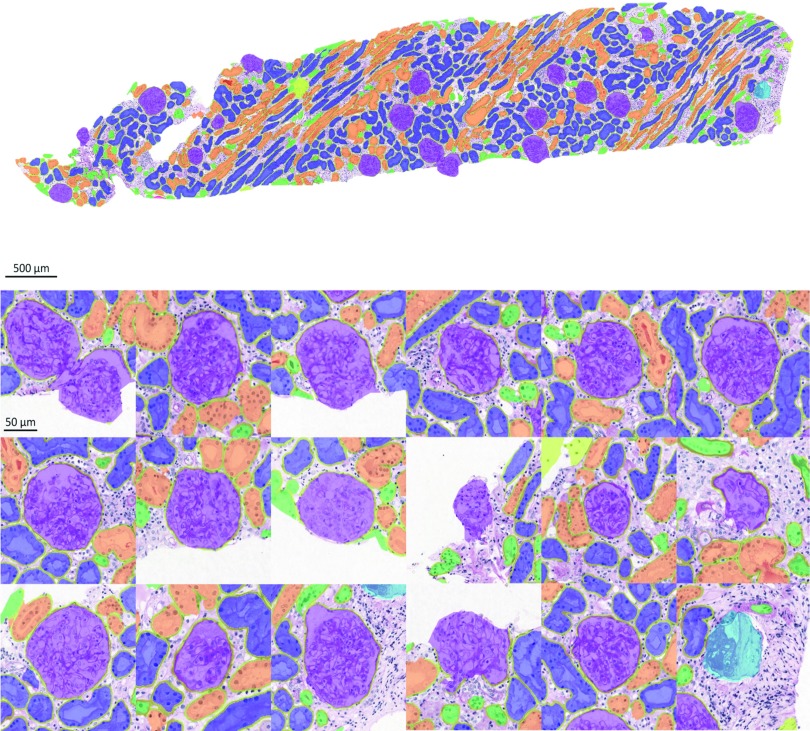
Significance Statement
Histopathologic assessment of kidney tissue currently relies on manual scoring or traditional image-processing techniques to quantify and classify tissue features, time-consuming approaches that have limited reproducibility. The authors present an alternative approach, featuring a convolutional neural network for multiclass segmentation of kidney tissue in sections stained by periodic acid–Schiff. Their findings demonstrate applicability of convolutional neural networks for tissue from multiple centers, for biopsies and nephrectomy samples, and for the analysis of both healthy and pathologic tissues. In addition, they validated the network’s results with components from the Banff classification system. Their convolutional neural network may have utility for quantitative studies involving kidney histopathology across centers and potential for application in routine diagnostics.
Keywords: deep learning, kidney transplantation, histopathology, Banff classification
Visual Abstract
Abstract
Background
The development of deep neural networks is facilitating more advanced digital analysis of histopathologic images. We trained a convolutional neural network for multiclass segmentation of digitized kidney tissue sections stained with periodic acid–Schiff (PAS).
Methods
We trained the network using multiclass annotations from 40 whole-slide images of stained kidney transplant biopsies and applied it to four independent data sets. We assessed multiclass segmentation performance by calculating Dice coefficients for ten tissue classes on ten transplant biopsies from the Radboud University Medical Center in Nijmegen, The Netherlands, and on ten transplant biopsies from an external center for validation. We also fully segmented 15 nephrectomy samples and calculated the network’s glomerular detection rates and compared network-based measures with visually scored histologic components (Banff classification) in 82 kidney transplant biopsies.
Results
The weighted mean Dice coefficients of all classes were 0.80 and 0.84 in ten kidney transplant biopsies from the Radboud center and the external center, respectively. The best segmented class was “glomeruli” in both data sets (Dice coefficients, 0.95 and 0.94, respectively), followed by “tubuli combined” and “interstitium.” The network detected 92.7% of all glomeruli in nephrectomy samples, with 10.4% false positives. In whole transplant biopsies, the mean intraclass correlation coefficient for glomerular counting performed by pathologists versus the network was 0.94. We found significant correlations between visually scored histologic components and network-based measures.
Conclusions
This study presents the first convolutional neural network for multiclass segmentation of PAS-stained nephrectomy samples and transplant biopsies. Our network may have utility for quantitative studies involving kidney histopathology across centers and provide opportunities for deep learning applications in routine diagnostics.
Quantification and classification of tissue features are important elements of the histopathologic assessment of renal tissue. In routine diagnostics for example, biopsy quality is assessed by glomerular counting, and kidney transplant biopsies are scored extensively with the Banff classification system. Likewise, chronic damage is usually assessed by visual estimation of the extent of interstitial fibrosis and the fraction of atrophic tubuli.1,2 Visual estimation of tissue features can be strengthened by application of digital image analysis techniques. Computer algorithms can increase reproducibility, which is less optimal for human observers, and may increase discriminative power to detect subtle yet relevant pathologic changes. Automated assessment can also overcome the tedious nature of visual assessment, which can be a limiting factor in large studies.
Digital image analysis has been studied widely to enable high-throughput, accurate, and reproducible assessment of digitized microscopic images of kidney tissue sections. Most research in this area has been performed with “traditional” image-processing techniques.3–9 Although these traditional techniques are valuable for studies that are limited in scope and size, the generalization to larger-scale applications and to multicenter data sets with inherently more variation in terms of tissue quality, staining and digitization is problematic because these techniques are insufficiently robust to these variations and therefore require manual intervention (e.g., thresholding).10
Advances in machine learning (mainly the emergence of deep neural networks, collectively called “deep learning”) combined with the possibility to digitize entire tissue sections at microscopic resolution within minutes (whole-slide images [WSIs]) have paved the way for more advanced digital analysis of histopathologic images.11–13 Deep learning techniques allow autonomous learning of increasingly complex structures during the transformation from input (WSI) to desired output (e.g., structure detection). The most widely applied deep learning models for analysis of images are so-called convolutional neural networks (CNNs). CNNs have only recently been introduced in kidney histopathology and focus merely on the detection of glomeruli, leaving other relevant structures unaddressed.14,15
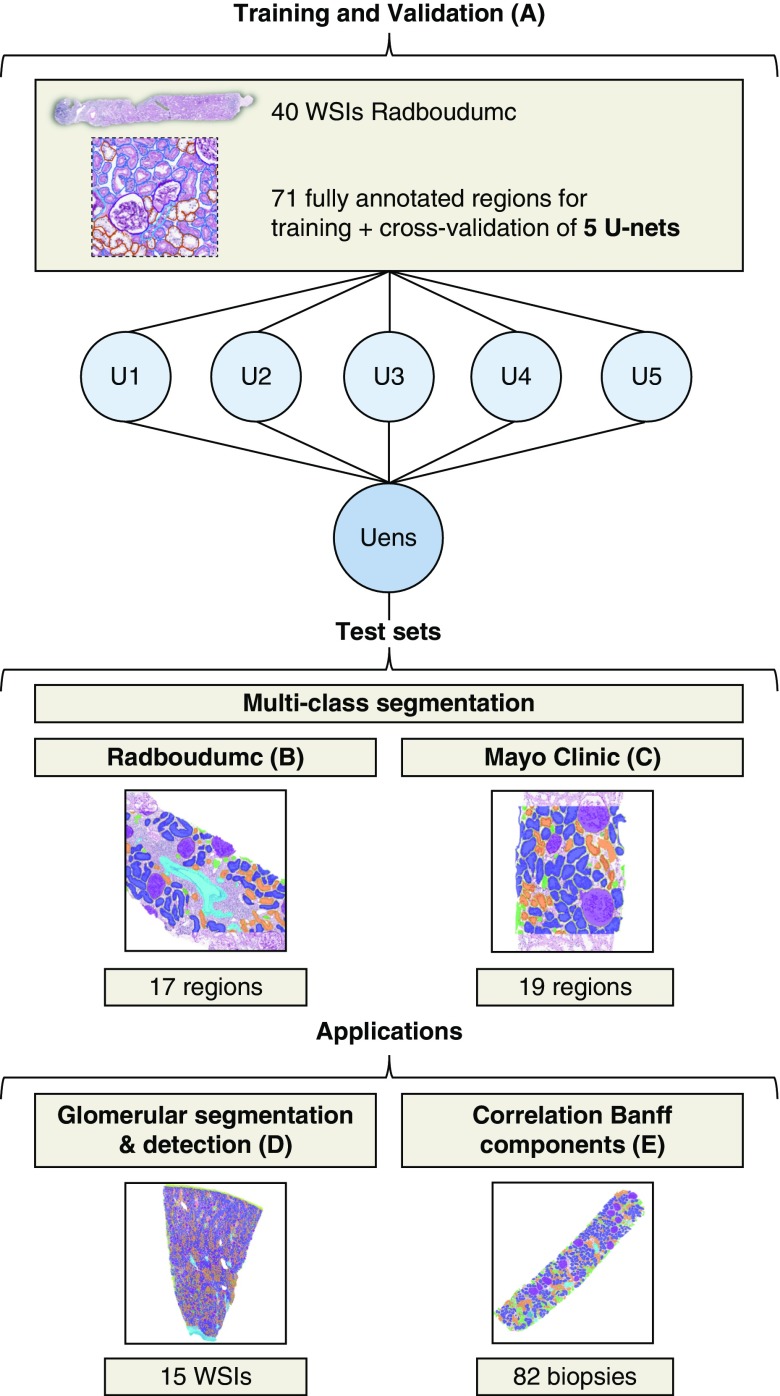
The aim of this study was to develop and validate a CNN for histologic analysis in renal tissue stained with periodic acid–Schiff (PAS). To achieve this goal, we addressed four major objectives (Figure 1). First, the CNN should be able to accurately segment cortical regions of both healthy and pathologic renal tissue biopsies into multiple tissue classes, e.g., (sclerotic) glomeruli, (proximal, distal, and atrophic) tubuli, and interstitium (Figure 1B). Second, digital pathology facilitates large-scale, multicentric studies and intercollegial consultations by telepathology.16 Therefore, the CNN should have comparable multiclass segmentation performance on tissue that is processed, stained, and scanned at an external center (Figure 1C). Third, although the network was trained on biopsy material, the application of the CNN should not be limited to biopsies only. Therefore, we aimed to fully segment tumor nephrectomy samples containing both cortex and medulla. As a proof of concept, we report the detection rate and segmentation performance for (sclerotic) glomeruli by the CNN on these larger tissue sections (Figure 1D). Finally, we assessed the applicability of deep learning in kidney transplant pathology by comparing quantifications of our CNN to manually scored elements of the Banff classification system by multiple renal pathologists (Figure 1E).
Figure 1.
A summarizing overview of the image sets used and their corresponding objectives. (A) Five U-nets were trained using kidney biopsies from Radboudumc and applied as an ensemble (Uens) on several data sets. The multiclass segmentation performance was assessed on (B) ten kidney transplant biopsies from Radboudumc, and (C) ten kidney transplant biopsies from Mayo Clinic as an external data set. Data set D was used to assess the network’s ability to segment and detect glomeruli on WSI level in 15 large tissue specimens obtained after nephrectomies. Data set E served to assess the CNN’s routine examination of 82 kidney transplant biopsies using the Banff classification.
Methods
Tissue Samples
Transplant Biopsies from Radboudumc
We used Bouin-fixed, paraffin-embedded needle-core biopsies that were obtained on indication from 101 patients who underwent a kidney transplantation between 2008 and 2012 in the Radboud University Medical Center, Nijmegen, The Netherlands (Radboudumc). PAS-stained slides (n=132) were collected from the Radboudumc pathology archives. The majority of the slides were stained between 2008 and 2012, whereas for 15 cases 3-µm-thick slides were newly cut and stained because the original PAS-stained slides were not available. Slide digitization was performed using a Pannoramic 250 Flash II digital slide scanner (3DHistech, Budapest, Hungary) with a 20× objective at a resolution of 0.24 µm/pixel. A total of 40 WSIs were used for training and validation of the CNN (Figure 1A), and ten WSIs for testing the multiclass segmentation performance (Figure 1B). Validation with elements of the Banff classification system, manually scored by multiple experienced renal pathologists, was conducted on 82 glass slides and their corresponding WSIs (Figure 1E). The need for approval for use of any of the Radboudumc tissues used in this study was waived by the local Institutional Review Board (IRB; #2016-2269).
Transplant Biopsies from External Center
Needle-core biopsies were taken at the time of transplantation from living donor kidneys at the Mayo Clinic (Rochester, MN). Biopsy specimens were formalin fixed, paraffin embedded, cut in to 3-µm sections, and stained using PAS reagent. Digital images were created using Aperio’s ScanScope XT System scanner (Leica Biosystems, Wetzlar, Germany) with a 20× objective at a resolution of 0.49 µm/pixel as part of one of the studies on renal aging by Denic et al.17 Ten WSIs from ten biopsies obtained between 2002 and 2008 were used for validation of the CNN trained at Radboudumc (Figure 1C). Biopsy slides were scanned with IRB approval (#17-002391 and 10-004644). Furthermore, external file transfer after all slides were de-identified was approved under IRB #18-005592.
Nephrectomy Specimens
Macroscopically normal kidney tissue was obtained from surgically removed kidneys of nine patients with renal cell carcinoma. We selected 15 formalin-fixed, paraffin-embedded tissue blocks that were sectioned at 3 µm and stained using PAS reagent. Absence of malignant lesions was confirmed by microscopic assessment. Glass slides were digitized as described above (see Transplant Biopsies from Radboudumc).
CNN Development and Design
Ground Truth Training and Test Sets
In 50 WSIs (Radboudumc) and ten WSIs (Mayo Clinic) of PAS-stained transplant biopsies, a human observer randomly selected one or two rectangular regions of approximately 720×960 µm2 (3000×4000 pixels). These regions were subsequently exhaustively annotated, using an automated slide analysis platform software (ASAP; version 1.8, available as open-source software from https://github.com/computationalpathologygroup/ASAP). The following predefined classes were applied: “glomeruli,” “sclerotic glomeruli,” “empty Bowman’s capsules,” “proximal tubuli,” “distal tubuli,” “atrophic tubuli,” “undefined tubuli,” “capsule,” “arteries,” and “interstitium.” Globally sclerosed glomeruli were labeled as sclerotic glomeruli. All nonglobally sclerotic glomeruli, thus healthy and segmentally sclerotic glomeruli, were labeled as glomeruli. The undefined-tubuli class was used for tubuli that could not be classified. Thin ascending limbs of Henle, convoluted distal tubuli, and cortical collecting ducts were collectively labeled as distal tubuli. All remaining unannotated tissue, including smaller vessels, was labeled as interstitium. To facilitate accurate detection of individual objects, single structure delineation is needed. An additional class, representing the border of all structures, was therefore included. The outer rim of every annotated object (measuring on average 4 pixels) was automatically determined and assigned to this additional “border” class. All annotations were checked and corrected where necessary by an experienced renal pathologist. The 50 annotated WSIs from Radboudumc were divided into a set for training and validation (n=40) and testing (n=10). The ten annotated WSIs from the Mayo Clinic were used as an additional external test set (Table 1).
Table 1.
Number of annotations per class used in the training, validation, and test sets of the CNN
| Class | Training | Validation 1 | Validation 2 | Validation 3 | Validation 4 | Validation 5 | Test Radboudumc | Test Mayo Clinic |
|---|---|---|---|---|---|---|---|---|
| Glomeruli | 84 | 12 | 10 | 12 | 19 | 6 | 39 | 37 |
| Sclerotic glomeruli | 7 | 1 | 1 | 1 | 1 | 2 | 5 | 1 |
| Empty Bowman’s capsules | 5 | 2 | 1 | 1 | 3 | 1 | 5 | 3 |
| Proximal tubuli | 1941 | 433 | 253 | 139 | 346 | 145 | 636 | 1060 |
| Distal tubuli | 1321 | 274 | 224 | 141 | 248 | 156 | 374 | 579 |
| Atrophic tubuli | 1160 | 88 | 124 | 198 | 180 | 232 | 328 | 12 |
| Undefined tubuli | 883 | 186 | 42 | 174 | 222 | 73 | 495 | 69 |
| Arteries | 43 | 4 | 1 | 5 | 1 | 1 | 11 | 14 |
| Interstitium | 44 | 6 | 5 | 6 | 5 | 5 | 17 | 19 |
| Capsule | 4 | 2 | 1 | 1 | 1 | 1 | 4 | —a |
The Mayo Clinic WSIs did not contain any capsular structures.
CNN Design
For our CNN design, we chose a U-net architecture because this has been proven to be specifically powerful for tissue segmentation.18 To train the U-net architecture, we subdivided the 40 WSIs in the training subset into five sets for crossvalidation, each consisting of training (n=37) and validation (n=3) WSIs. Crossvalidation is often applied in smaller data sets to account for variations that naturally occur in the training data, preventing over-fitting on a specific validation subset. On each of the folds, a U-net was independently trained for 100 epochs, at 300 iterations per epoch with batch sizes of six patches (412×412 pixels at a resolution of 0.96 µm/pixel). Spatial (rotation, flipping, elastic deformation, zooming) and color (brightness, contrast, saturation, hue shifting, Gaussian noise, Gaussian blur) augmentation techniques were applied to improve the algorithm’s robustness for variation in tissue morphology and staining.19 Adam was used as learning rate optimization algorithm20 and categorical cross entropy as loss function. The five U-net models were applied as an ensemble for segmentation of all image sets. The probability per pixel for all five U-nets was averaged per class, and the class with the subsequent highest probability was assigned as predicted label, defined as:
 |
Postprocessing
The CNN provides a segmentation mask where individual pixels are assigned to one of the predefined classes. To convert labeled pixels into meaningful objects (e.g., a glomerulus), all pixels within an area that was entirely surrounded by pixels labeled as either border or intersitium were considered to form one object. As a first postprocessing step, objects smaller than 300 pixels (the size of the smallest annotated tubuli) were assigned to the interstitium class. Second, if an object consisted of multiple class labels, the most predominant class, the class exceeding at least 35% of the area, was assigned to the entire object.
Assessments of CNN Performance
Multiclass Segmentation Performance
The CNN’s multiclass segmentation performance in kidney transplant biopsies was assessed using the Dice coefficient (DC) on ten WSIs from Radboudumc and ten WSIs from the Mayo Clinic. Ground truth annotations were produced as reported above (see Ground Truth Training and Test Sets). The DC measures the spatial overlap between ground truth (A) and segmentation result (B) and is defined as DC(A, B)=2(|A|∩|B|)/(|A|+|B|), where ∩ is the intersection. DC ranges from zero (no pixels in common between ground truth and segmentation result) to one (perfect agreement). DCs are presented per class and as a weighted mean DC (correcting for unequal class representation). We also report the DC for all tubuli classes combined.
Glomerular Segmentation and Detection on Nephrectomy Specimens
All healthy, segmentally and globally sclerotic glomeruli in 15 WSIs of PAS-stained nephrectomy sections were annotated by a human observer using ASAP. In addition to segmentation performance (expressed in the DC), we assessed the network’s ability to detect the glomeruli inside of these large specimens. All annotated glomeruli with overlap with the CNN’s segmentation were considered to be detected objects. Segmentations located entirely outside an annotated glomerulus were noted as false positive detections.
CNN for Scoring Components of the Banff Classification System
The CNN was validated for diagnostic application by comparing the network’s output with histologic components visually scored by multiple pathologists. In 82 WSIs of PAS-stained transplant biopsies, one cortical biopsy specimen was selected and analyzed using our CNN. For this part of the study, we derived quantitative and morphometric data from the segmentation mask of several classes. The sum of the objects labeled as glomeruli and sclerotic glomeruli was used as glomerular count. The number of pixels labeled as interstitium was divided by the total number of segmented pixels to calculate the area percentage of interstitium. The number of objects labeled as atrophic tubuli was divided by the sum of objects labeled as one of the four tubuli classes, to determine the proportion of atrophic tubuli. The identical biopsy specimen was marked on each of the 82 corresponding glass slides and visually scored by multiple pathologists. Glomeruli were counted (E.J.S., J.K., and J.T.H.R.) and the intertubular area percentage was estimated in steps of ten percent (J.K. and J.T.H.R.). Also, the extent of interstitial fibrosis (ci score), total inflammation (ti score), and tubular atrophy (ct score) was scored (E.J.S., J.K., and S.F.). Interstitial fibrosis and tubular atrophy (IFTA) grades were derived from the pathologists’ ci and ct scores, following the Banff reference guide 2018.21
For glomerular counting, the intraclass correlation coefficients (ICCs) among the pathologists and the CNN were calculated. For each case, the pathologists’ average intertubular area percentage, ci score, ti score, ct score, and IFTA grading were compared with the percentage for interstitium and atrophic tubuli, calculated by the CNN, respectively. This correlation was assessed by calculating the Spearman correlation coefficient and the coefficient of determination (R2).
Interobserver agreement between pathologists was assessed by calculating linear weighted κ values for ci, ti, and ct scores and by calculating the ICCs for intertubular area percentage. We will report the average κ values and ICCs. No κ values were calculated for IFTA grading as these were derived from the pathologists’ ci and ct score, and not given by the pathologists themselves.
Results
Multiclass Segmentation Performance in the Radboudumc Test Set
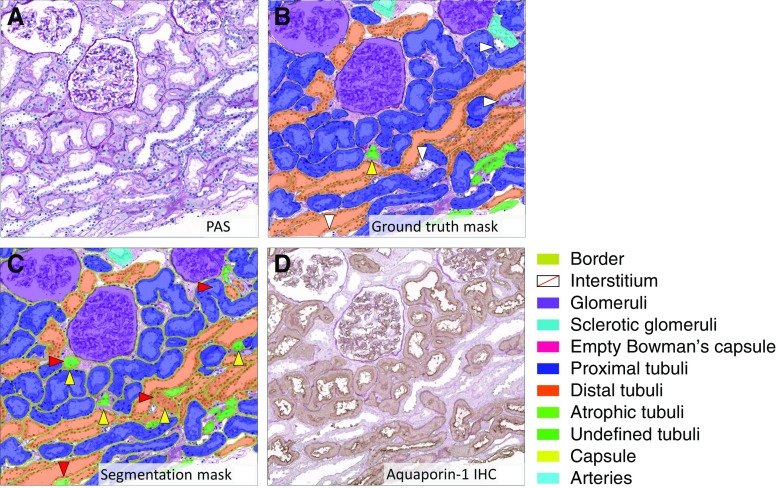
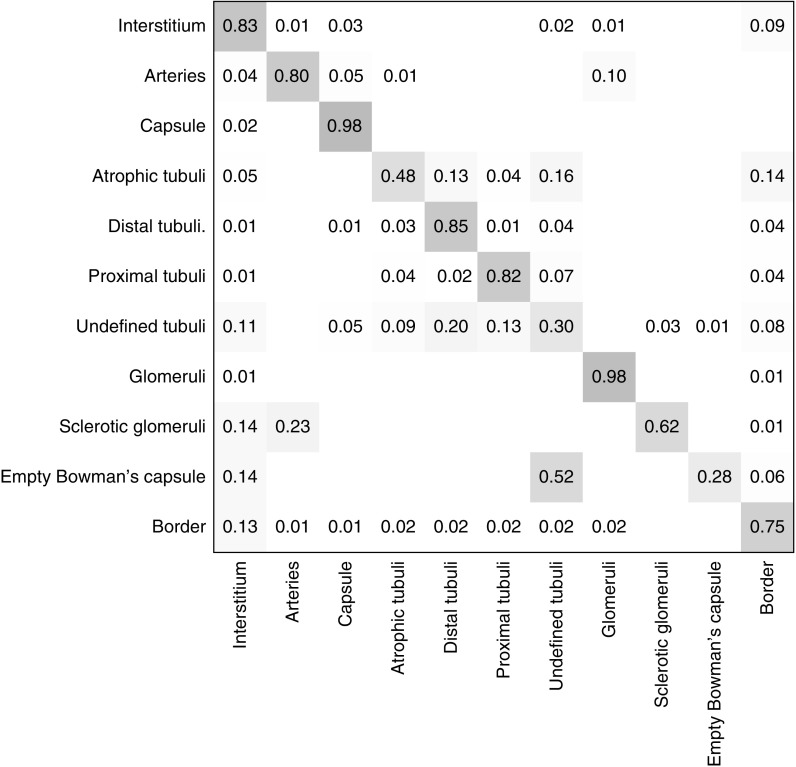
A representative example of a ground truth and segmentation mask as used in the test set is depicted in Figure 2. The CNN’s multiclass segmentation performance was assessed on ten WSIs from Radboudumc kidney transplant biopsies and calculated using the DC. The highest DC was obtained for the segmentation of healthy and segmentally sclerotic glomeruli, represented in the glomeruli class, followed by the interstitium, capsule, and proximal tubuli classes (Table 2). Lower DCs were observed for empty Bowman’s capsules, undefined tubuli, and atrophic tubuli. The test set’s confusion matrix illustrates how misclassified tubuli are often segmented as one of the other classes of tubuli (Figure 3). When misclassification of tubuli subtypes was disregarded, the tubuli were the second-best segmented structures (tubuli combined, Table 2). The overall performance of the CNN was assessed by calculating the weighted mean DC, which remained unaltered after postprocessing (Table 2).
Figure 2.
Region of PAS-stained slide with ground truth, segmentation by the CNN, and immunohistochemical staining (Aquaporin-1). (A) Represents regions that were used for testing of the CNN (PAS, Radboudumc). (B) The mask of the manually produced annotations (ground truth). (C) The CNN’s result. (D) For illustrative purposes, the PAS slide was restained using anti-Aquaporin-1 antibody, highlighting proximal tubuli. Red arrowhead highlights inconsistency between CNN and ground truth; yellow arrowhead highlights inconsistency with the anti-Aquaporin-1 staining; white arrowhead highlights annotation error. The ground truth and the output of the network overlap largely with the immunohistochemical staining, illustrating the high quality of both.
Table 2.
DCs per class and weighted mean DC on ten WSIs of kidney biopsies from Radboudumc and the Mayo Clinic
| Feature | Radboudumc | Mayo Clinic | ||
|---|---|---|---|---|
| DC Before Postprocessing | DC After Postprocessing | DC Before Postprocessing | DC After Postprocessing | |
| Class | ||||
| Glomeruli | 0.95 | 0.95 | 0.94 | 0.94 |
| Sclerotic glomeruli | 0.63 | 0.62 | 0.00 | 0.00 |
| Empty Bowman’s capsules | 0.52 | 0.37 | 0.44 | 0.00 |
| Proximal tubuli | 0.86 | 0.87 | 0.91 | 0.92 |
| Distal tubuli | 0.82 | 0.81 | 0.72 | 0.71 |
| Atrophic tubuli | 0.48 | 0.49 | 0.10 | 0.11 |
| Undefined tubuli | 0.32 | 0.30 | 0.12 | 0.10 |
| Tubuli combined | 0.93 | 0.92 | 0.92 | 0.92 |
| Arteries | 0.69 | 0.70 | 0.47 | 0.55 |
| Interstitium | 0.88 | 0.88 | 0.76 | 0.76 |
| Capsule | 0.89 | 0.84 | —a | —a |
| Weighted mean | 0.80 | 0.80 | 0.84 | 0.84 |
| Weighted mean with tubuli combined | 0.88 | 0.88 | 0.87 | 0.88 |
The Mayo Clinic WSIs did not contain any capsular structures.
Figure 3.
Confusion matrix for the U-net ensemble on the Radboudumc test set for multiclass segmentation performance in kidney transplant biopsies. Confusion matrices provide insight on how predictions are distributed over the different classes. In this figure, the ground truth labels are given vertically and the predicted labels by the CNN are written on the horizontal axis. Here can be seen that, e.g., 98% of all pixels with ground truth label glomeruli, were classified as glomeruli by the CNN ensemble.
Multiclass Segmentation Performance in Mayo Clinic Test Set
To study the CNN’s multiclass segmentation performance on material that was processed, stained, and scanned at an external center, we used ten WSIs from Mayo Clinic kidney transplant biopsies (Table 2). The weighted mean DC was slightly higher than that in the Radboudumc test set, but also here the highest DCs were obtained for glomeruli, proximal tubuli, and interstitium. In this test set of healthy donor kidney biopsies only one globally sclerotic glomerulus and one empty Bowman’s capsule were annotated. Both were not correctly segmented by the network, leading to a DC of zero for these classes. The combination of tubuli subtypes into one class resulted in a weighted average DC of 0.88, equal to the performance of the CNN on the data set originating from Radboudumc.
Glomerular Segmentation and Detection in Nephrectomy Sections
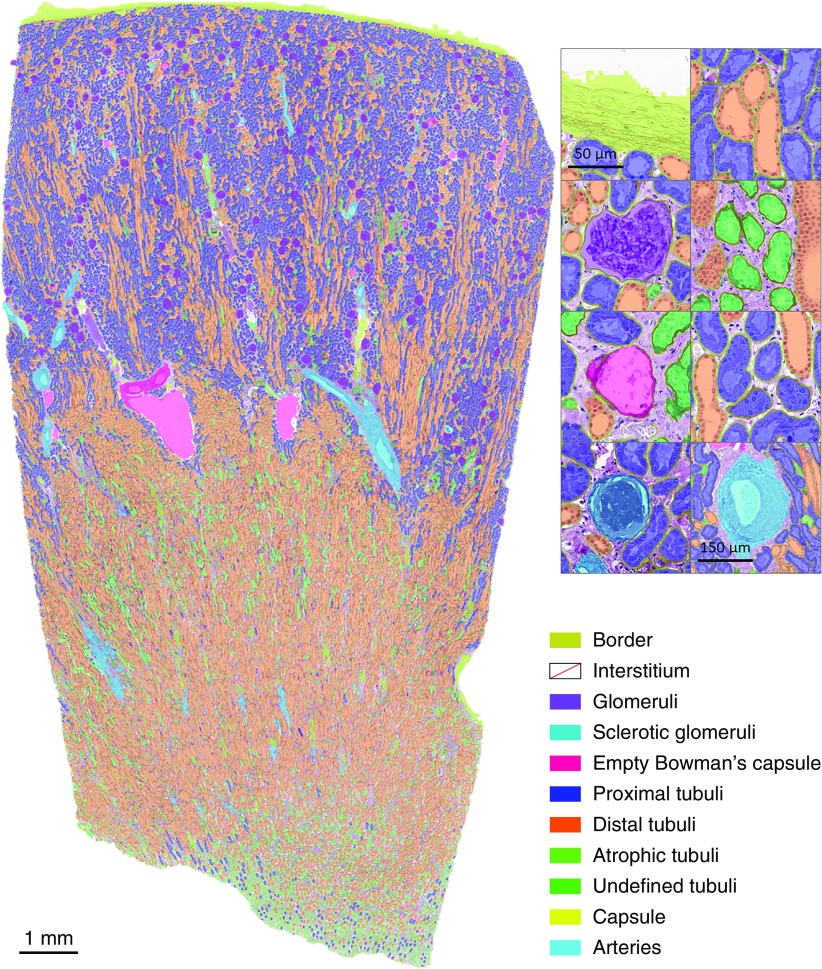
To assess the utility of the CNN for the assessment of kidney specimens other than biopsies, we applied the network on WSIs of 15 tumor nephrectomy specimens. In total, 1747 healthy or segmentally sclerotic glomeruli (glomeruli) and 72 globally sclerotic glomeruli (sclerotic glomeruli) were labeled in the WSIs. The CNN’s ability to fully segment the nephrectomy specimens can be appreciated from Figure 4. The segmentation mask nicely depicts the distinct representation of the cortical and medullar compartment and in higher magnification the accurate delineation of multiple tissue structures can be seen. Because the glomerulus is one of the most extensively studied components of the kidney, we focused our proof of concept on the glomeruli and sclerotic glomeruli classes. For these classes, we found an average DC of 0.90 (glomeruli) and 0.59 (sclerotic glomeruli), respectively, which did not change after postprocessing. The CNN was able to detect 92.7% of all 1819 annotated glomeruli in the nephrectomy samples. Specifically, the network detected 93.4% of all objects labeled as glomeruli (1632 out of 1747) and 76.4% of the objects labeled as sclerotic glomeruli (55 out of 72). There were 149 and 46 false positive detections for glomeruli and sclerotic glomeruli, respectively.
Figure 4.
Full segmentation of a tumor nephrectomy specimen by the CNN on WSI level. Left: segmentation result on low magnification. Top right: segmentation result depicted for specific structures on high magnification.
CNN versus Banff Classification System
The network’s applicability for routine diagnostic tasks was assessed by comparing the CNN’s quantification of a selection of structures to visually scored histologic (Banff) components in 82 PAS-stained transplant biopsies. Visual scoring was performed on glass slides by multiple pathologists. Automated quantification was performed using the CNN’s segmentation mask of the corresponding WSI.
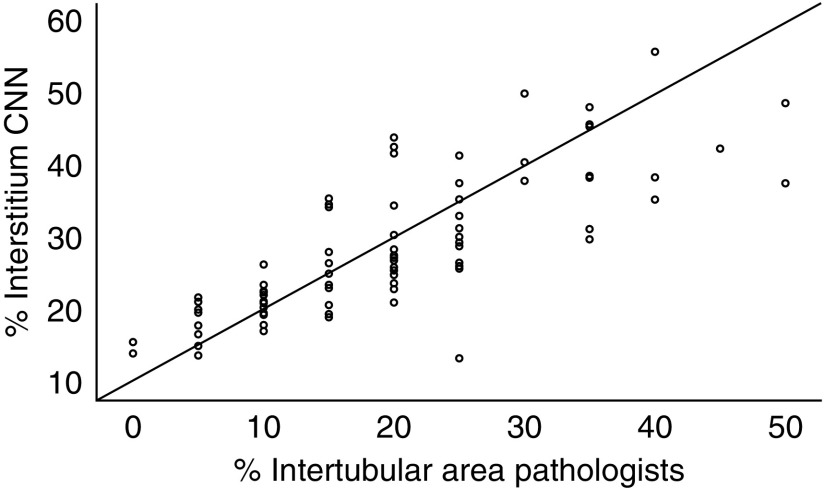
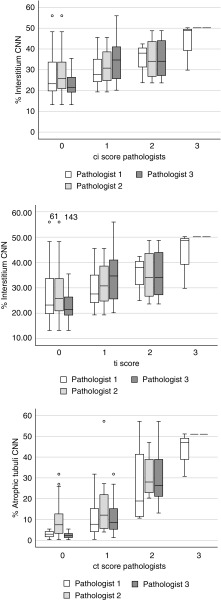
An example of a fully segmented transplant biopsy is depicted in Figure 5. The ICCs for glomerular counting by the CNN and the pathologists ranged from 0.93 to 0.96 (Figure 6, Table 3). The Spearman correlation coefficient for the average intertubular area percentage visually estimated by two pathologists and the interstitium area percentage calculated by our CNN was 0.81 (R2=0.66, P<0.001) (Figure 7, Table 4). Several pathologic processes can cause expansion of the interstitial compartment of the kidney. Next to edema, influx of inflammatory cells and fibrosis are the main reasons for an increased interstitial area. We assessed the relation between the area percentage interstitium and the average ci and ti lesion scores of three pathologists, scored according to the Banff reference guide. The Spearman correlation coefficient for these analyses were 0.55 (R2=0.30, P<0.001) and 0.71 (R2=0.50, P<0.001), respectively (Figure 8, Table 4). In a similar analysis, to assess the relationship between the percentage atrophic tubuli and the average ci and ct score of the three pathologists, the Spearman correlation coefficient was 0.62 (R2=0.38, P<0.001) and 0.58 (R2=0.34, P<0.001) (Figure 8, Table 4). The Spearman correlation coefficients for interstitium and atrophic tubuli percentage and average IFTA grading were 0.33 (R2=0.11, P<0.01) and 0.58 (R2=0.30, P<0.001), respectively (Table 4).
Figure 5.
Full segmentation of a transplant biopsy on whole-biopsy level. The (sclerotic) glomeruli segmentations by the CNN are depicted in high magnification in the lower panel; all are correct. The CNN could not separate the two closely adjacent glomeruli (top left), leading to a count of 17 nonsclerotic glomeruli and one sclerotic glomerulus (bottom right).
Figure 6.
Bland–Altman plots representing the glomerular counts per WSI by three nephropathologists and the glomerular count by the CNN.
Table 3.
ICCs for glomerular counting by three pathologists (P1–P3) and the CNN
| Pathologist | CNN |
|---|---|
| P1 | 0.94 |
| P2 | 0.96 |
| P3 | 0.93 |
Figure 7.
Scatterplot visualizing the correlation between CNN-based area percentage interstitium and the average intertubular area percentage estimated by two pathologists.
Table 4.
The Spearman correlation coefficients for quantifications by the CNN and the average visual scores of multiple pathologists for relevant (Banff) components
| CNN | Visual Scoring Pathologists | ||||
|---|---|---|---|---|---|
| Intertubular Area (% Total Cortical Area) | ci Score | ti Score | IFTA Grade | ct Score | |
| Interstitium (% total cortical area) | 0.81 | 0.55 | 0.71 | 0.33 | — |
| Atrophic tubuli area (% of total n tubuli) | — | 0.62 | — | 0.58 | 0.58 |
– , these analyses were not performed.
Figure 8.
Box plots visualizing the CNN’s quantification of interstitium and atrophic tubuli and the ci, ti, and ct lesion scores per pathologist. Top: percentage area of interstitium scored by the CNN and the ci score per pathologist. Middle: percentage area of interstitium scored by the CNN and the ti score per pathologist. Bottom: percentage of atrophic tubuli scored by the CNN and the ct score per pathologist. Each bar represents one pathologist.
The interobserver agreement among the pathologists for glomerular counting and estimation of intertubular area percentage was expressed in ICCs. The interobserver agreement for Banff lesion scoring was expressed in linear weighted κ values. The average ICCs and κ values of the pathologists are listed in Table 5.
Table 5.
Interobserver agreement between pathologists on histologic (Banff) components scoring
| Measure | ICC | κ |
|---|---|---|
| Glomerular counting | 0.97 | — |
| Intertubular area % | 0.59 | — |
| ci score | — | 0.50 |
| ti score | — | 0.59 |
| ct score | — | 0.23 |
The average ICCs and average linear weighted κ values are presented.
– , these analyses were not performed.
Discussion
In this study we developed a CNN for the multiclass segmentation of renal tissue in routinely PAS-stained sections. Our main findings are that the CNN achieves accurate segmentation of glomeruli, tubuli, and interstitium in kidney transplant biopsies; living donor kidney biopsies from an external center; and nephrectomy samples.
The number of studies aiming to develop deep learning applications for nephropathology has increased rapidly over the past years.11,14,15 Pretrained neural networks have successfully been applied for the distinction between glomerular and nonglomerular regions. Pedraza et al.14 trained a CNN for the detection of glomeruli in preselected areas on PAS-stained human renal biopsies. The glomerulus localizer of Bukowy et al.15 detected healthy and injured glomeruli in whole rat kidney sections stained with either Gömöri or Masson Trichrome using a CNN. Both groups did not further classify the detected glomeruli and aimed for detection of the glomeruli rather than segmentation. Gadermayr et al.11 investigated two cascades where two U-nets are combined with a sliding window CNN: 80% of glomerular objects where found with a DC of 0.90 or higher when detection and segmentation were combined.11 As a limitation, this work focused only on segmentation of glomeruli, whereas the simultaneous segmentation and classification of the whole renal cortex can be of great added value in kidney diagnostics and research. Our first objective was therefore to train a CNN for the segmentation of PAS-stained kidney sections into ten significant tissue classes. Adding a border class to the segmentation network, representing the structure’s basal membranes (visible in Figure 2), allowed us to separate touching structures and to identify individual objects. The best segmentation performance was achieved for the class glomeruli, containing healthy and segmentally sclerotic glomeruli (mean DC 0.95). The biopsies included in our training set displayed a wide range of inflammation and/or chronic damage, which facilitated us to include globally sclerotic glomeruli in the training data as well. The CNN’s segmentation performance for this class was reasonable (DC 0.62) but requires improvement. The number of annotations for this class was limited (Table 1) and additional training with more sclerotic glomeruli annotations will probably improve the segmentation results for this class. The potential of the CNN to discriminate between different types of tubuli was nicely illustrated by the high DCs for segmentation of proximal (0.87) and distal tubuli (0.81) (Figure 2, Table 2). Tubular atrophy is a continuous process resulting in the presence of mildly injured to severe atrophic tubuli. Therefore, tubuli could not always be indisputably classified during ground truth development. The difficulty of this annotation task is emphasized by the low κ values for ct scoring by three pathologists. Correct classification of tubuli with different extents of tubular atrophy by the CNN will benefit from more training data. Crowd-sourcing experiments, where groups of nonexpert annotators collectively generate a high number of annotations, have shown how the size of a data set can compensate for having a “noisy” ground truth.22 Without differentiation in subtypes, the CNN classified tubuli very well (DC 0.92). Combined with the DC of 0.88 for the segmentation of interstitium, this led to a high-quality segmentation of the tubulo-interstitial compartment. Although the CNN was not optimized for speed, segmentation of one kidney biopsy WSI took under 2 minutes using a standard desktop configuration with 20 gigabytes of RAM, using two CPU cores and a single NVIDIA GTX 1080 GPU.
Large-scale, histopathologic studies often include material from multiple centers. Differences in tissue processing techniques, staining protocols, and slide scanners cause heterogeneous data sets, and WSIs of different file formats and resolutions. With digital pathology gaining territory in diagnostic settings, peer consultation using telepathology platforms will happen more often. Therefore, our second objective was to test the CNN’s robustness to the abovementioned variations. Our CNN was trained using slides produced and digitized at Radboudumc. The data augmentation techniques applied during training should make the CNN robust to variances in, e.g., color and morphology.19 We applied the Radboudumc CNN to an external set of biopsy material. This external tissue was fixed using formalin, leading to subtle morphologic differences in the tissue when compared with Radboudumc tissue (fixed with Bouin fixative). The use of a different staining protocol and tissue scanner resulted in color variance and lower image resolution. Nevertheless, the performance of the CNN on this external data set was quite comparable to that on Radboudumc tissue (weighted mean DC 0.84 versus 0.80, respectively), which obviates the need for additional, external training data.
Our network appeared to be very capable of segmenting WSIs of nephrectomy specimens, even though it was not trained on this type of material. A clear distinction between capsule, cortex, and medulla could be derived from the segmentation mask, as visualized in Figure 4. To provide a proof of concept for the applicability of our CNN for glomerular assessment in research settings, we calculated the segmentation performance for glomeruli and sclerotic glomeruli in the nephrectomy samples. The DCs calculated for the nephrectomy samples were comparable to those calculated for the kidney transplant biopsies (0.90 versus 0.95 and 0.59 versus 0.62, respectively). Additionally, we have demonstrated that our segmentation masks can be used for detection of glomeruli. Of all glomeruli (healthy, segmentally and globally sclerotic), 92.7% were detected by the CNN. The high false positive rate in the sclerotic glomeruli class emphasizes that more training data are crucial for correct segmentation of this class.
Finally, we compared quantifications based on CNN segmentation data to visually scored components of the Banff classification system in whole transplant biopsies. Biopsy quality assessment is generally performed by glomerular counting. We observed high correlations for glomerular counting performed by the network and the renal pathologists. The rules for counting glomeruli are established in the Banff reference guide. Nevertheless, the pathologists did not fully agree for every case, indicating room for interpretation and, thus, subjectivity. Using a CNN for counting glomeruli will eliminate this subjectivity and could take over this tedious task from the pathologist.
An additional high correlation was observed for the area percentage of interstitium generated from the CNN’s segmentation mask and the intertubular area percentage estimated by two renal pathologists. Next to edema, inflammation and fibrosis are some of the main causes for interstitium expansion. In this cohort, we found a high correlation between the area percentage interstitium and the average total inflammation score of three pathologists. This was lower for the average interstitial fibrosis score. These correlations will depend highly on the composition of the data set. Without an additional technique for the automated segmentation of fibrosis or detection of inflammatory cells, the reason for interstitial expansion cannot solely be based on the interstitium segmentation.
The atrophic tubuli percentage derived from the CNN’s segmentation mask showed significant correlation to parameters of chronic damage, expressed by the average tubular atrophy score or by IFTA grading of three pathologists. After more extensive training of the CNN for this heterogeneous class, the atrophic tubuli percentage could potentially become an even more reliable indicator of chronic injury.
The number of neural networks trained for the analysis of histopathologic slides has increased tremendously in the past decade, showing exciting results for tumor detection and tumor grading. The delicate histologic changes in renal tissue, which are connected to a wide range of kidney diseases or types of allograft rejection, make renal pathology one of the most complicated specializations in pathology. Nevertheless, the renal field could benefit equally from this digitization of pathology with a robust segmentation algorithm. We present a CNN for the multiclass segmentation of renal tissue in routinely PAS-stained sections. Our CNN is applicable on material from multiple centers and we sustained a high segmentation performance, despite differences in fixation, staining protocol, and scanning resolution. The CNN is capable of segmenting biopsies and nephrectomy samples, and can be used for the analysis of healthy and pathologic tissue. Significant relations were found between the CNN’s quantification of the glomeruli, interstitium, and atrophic tubuli tissue classes and visually scored glomerular count, intertubular area percentage, interstitial lesions, and chronic damage parameters. This is the first validation of quantitative results obtained by a CNN with components of the Banff classification system. The results of these analyses are encouraging for the application of deep learning in renal (transplantation) pathology.
Disclosures
Prof. van der Laak reports grants from ZonMw (The Netherlands) during the conduct of the study, personal fees from Philips (The Netherlands), personal fees from ContextVision, grants from Philips (The Netherlands), and grants from Sectra (Sweden), outside of the submitted work. All of the remaining authors have nothing to disclose.
Funding
This work was supported by ERACoSysMed’s SysMIFTA project, as part of the European Union’s Horizon 2020 Framework Programme (grant number 9003035004). Dr. Kers received financial support from the Dutch Kidney Foundation (Nierstichting) (project DEEPGRAFT, grant number 17OKG23).
Acknowledgments
The authors thank Milly van den Warenburg and Jimmy Knuiman for their contributions to the CNN’s ground truth development.
Ms. Hermsen, Prof. van der Laak, Prof. Hilbrands and Prof. Smeets designed the study. Dr. Steenbergen and Prof. Alexander checked the manual annotations. Mr. de Bel and Ms. den Boer designed and trained the CNN. Dr. Steenbergen, Prof. Florquin, Dr. Kers and Dr. Roelofs performed the manual glomerular counting and/or provided the ci, ti and ct scores and intertubular area percentages. Prof. Stegall, Prof. Alexander and Dr. Smith provided the external material. Ms. Hermsen and Ms. den Boer validated the CNN. Ms. Hermsen, Prof. van der Laak, Prof. Hilbrands and Prof. Smeets analyzed the data and Ms. Hermsen made the figures. Ms. Hermsen, Prof. van der Laak, Prof. Hilbrands and Prof. Smeets drafted the paper. The final version of the manuscript was revised and approved by all authors.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al.: The Banff 97 working classification of renal allograft pathology. Kidney Int 55: 713–723, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al.: The Banff 2015 kidney meeting report: Current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant 17: 28–41, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Servais A, Meas-Yedid V, Noël LH, Martinez F, Panterne C, Kreis H, et al.: Interstitial fibrosis evolution on early sequential screening renal allograft biopsies using quantitative image analysis. Am J Transplant 11: 1456–1463, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Grimm PC, Nickerson P, Gough J, McKenna R, Stern E, Jeffery J, et al.: Computerized image analysis of Sirius Red-stained renal allograft biopsies as a surrogate marker to predict long-term allograft function. J Am Soc Nephrol 14: 1662–1668, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Kato T, Relator R, Ngouv H, Hirohashi Y, Takaki O, Kakimoto T, et al.: Segmental HOG: New descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics 16: 316–332, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klapczynski M, Gagne GD, Morgan SJ, Larson KJ, LeRoy BE, Blomme EA, et al. : Computer-assisted imaging algorithms facilitate histomorphometric quantification of kidney damage in rodent renal failure models. J Pathol Inform 3: 20, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadermayr M, Klinkhammer KM, Boor P, Merhof D: Do we need large annotated training data for detection applications in biomedical imaging? A case study in renal glomeruli detection. In: Machine Learning in Medical Imaging. MLMI 2016 (Lecture Notes in Computer Science, Vol. 10019), edited by Wang L, Adeli E, Wang Q, Shi Y, Suk HI, Cham, Switzerland, Springer, 2016, pp 18–26 [Google Scholar]
- 8.Ginley BG, Tomaszewski JE, Yacoub R, Chen F, and Sarder P: Unsupervised labeling of glomerular boundaries using Gabor filters and statistical testing in renal histology. J Med Imaging (Bellingham) 16: 021102, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginley BG, Tomaszewski JE, Jen K, Fogo A, Jain S, and Sarder P: Computational analysis of the structural progression of human glomeruli in diabetic nephropathy. Medical Imaging: Digital Pathology, 10581, 2018
- 10.Tadrous PJ: On the concept of objectivity in digital image analysis in pathology. Pathology 42: 207–211, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Gadermayr M, Dombrowski AK, Klinkhammer BM, Boor P, Merhof D: CNN cascades for segmenting sparse objects in gigapixel whole slide images. Comput Med Imaging Graph 71: 40–48, 2019 [DOI] [PubMed] [Google Scholar]
- 12.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al.: A survey on deep learning in medical image analysis. Med Image Anal 42: 60–88, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, et al. : Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6: 26286, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedraza A, Gallego J, Lopez S, Gonzalez L, Laurinavicius A, Bueno G: Glomerulus classification with convolutional neural networks. In: Medical image understanding and analysis. MIUA 2017 (Communications in Computer and Information Science, Vol. 723), edited by Valdés Hernández M, González-Castro V, Cham, Switzerland, Springer, 2017, pp 839–849 [Google Scholar]
- 15.Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, et al.: Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 29: 2081–2088, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barisoni L, Nast CC, Jennette JC, Hodgin JB, Herzenberg AM, Lemley KV, et al.: Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin J Am Soc Nephrol 8: 1449–1459, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denic A, Lieske JC, Chakkera HA, Poggio ED, Alexander MP, Singh P, et al.: The substantial loss of nephrons in healthy human kidneys with aging. J Am Soc Nephrol 28: 313–320, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronneberger O, Fischer P, Brox T: U-Net: Convolutional networks for biomedical image segmentation. In: Medical image Computing and Computer-Assisted Intervention – MICCAI 2015 (Lecture Notes in Computer Science, Vol. 9351), edited by Navab N, Hornegger J, Wells W, Frangi A, Cham, Switzerland, Springer, 2015, pp 234–241 [Google Scholar]
- 19.Tellez D, Balkenhol M, Otte-Höller I, van de Loo R, Vogels R, Bult P, et al.: Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans Med Imaging 37: 2126–2136, 2018 [DOI] [PubMed] [Google Scholar]
- 20.Kingma DP, Ba JL: Adam: a method for stochastic optimization. Presented at the Third International Conference on Learning Representations, San Diego, CA, May 7–9, 2015. 29994086 [Google Scholar]
- 21.Roufosse C, Simmonds N, Clahsen-van Groningen M, Haas M, Henriksen KJ, Horsfield C, et al.: A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation 102: 1795–1814, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitry D, Zukis K, Dhillon B, Peto T, Hayat S, Khaw KT, et al. : The accuracy and reliability of crowdsource annotations of digital retinal images [published correction appears in Trans Vis Sci Technol 2016; 5: 9]. Transl Vis Sci Technol 5: 6, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]