Significance Statement
Pathologists usually classify diabetic nephropathy on the basis of a visual assessment of glomerular pathology. Although diagnostic guidelines are well established, results may vary among pathologists. Modern machine learning has the potential to automate and augment accurate and precise classification of diabetic nephropathy. Digital algorithms may also be able to extract novel features relevant to disease progression and prognosis. The authors used image analysis and machine learning algorithms to digitally classify biopsy samples from 54 patients with diabetic nephropathy and found substantial agreement between digital classifications and those by three different pathologists. The study demonstrates that digital processing of renal tissue may provide useful information that may augment traditional clinical diagnostics.
Keywords: Computational renal pathology, diabetic nephropathy, glomerulus, Tervaert's classification, Digital pathology, Image analysis
Visual Abstract
Abstract
Background
Pathologists use visual classification of glomerular lesions to assess samples from patients with diabetic nephropathy (DN). The results may vary among pathologists. Digital algorithms may reduce this variability and provide more consistent image structure interpretation.
Methods
We developed a digital pipeline to classify renal biopsies from patients with DN. We combined traditional image analysis with modern machine learning to efficiently capture important structures, minimize manual effort and supervision, and enforce biologic prior information onto our model. To computationally quantify glomerular structure despite its complexity, we simplified it to three components consisting of nuclei, capillary lumina and Bowman spaces; and Periodic Acid-Schiff positive structures. We detected glomerular boundaries and nuclei from whole slide images using convolutional neural networks, and the remaining glomerular structures using an unsupervised technique developed expressly for this purpose. We defined a set of digital features which quantify the structural progression of DN, and a recurrent network architecture which processes these features into a classification.
Results
Our digital classification agreed with a senior pathologist whose classifications were used as ground truth with moderate Cohen’s kappa κ = 0.55 and 95% confidence interval [0.50, 0.60]. Two other renal pathologists agreed with the digital classification with κ1 = 0.68, 95% interval [0.50, 0.86] and κ2 = 0.48, 95% interval [0.32, 0.64]. Our results suggest computational approaches are comparable to human visual classification methods, and can offer improved precision in clinical decision workflows. We detected glomerular boundaries from whole slide images with 0.93±0.04 balanced accuracy, glomerular nuclei with 0.94 sensitivity and 0.93 specificity, and glomerular structural components with 0.95 sensitivity and 0.99 specificity.
Conclusions
Computationally derived, histologic image features hold significant diagnostic information that may augment clinical diagnostics.
In the United States, an estimated 9.4% of the population has diabetes mellitus (DM), and over one-third will develop diabetic nephropathy (DN), making diabetes the leading cause of CKD and ESKD. The effect of diabetes and DN on public health will only intensify, because the Centers for Disease Control predicts one in three Americans will have diabetes by 2050 if current trends continue.1
Although confirmatory biopsies are rarely performed to definitively establish the diagnosis of DN, there is typically good correlation between the clinical stages of DN and the renal morphologic changes seen on biopsy. A consensus pathologic classification system has been developed which divides DN into four hierarchic categories on the basis of glomerular morphologic findings. An important strength for this DN classification system is that the morphologic variables are well defined, which results in improved interobserver reproducibility. However, in practice, reproducibility remains an issue, and a broad morphologic spectrum of glomerular lesions can still be seen within each class.
In this study, our goal was to engineer an automated computational pipeline to classify digitized human renal biopsy samples according to the scheme by Tervaert and collegues.2 We also subsequently validated the pipeline for an alternative DN classification scheme defined by A.B.F. and also for a streptozotocin (STZ) mouse model of DM. This pipeline has four subtasks: (1) identify glomerular locations in digitized biopsy sample slides, (2) identify and discretize glomerular components, (3) quantify glomerular components, and (4) classify glomerular features. To accomplish these tasks, we integrated several computational methods, combining traditional image analysis and machine learning with modern machine learning. We have done this for two distinct reasons: (1) to superimpose biologic prior information onto our modeling, and (2) to capture important glomerular structures and their associated changes while minimizing the burden of human annotation. Our most important contributions to this pipeline are a set of computationally defined glomerular features which reflect DN glomerulopathic structural alteration, and a recurrent neural network (RNN) architecture which analyzes sequences of these glomerular features to yield the final biopsy diagnosis. Moreover, the output of the network is a continuous estimate between 1 and 5, which can be rounded to conform to the discrete Tervaert classes, but can also motivate the shift of diagnoses from discrete stages to continuous risk models. We compared this technique against the staging of three renal pathologists for 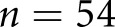 patients, and found that it was reaching human levels of agreement with pathologist annotations. With the most experienced pathologist set as ground truth, the computational method agreed with
patients, and found that it was reaching human levels of agreement with pathologist annotations. With the most experienced pathologist set as ground truth, the computational method agreed with 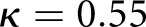 (
( confidence interval [95% CI], 0.50 to 0.60), as compared with two renal pathologists (
confidence interval [95% CI], 0.50 to 0.60), as compared with two renal pathologists (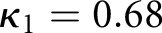 ;
; 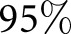 CI, 0.50 to 0.86; and
CI, 0.50 to 0.86; and 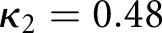 ; 95% CI, 0.32 to 0.64). When splitting multiple sections of patients into separate cases, the computational κ rose to
; 95% CI, 0.32 to 0.64). When splitting multiple sections of patients into separate cases, the computational κ rose to 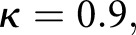 (
(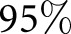 CI, 0.87 to 0.93). The result suggests that the computational approach has the potential to improve precision in a clinical workflow assisting humans. The framework we apply is exceptionally flexible and could be extended for application to many other glomerular diseases with histologic changes, such as FSGS, lupus nephritis, and IgA nephropathy, as well as other histologic studies where a sparse set of compartments are quantified. Our study suggests that image-based features derived computationally from histologic images hold significant diagnostic information that can be further explored for clinical applications.
CI, 0.87 to 0.93). The result suggests that the computational approach has the potential to improve precision in a clinical workflow assisting humans. The framework we apply is exceptionally flexible and could be extended for application to many other glomerular diseases with histologic changes, such as FSGS, lupus nephritis, and IgA nephropathy, as well as other histologic studies where a sparse set of compartments are quantified. Our study suggests that image-based features derived computationally from histologic images hold significant diagnostic information that can be further explored for clinical applications.
Methods
Human data collection followed protocols approved by the Institutional Review Board at the University at Buffalo (UB) and Vanderbilt University. All methods were performed according to federal guidelines and regulations. All animal studies were performed according to protocols approved by the UB Animal Studies Committee Procedures and the Institutional Animal Care and Use Committee.
Image Data
Image data consisted of whole-slide images (WSIs) of renal tissue sections from 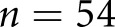 human patients and
human patients and 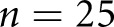 mice. Human tissues consisted of needle biopsy samples from patients with DN as disease data and nontumoral parenchyma of carcinoma nephrectomies as control. Although nephrectomy samples were rapidly processed after resection, we cannot measure significant differences due to biopsy or nephrectomy procedures in our results. Further, there is no readily available normal kidney biopsy tissue that can be ethically obtained from healthy volunteers or living donors. Our patients were sourced from two different institutions, some from the Kidney and Translational Research Core (KTRC) at Washington University in Saint Louis School of Medicine, and some from the Vanderbilt University Medical Center (VUMC). We will refer to the institutes anonymously as institute-1 and -2 throughout the manuscript. The tissues were also prepared in three separate laboratories, some at UB’s Pathology and Anatomical Science department’s histology core, some at the histology core in the local Roswell Park Cancer Institute, and some at VUMC. We chose to prepare the tissues separately in order to model stain variation. We will anonymously refer to these preparations as preparation-1, -2, and -3. All tissues were scanned using a whole-slide scanner (Aperio; Leica) at ×40 apparent magnification, resulting in images with resolution 0.25 µm per pixel. The analysis presented in this work is only valid for images which are scanned at a similar resolution.
mice. Human tissues consisted of needle biopsy samples from patients with DN as disease data and nontumoral parenchyma of carcinoma nephrectomies as control. Although nephrectomy samples were rapidly processed after resection, we cannot measure significant differences due to biopsy or nephrectomy procedures in our results. Further, there is no readily available normal kidney biopsy tissue that can be ethically obtained from healthy volunteers or living donors. Our patients were sourced from two different institutions, some from the Kidney and Translational Research Core (KTRC) at Washington University in Saint Louis School of Medicine, and some from the Vanderbilt University Medical Center (VUMC). We will refer to the institutes anonymously as institute-1 and -2 throughout the manuscript. The tissues were also prepared in three separate laboratories, some at UB’s Pathology and Anatomical Science department’s histology core, some at the histology core in the local Roswell Park Cancer Institute, and some at VUMC. We chose to prepare the tissues separately in order to model stain variation. We will anonymously refer to these preparations as preparation-1, -2, and -3. All tissues were scanned using a whole-slide scanner (Aperio; Leica) at ×40 apparent magnification, resulting in images with resolution 0.25 µm per pixel. The analysis presented in this work is only valid for images which are scanned at a similar resolution.
Glomerular Detection and Boundary Segmentation
To develop a robust glomerular detection and boundary segmentation model, we used our iterative convolutional learning interface, human-artificial-intelligence-loop (HAIL4), which uses the DeepLab V2 ResNet3 network to detect and segment glomerular boundaries on WSIs. We applied HAIL using a multipass approach, which uses a low- and high-resolution network in tandem to improve speed and accuracy of glomerular acquisition, described more in depth in our previous work.4 The glomerular detection models trained in that work were used as pretrained starting models. These models were trained on WSIs from 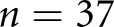 human patients with DN,
human patients with DN, 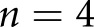 human control patients,
human control patients, 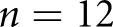 STZ mice, and
STZ mice, and 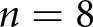 control mice. WSIs from
control mice. WSIs from 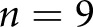 human patients with DN,
human patients with DN, 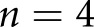 human control patients,
human control patients,  STZ mice, and
STZ mice, and 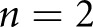 control mice were reserved for holdout testing. The high-resolution network was trained for
control mice were reserved for holdout testing. The high-resolution network was trained for 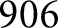 K steps, and the low-resolution network was trained for
K steps, and the low-resolution network was trained for  K steps, at which point the training loss curves for both networks had plateaued. For both networks, initial learning rate was
K steps, at which point the training loss curves for both networks had plateaued. For both networks, initial learning rate was 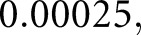 batch size was
batch size was  and input image size was
and input image size was 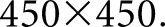 Remaining hyper-parameter values were left at DeepLab default.
Remaining hyper-parameter values were left at DeepLab default.
Glomerular Component Detection
To simplify glomerular compartmentalization, glomerular structures were assigned to one of three components on the basis of their appearance in periodic acid–Schiff (PAS) stains: (1) a nuclear component; (2) a PAS-positive (PAS+) component consisting of mesangium, glomerular basement membranes, and Bowman’s capsule; and (3) a luminal component consisting of Bowman’s space and capillary lumina.
Nuclear Component
For the detection of nuclei in human tissues, the DeepLab V2 ResNet convolutional neural network (CNN) was trained on three distinct sets of annotated images pooled into one. The first set of images contained  glomeruli, which had the majority of nuclei annotated with computational assistance by automatic thresholding5,6 of stain deconvolution7 for hematoxylin. Missed nuclei were added by manual annotation, and clumped nuclei split using a distance transform–based marker-controlled watershed8 (described in the Supplemental Material). We used computational assistance so that we could annotate a large number of images cheaply, and used them to form the bulk of network training. To improve the network’s identification of exact nuclear boundary as perceived by the human eye, which was not always identified by color deconvolution, we added another set of
glomeruli, which had the majority of nuclei annotated with computational assistance by automatic thresholding5,6 of stain deconvolution7 for hematoxylin. Missed nuclei were added by manual annotation, and clumped nuclei split using a distance transform–based marker-controlled watershed8 (described in the Supplemental Material). We used computational assistance so that we could annotate a large number of images cheaply, and used them to form the bulk of network training. To improve the network’s identification of exact nuclear boundary as perceived by the human eye, which was not always identified by color deconvolution, we added another set of  glomerulus images which were annotated entirely by a manual annotator. The third dataset consisted of six rectangular images with dimensions between
glomerulus images which were annotated entirely by a manual annotator. The third dataset consisted of six rectangular images with dimensions between 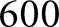 and
and 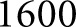 pixels, selected to include large patches of inflammation. These images were selected so that the network had more diverse examples to learn nuclear splitting, and were annotated manually. From the second dataset (manual annotations of whole glomeruli),
pixels, selected to include large patches of inflammation. These images were selected so that the network had more diverse examples to learn nuclear splitting, and were annotated manually. From the second dataset (manual annotations of whole glomeruli), 
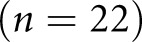 of the images were reserved, randomly, for holdout performance testing. We split the training and testing sets at the patch level because the annotation cost for nuclei is very high and we only had a limited amount of annotations for training use.
of the images were reserved, randomly, for holdout performance testing. We split the training and testing sets at the patch level because the annotation cost for nuclei is very high and we only had a limited amount of annotations for training use.
All remaining training images were chopped into  -pixel overlapping patches (
-pixel overlapping patches ( overlap), resulting in close to
overlap), resulting in close to 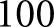 K images. We initialized our nuclear detection network training on the trained network model used for high-resolution glomerular detection. This was done with the assumption that the glomerular segmentation network had already been extensively exposed to nuclei, and would likely have developed some form of filters used to detect their presence. Starting with this pretrained model should theoretically improve convergence time of the network. We trained with a batch size of
K images. We initialized our nuclear detection network training on the trained network model used for high-resolution glomerular detection. This was done with the assumption that the glomerular segmentation network had already been extensively exposed to nuclei, and would likely have developed some form of filters used to detect their presence. Starting with this pretrained model should theoretically improve convergence time of the network. We trained with a batch size of 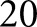 for
for 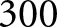 K steps of training (
K steps of training (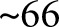 epochs) using an initial learning rate of
epochs) using an initial learning rate of 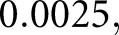 and remaining hyper-parameters were left at DeepLab default values. The resulting output may have still contained clumped nuclei because there was physically very little or no space between them at the
and remaining hyper-parameters were left at DeepLab default values. The resulting output may have still contained clumped nuclei because there was physically very little or no space between them at the  -µm resolution. Therefore, we split the remaining clumped nuclei via a morphologic postprocessing step. First, the output of the neural network segmentation was processed by estimating and removing single object clumps with the distance transform.9–11 Single objects were defined as binary regions that contained only one estimated peak region (see watershed splitting in the Supplemental Material). Then, remaining clumps were split using a distance transform–based marker-controlled watershed.8 Other groups have shown success using the watershed technique for histopathologic nucleus splitting in the past.12
-µm resolution. Therefore, we split the remaining clumped nuclei via a morphologic postprocessing step. First, the output of the neural network segmentation was processed by estimating and removing single object clumps with the distance transform.9–11 Single objects were defined as binary regions that contained only one estimated peak region (see watershed splitting in the Supplemental Material). Then, remaining clumps were split using a distance transform–based marker-controlled watershed.8 Other groups have shown success using the watershed technique for histopathologic nucleus splitting in the past.12
To modulate and investigate the sensitivity and specificity of the network, we applied weighting to the network’s raw output probabilities. The raw network output is a vector at each pixel with length equal to the number of classes, containing probabilities that correspond to the likelihood of each respective class assignment. We used a normalized weight, w  for the nuclear class and 1-w for the background class, and computed a normalized weighted average probability to bias the network probabilities toward the nuclear class or background class. At each pixel, the class with maximum weighted probability value was selected as the class assignment. Weight values were sampled uniformly in intervals of
for the nuclear class and 1-w for the background class, and computed a normalized weighted average probability to bias the network probabilities toward the nuclear class or background class. At each pixel, the class with maximum weighted probability value was selected as the class assignment. Weight values were sampled uniformly in intervals of 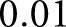 in the range (0, 1). We plotted the results of this weighting as a receiver operating curve describing sensitivity and specificity for a holdout set of glomeruli; see Figure 3G.
in the range (0, 1). We plotted the results of this weighting as a receiver operating curve describing sensitivity and specificity for a holdout set of glomeruli; see Figure 3G.
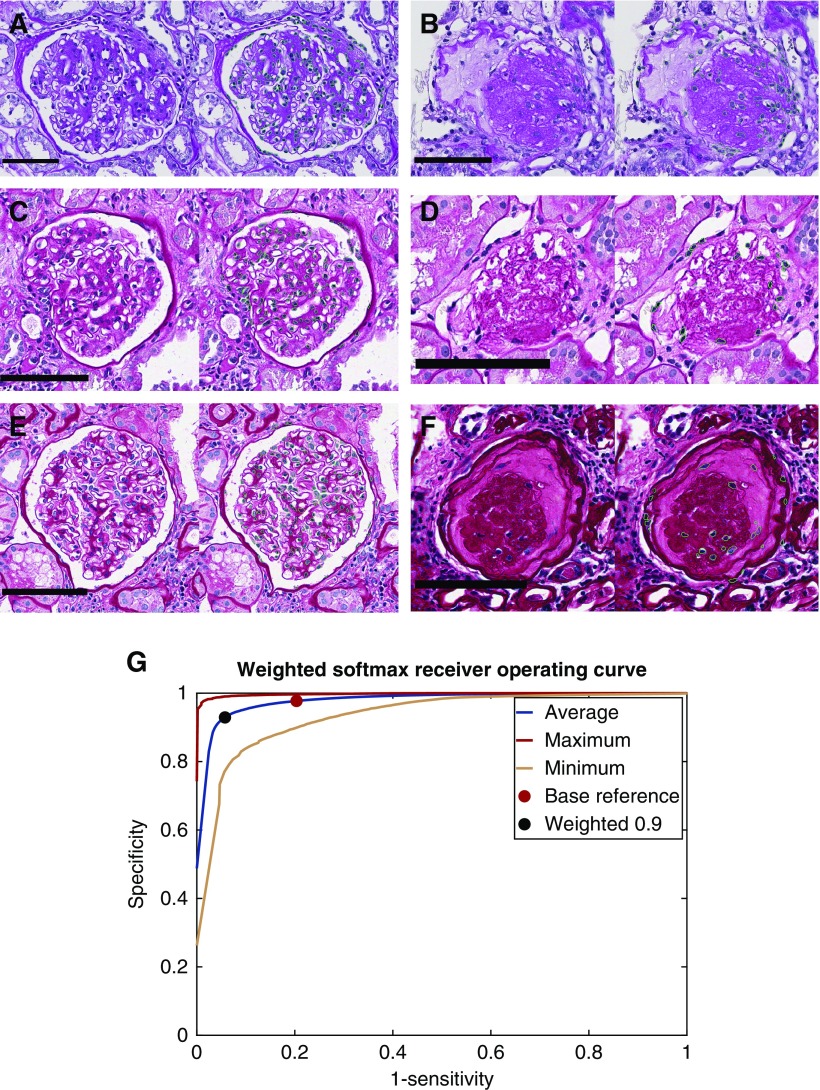
Figure 3.
Nuclear boundaries detected from varied, PAS stained glomerulus images with high accuracy. (A) Detection of nuclei from institute- 1 and preparation-1 in a glomerulus. Green boundaries in images indicate the perimeter of the detected nuclear region. (B) Detection of nuclei from institute-1, preparation-1, in a sclerotic glomerulus. (C and D) Detection of nuclei in glomeruli from institute-2, preparation-2, for a glomerulus and a sclerotic glomerulus. (E and F) Detection of nuclei in glomeruli from institute-2, preparation-3, for a glomerulus and a sclerotic glomerulus. (G) Receiver operating curve for the nuclear detection method calculated as the average, minimum, and maximum of a 22-image holdout set. Red dot indicates the network’s performance without any weighting on the network’s output. Black dot indicates the network’s performance when the network’s output is weighted toward the nuclear class with weight 0.9 (a weight of 1 would result in every pixel in the image detected as nuclear). Scale bars, 100 µm.
PAS+ and Luminal Components
The luminal and PAS+ components were each identified using a two-step procedure. First, a rough mask of each component was defined by thresholding grayscale images. To identify a grayscale image that reflected PAS+ regions, we used the HSV (hue, saturation, value) transformation.13 Specifically, we divided the saturation channel by the value channel, which resulted in an image that had high intensity where there was darkly saturated material (such as mesangium). This image was thresholded at a fixed value of 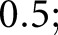 we will call it the PAS+ precursor mask. To identify a grayscale image that reflected luminal regions, the L* component of the L*a*b color space14 was used. This grayscale image was thresholded with Otsu’s method5 to yield the luminal precursor mask. In the combined precursor mask, some pixels were unlabeled, and some had a double label. To achieve a final mask without double-labeled or unlabeled pixels, the Red-Green-Blue (RGB) values that underlie the precursor masks were used to train a two-class naïve Bayesian classifier15 (MathWorks, Natick, MA). This trained classifier was then used to predict a label for all pixels in the glomerular region.
we will call it the PAS+ precursor mask. To identify a grayscale image that reflected luminal regions, the L* component of the L*a*b color space14 was used. This grayscale image was thresholded with Otsu’s method5 to yield the luminal precursor mask. In the combined precursor mask, some pixels were unlabeled, and some had a double label. To achieve a final mask without double-labeled or unlabeled pixels, the Red-Green-Blue (RGB) values that underlie the precursor masks were used to train a two-class naïve Bayesian classifier15 (MathWorks, Natick, MA). This trained classifier was then used to predict a label for all pixels in the glomerular region.
Detection Performance Analysis
All object detection performances were assessed at the pixel level. Performance for glomerular detection in WSIs was calculated against manual annotation of holdout WSIs. We report sensitivity and specificity, but also balanced accuracy, because the number of glomerular class pixels is extremely imbalanced with respect to the number of background class pixels. Performance for glomerular nucleus detection was assessed against manual annotation of nuclei from holdout glomerulus images. PAS+ and lumina detection was assessed against sparse manual annotation of 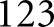 holdout images, sampled to represent each Tervaert disease stage I–IV as well as each category of the DM mouse model described in the Supplemental Material.
holdout images, sampled to represent each Tervaert disease stage I–IV as well as each category of the DM mouse model described in the Supplemental Material.
Feature Extraction
We defined six types of features for classification of glomerular structures: color, textural, morphologic, containment, interstructural distance, and intrastructural distance. Color features included the mean and SDs of R, G, and B values observed in PAS+, luminal, and nuclear regions. Textural features (entropy, contrast, correlation, homogeneity), which reflect subvisual compartment changes, were computed via gray-level co-occurrence16 computed on each respective component. Morphologic features included number, area, and convexity of component objects (measures expansion or reduction of components). Containment features quantified the relative amount of one component within another and describe how components are expanding or reducing relative to each other (e.g., nuclear area contained within a mesangial segment). To calculate containment, first, each unique object in each component was identified. Then, for each identified object, the convex hull of the region was taken. The containment feature was defined as the area in this convex hull, divided by the area of other components contained within its boundaries (e.g., nuclear area). Interstructural distance features assessed the distance between same-class objects (e.g., nuclei) and their distance to glomerular landmarks (e.g., distance to glomerular boundary), describing a measure of how components move relative to one another. Intrastructural distance features assessed the thickness of objects, such as mesangial width. They were assessed by taking histogram data on distance-transformed glomerular components, such as the PAS+ precursor mask (see Figure 5). The total number of features we defined was 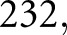 and a full list of exact names of these features is available as Supplemental Table 1.
and a full list of exact names of these features is available as Supplemental Table 1.
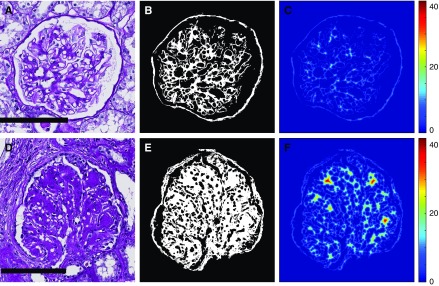
Figure 5.
Intracompartmental distance features continuously quantitate mesangial thickening on PAS+ regions of DN glomeruli. (A) Example of glomerulus with mild mesangial thickening. (B) PAS+ precursor mask for glomerulus in image (A), used as an estimate for mesangial regions. (C) Distance transform of the image shown in (B), resulting in maximum value calculated at 18.4. (D) Example of glomerulus with extensive mesangial thickening, including Kimmelstiel–Wilson nodules. (E) PAS+ precursor mask for glomerulus in (D). (F) Distance transform of the image shown in (E), with maximum value calculated at 42. Compared with (C), it can be seen that this analysis yields higher values for thicker PAS+ structures, and lower values for thinner ones. Scale bars, 100 µm.
Recurrent DN Classification
We computationally classified DN biopsy samples according to two different staging systems, one being the Tervaert classification scheme,2 and the other being a classification scheme defined by coauthor A.B.F.; the latter scheme has the following classes: normal, no significant DN-related changes; mild, up to 25% tubulointerstitial fibrosis/glomerular sclerosis; moderate, 25%–50%; and severe, >50%. We have also compared our technique against the grading of three renal pathologists for the Tervaert scheme. For grading, all glomeruli with sufficient cross-sectional area of the glomerular tuft (two or more visualized mesangial regions) were evaluated.
Although the tubulointerstitial compartment is an important component of both of these grading systems, and is a key prognostic indicator in many glomerular diseases, we did not utilize this information in our computational grading system. Identification of this compartment requires extensive work in its own right and we will be pursuing its quantification in a separate work.
Training
We computationally classified tissues of  patients,
patients, 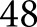 of which were biopsy samples taken with the suspicion of DN, and six control tissues. Control tissues were not classed by pathologists because they were nephrectomies, containing an extensive portion of a whole kidney section. Such an extent of tissue would make it very difficult to blind the annotators as to the source of the tissue. These tissues were independently validated by the pathology core at KTRC for use as control tissue. Contained within these patient data were
of which were biopsy samples taken with the suspicion of DN, and six control tissues. Control tissues were not classed by pathologists because they were nephrectomies, containing an extensive portion of a whole kidney section. Such an extent of tissue would make it very difficult to blind the annotators as to the source of the tissue. These tissues were independently validated by the pathology core at KTRC for use as control tissue. Contained within these patient data were  glomerulus images from patients with DN, and, in the control data, another
glomerulus images from patients with DN, and, in the control data, another 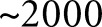 glomerulus images.
glomerulus images.
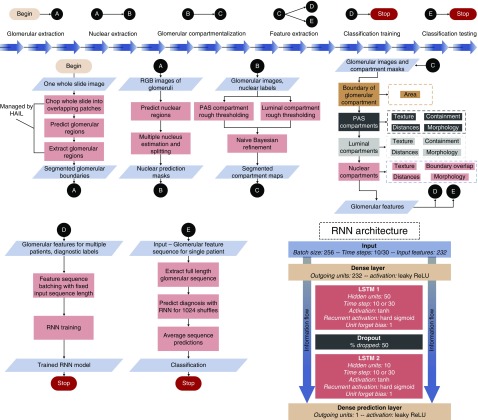
To transform the glomerular features that we crafted into the final diagnosis, we designed a simple RNN architecture17 (Figure 1) which treats a sequence of glomerular features from a renal biopsy sample as its temporal data. The quantified glomerular features were normalized by mean and SD before input to the network. The network architecture can be described as follows: First, input features were passed to a densely connected layer with output size equal to the number of input features. This layer was connected to a long short-term memory (LSTM17) unit with 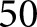 hidden features, which was connected to a second LSTM unit with
hidden features, which was connected to a second LSTM unit with  hidden features. For experiments run using individual patients as cases, both LSTMs had an input sequence length of
hidden features. For experiments run using individual patients as cases, both LSTMs had an input sequence length of 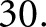 For experiments run using individual sections as separate cases, there was a smaller pool of glomeruli in each case, so the LSTMs had input sequence length of
For experiments run using individual sections as separate cases, there was a smaller pool of glomeruli in each case, so the LSTMs had input sequence length of 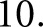 There was a
There was a  dropout between the LSTM units. The output of the second LSTM unit was fed into a dense layer with one output, which is the predicted diagnostic value, a continuous number between 1 and the number of classes. The activation function for both dense layers was a leaky rectified linear unit,18 and for both LSTM units, a hyperbolic tangent. We defined our loss function as the absolute distance of the predicted labels from the true labels. This loss function, combined with the single-valued output structure, allowed us to enforce the continuous nature of the task onto the network. Namely, classifying a class IV biopsy sample as class I is considerably more erroneous than classifying a class IIa biopsy sample as class IIb (and, therefore, the latter should be rewarded more than the former). An Adam optimizer19 was used to schedule the learning rate. As previously mentioned, the network accepts fixed-length sequences of glomerular features drawn from a single biopsy sample. A single batch of training data consisted of
dropout between the LSTM units. The output of the second LSTM unit was fed into a dense layer with one output, which is the predicted diagnostic value, a continuous number between 1 and the number of classes. The activation function for both dense layers was a leaky rectified linear unit,18 and for both LSTM units, a hyperbolic tangent. We defined our loss function as the absolute distance of the predicted labels from the true labels. This loss function, combined with the single-valued output structure, allowed us to enforce the continuous nature of the task onto the network. Namely, classifying a class IV biopsy sample as class I is considerably more erroneous than classifying a class IIa biopsy sample as class IIb (and, therefore, the latter should be rewarded more than the former). An Adam optimizer19 was used to schedule the learning rate. As previously mentioned, the network accepts fixed-length sequences of glomerular features drawn from a single biopsy sample. A single batch of training data consisted of  of these feature sequence sets. We used identical hyper-parameters for all experiments within this work; namely, we used initial learning rate
of these feature sequence sets. We used identical hyper-parameters for all experiments within this work; namely, we used initial learning rate 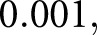 batch size
batch size 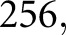 and
and  training steps. Reported agreement statistics were obtained by taking the accumulated prediction results of
training steps. Reported agreement statistics were obtained by taking the accumulated prediction results of  trials of
trials of 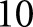 -fold crossvalidation.
-fold crossvalidation.
Figure 1.
General overview of our computational pipeline and recurrent architecture. ReLU, rectified linear unit.
Prediction
One reason recurrent networks are advantageous is because they do not require fixed-length input sequences for prediction as they do for training. They can predict on sequences of any length, and, therefore, each prediction sequence had length equal to the number of observed glomeruli in the case. To prevent the resultant prediction from being dependent on the particular order in which the glomeruli were extracted, each full-length prediction sequence was shuffled 1024 times. This block of 1024 shuffled sequences of glomeruli represented one set of sequences to be predicted for a single case. The final classification for that particular case was made by taking the average of predicted values on the 1024 sequences and rounding this to the closest whole number.
Feature Ranking
To investigate which features the network depended on most for its diagnostic decisions, we sequentially dropped features from the network input, and recorded how much the network’s prediction changed when each feature was removed. The model for this experiment was trained on  cases. Four cases were reserved as holdout (one case each from DN classes I, IIb, III, and IV) to ensure the model was not overfitting. Note that class IIa was excluded because there were only two cases in this class. The model was trained for
cases. Four cases were reserved as holdout (one case each from DN classes I, IIb, III, and IV) to ensure the model was not overfitting. Note that class IIa was excluded because there were only two cases in this class. The model was trained for 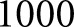 iterations with batch size
iterations with batch size  and learning rate
and learning rate  After training, the network was run for prediction once on all 54 cases, to get a base reference of how the network was functioning without losing any features. After this, the network was run for prediction
After training, the network was run for prediction once on all 54 cases, to get a base reference of how the network was functioning without losing any features. After this, the network was run for prediction  times (which is equal to the total number of features), and, on each run, a different feature value was set to zero before prediction. The predicted output value is likely to shift when a feature is lost, and the amount that this value changes is a reflection of how dependent the network was on that feature. We measured this prediction drift for all features, and for all of the cases
times (which is equal to the total number of features), and, on each run, a different feature value was set to zero before prediction. The predicted output value is likely to shift when a feature is lost, and the amount that this value changes is a reflection of how dependent the network was on that feature. We measured this prediction drift for all features, and for all of the cases  The final score of dependence for each feature was measured as the average prediction drift across all 54 cases. Mathematically, the function we used can be described as Equation 1 below:
The final score of dependence for each feature was measured as the average prediction drift across all 54 cases. Mathematically, the function we used can be described as Equation 1 below:
 |
(1) |
where 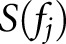 is the score for the dropped feature
is the score for the dropped feature  such that
such that 
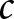 is the number of cases,
is the number of cases,  is the network’s predicted label for case
is the network’s predicted label for case  given feature
given feature 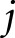 has been dropped,
has been dropped, 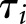 is the true label for case
is the true label for case 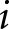 , and
, and 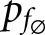 is the network’s predicted label given no features have been dropped. This function is defined in such a way that a large negative score would indicate a feature with an obfuscating influence on the network’s classification ability. Conversely, a large positive score would mean that the network was highly dependent on that feature to get the correct diagnosis, and suffered from its loss.
is the network’s predicted label given no features have been dropped. This function is defined in such a way that a large negative score would indicate a feature with an obfuscating influence on the network’s classification ability. Conversely, a large positive score would mean that the network was highly dependent on that feature to get the correct diagnosis, and suffered from its loss.
Statistical Analyses
To compare the performance of our algorithm against ground truth, we calculated the agreement between our method and human pathologists using a linear weighted κ, which is most useful to describe agreement when order is important. κ
 indicates no agreement,
indicates no agreement, 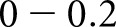 slight agreement,
slight agreement, 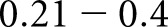 fair agreement,
fair agreement, 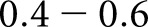 moderate agreement,
moderate agreement, 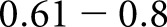 substantial agreement, and
substantial agreement, and 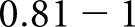 almost perfect agreement.20 For all comparisons, we computed the conditional probability of class assignment given ground truth, the proportion of agreement, and the
almost perfect agreement.20 For all comparisons, we computed the conditional probability of class assignment given ground truth, the proportion of agreement, and the  CIs for both of these sets of statistics. For calculation of 95% CIs on conditional probabilities, we used the Clopper–Pearson exact method.21
CIs for both of these sets of statistics. For calculation of 95% CIs on conditional probabilities, we used the Clopper–Pearson exact method.21
Data Sharing
Codes used to perform this study are made openly available at GitHub at https://github.com/SarderLab/DN_classification. We also provide our WSIs used for this analysis at https://buffalo.box.com/s/e40wzg2flb3p0r73zyhelhqvhle46vvr.
Results
An overview of the computational scheme is provided in Figure 1. The pipeline consisted of: (1) glomerular boundary detection (Begin-A), (2) glomerular nucleus boundary detection (A–B), (3) detection of glomerular components (B–C), (4) glomerular feature extraction (C–D, C–E), and (5) biopsy sample classification (D-stop, E-stop). Classification of an STZ DM mouse model is available in our Supplemental Material. See Supplemental Figure 1 and Supplemental Tables 2 and 3.
Glomerular Boundary Identification
The first step in automating analysis of glomerular disease is to automate glomerular identification. Much work has already been done in this area, and other groups have shown detection of glomeruli using CNNs.22,23 We developed the glomerular detection network in this work using our iterative whole-slide CNN training interface, HAIL. HAIL enables image data transmission to and from the DeepLab network by directly annotating and displaying predictions on WSIs.
High performance was achieved for glomerular boundary detection on a set of 11 holdout biopsy samples. The network scored balanced accuracy  (sensitivity
(sensitivity 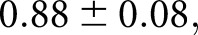 specificity
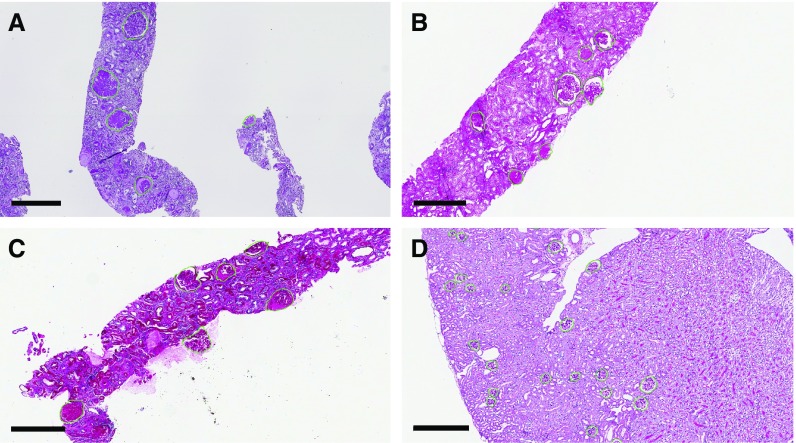
specificity  ). We reported balanced accuracy because the number of pixels in each class was extremely imbalanced. The network’s predicted glomerular boundaries are displayed in green in Figure 2. The network has learned an efficient representation of glomeruli such that it can recognize glomeruli from different disease stages (sclerotic and not), different source institutions (institute-1, Figure 2A, versus institute-2, Figure 2, B and C), staining preparations (preparation-1, Figure 2A, preparation-2, Figure 2B, preparation-3, Figure 2C), and species (human, Figure 2, A–C, mouse, Figure 2D).
). We reported balanced accuracy because the number of pixels in each class was extremely imbalanced. The network’s predicted glomerular boundaries are displayed in green in Figure 2. The network has learned an efficient representation of glomeruli such that it can recognize glomeruli from different disease stages (sclerotic and not), different source institutions (institute-1, Figure 2A, versus institute-2, Figure 2, B and C), staining preparations (preparation-1, Figure 2A, preparation-2, Figure 2B, preparation-3, Figure 2C), and species (human, Figure 2, A–C, mouse, Figure 2D).
Figure 2.
Accurate detection of glomerular boundaries from WSIs depicting PAS stained renal tissue. (A) Detection of glomeruli in human biopsy sample sourced and prepared in institute-2, with a purple appearance. (B) Detection of human glomeruli sourced from institute-2, prepared in a different institute than (A and C), with a pink appearance. Occasionally, two closely abutting glomeruli will be identified as one doublet object. (C) Detection of human glomeruli sourced from institute-2, prepared in a different institute than (A and B), with reddish-pink appearance. (D) Detection of glomeruli in mouse kidney sections. Scale bars, 400 µm.
Glomerular Component Analysis
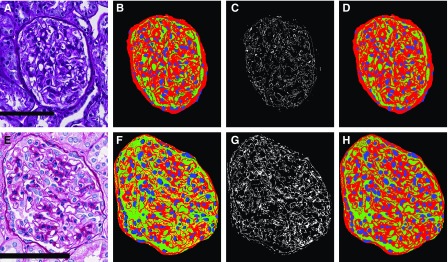
We developed a glomerular component analysis technique to describe glomerular structure; see Figures 3 and 4 for a demonstration on select glomeruli. We simplified the glomerulus into three components: (1) the nuclear component, (2) the PAS+ component, and (3) the luminal component. These components were selected because they simplify the complex glomerular system, facilitating computational detection in widely varying phenotypes and stain presentations (see Figure 4, D and H). Although we are sacrificing some technical accuracy, we will still show that the delineation of exact glomerular compartments is not required to classify glomerular structure.
Figure 4.
Glomerular components are detected consistently in images with varying presentation. (B) Glomerular component precursor mask. Red, PAS+ precursor mask; green, luminal precursor mask; blue, nuclear detections from DeepLab V2. (C) White pixels indicate regions from the glomerular component precursor mask which either have no detected label or are detected as both luminal and PAS+. (D) Naïve Bayesian classification correction of the glomerular component precursor mask, where every pixel has specifically one class label belonging to one of PAS+ (red), lumina (green), or nuclei (blue). (E–H) Identical computation as that shown in (A–D), but with a glomerulus from institute-2, preparation-3. Scale bars, 100 µm.
Nuclear Detection
Figure 3 shows nuclei segmentation performance for different institutes and preparations. We assessed network performance on  holdout glomeruli. A receiver operator curve for this strategy is shown in Figure 3G, along with examples of the network’s predictions. The red star marks the network’s default unweighted predictions,
holdout glomeruli. A receiver operator curve for this strategy is shown in Figure 3G, along with examples of the network’s predictions. The red star marks the network’s default unweighted predictions,  sensitivity and
sensitivity and 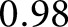 specificity, and the black star marks a weighting of
specificity, and the black star marks a weighting of  which optimized the network’s sensitivity and specificity to
which optimized the network’s sensitivity and specificity to 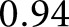 and
and  , respectively. As demonstrated, the network efficiently identified nuclei in widely varying stains and disease phenotypes.
, respectively. As demonstrated, the network efficiently identified nuclei in widely varying stains and disease phenotypes.
Luminal and PAS+ Image Analysis
Figure 4 shows detection of glomerular components. The luminal component identified capillaries and Bowman’s space (shown in green). The PAS+ component identified mesangium and basement membranes (red). Combining these with nuclear predictions from DeepLab resulted in the precursor glomerular component maps shown in Figure 4, B and F, which were corrupted with double predictions (yellow pixels, estimated as both luminal and PAS+) or missing predictions (black pixels, no predicted label). Corrupted pixels are shown as a binary mask in Figure 4, C and G. A naïve Bayesian classifier was trained on the RGB pixels which underlie the luminal and PAS+ pixels of the precursor masks, and was used to predict the pixel labels of the final masks (Figure 4, D and H). This method identified PAS+ components with average sensitivity/specificity 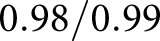 and luminal components
and luminal components 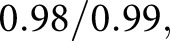 against sparse manual annotations of
against sparse manual annotations of 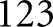 glomeruli.
glomeruli.
Glomerular Feature Extraction
We designed hand-crafted features to target pathologic progression of glomerular structure in DN, and allowed a neural network to combine these features and determine the final diagnosis. This allowed us to enforce biologic prior information onto the model without sacrificing the state-of-the-art performance of network learning. Supplemental Table 1 lists the  features explicitly. Our features can be categorized into six types: color, textural, morphologic, containment, interstructural distance, and intrastructural distance. One feature class which is easily interpretable is the intrastructural distance features. These were computed by performing a distance transform on a glomerular component and then obtaining a histogram of the result. A distance transform assigns each pixel in a binary image with the distance it has from the background. Examples are shown in Figure 5, C and F, where blue regions are low valued (close to background), and red are high valued (far from background). The glomerulus in Figure 5A, having only mild mesangial thickening, had maximum value in distance transform of
features explicitly. Our features can be categorized into six types: color, textural, morphologic, containment, interstructural distance, and intrastructural distance. One feature class which is easily interpretable is the intrastructural distance features. These were computed by performing a distance transform on a glomerular component and then obtaining a histogram of the result. A distance transform assigns each pixel in a binary image with the distance it has from the background. Examples are shown in Figure 5, C and F, where blue regions are low valued (close to background), and red are high valued (far from background). The glomerulus in Figure 5A, having only mild mesangial thickening, had maximum value in distance transform of 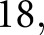 but the glomerulus in Figure 5D, where significantly more thickening was present, had maximum value
but the glomerulus in Figure 5D, where significantly more thickening was present, had maximum value  Further, clusters of high values corresponded to the thickest structures, such as Kimmelstiel–Wilson nodules in the glomerulus of Figure 5D. As mesangium progressively thickens over disease course, it is slowly reflected by shifts in the values of the distance transform.
Further, clusters of high values corresponded to the thickest structures, such as Kimmelstiel–Wilson nodules in the glomerulus of Figure 5D. As mesangium progressively thickens over disease course, it is slowly reflected by shifts in the values of the distance transform.
Classification of Biopsy Sample Structure
To incorporate all glomerular features of a single patient into a single output value, we used an RNN. RNNs are used to process sequential data, such as a time-series24–26 or written language.27–29 In our case, we used an RNN to process the glomerular features of a patient as a sequence, and funneled the output into a single, continuous number. We found that this method works exceptionally well to classify human DN biopsy samples. We compared this method against three pathologists, for  patients with DN (
patients with DN (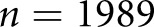 individual glomeruli) and
individual glomeruli) and  control tissues, using multiple schema. We also classified STZ mouse tissues in our Supplemental Material.
control tissues, using multiple schema. We also classified STZ mouse tissues in our Supplemental Material.
To train the algorithm, we took the renal pathologist with most experience as ground truth, and compared against two other renal pathologists. The second renal pathologist agreed with the ground truth with Cohen’s κ 0.48 (0.32 to 0.64) (brackets indicate  CI), and the third renal pathologist agreed with Cohen’s κ 0.68 (0.50 to 0.86) (Table 1). We first assessed a baseline classifier which selected predictions randomly with probability equal to the provided ground truth label distributions (
CI), and the third renal pathologist agreed with Cohen’s κ 0.68 (0.50 to 0.86) (Table 1). We first assessed a baseline classifier which selected predictions randomly with probability equal to the provided ground truth label distributions ( for classes I, IIa, IIb, III, and IV, respectively). The Cohen’s κ for this baseline classifier was less than zero; that is, no agreement over random chance. This proves that random guessing cannot agree with pathologist classification when continuity is taken into account. On the other hand, our RNN strategy agreed with the ground truth renal pathologist with Cohen’s κ 0.55 (0.50 to 0.6). This result was almost exactly between the other two human renal pathologists, suggesting that the network developed its own opinion on the interpretation of cases, without overfitting to the particular interpretation provided by the ground truth. Further, the 95% CIs suggest that the computational method was more precise when averaging over many trials (something not reasonable to request of a time-pressed clinician). To further validate, we computed two statistics describing per-class agreement, the proportions of agreement per class, and the conditional probability of class assignment given the ground truth assignment. We computed these measures between human annotators and ground truth as well as the computational method and ground truth (Table 2). Because our data were limited, we lacked sufficient data in some classes, making them difficult to learn and resulting in poor performance. The only class convincingly concurring to human levels with the ground truth was class III, which, not coincidentally, was the class with most data points. To increase our dataset’s size, we next investigated taking each available section of a given patient as a single data point. The biopsy samples contained anywhere between 1 and 4 adjacent sections that could be split into separate cases. We acknowledge that assuming each section as a separate case is biased, but it is motivated on the basis of a similar ideology in a recent work done by colleagues.30 Computational renal pathology is still in its genesis, and large, well curated image datasets for specific diseases are not available. In other computational histopathology studies,31 authors have shown excellent κ values for network approaches when using larger datasets (
for classes I, IIa, IIb, III, and IV, respectively). The Cohen’s κ for this baseline classifier was less than zero; that is, no agreement over random chance. This proves that random guessing cannot agree with pathologist classification when continuity is taken into account. On the other hand, our RNN strategy agreed with the ground truth renal pathologist with Cohen’s κ 0.55 (0.50 to 0.6). This result was almost exactly between the other two human renal pathologists, suggesting that the network developed its own opinion on the interpretation of cases, without overfitting to the particular interpretation provided by the ground truth. Further, the 95% CIs suggest that the computational method was more precise when averaging over many trials (something not reasonable to request of a time-pressed clinician). To further validate, we computed two statistics describing per-class agreement, the proportions of agreement per class, and the conditional probability of class assignment given the ground truth assignment. We computed these measures between human annotators and ground truth as well as the computational method and ground truth (Table 2). Because our data were limited, we lacked sufficient data in some classes, making them difficult to learn and resulting in poor performance. The only class convincingly concurring to human levels with the ground truth was class III, which, not coincidentally, was the class with most data points. To increase our dataset’s size, we next investigated taking each available section of a given patient as a single data point. The biopsy samples contained anywhere between 1 and 4 adjacent sections that could be split into separate cases. We acknowledge that assuming each section as a separate case is biased, but it is motivated on the basis of a similar ideology in a recent work done by colleagues.30 Computational renal pathology is still in its genesis, and large, well curated image datasets for specific diseases are not available. In other computational histopathology studies,31 authors have shown excellent κ values for network approaches when using larger datasets (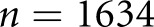 WSIs). We do not have the same magnitude of dataset available; however, by enforcing our biologic prior information, we were able to achieve a moderate κ in a small dataset nonetheless. Further, improved performance obtained as a result of case splitting would help us confirm that our technique was valid and that the patient-level analysis was not obtained by chance. Splitting the data increased the size to
WSIs). We do not have the same magnitude of dataset available; however, by enforcing our biologic prior information, we were able to achieve a moderate κ in a small dataset nonetheless. Further, improved performance obtained as a result of case splitting would help us confirm that our technique was valid and that the patient-level analysis was not obtained by chance. Splitting the data increased the size to 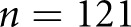 cases. Looking again to Tables 1 and 2, it can be observed that increasing the number of cases via sectioning enabled the network to learn a much higher level of agreement.
cases. Looking again to Tables 1 and 2, it can be observed that increasing the number of cases via sectioning enabled the network to learn a much higher level of agreement.
Table 1.
Agreement statistics for human annotators and computational technique
| Comparison | n | Observed κ | Lower 95% CI | Upper 95% CI |
|---|---|---|---|---|
| RP1 versus GT, scheme: T, data: patients | 48 | 0.48 | 0.32 | 0.64 |
| RP2 versus GT, scheme: T, data: patients | 48 | 0.68 | 0.50 | 0.86 |
| C versus GT, scheme: T, data: patients | 54 | 0.55 | 0.50 | 0.60 |
| C versus GT, scheme: T, data: sections | 121 | 0.9 | 0.87 | 0.93 |
| C versus GT, scheme: F, data: patients | 33 | 0.31 | 0.17 | 0.45 |
| C versus GT, scheme: F, data: sections | 85 | 0.98 | 0.94 | 1 |
| Baseline versus GT, scheme: T, data: patients | 54 | <0 | n/a | n/a |
Reported values include linear weighted Cohen’s κ and upper and lower 95% CIs. T refers to classifications according to the Tervaert scheme; F refers to classifications according to the Fogo scheme. Data description identifies whether the experiment was performed using separate patients or separate sections as individual data. RP, renal pathologist; GT, ground truth; C, computer; n/a, not applicable.
Table 2.
Conditional probability of class assignment, proportions of agreement, and 95% CIs for reported experiments
| Experiment Description | Comparison | Proportions of Agreement (95% CIs) | ||||
|---|---|---|---|---|---|---|
| I (n=11) | IIa (n=2) | IIb (n=12) | III (n=23) | IV (n=6) | ||
| RP1 versus GT, scheme: T, data: patients | Proportion of agreement | 1 (0.03 to 1) | 0.07 (0.004 to 0.34) | 0.26 (0.13 to 0.45) | 0.63 (0.44 to 0.78) | 0.13 (0.007 to 0.53) |
| P(RP1|GT) | 1 (0.29 to 1) | 0.5 (0.01 to 0.99) | 0.4 (0.19 to 0.64) | 0.71 (0.51 to 0.87) | 0.13 (0.003 to 0.53) | |
| RP2 versus GT, scheme: T, data: patients | Proportion of agreement | 1 (0.31 to 1) | 0.33 (0.02 to 0.87) | 0.64 (0.44 to 0.81) | 0.80 (0.61 to 0.92) | 0.33 (0.09 to 0.69) |
| P(RP2|GT) | 1 (0.29 to 1) | 0.50 (0.01 to 0.99) | 0.90 (0.68 to 0.99) | 0.86 (0.67 to 0.96) | 0.38 (0.09 to 0.76) | |
| C versus GT, scheme: T, data: patients | Proportion of agreement | 0.67 (0.58 to 0.76) | 0.03 (0.005 to 0.12) | 0.11 (0.07 to 0.17) | 0.47 (0.42 to 0.52) | 0.04 (0.01 to 0.13) |
| P(C|GT) | 0.67 (0.58 to 0.76) | 0.10 (0.01 to 0.32) | 0.18 (0.12 to 0.26) | 0.76 (0.66 to 0.81) | 0.05 (0.01 to 0.14) | |
| I (n=18) | IIa (n=6) | IIb (n=27) | III (n=58) | IV (n=12) | ||
| C versus GT, scheme: T, data: sections | Proportion of agreement | 0.90 (0.80 to 0.96) | 0.69 (0.48 to 0.85) | 0.72 (0.63 to 0.79) | 0.9 (0.85 to 0.93) | 0.70 (0.60 to 0.85) |
| P(C|GT) | 0.90 (0.81 to 0.96) | 0.75 (0.53 to 0.90) | 0.84 (0.76 to 0.91) | 0.97 (0.94 to 0.99) | 0.83 (0.70 to 0.93) | |
| Normal (n=2) | Mild (n=18) | Moderate (n=13) | Severe (n=0) | |||
| C versus GT, scheme: F, data: patients | Proportion of agreement | 0 (0 to 0.37) | 0.52 (0.42 to 0.62) | 0.43 (0.31 to 0.55) | X | |
| P(C|GT) | 0 (0 to 0.26) | 0.75 (0.66 to 0.83) | 0.60 (0.49 to 0.71) | |||
| Normal (n=6) | Mild (n=46) | Moderate (n=33) | Severe (n=0) | |||
| C versus GT, scheme: F, data: sections | Proportion of agreement | 1 (0.78 to 1) | 0.97 (0.93 to 0.99) | 0.96 (0.89 to 0.99) | X | |
| P(C|GT) | 1 (0.84 to 1) | 1 (0.97 to 1) | 0.96 (0.90 to 0.99) | |||
T refers to classifications according to the Tervaert scheme; F refers to classifications according to the Fogo scheme. Data description identifies whether the experiment was performed using separate patients or separate sections as individual data. Each value is reported with the format observed value (lower 95% CI to upper 95% CI). Columns also indicate the number of samples in each class. GT, ground truth; RP, renal pathologist; P(RP1|GT), conditional probability of the class selection by the RP1 given the GT class; P(C|GT), conditional probability of the class prediction by the C given the GT class; X, not applicable.
As secondary validation we also tested our method to classify data on the basis of a classification scheme defined by coauthor A.B.F. (see Methods). We did not have any cases classed as severe. Using patient-level holdout, the computational method agreed with κ 0.31 (0.17 to 0.45); however, we only had 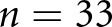 biopsy samples available which were classified according to this scheme. When we split the cases, the performance rose significantly to 0.98 (0.94 to 1), which was likely a result of increased homogeneity between data preparation and sections, and the reduced number of total classes (3).
biopsy samples available which were classified according to this scheme. When we split the cases, the performance rose significantly to 0.98 (0.94 to 1), which was likely a result of increased homogeneity between data preparation and sections, and the reduced number of total classes (3).
Table 3 demonstrates the network’s continuous nature of prediction by measurement of distance from true label. It was rare for either of the pathologists to overcall as compared with the ground truth, despite overall levels of agreement being quite different. The network took a more balanced approach of overcalls and undercalls. This was likely because pathologists have a conceptualization of the diagnostic tradeoff between undercalling and overcalling a case, and prefer to err on the lower side, whereas the network does not. This provided evidence that the network was learning a balanced approach to prediction of class, and a continuous progression through the data.
Table 3.
Distance metrics for human annotators and computational technique
| Comparison | Case Number | Fraction=GT | Average Distance <GT | Average Distance >GT |
|---|---|---|---|---|
| RP1 versus GT, scheme: T, data: patients | 48 | 0.56 | −1.19±0.48 | 2±0 (1 case) |
| RP2 versus GT, scheme: T, data: patients | 48 | 0.8 | −1.4±0.52 | 2±0 (2 cases) |
| C versus GT, scheme: T, data: patients | 54 | 0.5 | −0.61±0.57 | 0.57±0.60 |
| C versus GT, scheme: T, data: sections | 121 | 0.52 | −0.65±0.63 | 0.66±0.71 |
| C versus GT, scheme: F, data: patients | 33 | 0.65 | −0.36±0.4 | 0.40±0.47 |
| C versus GT, scheme: F, data: sections | 85 | 0.98 | −0.08±0.2 | 0.01±0.04 |
| Baseline versus GT, scheme: T, data: patients | 54 | 0.27 | −1.79±0.88 | 1.9±0.96 |
Distance is defined as the difference of the assigned label minus the ground truth label. Negative distances indicate undercalling; positive distances indicate overcalling. T refers to classifications according to the Tervaert scheme; F refers to classifications according to the Fogo scheme. Values are reported as mean±SD taken over all of the cases. Data description identifies whether the experiment was performed using separate patients or separate sections as individual data. GT, ground truth; RP, renal pathologist; C, computer.
Feature Relevance
We defined a study to identify which features were most important for classification, by sequentially removing features from the network at prediction time and comparing how the output changed at each step. The scoring function  defined in Equation 1 indicates with what magnitude the network’s predictions were deviated from baseline (baseline meaning no features dropped), after each feature had been dropped. That is, when looking at the average of diagnostic decisions across all of the patient data in this study, Equation 1 measured whether the loss of a feature improved the ability of the network to get the correct answer. Negative values would imply that the loss of a feature improved the network’s average performance, and positive values would imply the opposite. The raw numeric output from Equation 1 was difficult to interpret, so we standardized the features according to their mean and SD, and also normalized them by minimum and maximum. These numbers are available in Supplemental Table 1. The most important feature was the SD of red values in PAS+ regions (likely targets variability in staining intensity), with standardized value 9 deviations greater than average. The next two most important features, each with a standardized score near 5 deviations greater than average, were the mean nuclear blue values (likely decreases as nuclei become obscured by mesangium) and the deviation of PAS+ blue values (likely increases with increased nuclear stain in mesangium). A scatterplot of the raw deviation values (
defined in Equation 1 indicates with what magnitude the network’s predictions were deviated from baseline (baseline meaning no features dropped), after each feature had been dropped. That is, when looking at the average of diagnostic decisions across all of the patient data in this study, Equation 1 measured whether the loss of a feature improved the ability of the network to get the correct answer. Negative values would imply that the loss of a feature improved the network’s average performance, and positive values would imply the opposite. The raw numeric output from Equation 1 was difficult to interpret, so we standardized the features according to their mean and SD, and also normalized them by minimum and maximum. These numbers are available in Supplemental Table 1. The most important feature was the SD of red values in PAS+ regions (likely targets variability in staining intensity), with standardized value 9 deviations greater than average. The next two most important features, each with a standardized score near 5 deviations greater than average, were the mean nuclear blue values (likely decreases as nuclei become obscured by mesangium) and the deviation of PAS+ blue values (likely increases with increased nuclear stain in mesangium). A scatterplot of the raw deviation values (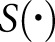 ) is also provided in Supplemental Figure 2, demonstrating that the network was most dependent on the color features (feature indices 215–232).
) is also provided in Supplemental Figure 2, demonstrating that the network was most dependent on the color features (feature indices 215–232).
Time and Space Complexity Analysis
Experiments were performed on a Linux distribution (Ubuntu 16.04) computer with an Intel Xeon E5–2630 CPU with 40 cores at 2.20 GHz, 64 GB of RAM, and 64 GB of swap memory, and an NVIDIA Titan X GPU with 12 GB of memory.
WSI chopping and prediction scaled with complexity 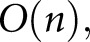 where
where 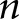 was number of pixels contained in a WSI. Glomerular component detection and feature extraction also scaled with
was number of pixels contained in a WSI. Glomerular component detection and feature extraction also scaled with  ; however, here
; however, here 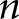 was number of glomeruli. The pairwise distance feature calculations scaled with
was number of glomeruli. The pairwise distance feature calculations scaled with  with
with  as number of objects to be compared; however, the number of glomerular compartments is assumed to be bounded finitely. The amount of time taken to provide a diagnosis for a biopsy sample took approximately 2 minutes for the smallest WSI, and up to 15 minutes for the largest. Memory use was most dictated by the size of image regions used to chop the WSIs, as can be seen by comparing the plots in Supplemental Figure 3, A and C (chopping image size
as number of objects to be compared; however, the number of glomerular compartments is assumed to be bounded finitely. The amount of time taken to provide a diagnosis for a biopsy sample took approximately 2 minutes for the smallest WSI, and up to 15 minutes for the largest. Memory use was most dictated by the size of image regions used to chop the WSIs, as can be seen by comparing the plots in Supplemental Figure 3, A and C (chopping image size  , maximum memory 25 and 15 GB, respectively) against Supplemental Figure 3B (chopping size
, maximum memory 25 and 15 GB, respectively) against Supplemental Figure 3B (chopping size  maximum memory 40 GB). Altering the chopping image size provided moderate time savings for increased memory usage, although we did not find this significant enough to warrant complete adoption of large chopping blocks.
maximum memory 40 GB). Altering the chopping image size provided moderate time savings for increased memory usage, although we did not find this significant enough to warrant complete adoption of large chopping blocks.
Comparing these results with humans, the ground truth renal pathologist took an average of  seconds to classify a biopsy sample case, the second renal pathologist
seconds to classify a biopsy sample case, the second renal pathologist  seconds, and the third
seconds, and the third 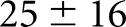 seconds. However, we do not believe we need to achieve these speeds to be effective. As an example, many clinical facilities are understaffed with respect to renal pathologists, especially on weekends and nights, and on-call pathologist readings are not as accurate or prognostic as those done by a renal pathologist.32 We envision one potential effect for our algorithm would be providing renal-pathologist–calibrated structural analysis to on-call pathologists to improve their structural readings. On a separate note, often, pathologists do not read digitized images the moment they are prepared. There is typically a turnaround time between specimen collection, preparation, and image digitization. The algorithm could easily be run on the digitized image during this turnaround time, and the prediction would be available before the pathologist reads the case. Therefore, the amount of space and time used by the algorithm is reasonable with respect to the computational resources accessible by large medical centers, and the relatively cheap cost of memory and computing power of modern hardware.
seconds. However, we do not believe we need to achieve these speeds to be effective. As an example, many clinical facilities are understaffed with respect to renal pathologists, especially on weekends and nights, and on-call pathologist readings are not as accurate or prognostic as those done by a renal pathologist.32 We envision one potential effect for our algorithm would be providing renal-pathologist–calibrated structural analysis to on-call pathologists to improve their structural readings. On a separate note, often, pathologists do not read digitized images the moment they are prepared. There is typically a turnaround time between specimen collection, preparation, and image digitization. The algorithm could easily be run on the digitized image during this turnaround time, and the prediction would be available before the pathologist reads the case. Therefore, the amount of space and time used by the algorithm is reasonable with respect to the computational resources accessible by large medical centers, and the relatively cheap cost of memory and computing power of modern hardware.
Discussion
Predictive capacity of automated digital quantitation is dictated first by preanalytic variance contributing to structural differences from image to image, and second by computational model. The mutability of algorithmic pipelines can be leveraged to conform to the amount of control one has over preanalytic variance. Given the extreme amount of heterogeneity embedded within the field of pathology, it is unlikely that any one-size-fits-all approaches will be fruitful, and selection of algorithmic pipelines to efficiently acquire and explore data will become increasingly important. With this theme in mind, we fused traditional and modern machine learning and image analysis to acquire glomerular morphometric data efficiently, and applied this strategy to evaluate DN classification as a proof of concept. Given the complexity of glomerular compartment distinction in PAS+ images, we simplified the glomerulus into three components on the basis of their appearance in PAS stains. In future work with more data, possibly annotated by modern imaging techniques such as multicolor immunofluorescence, we will be able to extend our method to detect actual glomerular compartments, which will likely increase performance and disease-relevant feature discovery.
Our developed pipeline is flexible for extension to any glomerular disease that is interpreted histologically, such as IgA nephropathy or lupus nephritis, and can also be trained to predict any numeric outcome label, e.g., proteinuria. This work was only a pilot study to understand whether computational diagnosis of renal tissue is possible as compared with a specific pathologist. However, our ultimate goal is to use pooled annotations to train a generalized network that represents the balanced opinion of many renal professionals.
Our work motivates the shift of pathologic diagnoses from discrete categories extrapolated from visual characterizations to continuous risk models derived from structural quantification. Continuous risk models could improve the precision with which disease is described, prompting improved prognostications. Combining high-level structural analysis with molecular or genomic information could help create next-generation individualized renal therapy, not only in detecting those with diabetes who are at risk of developing DN, but possibly also for many other difficult-to-treat diseases such as allograft nephropathy.
Disclosures
Prof. Sarder, Dr. Tomaszewski, Mr. Ginley, and Mr. Lutnick report Diabetic Complications Consortium grant DK076169 and grant R01DK114485 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), during the conduct of the study. Dr. Tomaszewski reports a grant from Neurovascular Diagnostics Inc. and from Inspirata Inc., outside the submitted work. In addition, Dr. Tomaszewski has a patent (1) “Malignancy Diagnosis Using Content-Based Image Retrieval of Tissue Histopathology,” Anant Madabhushi, Michael D. Feldman, John Tomaszewski, Scott Doyle, International Publication Number: WO 2009/017483 A1 issued; a patent (2) “Systems and Methods for Automated Detection of Cancer,” Anant Madabhushi, Michael D. Feldman, Jianbo Shi, Mark Rosen, John Tomaszewski, United States Serial Number (USSN): 60/852,516 issued; a patent (3) “System and Method for Image Registration,” Anant Madabhushi, Jonathan Chappelow, Mark Rosen, Michael Feldman, John Tomaszewski, USSN: 60/921 issued; and a patent (4) “Computer Assisted Diagnosis (CAD) of cancer using Multi-Functional Multi-Modal in vivo Magnetic Resonance Spectroscopy (MRS) and Imaging (MRI),” by Anant Madabhushi, Satish Viswanath, Pallavi Tiwari, Robert Toth, Mark Rosen, John Tomaszewski, Michael D. Feldman, PCT/US08/81656, Oct 2008 issued.
Funding
The project was supported by the faculty startup funds from the Jacobs School of Medicine and Biomedical Sciences, University at Buffalo; the https://doi.org/10.13039/100008209 Innovative Micro-Programs Accelerating Collaboration in Themes (IMPACT) award; National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Diabetic Complications Consortium grant DK076169; and NIDDK grant R01 DK114485.
Supplementary Material
Acknowledgments
We thank NVIDIA Corporation for the donation of the Titan X Pascal GPU used for this research (NVIDIA, Santa Clara, CA). We thank Diane Salamon for assistance in gathering diabetic nephropathy samples from the Kidney and Translational Research Core at Washington University in Saint Louis School of Medicine. We thank Ellen Donnert for her assistance in selecting diabetic nephropathy biopsy samples from the Vanderbilt University Medical Center collection. We acknowledge the assistance of the Histology Core Laboratory and Multispectral Imaging Suite in the Department of Pathology and Anatomical Sciences, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo.
Mr. Ginley designed, implemented, and conceptualized the computational detections; defined the glomerular pathobiologic computational features; designed network architecture; analyzed the results; and wrote the manuscript. Mr. Lutnick assisted in conceptualizing and designing the architecture and nuclear segmentation weighting scheme. Dr. Jen provided diabetic nephropathy diagnoses, critically assessed the manuscript with respect to the renal pathology domain, and conceptualized the study to maximize the effect for point of care. Dr. Fogo and Dr. Jain organized and curated the human data. Dr. Rosenberg, Dr. Rossi, and Dr. Walavalkar provided diabetic nephropathy diagnoses. Dr. Wilding assisted with statistical analysis. Dr. Tomaszewski mentored the team on diabetic nephropathy glomerular pathobiology. Dr. Yacoub implemented the streptozotocin mouse model. Prof. Sarder conceived the overall research scheme, coordinated with the study team, conceptualized the overall study design and the statistical performance analysis, supervised the computational implementation, critically analyzed the results, and assisted in manuscript preparation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018121259/-/DCSupplemental.
Supplemental Figure 1. Glomerular component maps for murine glomeruli.
Supplemental Figure 2. Deviation of network predictions as a function of dropped features.
Supplemental Figure 3. Memory as a function of run time for application of our algorithm in select cases.
Supplemental Table 1. Feature list and associated network impact scores.
Supplemental Table 2. Linear weighted Cohen’s kappa and confidence limits for mouse experiments.
Supplemental Table 3. Conditional probabilities for class assignment in mouse experiments given the ground truth class assignment.
References
- 1.CDC : Diabetes report card. 2014. Available at: https://www.cdc.gov/diabetes/library/reports/congress.html
- 2.Tervaert TW, Mooyaart AL, Amann K, Cohen AH, Cook HT, Drachenberg CB, et al.: Renal Pathology Society : Pathologic classification of diabetic nephropathy. J Am Soc Nephrol 21: 556–563, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL: DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 40: 834–848, 2018 [DOI] [PubMed] [Google Scholar]
- 4.Lutnick B, Ginley B, Govind D, McGarry SD, LaViolette PS, Yacoub R, et al. : An integrated iterative annotation technique for easing neural network training in medical image analysis. Nat Mach Intell 1: 112–119, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otsu N: A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern Syst 9: 62–66, 1976 [Google Scholar]
- 6.Bradley D, Roth G: Adaptive thresholding using the integral image. J Graphics Tools 12: 13–21, 2007 [Google Scholar]
- 7.Ruifrok AC, Johnston DA: Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23: 291–299, 2001 [PubMed] [Google Scholar]
- 8.Meyer F, Beucher S: Morphological segmentation. J Vis Commun Image Represent 1: 21–46, 1990 [Google Scholar]
- 9.Maurer C: A linear time algorithm for computing exact euclidean distance transforms of binary images in arbitrary dimensions. IEEE Trans Pattern Anal Mach Intell 25: 265–270, 2003 [Google Scholar]
- 10.Paglieroni DW: Distance transforms - properties and machine vision applications. Cvgip-Graph Model Im 54: 56–74, 1992 [Google Scholar]
- 11.Rosenfel A, Pfaltz JL: Sequential operations in digital picture processing. J Assoc Comput Mach 13: 471–491, 1966 [Google Scholar]
- 12.Veta M, Huisman A, Viergever MA, van Diest PJ, Pluim JPW: Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images. Presented at the 2011 8th IEEE International Symposium on Biomedical Imaging, Chicago, IL, 2011, pp 618–621 [Google Scholar]
- 13.Smith AR: Color gamut transform pairs. Comput Graph 12: 12–19, 1978 [Google Scholar]
- 14.McLAREN K. XIII—The Development of the CIE : 1976 (L* a* b*) uniform colour space and colour-difference formula. J Soc Dyers Colour 92: 338–341, 1976 [Google Scholar]
- 15.Hastie T, Tibshirani R, Friedman J: The Elements of Statistical Learning, 2nd Ed., Berlin, Germany, Springer, 2008 [Google Scholar]
- 16.Haralick RM, Shanmugam K, Dinstein I: Textural features for image classification. IEEE T Syst Man Cyb 3: 610–621, 1973 [Google Scholar]
- 17.Hochreiter S, Schmidhuber J: Long short-term memory. Neural Comput 9: 1735–1780, 1997 [DOI] [PubMed] [Google Scholar]
- 18.Mass AL, Haas AYH, Ng AY: Rectifier Nonlinearities Improve Neural Network Acoustic Models, Atlanta, GA, ICML; 2013 [Google Scholar]
- 19.Diederik P, Kingma JLB: Adam: A method for stochastic optimization. Presented at the International Conference on Learning Representations, 2015 [Google Scholar]
- 20.Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 33: 159–174, 1977 [PubMed] [Google Scholar]
- 21.Clopper CJ, Pearson ES: The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26: 404–413, 1934 [Google Scholar]
- 22.Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, et al. : Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 29: 2081–2088, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedraza A, Gallego J, Lopez S, Gonzalez L, Laurinavicius A, Bueno G: Glomerulus classification with convolutional neural networks. In: Medical Image Understanding and Analysis. MIUA 2017, edited by Valdés Hernández M, González-Castro V, Cham, Switzerland, Springer International Publishing, 2017, pp 839–849 [Google Scholar]
- 24.Cheng H, Tan P-N, Gao J, Scripps J, editors: Multistep-Ahead Time Series Prediction. Advances in Knowledge Discovery and Data Mining, Berlin, Springer, 2006 [Google Scholar]
- 25.Ho SL, Xie M, Goh TN: A comparative study of neural network and Box-Jenkins ARIMA modeling in time series prediction. Comput Ind Eng 42: 371–375, 2002 [Google Scholar]
- 26.Che Z, Purushotham S, Cho K, Sontag D, Liu Y: Recurrent neural networks for multivariate time series with missing values. Sci Rep 8: 6085, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikolov T, Zweig G: Context dependent recurrent neural network language model. Presented at the 2012 IEEE Spoken Language Technology Workshop (SLT), 2–5 Dec, 2012 [Google Scholar]
- 28.Mikolov T, Kombrink S, Burget L, Černocký J, Khudanpur S: Extensions of recurrent neural network language model. Presented at the 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 22–27 May, 2011 [Google Scholar]
- 29.Young T, Hazarika D, Poria S, Cambria E: Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 13: 55–75, 2018 [Google Scholar]
- 30.Kolachalama VB, Singh P, Lin CQ, Mun D, Belghasem ME, Henderson JM, et al. : Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 3: 464–475, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. : Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24: 1559–1567, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas M: Donor kidney biopsies: Pathology matters, and so does the pathologist. Kidney Int 85: 1016–1019, 2014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.