 strain) 3CLpro (PDB accession number: 4ZRO) as templates. The 3CLpros of TGEV and FIPV have high structural homology with 92.05–93.71% amino acid sequence identity. The crystal structure of TGEV 3CLpro bound with GC376 [Protein Data Bank number (PDB): 4F49] was previously reported by us (Kim et al., 2012a). Therefore, the modelled 3CLpro structures were superimposed with the crystal structures of TGEV 3CLpro-GC376 and feline coronavirus 3CLpro for comparative purposes (Kim et al., 2012b). The Ring 2.0 web server (Piovesan et al., 2016) was used to study the interactions between the residues at positions 25, 252 and 260 and other residues in feline coronavirus 3CLpro.
3. Results
3.1. The effects of amino acid changes in 3CLpro against GC376
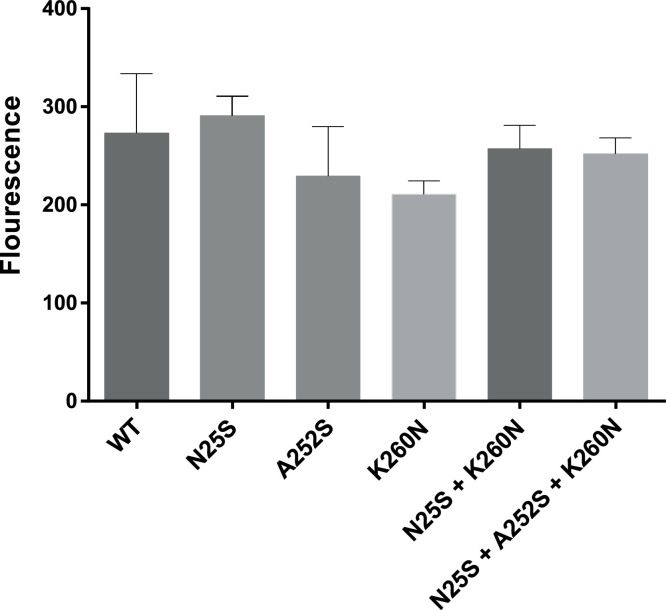
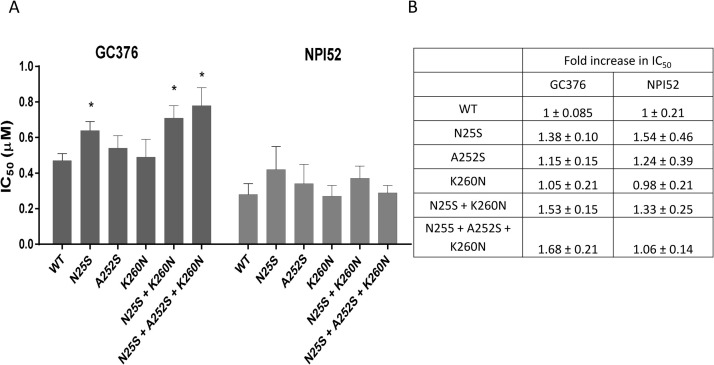
The activities of the recombinant WT and mutant 3CLpros in the FRET assay are shown in Fig. 2 . There was no statistically significant difference in the activity among the 3CLpros, which indicates that the amino acid changes do not affect the function of 3CLpro compared to that of WT 3CLpro (Fig. 2). When the inhibitory activity of GC376 was determined against the WT and mutant 3CLpros, it was found that single amino acid change of N25S, but not A252S or K260 N, led to a statistically significant increase in the IC50 value of GC376, compared to that of WT 3CLpro (Fig. 3 A and B). However, the increase in the IC50 value was marginal with a fold-change of 1.38 in IC50. The 3CLpro carrying a double mutation (N25S and N260 N) or a triple mutation (N25S, A252S and K260 N) also significantly increased the IC50 value of GC376 by 1.53 and 1.68-fold, respectively, compared to WT 3CLpro. However, the IC50 values of GC376 against 3CLpro with a double or a triple mutation were not statistically different from that of 3CLpro carrying a single mutation of N25S (Fig. 3A and B). These findings indicate that A252S and K260 N mutations do not affect the susceptibility of 3CLpro against GC376. NPI52 is a tripeptidyl compound that shares a similar backbone structure with GC376 and has a similarly potent activity against feline coronavirus as GC376 in the FRET assay and cell culture (Kim et al., 2015). However, unlike those of GC376, there was no difference in the IC50 values of 3CLpro with WT, single, double or triple mutations against NPI52 (Fig.3A and B).
Fig. 2.
Activity of the recombinant wild-type and mutant 3CLpros in the FRET assay. Each 3CLpro was mixed with fluorogenic substrate in assay buffer in a 96-well imaging microplate. After incubation at 37 °C for 30 min, fluorescence was measured on a fluorescence microplate reader. Each bar represents the mean ± standard error of mean of fluorescence units from at least four independent FRET assays. There was no statistical difference (P < 0.05) between the activities of recombinant 3CLpros. WT indicates wild-type (pre-treatment) 3CLpro. N25S, A252S and K260 N indicates 3CLpros carrying a single mutation. N25S + K260 N and N25S + A252S + K260 N indicate double and triple mutation, respectively.
Fig. 3.
Effects of GC376 and NPI52 on recombinant wild-type and mutant 3CLpros in the FRET assay. Each 3CLpro was incubated with serial dilutions of GC376 or NPI52 for 30 min at 37 °C, and then fluorogenic substrate was added to the mixture. Following incubation for 30 min, fluorescence readings were measured on a fluorescence microplate reader for the determination of IC50 values. (A) Each bar represents the mean ± standard error of the means from at least three independent FRET assays. (B) Fold increases in IC50 values of mutant 3CLpro over those of WT 3CLpro are shown in the table. WT indicates wild-type (pre-treatment) 3CLpro. N25S, A252S and K260 N indicates 3CLpros carrying a single mutation. N25S + K260 N and N25S + A252S + K260 N indicate double and triple mutations, respectively. Asterisks indicate statistically significant difference compared to WT (P < 0.05).
3.2. Multiple sequence alignment and three-dimensional homology structure of feline coronavirus 3CLpro
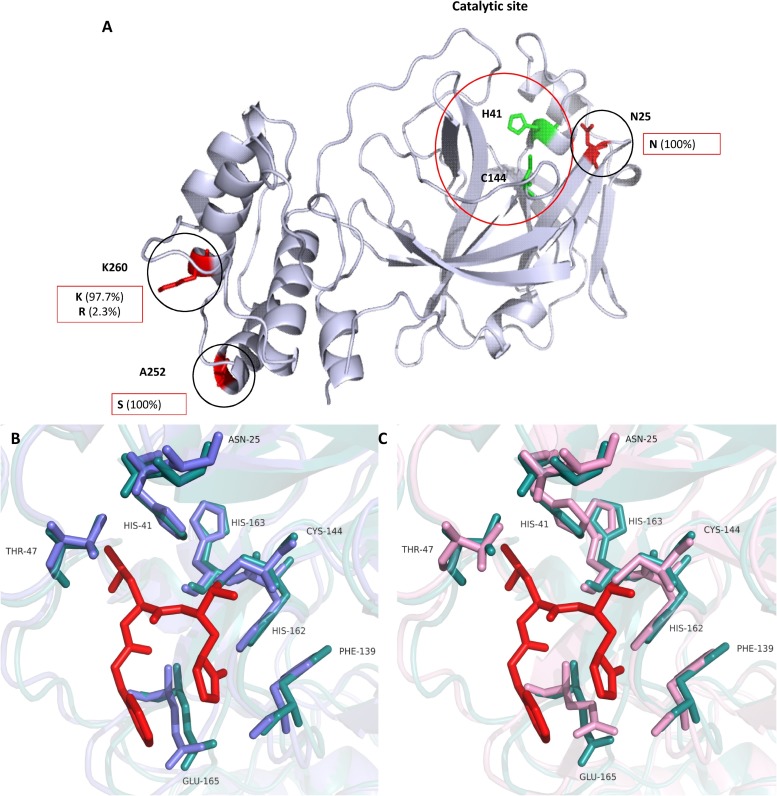
The crystal structure of feline coronavirus 3CLpro (PDB number: 4ZRO) modified to show N25S, A252S and K260 N was analyzed using the PyMol molecular graphics system, Version 1.8 (Schrodinger LLC, Cambridge, MA) (DeLano, 2010) (Fig.4 A). The N25 is adjacent to the catalytic site, while A252 and K260 are located in the domain III near the C terminus. Multiple sequence analysis of feline coronavirus strains whose 3CLpro sequences are available in the PubMed database showed that N25 and A252 are invariably conserved in all feline coronavirus strains and K260 is also highly conserved with only one strain having Arg (R) at the same position (Fig. 4A). Multiple sequence alignment of feline coronavirus 3CLpro also revealed that the residues that form hydrogen bond interactions with GC376 in TGEV 3CLpro (T47, F139 H162, H163 and E165) (Kim et al., 2012a) are conserved in all feline coronaviruses (Data not shown). Superposition of the homology structure of CT10 3CLpro (WT) and the crystal structures of TGEV-GC376 (PDB accession number: 4F49) and feline coronavirus 3CLpro (PDB accession number: 4ZRO) showed that these residues (T47, F139 H162, H163 and E165) are aligned closely in the catalytic site of 3CLpro (Fig. 4B and C), suggesting that these residues likely form hydrogen bonds with GC376 in feline coronavirus 3CLpro. In addition, N25 does not seem to form interactions with GC376 but is located close to H41 (Fig. 4B and C). C144 and H41 are the catalytic residues in the active site of 3CLpro.
Fig. 4.
The structure of feline coronavirus 3CLpro and amino acid substitutions. (A) A ribbon representation of a feline coronavirus 3CLpro (PDB accession: 4ZRO) was prepared using the PyMol program. The catalytic residues (H41 and C144) are shown in green, and N25, A252 and K260 are shown in red. The amino acids found at the position of 25, 252 and 260 in 3CLpro among feline coronavirus strains whose 3CLpro amino acid sequence are available in the PubMed database are shown in red boxes. (B) Active site of the superimposed crystal structures of TGEV 3CLpro-GC376 (PDB accession: 4F49) (teal) and feline coronavirus 3CLpro (PDB accession: 4ZRO) (slate). (C) Active site of the superimposed crystal structure of TGEV 3CLpro-GC376 (PDB accession: 4F49) (teal) and a homology model of CT10 WT 3CLpro (pink). GC376 (red) is shown as a red stick (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
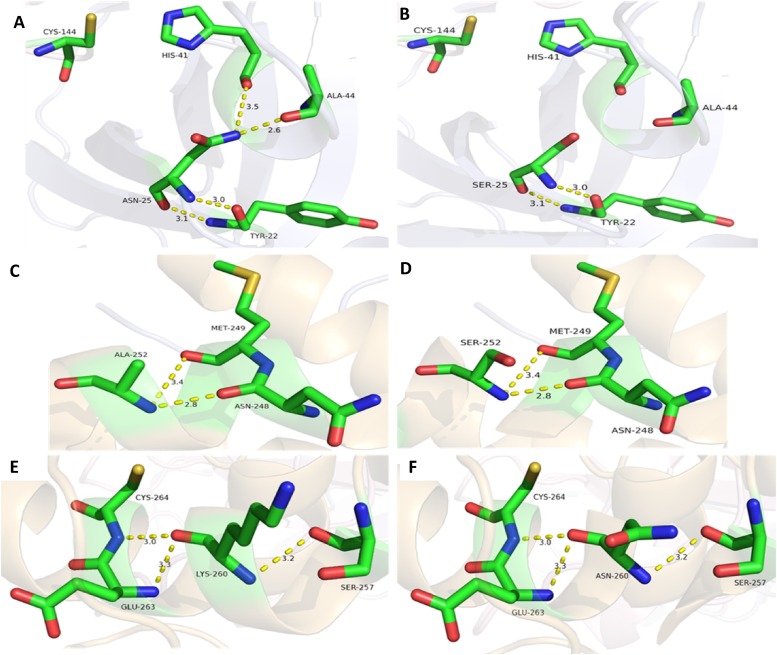
We then explored the potential interactions of N25, A252 and K260 with neighboring residues using a crystal structure of feline coronavirus 3CLpro (PDB: 4ZRO) with the Ring web server. N25 is predicted to form hydrogen bonds with the main chains of Y22, H41 (a catalytic residue) and A44 with hydrogen bond lengths of 3.029, 3.131, 3.486 and 2.615 Å, respectively (Fig. 5 A). Substitution of Asn with Ser at position 25 may disrupt the formation of the putative hydrogen bonds with H41 and A44, while retaining the hydrogen bonds with Y22 (Fig. 5B). A252 is predicted to form hydrogen bonds with the main chains of N248 and M249 with hydrogen bond lengths of 2.810 and 3.432 Å, respectively (Fig. 5C). Substitution of Ala with Ser at position 252 does not seem to affect those hydrogen bond formations (Fig. 5D). K260 is predicted to form hydrogen bonds with the main chains of S257, G263 and C264 with hydrogen bond lengths of 3.226, 3.267 and 3.037 Å, respectively (Fig. 5E). Substitution of Lys with Asn at position 260 does not seem to affect those hydrogen bond formations (Fig. 5F).
Fig. 5.
The effects of amino acid changes on the putative hydrogen bond formation by N25, A252 and K260 in feline coronavirus 3CLpro. The active site of wild type 3CLpro with predicted hydrogen bonds are shown in panels A, C and E. (A and B) The N25S substitution is predicted to result in the loss of hydrogen bonds (dashed lines) formed between N25 and H41 and A44, but not with Y22. (C to F) The A252S (C and D) and K260 N (E and F) substitutions are not predicted to affect the hydrogen bonds. The hydrogen bonds between N25 (A), A252 (C) and K260 (E) with neighboring amino acids in feline coronavirus 3CLpro are shown with the distance measurements in Å.
4. Discussion
Drug resistance is an increasing concern in antiviral therapy with prolonged drug exposure and high viral mutation rates because greatly reduced susceptibility to a drug can compromise clinical efficacy of antiviral treatment. Emergence of drug resistance is influenced by multiple factors, such as patient compliance, immune status of patients, suboptimal dosage, drug potency, genetic barrier of a drug to resistance, viral fitness or the biology of virus (reviewed in (Gotte, 2012; Strasfeld and Chou, 2010)). Drug resistance is caused by the development of viral mutations that arise during antiviral treatment or by pre-existing virus variants that carry such mutations. Viral mutations can confer various levels of reduced susceptibility or resistance to a drug, and clinically meaningful drug resistance can arise when viral replication is insufficiently suppressed by plasma drug concentrations. Antiviral compounds have varying genetic barriers to resistance. Those with low genetic barriers relatively readily lead to the emergence of mutations associated with clinically meaningful resistance and may not be suitable for monotherapy or longer duration of treatment. In contrast, those with high genetic barriers have reduced risks of developing resistance during antiviral treatment (reviewed in (Gotte, 2012; Strasfeld and Chou, 2010)). Therefore, identifying and understanding drug resistance of an antiviral compound can help effective treatment regime to be devised.
Feline coronaviruses are divided into serotypes I and II based on antigenicity (Addie et al., 2003; Benetka et al., 2004; Kummrow et al., 2005; Pedersen, 2009; Wang et al., 2014). Serotype I feline coronaviruses are the predominant type in the field, but they hardly grow in cell culture. In our previous study, we reported that serial passages of a serotype II feline coronavirus in the presence of GC376 in cell culture did not lead to emergence of resistant viruses for up to 20 passages, although a tripeptidyl compound (NPI52) that shares a similar backbone structure with GC376 readily led to resistant viruses (Kim et al., 2016). In the field trial, we identified amino acid changes in feline coronavirus 3CLpro from a feline patient that received intermittent treatment for the duration of about 8 months (Fig. 1) (Pedersen et al., 2018). This patient did not show evidence of clinical drug resistance during treatment. However, we set out to study the nature of those amino acid changes in 3CLpro. The feline coronavirus from the patient is a serotype I strain based on the sequence analysis of the regions spanning the spike gene. To examine the effects of the amino acid changes on the susceptibility to GC376, we generated the recombinant 3CLpro carrying the amino acid changes and determined the changes in their susceptibility to GC376 and NPI52 in the FRET assay (Fig. 2, Fig. 3). Among single (N25S, A252S or K260 N), double (N25S + K260 N) or triple (N25S + A252S + K260 N) amino acid changes, only those containing N25S led to a statistically significant reduction in susceptibility to GC376, but the reduction was marginal (up to a 1.68-fold increase in IC50 compared to that of WT) (Fig. 3A). Interestingly, A252S or K260 N single mutations did not change the susceptibility of 3CLpro to GC376, nor did further potentiate the effect of N25S in 3CLpro in the FRET assay (Fig. 3A). Considering the fact that the plasma drug concentrations of GC376 are 205˜834-fold over the 50% effective concentration (EC50) in cats receiving the drug (Kim et al., 2016), this result, combined with the previous finding of observed difficulty of raising drug resistance viruses in cell culture, may help explain the absence of clinical drug resistance in the patients that received treatment for an extended period in the field trial.
NPI52 is a tripeptidyl compound that shares the core structure with GC376 but has an additional residue that corresponds to the P3 position. In our previous report, serial passages of serotype II feline coronavirus in the presence of NPI52 readily led to emergence of viruses with NPI52-resistant phenotype with an increase of EC50 by 15-fold compared to WT (Kim et al., 2016). NPI52-resistant viruses have a single S131C mutation and this mutation is away from the catalytic site of 3CLpro. Interestingly, NPI52-resistant viruses retained susceptibility to GC376 in cell culture (Kim et al., 2016). In this study, the FRET assay showed that the amino acid substitutions of N25S, A252S and K260N did not affect the susceptibility of 3CLpro to NPI52 (Fig. 3A and B), confirming that GC376 and NPI52, although sharing a similar chemical structure, have disparate mechanisms of inducing mutations in feline coronavirus 3CLpro.
There are only a few reports available on coronavirus 3CLpro inhibitors and their resistance. Deng et al (Deng et al., 2014) reported single mutation (T26I and D65 G) and double mutations (T26I/D65G and T26I/A298D) that are responsible for resistance of a murine hepatitis virus (MHV), a coronavirus, to a 3CLpro inhibitor GRL-001 in cell culture. MHV carrying a single mutation was partially resistant to the inhibitor, compared to WT virus, but double mutations further increased viral resistance in cells. Although these mutations arose rapidly in cell culture, indicating that this specific inhibitor has low-genetic barrier to resistance, the mutated viruses had reduced viral fitness in cells and were highly attenuated in the host (mice). In that study, T26I and D65 located in and away from the active site of MHV 3CLpro, respectively, was speculated to affect inhibitor binding indirectly or directly, although the role of A298D in the domain III was unclear (Deng et al., 2014). The N25S in feline coronavirus 3CLpro is in the active site of 3CLpro, and our 3D modelling study showed that the loss of a side chain from N25S substitution is predicted to partially lose hydrogen bonds with residues in the active site (Fig. 5A and B). This partial loss of bonds may explain the marginal reduction in susceptibility of 3CLpro to GC376 by N25S substitution. The modelling study also predicted that A252S and K260 N substitutions in domain III (Fig. 5C-F) have no changes in the interactions with neighboring residues, which may explain the findings from the FRET assay where these mutations do not seem to affect the proteolytic activity of 3CLpro.
5. Conclusions
Emergence of viral resistance to antivirals during treatment, especially long-term treatment, is a concern. Amino acid changes of N25S, A252S and K260N were found in the 3CLpro of feline coronavirus obtained from a cat patient that received multiple rounds of antiviral treatment but did not show clinical resistance. The N25S substitution in the 3CLpro led to a statistically significant but slight reduction in susceptibility to GC376 in the FRET assay (up to 1.68-fold in IC50), but no change in susceptibility was observed with A252S and K260N substitutions. The results of this study are consistent with the previous findings showing high genetic barriers of feline coronavirus 3CLpro for selecting resistance against GC376. Further studies, including X-ray crystallography on mutant 3CLpro and generation of recombinant viruses carrying those mutations, will help us to gain better understanding of the roles of those substitutions.
Funding statement
This research was funded by the National Institutes of Health, grant number R01 AI130092, and Morris Animal Foundation, grant numbers D14FE-012 and D16FE-512.
Declaration of Competing Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. Y.K. K.O. and W.C.G. have patent claims on the protease inhibitors in the manuscript.
Acknowledgment
We thank David George for technical assistance.
References
- Addie D., Houe L., Maitland K., Passantino G., Decaro N. Effect of cat litters on feline coronavirus infection of cell culture and cats. J. Feline Med. Surg. 2019 doi: 10.1177/1098612X19848167. 1098612X19848167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addie D.D., Schaap I.A., Nicolson L., Jarrett O. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 2003;84:2735–2744. doi: 10.1099/vir.0.19129-0. [DOI] [PubMed] [Google Scholar]
- Barker E.N., Tasker S., Gruffydd-Jones T.J., Tuplin C.K., Burton K., Porter E., Day M.J., Harley R., Fews D., Helps C.R., Siddell S.G. Phylogenetic analysis of feline coronavirus strains in an epizootic outbreak of feline infectious peritonitis. J. Vet. Intern. Med. 2013;27:445–450. doi: 10.1111/jvim.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetka V., Kubber-Heiss A., Kolodziejek J., Nowotny N., Hofmann-Parisot M., Mostl K. Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet. Microbiol. 2004;99:31–42. doi: 10.1016/j.vetmic.2003.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.W., Egberink H.F., Halpin R., Spiro D.J., Rottier P.J. Spike protein fusion Peptide and feline coronavirus virulence. Emerg Infect Dis. 2012;18:1089–1095. doi: 10.3201/eid1807.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. DeLano Scientific LLC; San Carlos, CA: 2010. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- Deng X., StJohn S.E., Osswald H.L., O’Brien A., Banach B.S., Sleeman K., Ghosh A.K., Mesecar A.D., Baker S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte M. The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance. Curr. Opin. Virol. 2012;2:644–650. doi: 10.1016/j.coviro.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Mandadapu S.R., Groutas W.C., Chang K.O. Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antiviral Res. 2013;97:161–168. doi: 10.1016/j.antiviral.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Scott L., Kok-Chuan T., Sivakoteswara Rao M., Kevin R.A., Kevin P.B., William C.G., Kyeong-Ok C. Broad-spectrum antivirals against 3C or 3C-Like proteases of Picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Shivanna V., Narayanan S., Prior A.M., Weerasekara S., Hua D.H., Kankanamalage A.C., Groutas W.C., Chang K.O. Broad-spectrum inhibitors against 3C-Like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89:4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M., Lefkowitz E., Adams M.J., Carstens E.B. Elsevier; 2011. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- Kummrow M., Meli M.L., Haessig M., Goenczi E., Poland A., Pedersen N.C., Hofmann-Lehmann R., Lutz H. Feline coronavirus serotypes 1 and 2: seroprevalence and association with disease in Switzerland. Clin. Diagn. Lab. Immunol. 2005;12:1209–1215. doi: 10.1128/CDLI.12.10.1209-1215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntal B.K., Aparoy P., Reddanna P. EasyModeller: a graphical interface to MODELLER. BMC Res. Notes. 2010;3:226. doi: 10.1186/1756-0500-3-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19:1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963-2008. J. Feline Med. Surg. 2009;11:225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. An update on feline infectious peritonitis: virology and immunopathogenesis. Vet. J. 2014;201:123–132. doi: 10.1016/j.tvjl.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Allen C.E., Lyons L.A. Pathogenesis of feline enteric coronavirus infection. J. Feline Med. Surg. 2008;10:529–541. doi: 10.1016/j.jfms.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Kim Y., Liu H., Galasiti Kankanamalage A.C., Eckstrand C., Groutas W.C., Bannasch M., Meadows J.M., Chang K.O. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 2018;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Liu H., Dodd K.A., Pesavento P.A. Significance of coronavirus mutants in feces and diseased tissues of cats suffering from feline infectious peritonitis. Viruses. 2009;1:166–184. doi: 10.3390/v1020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Liu H., Scarlett J., Leutenegger C.M., Golovko L., Kennedy H., Kamal F.M. Feline infectious peritonitis: role of the feline coronavirus 3c gene in intestinal tropism and pathogenicity based upon isolates from resident and adopted shelter cats. Virus Res. 2012;165:17–28. doi: 10.1016/j.virusres.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D., Minervini G., Tosatto S.C. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44:W367–374. doi: 10.1093/nar/gkw315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasfeld L., Chou S. Antiviral drug resistance: mechanisms and clinical implications. Infect. Dis. Clin. North Am. 2010;24:809–833. doi: 10.1016/j.idc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Wang Y.T., Chueh L.L., Wan C.H. An eight-year epidemiologic study based on baculovirus-expressed type-specific spike proteins for the differentiation of type I and II feline coronavirus infections. BMC Vet. Res. 2014;10:186. doi: 10.1186/s12917-014-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]