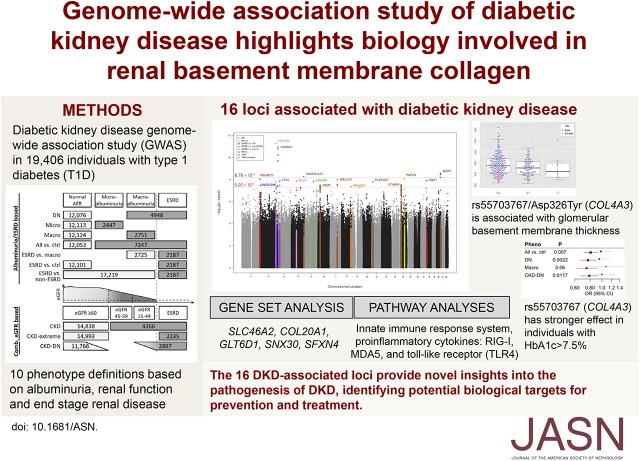
Significance Statement
Although studies show that diabetic kidney disease has a heritable component, searches for the genetic determinants of this complication of diabetes have had limited success. In this study, a new international genomics consortium, the JDRF funded Diabetic Nephropathy Collaborative Research Initiative, assembled nearly 20,000 samples from participants with type 1 diabetes, with and without kidney disease. The authors found 16 new diabetic kidney disease–associated loci at genome-wide significance. The strongest signal centers on a protective missense coding variant at COL4A3, a gene that encodes a component of the glomerular basement membrane that, when mutated, causes the progressive inherited nephropathy Alport syndrome. These GWAS-identified risk loci may provide insights into the pathogenesis of diabetic kidney disease and help identify potential biologic targets for prevention and treatment.
Keywords: diabetes, end-stage renal disease, genetic renal disease, kidney disease, human genetics, diabetic nephropathy
Visual Abstract
Abstract
Background
Although diabetic kidney disease demonstrates both familial clustering and single nucleotide polymorphism heritability, the specific genetic factors influencing risk remain largely unknown.
Methods
To identify genetic variants predisposing to diabetic kidney disease, we performed genome-wide association study (GWAS) analyses. Through collaboration with the Diabetes Nephropathy Collaborative Research Initiative, we assembled a large collection of type 1 diabetes cohorts with harmonized diabetic kidney disease phenotypes. We used a spectrum of ten diabetic kidney disease definitions based on albuminuria and renal function.
Results
Our GWAS meta-analysis included association results for up to 19,406 individuals of European descent with type 1 diabetes. We identified 16 genome-wide significant risk loci. The variant with the strongest association (rs55703767) is a common missense mutation in the collagen type IV alpha 3 chain (COL4A3) gene, which encodes a major structural component of the glomerular basement membrane (GBM). Mutations in COL4A3 are implicated in heritable nephropathies, including the progressive inherited nephropathy Alport syndrome. The rs55703767 minor allele (Asp326Tyr) is protective against several definitions of diabetic kidney disease, including albuminuria and ESKD, and demonstrated a significant association with GBM width; protective allele carriers had thinner GBM before any signs of kidney disease, and its effect was dependent on glycemia. Three other loci are in or near genes with known or suggestive involvement in this condition (BMP7) or renal biology (COLEC11 and DDR1).
Conclusions
The 16 diabetic kidney disease–associated loci may provide novel insights into the pathogenesis of this condition and help identify potential biologic targets for prevention and treatment.
The devastating diabetic complication of diabetic kidney disease (DKD) is the major cause of worldwide.1,2 Current treatment strategies at best slow the progression of DKD, and do not halt or reverse the disease. Although improved glycemic control influences the rate of diabetic complications, a large portion of the variation in DKD susceptibility remains unexplained: one third of people with type 1 diabetes (T1D) develop DKD despite adequate glycemic control, whereas others maintain normal renal function despite long-term severe chronic hyperglycemia.3
Though DKD demonstrates both familial clustering4–6 and single nucleotide polymorphism (SNP) heritability,7 the specific genetic factors influencing DKD risk remain largely unknown. Recent genome-wide association studies (GWAS) have only identified a handful of loci for DKD, albuminuria, or eGFR in individuals with diabetes.7–13 Potential reasons for the limited success include small sample sizes, modest genetic effects, and lack of consistency of phenotype definitions and statistical analyses across studies. Through collaboration within the JDRF Diabetes Nephropathy Collaborative Research Initiative, we adopted three approaches to improve our ability to find new genetic risk factors for DKD: (1) assembling a large collection of T1D cohorts with harmonized DKD phenotypes, (2) creating a comprehensive set of detailed DKD definitions, and (3) augmenting genotype data with low frequency and exome array variants.
Methods
Cohorts and Phenotype Definitions
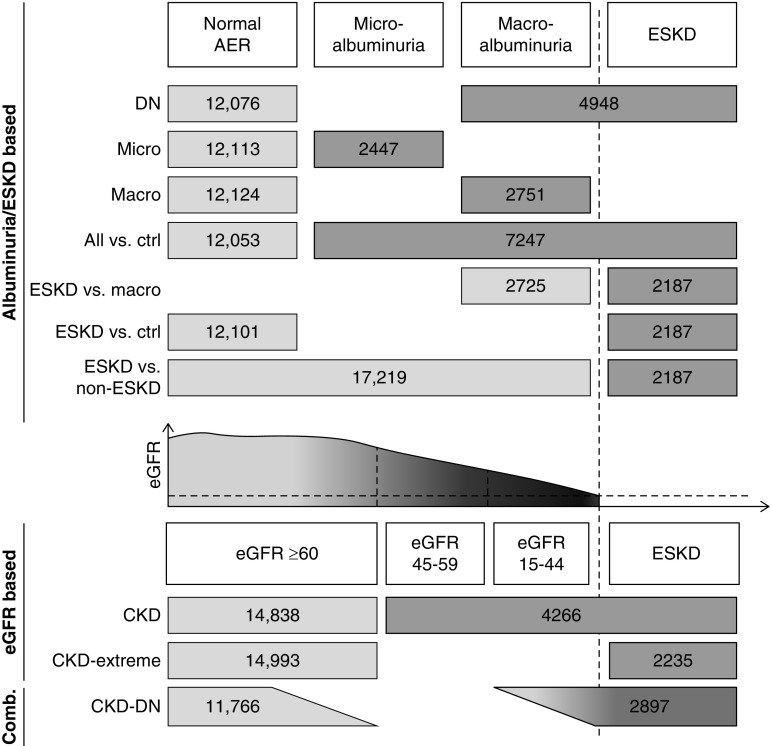
The GWAS meta-analysis included up to 19,406 patients with T1D of European origin from 17 cohorts (for study list and details see Supplemental Table 1). All participants gave informed consent and all studies were approved by ethics committees from participating institutions. We defined a total of ten different case-control outcomes to cover the different aspects of renal complications, using both albuminuria and eGFR (Figure 1). Five comparisons (“All versus control (ctrl),” “Micro,” “diabetic nephropathy [DN],” “Macro,” and “ESKD versus macro”) were on the basis of albuminuria, measured by albumin excretion rate (AER) from overnight or 24-hour urine collection, or by albumin-to-creatinine ratio. Two out of three consecutive collections were required (when available) to classify the renal status of patients as either normoalbuminuria, microalbuminuria, macroalbuminuria, or ESKD; for detailed thresholds, see Figure 1. Controls with normal AER were required to have a minimum diabetes duration of 15 years; participants with microalbuminuria/macroalbuminuria/ESKD were required to have minimum diabetes duration of 5, 10, and 10 years, respectively, to exclude renal complications of nondiabetic origins. Two comparisons (“ESKD versus ctrl” and “ESKD versus non-ESKD”) were on the basis of presence of ESKD as defined by eGFR<15 ml/min or dialysis or renal transplant. Two phenotypes (“CKD” and “CKD extreme”) were defined on the basis of eGFR estimated by the CKD Epidemiology Collaboration formula: controls had eGFR≥60 ml/min per 1.73 m2 for both phenotypes, and ≥15 years of diabetes duration; cases had eGFR<60 ml/min per 1.73 m2 for the CKD phenotype, and eGFR<15 ml/min per 1.73 m2 or dialysis or renal transplant for the CKD extreme phenotype, and ≥10 years of diabetes duration. For the “CKD-DN” phenotype that combined both albuminuria and eGFR data, controls were required to have both eGFR≥60 ml/min per 1.73 m2 and normoalbuminuria; cases had both eGFR<45 ml/min per 1.73 m2 and micro- or macroalbuminuria, or ESKD.
Figure 1.
Phenotypic analysis of DKD. Schematic diagram of outcomes analyzed in this study. Numbers indicate the total number of cases (darker gray) and controls (lighter gray) included in the meta-analyses for each phenotype. ESKD defined as eGFR<15 ml/min per 1.73 m2 or undergoing dialysis or having renal transplant.
GWAS Genotyping, Quality Control, and Imputation
All study samples underwent genotyping, quality control (QC), and imputation centrally at the University of Virginia. In brief, samples were genotyped on the HumanCore BeadChip array (Illumina, San Diego, CA), which contains approximately 250,000 genome-wide tag SNPs and >200,000 exome-focused variants. All samples were passed through a stringent QC protocol. Following initial genotype calling with Illumina software, all samples were recalled with zCall, a calling algorithm specifically designed for rare SNPs from arrays. Variant orientation and position were aligned to hg19 (Genome Reference Consortium Human Build 37, GRCh37). Variant names were updated using 1000 Genomes as a reference. The data were then filtered for low-quality variants (e.g., call rates <95% and excessive deviation from Hardy–Weinberg equilibrium) and samples (e.g., call rates <98%, sex mismatch, extreme heterozygosity). Principal component analysis was performed separately for each cohort to empirically detect and exclude outliers with evidence of non-European ancestry (see Supplemental Material for full QC details, and Supplemental Figure 1 for trait-specific Manhattan and QQ plots). Genotypes were expanded to a total of approximately 49 million by imputation, using the minimac imputation tool14,15 and 1000 Genomes Project (phase 3v5) as a reference.
GWAS Analysis
A genome-wide association analysis was performed for each of the case-control definitions under an additive genetic model, adjusting for age, sex, diabetes duration, study site (where applicable), and principal components. We conducted a second set of analyses, adjusting for body mass index and glycated hemoglobin (HbA1c), which we refer to as our fully adjusted covariate model. Allele dosages were used to account for imputation uncertainty. Inverse-variance fixed effects meta-analysis was performed using METAL and the following filters: INFO score >0.3, minor allele count >10 in both cases and controls, and presence of variant in at least two cohorts (Manhattan and QQ plots each trait and covariate model presented in Supplemental Figure 1). The X chromosome was similarly analyzed for men and women both separately and in a combined analysis, with the exception of using hard call genotypes in place of allele dosages.16 We estimated the percentage of variance explained for all genome-wide significant SNPs across all disease definitions using the McKelvey and Zavoina17 pseudo-R2 statistic predicting continuous latent variables underlying binary outcomes.
Glomerular Basement Membrane Measurement in Renin-Angiotensin System Study
In brief, the renin-angiotensin system study (RASS) was a double-blind, placebo-controlled, randomized trial of enalapril and losartan on renal pathology among 285 normoalbuminuric, normotensive participants with T1D and normal or increased measured GFR (>90 ml/min per 1.73 m2).18 Participants were followed for 5 years with percutaneous kidney biopsy completed prior to randomization and at 5 years. Structural parameters measured by electron microscopy on biopsy included glomerular basement membrane (GBM) width, measured by the electron microscopic orthogonal intercept method.18 All RASS participants contributed DNA for genotyping.
In Silico Replication in SUMMIT Consortium
The Surrogate Markers for Micro- and Macro-vascular Hard Endpoints for Innovative Diabetes Tools (SUMMIT) consortium included up to 5193 European Ancestry participants with type 2 diabetes (T2D), with and without kidney disease. In silico replication was performed on previously published GWAS on DKD with harmonized trait definitions for seven of our primary T1D analyses: “DN,” “Micro,” “Macro,” “ESKD,” “ESKD versus non-ESKD,” “CKD,” and “CKD-DN” under an additive model, adjusting for age, sex, and duration of diabetes.13
RNA-Sequencing and Microarray Profiling of Human Kidney Samples from the Pima Cohort
Kidney biopsy samples from the Pima Indian cohort were manually microdissected into 119 glomerular and 100 tubule-interstitial tissues to generate gene expression profiles.19 Expression profiling in the Pima Indian cohort kidney biopsies was carried out using Affymetrix GeneChip Human Genome U133 Array and U133Plus2 Array, as reported previously, and Affymetrix Human Gene ST GeneChip 2.1,20,21 and on RNA-seq (Illumina). The libraries were prepared using the ClonTech SMARTSeq v4 Ultra Low Input polyA selection kit. Samples were sequenced on a HiSeq4000 using single-end, 75 bp reads. Mapping to human reference genome GRCh38.7 was performed with STAR 2.5.2b (https://github.com/alexdobin/STAR). For annotation and quantification of mapping results we used cufflinks, cuffquant, and cuffnorm in version 2.2.1 (https://cole-trapnell-lab.github.io/cufflinks/). After mapping and quantification, principal component analysis and hierarchical clustering was used to identify outliers and reiterated until no more outliers could be identified.
RNA-Sequencing and cis Expression Quantitative Trait Loci Analysis in Human Kidney Samples from University of Pennsylvania Cohort
Human kidney samples were obtained from surgical nephrectomies for a total of 455 participants with pathologic data and were manually microdissected under a microscope in RNAlater for glomerular and tubular compartments (433 tubule and 335 glomerulus samples). The local renal pathologist performed an unbiased review of the tissue section by scoring multiple parameters, and RNA were prepared using RNAeasy mini columns (Qiagen, Valencia, CA) according to manufacturer’s instructions.
Whole kidney,22 tubularm and glomerular23 expression quantitative trait loci (eQTL) analyses have been described previously. Tubular and glomerular eQTL data sets were generated by 121 samples of tubules and 119 samples of glomeruli, respectively.23 The cis window was defined as 1 Mb up- and downstream of the transcriptional start site (±1 Mb). The whole kidney cis-eQTL (further referred to as just eQTL) data set was generated from 96 human samples obtained from The Cancer Genome Atlas (TCGA) through the TCGA Data portal.22
Mouse Kidney Single-Cell RNA-Sequencing
Kidneys were harvested from 4- to 8-week-old male mice with C57BL/6 background and dissociated into single-cell suspension as described in our previous study.24 The single-cell sequencing libraries were sequenced on an Illumina HiSeq with 2×150 paired-end kit. The sequencing reads were demultiplexed, aligned to the mouse genome (mm10) and processed to generate gene-cell data matrix using Cell Ranger 1.3 (http://10xgenomics.com).24
Genomic Features of Human Kidney
Human kidney-specific chromatin immunoprecipitation sequencing data can be found at Gene Expression Omnibus under accession numbers GSM621634, GSM670025, GSM621648, GSM772811, GSM621651, GSM1112806, and GSM621638. Different histone markers were combined into chromatin states using ChromHMM.25
Gene and Gene Set Analysis
PASCAL and MAGMA (v1.06) gene and pathway scores were conducted on all 20 sets of GWAS summary statistics using default pathway libraries from BioCarta, REACTOME, and KEGG. MAGENTA (vs2, July 2011) pathway analysis included 4725 pathways with a minimum of five genes within the gene set for the ten standard adjustment models. We conducted DEPICT individually on all 20 sets of GWAS summary statistics with P<10−5 and additional pooled analyses using genome-wide minimum P values from all 20 analyses (ten phenotypes and two covariate models) and 16 analyses of the eight most related phenotypes, which excluded ESKD versus Macro and Micro.
Transcriptome-Wide Association Study
A transcriptome-wide association study (TWAS) of kidney glomeruli and tubules was performed using MetaXcan with default parameters,26 on the basis of eQTL data for human glomerular and tubular cells.23
Results
Phenotypic Comparisons
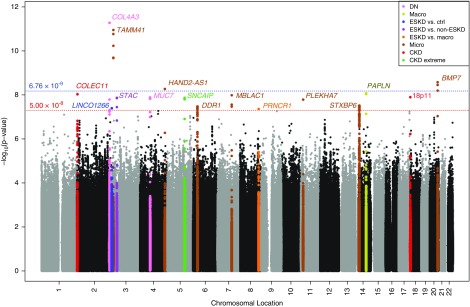
We investigated a broad spectrum of DKD definitions on the basis of albuminuria and renal function criteria, defining a total of ten different case-control comparisons to cover the different aspects of disease progression (Figure 1). Seven comparisons were on the basis of albuminuria and/or ESKD (including DN, defined as either macroalbuminuria or ESKD), two were defined on the basis of eGFR (used to classify severity of CKD), and one combined both albuminuria and eGFR data (CKD-DN). Each phenotypic definition was analyzed separately in GWAS; to account for the ten definitions each analyzed under two covariate adjustment models, we estimated16 the total effectively independent tests as 7.4, allowing us to compute a more conservative study-wide significance threshold (P<6.76×10−9), on the basis of genome-wide significance (P<5×10−8) and Bonferroni correction for 7.4 effective tests.
Top Genome-Wide Association Results Highlight COL4A3
GWAS meta-analysis included association results for up to 19,406 individuals with T1D of European descent from 17 cohorts for the ten case-control definitions (Supplemental Table 1). We identified 16 novel independent loci that achieved genome-wide significance (P<5×10−8) in either the minimal or fully adjusted models, in which four lead SNPs also surpassed our more conservative study-wide significance threshold (Figure 2, Manhattan plot; Table 1; Supplemental Figure 2, A–P, regional association and forest plots). None of the loci reaching genome-wide significance have been previously identified in GWAS or candidate gene studies for DKD or closely related traits. All SNPs with minor allele frequency (MAF) >1% explain 2.5% and 3.0% of the total variance (McKelvey and Zavoina17 pseudo-R2) of DN after adjusting for covariates in the minimal and full covariate models, respectively (Supplemental Table 2).
Figure 2.
Genome-wide association testing of all ten phenotypic comparisons. Multiphenotype Manhattan plot shows lowest P value at each marker for each of the ten phenotypic comparisons, under the standard and fully-adjusted model. Significance of SNPs (−log10[P value], y axis) is plotted against genomic location (x axis). Loci surpassing genome-wide significance (red line) and/or study-wide significance (blue line) are colored by phenotype.
Table 1.
Loci associated with DKD at study-wide (P<6.76×10−9, see bolding) and genome-wide (P<5×10−8) significance
| SNP | Chr:pos | Effect Allele | Other Allele | EAF | Notable Gene(s) | Phenotype | ORmin | P Valuemin | ORfull | P Valuefull |
|---|---|---|---|---|---|---|---|---|---|---|
| Common Variants | ||||||||||
| rs55703767 | 2:228121101 | T | G | 0.206 | COL4A3 (M, B, N) | DN | 0.79 | 5.34×10–12 | 0.78 | 8.19×10−11 |
| All versus ctrl | 0.83 | 3.88×10−10 | 0.84 | 9.68×10−9 | ||||||
| CKD+DN | 0.77 | 5.30×10−9 | 0.76 | 3.77×10−8 | ||||||
| Macro | 0.78 | 9.28×10−9 | 0.77 | 9.38×10−9 | ||||||
| rs12615970 | 2:3745215 | G | A | 0.133 | COLEC11 (B) ALLC (N, G) | CKD | 0.76 | 9.43×10−9 | 0.77 | 1.60×10−7 |
| rs142823282 | 3:11910635 | G | A | 0.011 | TAMM41 (N, B) | Micro | 6.73 | 8.32×10−10 | 9.18 | 1.13×10−11 |
| rs145681168 | 4:174500806 | G | A | 0.014 | HAND2-AS1 (N, G, B) | Micro | 5.53 | 2.06×10−7 | 7.47 | 5.40×10−9 |
| rs118124843 | 6:30887465 | T | C | 0.011 | DDR1 (B) VARS2 (G) | Micro | 3.79 | 4.42×10−8 | 3.99 | 3.37×10−8 |
| rs551191707 | 8:128100029 | CA | C | 0.122 | PRNCR1 (N) | ESKD versus macro | 1.70 | 4.39×10−8 | 1.71 | 3.15×10−6 |
| rs61983410 | 14:26004712 | T | C | 0.213 | STXBP6 (N) | Micro | 0.79 | 9.84×10−8 | 0.78 | 3.06×10−8 |
| rs144434404 | 20:55837263 | T | C | 0.011 | BMP7 (N, G, B) | Micro | 6.78 | 2.67×10−9 | 6.66 | 4.65×10−9 |
| Uncommon Variants | ||||||||||
| rs115061173 | 3:926345 | A | T | 0.014 | LINC01266 (N) | ESKD versus ctrl | 9.40 | 4.07×10−8 | 8.34 | 4.08×10−5 |
| rs116216059 | 3:36566312 | A | C | 0.016 | STAC (N, G) | ESKD versus non-ESKD | 8.73 | 1.37×10−8 | 11.78 | 1.41×10−4 |
| rs191449639 | 4:71358776 | A | T | 0.005 | MUC7 (N) | DN | 32.42 | 1.32×10−8 | 32.47 | 2.09×10−8 |
| rs149641852 | 5:121774582 | T | G | 0.012 | SNCAIP (N, G) | CKD extreme | 9.01 | 1.37×10−8 | — | — |
| rs77273076 | 7:99728546 | T | C | 0.008 | MBLAC1 (N) | Micro | 9.16 | 1.04×10−8 | 7.10 | 2.28×10−7 |
| rs183937294 | 11:16937846 | G | T | 0.007 | PLEKHA7 (N, G) | Micro | 17.22 | 1.65×10−8 | 23.62 | 2.10×10−6 |
| rs113554206 | 14:73740250 | A | G | 0.012 | PAPLN (N, G) | Macro | 4.60 | 5.39×10−7 | 10.42 | 8.46×10−9 |
| rs185299109 | 18:1811108 | T | C | 0.007 | intergenic | CKD | 20.75 | 1.28×10−8 | 44.75 | 4.99×10−7 |
Common variants and/or genes with relevant kidney biology are reported in the top half of the table. Uncommon variants (MAF<2%) with no known relevant kidney biology are reported in the bottom half of the table. Genes are annotated as follows: missense variant in the indicated gene (M); intronic, synonymous, or noncoding variant in the indicated gene (G); gene nearest to lead variant (N); gene has relevant kidney (B). Chr, chromosome; pos, position; EAF, effect allele frequency; min, minimally adjusted covariate model; full, fully adjusted covariate model.
The strongest signal was rs55703767 (MAF=0.21), a common missense variant (G>T; Asp326Tyr) in exon 17 of COL4A3. This SNP was associated with protection from DN (odds ratio [OR], 0.79; 95% Confidence Interval [95% CI], 0.73 to 0.84; P=5.34×10−12), any albuminuria (OR, 0.83; 95% CI, 0.79 to 0.88; P=3.88×10−10), the combined CKD-DN phenotype (OR, 0.77; 95% CI, 0.71 to 0.84; P=5.30×10−9), and macroalbuminuria (OR, 0.78; 95% CI, 0.72 to 0.85; P=9.28×10−9). Interestingly, we found that rs55703767 in COL4A3 was more strongly associated in men (OR, 0.73; 95% CI, 0.66 to 0.80; P=1.29×10−11) than in women (OR, 0.85; 95% CI, 0.76 to 0.94; P=1.39×10−3; Phet=1.58×10−2). COL4A3 encodes the α3 chain of collagen type IV, a major structural component of the GBM.27
COL4A3 Variation and Kidney Phenotypes
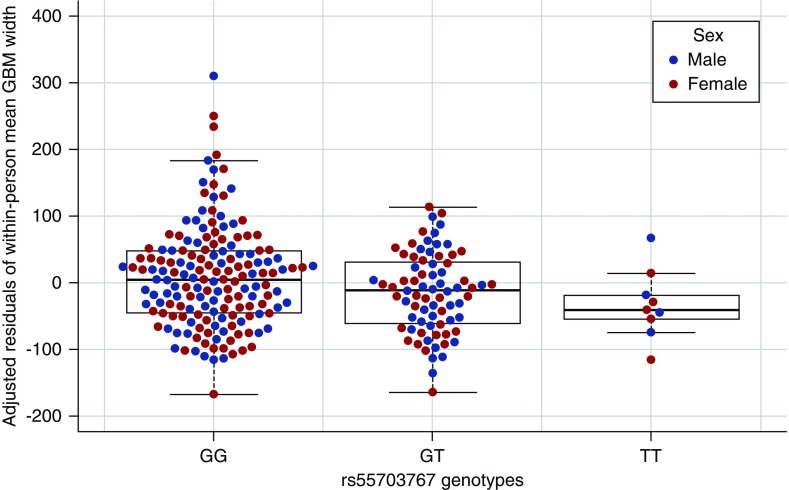
In persons with T1D and normoalbuminuria, GBM width predicts progression to proteinuria and ESKD independently of glycated hemoglobin (HbA1c).28 We examined the influence of the COL4A3 variant on GBM width in 253 RASS18 participants with T1D and normal AER, eGFR (>90 ml/min per 1.73 m2), and BP, who had biopsy and genetic data (Supplemental Table 3). The DKD-protective minor T allele was associated with 19.7 nm lower GBM width (SEM 8.2 nm; P=0.02), with the lowest mean GBM width among TT homozygotes (Figure 3, Supplemental Table 4), after adjusting for age, sex, and diabetes duration, and without detectable interactions with T1D duration or mean HbA1c. Thus, the protective T allele carriers had thinner GBM before any renal complications.
Figure 3.
Adjusted residuals of GBM width by rs55703767 genotype and sex. Box and whisker plot of residuals of mean GBM width after adjusting for age, sex, and diabetes duration, stratified by GG, GT, or TT genotype at rs55703767, with overlay of individual data points for both women (pink) and men (blue).
We did not detect any eQTL association between rs55703767 and COL4A3 expression in mouse glomeruli or in human tissues, and thus assume that the variant affects the COL4A3 structure rather than expression levels. Nevertheless in a Pima Indian cohort of 97 participants with DKD with morphometric and expression data from renal biopsies, COL4A3 expression was negatively correlated with the GBM surface density (filtration surface density) (β=−0.27; P=0.02), which is associated with eGFR in DKD in both T1D and T2D.29,30 Furthermore, in 335 microdissected human glomerulus samples, expression of COL4A3 was negatively correlated with glomerulosclerosis, potentially reflecting podocyte depletion in sclerotic glomeruli (correlation=0.16; P=4.8×10−3; Supplemental Figure 3). COL4A3 expression in glomeruli, but not in tubules, was also nominally correlated with eGFR (correlation=0.108; P=0.05; Supplemental Figure 3).
Evidence for Hyperglycemia Specificity
Hyperglycemia promotes the development of diabetic complications. If a genetic variant exerts a stronger effect in the setting of hyperglycemia, (1) it might not be detected in general CKD, (2) it may be detected whether hyperglycemia is conferred by T1D or T2D, (3) its effect may be stronger at higher glycemic strata, and (4) interventions that reduce glycemia may attenuate the association signal. COL4A3 rs55703767 was not associated with eGFR in a general population sample of 110,517 mainly nondiabetic participants of European ancestry31 (Supplemental Table 5). However, in a smaller cohort of 5190 participants with T2D and DKD phenotypes in the SUMMIT consortium, we detected a directionally consistent suggestive association of COL4A3 rs55703767 with DN (two-tailed P=0.08; Supplemental Table 6).
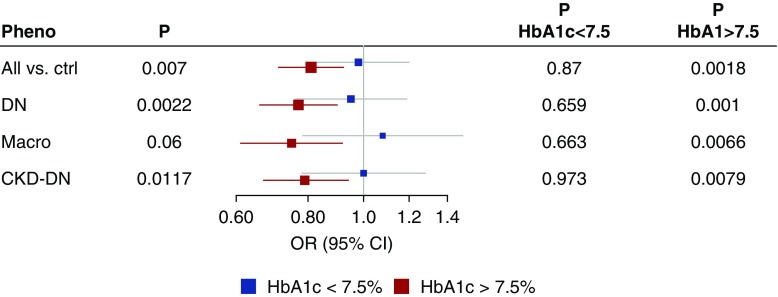
We further stratified the association analyses by HbA1c in the Finnish Diabetic Nephropathy (FinnDiane) Study, a T1D cohort study with extensive longitudinal phenotypic data.32 On the basis of the time-weighted mean of all available HbA1c measurements for each individual, 1344 individuals had mean HbA1c <7.5% (58 mmol/mol), and 2977 with mean HbA1c ≥7.5%. COL4A3 rs55703767 was nominally significant (P<0.05) only in individuals with HbA1c ≥7.5% (Figure 4, Supplemental Figure 4, Supplemental Table 7). However, the interaction between HbA1c and COL4A3 rs55703767 was not significant (P=0.83). Upon further examination, the genetic effect was diminished also in the highest HbA1c quartile (>9.3%), as the environmental effect of HbA1c seems to overwhelm any potential genetic effects at COL4A3 (test of heterogeneity nonsignificant). In a similar setting of individuals with T2D from the Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) study (n=3226),13,33 no difference was observed for COL4A3 rs55703767 between HbA1c strata below or above 7.5% (Supplemental Figure 5). We performed a similar HbA1c stratified analysis in the Diabetes Control and Complications Trial (DCCT), whose participants, all with T1D, continue to be followed in the Epidemiology of Diabetes Interventions and Complications (EDIC).34,35 In DCCT-EDIC the effect of COL4A3 rs55703767 was stronger among those recruited in the secondary cohort (mild retinopathy and longer diabetes duration at baseline) who were originally randomized to conventional treatment and therefore had higher HbA1c than the intensive treatment group (Supplemental Table 8). Taken together, these independent lines of evidence strongly suggest that the COL4A3 variant effect on DKD risk is amplified by poor glycemic control.
Figure 4.
Association at rs55703767 (COL4A3) stratified by HbA1c below or above 7.5%, for the phenotypes reaching genome-wide significance in the combined meta-analysis. Analysis included 1344 individuals with time-weighted mean HbA1c <7.5% (58 mmol/mol), and 2977 with mean HbA1c ≥7.5% from the FinnDiane study; the individuals had median 19 HbA1c measurements (range 1–129).
Other Association Signals
A comprehensive list of all loci that achieved genome-wide significance from either the minimal or the fully adjusted covariate models is reported in Table 1. The fewer covariates required for the minimal model results in improved statistical power because of fewer participants with missing data, whereas the fully adjusted model allows the identification of associations potentially mediated by covariates. Comparison of the adjustment models revealed strong consistency between the two models (Supplemental Figure 6). Table 1 is stratified into two sets of loci: common and/or known kidney biology loci (top half, n=7) and uncommon and no known kidney biology loci (bottom half, n=9). We focus on loci in the top half of the table, common and/or in/near genes with relevant kidney biology.
Two other genome-wide significant signals were near genes encoding proteins related to collagen. Variant rs12615970 (MAF=0.13), located 53 kb downstream of COLEC11, was associated with CKD (OR, 1.31; 95% CI, 1.19 to 1.45; P=9.43×10−9), and rs116772905 (MAF=0.011) in exon 14 of DDR1 was associated with microalbuminuria (OR, 3.78; 95% CI, 2.35 to 6.11; P=4.40×10−8). rs116772905 is in perfect linkage disequilibrium with rs118124843, the lead association with microalbuminuria for this locus under the full adjustment model (taking into account both body mass index and HbA1c), located 29 kb downstream of DDR1 (OR, 3.97; 95% CI, 2.44 to 6.52; P=3.37×10−8). COLEC11 encodes a collectin protein containing both a collagen-like domain and a carbohydrate recognition domain for binding sugars, and DDR1 encodes the discoidin domain-containing receptor 1, which binds collagens including type IV collagen.
In addition to COL4A3 rs55703767, three other low-frequency variants achieved study-wide significance (P<6.76×10−9), each associated with microalbuminuria: rs142823282 (MAF=0.017), 22 kb upstream of TAMM41 encoding a mitochondrial translocator assembly and maintenance protein36,37 (OR, 6.75; 95% CI, 3.66 to 12.38; P=1.13×10−11); rs144434404 (MAF=0.011), in intron 1 of BMP7 encoding the bone morphogenetic protein 7 previously implicated in DKD38 (OR, 6.75; 95% CI, 3.61 to 12.74; P=2.67×10−9); and rs145681168 (MAF=0.014), in intron 3 of two transcripts of HAND2 antisense RNA 1 (HAND2-AS1; OR, 5.53; 95% CI, 2.90 to 10.53; P=5.40×10−9) and 50 kb upstream of HAND2, encoding a heart and neural crest derivatives transcription factor.
Two additional common variants achieved genome-wide significance: rs551191707 (MAF=0.122) in PRNCR1 associated with ESKD when compared with macroalbuminuria (OR, 1.70; 95% CI, 1.40 to 2.04; P=4.39×10−8); and rs61983410 (MAF=0.213) in an intergenic region on chromosome 14 associated with microalbuminuria (full model OR, 0.78; 95% CI, 0.71 to 0.85; P=3.06×10−8). The remaining eight variants associated with features of DKD had lower allele frequencies (four with 0.01≤MAF≤0.05 and four with MAF<0.01) and did not achieve study-wide significance.
As we had done for COL4A3 rs55703767, we tested whether the associations of the 15 other variants were amplified by hyperglycemia. None of the 15 variants were significantly associated with eGFR in the general population (Supplemental Table 5). In the smaller SUMMIT T2D cohort13 we were able to interrogate seven loci with comparable trait definitions. The ORs were directionally consistent in six of them (binomial sign test: P=0.06; Supplemental Table 6). In FinnDiane seven of the remaining 15 loci were observed with sufficient frequency (minor allele counts >10) to allow subgroup analysis. Two additional SNPs (rs149641852 in SNCAIP and rs12615970 near COLEC11) were nominally significant (P<0.05) only in individuals with HbA1c ≥7.5%; however, the genotype-by-HbA1c interaction term was nonsignificant (Supplemental Figure 4, Supplemental Table 7).
Variants Previously associated with DKD
We investigated the effect of variants previously associated at genome-wide significance with renal complications in individuals with diabetes.8–13,39 Across the ten subphenotypes in our meta-analysis, we found evidence of association for seven of nine examined loci (P<0.05; Supplemental Figure 7): We replicated two loci that were previously discovered without overlapping individuals with the current study: SCAF8/CNKSR3 rs12523822, originally associated with DKD (P=6.8×10−4 for “All versus ctrl”)8; and UMOD rs77924615, originally associated with eGFR in both individuals with and without diabetes (P=5.2×10−4 for “CKD”).31 Associations at the AFF3, RGMA-MCTP2, and ERBB4 loci, identified in the Genetics of Nephropathy–an International Effort consortium12 comprising a subset of studies included in this current effort, remained associated with DKD, although the associations were attenuated in this larger dataset (RGMA-MCTP2 rs12437854 P=2.97×10−5; AFF3 rs7583877 P=5.97×10−4; ERBB4 rs7588550 P=3.53×10−5; Supplemental Figure 8). Associations were also observed at the CDCA7/SP3 (rs4972593, P=0.02 for “CKD-DN,” originally for ESKD exclusively in women11) and GLRA3 (rs1564939, P=0.02 for “CKD extremes,” originally for AER10,39), but these analyses also include individuals that overlap with the original studies. Apart from the UMOD locus, none of the 63 loci associated with eGFR in the general population31 were associated with DKD after correction for multiple testing (P<7.0×10−4; Supplemental Figure 9).
Gene and Gene Set Analysis
We conducted gene-level analyses by using two methods that aggregate SNP summary statistics over a gene region while accounting for linkage disequilibrium: MAGMA and PASCAL.40,41 MAGMA identified five genes at a Bonferroni-corrected threshold (P<0.05/18,222 genes tested =2.74×10−6): the collagen gene COL20A1 associated with “CKD extreme” (full model P=5.77×10−7) and “ESKD versus non-ESKD” (full model P=9.53×10−7), SLC46A2 associated with “All versus ctrl” (P=7.38×10−7), SFXN4 associated with “Macro” (full model P=1.65×10−7), GLT6D1 associated with “ESKD versus macro” (P=1.49×10−6), and SNX30 associated with “All versus ctrl” (P=2.49×10−6) (Supplemental Table 9). Although PASCAL did not identify any significant gene-level associations, the five MAGMA-identified genes had P<5.0×10−4 in PASCAL (Supplemental Table 10). Both SFXN4 and CBX8 have been reported to be differentially methylated in patients with diabetes with and without nephropathy.42,43
Additionally, we used MAGMA, PASCAL, DEPICT, and MAGENTA to conduct gene-set analysis in our GWAS dataset. The four methods identified 12 significantly enriched gene sets (Supplemental Table 11). One gene set, “negative regulators of RIG-I MDA5 signaling” was identified in two different pathway analyses (MAGMA and PASCAL) of our fully adjusted GWAS of ESKD versus Macro. Several additional related and overlapping gene sets were identified, including “RIGI MDA5 mediated induction of IFN alpha beta pathways,” “TRAF3 dependent IRF activation pathway,” and “TRAF6 mediated IRF activation” (PASCAL) and “activated TLR4 signaling” (MAGENTA). RIG-I, MDA5 and the toll-like receptor TLR4 are members of the innate immune response system that respond to both cellular injury and infection44,45 and transduce highly intertwined signaling cascades. These include the signaling molecules TRAF3 and TRAF6, which induce expression of type 1 IFN and proinflammatory cytokines implicated in the progression of DKD.46,47 Specifically, the TLR4 receptor and several of its ligands and downstream cytokines display differential levels of expression in DKD renal tubules versus normal kidneys and versus non-DKD controls,48 and TLR4 knockout mice are protected from DKD and display marked reductions in interstitial collagen deposition in the kidney.49 Other pathways of interest include “other lipid, fatty acid and steroid metabolism,” “nitric oxide signaling in the cardiovascular system,” and “TNF family member,” with both nitric oxide and TNF-α implicated in DKD.50,51,52
Expression and Epigenetic Analyses
We interrogated gene expression datasets in relevant tissues to determine whether our top signals underlie eQTL. We first analyzed genotype and RNA-sequencing gene expression data from 96 whole human kidney cortical samples22 and microdissected human kidney samples (121 tubule and 119 glomerular samples)23 from participants of European descent without any evidence of renal disease (Supplemental Figure 10). No findings in this data set achieved significance after correction for multiple testing. In the GTEx and eQTLgen datasets, COL4A3 rs55703767 had a significant eQTL (P=5.63×10−38) with the MFF gene in blood, but is most likely due to modest LD with other nearby strong eQTLs in the region. rs118124843 near DDR1 and VARS2 had multiple significant eQTLs in blood besides VARS2 (P=1.71×10−5; Supplemental Table 12). Interestingly, rs142823282 near TAMM41 was a cis-eQTL for PPARG (P=4.60×10−7), a transcription factor regulating adipocyte development and glucose and lipid metabolism; PPARγ agonists have been suggested to prevent DKD.53
To ascertain the potential functional role of our top noncoding signals, we mined chromatin immunoprecipitation sequencing data derived from healthy adult human kidney samples.25 SNP rs142823282 near TAMM41 was located close to kidney histone marks H3K27ac, H3K9ac, H3K4me1, and H3K4me3, suggesting that this is an active regulator of TAMM41 or another nearby gene (Supplemental Figure 11). Interestingly, in recent work we have shown that DNA methylation profiles in participants with T1D with/without kidney disease show the greatest differences in methylation sites near TAMM41.54
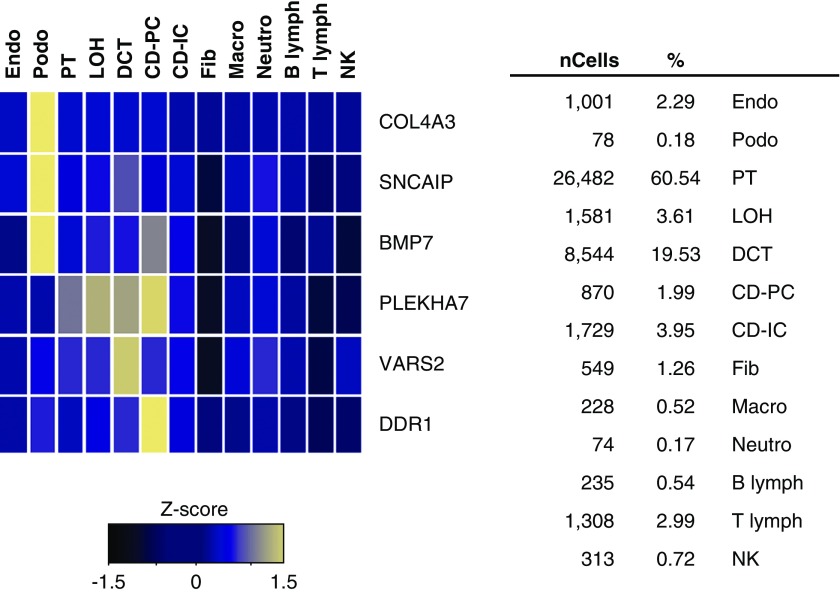
To establish whether the expression of our top genes shows enrichment in a specific kidney cell type, we queried an expression dataset of approximately 50,000 single cells obtained from mouse kidneys.24 Expression was detected for six genes in the mouse kidney atlas: three (COL4A3, SNCAIP, and BMP7) were almost exclusively expressed in podocytes (Figure 5), supporting the significant role for podocytes in DKD.
Figure 5.
Single-cell RNA-sequencing in mouse kidney shows COL4A3, SNCAIP, and BMP7 are specifically expressed in podocytes. Mean expression values of the genes were calculated in each cluster. The color scheme is on the basis of z-score distribution; the map shows genes with z-score >2. In the heatmap, each row represents one gene and each column is a single cell type. Percentages of assigned cell types are summarized in the right panel. CD-IC, collecting duct intercalated cell; CD-PC, collecting duct principal cell; CD-Trans, collecting duct transitional cell; DCT, distal convoluted tubule; Endo, containing endothelial, vascular, and descending loop of Henle; Fib, fibroblast; LOH, ascending loop of Henle; Lymph, lymphocyte; Macro, macrophage; Neutro, neutrophil; NK, natural killer cell; Podo, podocyte; PT, proximal tubule.
Gene expression levels in kidneys in cases versus controls were predicted with TWAS on the basis of the GWAS summary statistics and eQTL data of kidney glomeruli and tubuli.23 While none of the genes survived correction for multiple testing, analysis suggested 18 genes with differential expression in cases and controls with P<1×10−4, including the NPNT, PRRC2C, and VPS33B genes (Supplemental Table 13). NPNT encodes for nephronectin, an extracellular matrix protein on GBM. Knocking out NPNT or decreasing NPNT expression levels have been shown to induce podocyte injury related to GBM.55 On the contrary, TWAS predicted higher NPNT expression within DN cases versus normal AER. In line with our TWAS finding, NPNT is significantly upregulated in glomeruli of DN mouse model versus nondiabetic mouse (P=6.4×10−4; fold change 1.3, in top 2%, accessed through www.neproseq.org).56 Furthermore, a variant near PRRC2C was recently associated with albuminuria in the UK Biobank,57 and rare mutations in VPS33B gene cause arthrogryposis, renal dysfunction, and cholestasis-1 syndrome involving proximal–tubular dysfunction and usually death by 1 year of age.58
Discussion
Our genome-wide analysis of 19,406 participants with T1D identified 16 genome-wide significant loci associated with DKD, four of which remained significant after a conservative correction for multiple testing. Four of the 16 genome-wide significant signals are in or near genes with known or suggestive biology related to renal function/collagen (COL4A3, BMP7, COLEC11, and DDR1), but this is the first time that naturally occurring variation (MAF>1%) in these loci has been associated with DKD. Our most significant signal was a protective missense variant in COL4A3, rs55703767, reaching both genome-wide and study-wide significance with multiple definitions of DKD. Moreover, this variant demonstrated a significant association with GBM width such that protective allele carriers had thinner GBM before any signs of kidney disease, and its effect was dependent on glycemia.
COL4A3, with COL4A4 and COL4A5, make up the so-called “novel chains” of type IV collagen,59 which together play both structural and signaling roles in the GBM. Specifically, COL4A3 is known to bind a number of molecules including integrins, heparin, and heparin sulfate proteoglycans, and other components of the GBM, such as laminin and nidogen. These interactions mediate the contact between cells and the underlying collagen IV basement membrane, and regulate various processes essential to embryonic development and normal physiology, including cell adhesion, proliferation, survival, and differentiation. Dysregulation of these interactions has been implicated in several pathologic conditions, including CKD.60
Mutations in COL4A3 are responsible for the autosomal recessive form of Alport syndrome, a progressive inherited nephropathy, as well as benign familial hematuria, characterized by thin (or variable width) GBM, and thought to be a milder form of Alport syndrome.61 Furthermore, mutations in COL4A3 have also been identified in patients with FSGS, often leading to proteinuria and renal failure. Some of these patients with FSGS presented with segmental GBM thinning.62 Of note, the common rs55703767 (COL4A3 Asp326Tyr) variant, protecting from DKD, was also associated with thinner GBM in individuals with diabetes but without renal complications, a feature that seems to be beneficial in the context of diabetes. The rs55703767 SNP is predicted to alter the third amino acid of the canonical triple-helical domain sequence of Glycine (G)-X-Y (where X and Y are often proline [P] and hydroxyproline [Y], respectively) from G-E-D (D=Aspartic) to G-E-Y,63 potentially affecting the structure of the collagen complex. In addition, a recent study64 of candidate genes involved in renal structure reported rs34505188 in COL4A3 (not in linkage disequilibrium with rs55703767, r2<0.01) to be associated with ESKD in black Americans with T2D (MAF=2%; OR, 1.55; 95% CI, 1.22 to 1.97; P=5×10−4). Together with the trend toward association we have seen in SUMMIT and the glycemic interaction we have reported here, these findings suggest variation in COL4A3 may be associated with DKD in T2D as well.
Given its association as a protective SNP, we can speculate that the rs55703767 variant may confer tensile strength or flexibility to the GBM, which may be of particular relevance in the glomerular hypertension associated with DKD. Alternatively, COL4A3 may regulate the rates of production and/or turnover of other GBM components, affecting GBM width changes in diabetes. How these effects might confer protection in a manner dependent on ambient glucose concentrations is unknown. Future mechanistic studies will be required to determine the precise role of this variant in DKD; elucidation of its interaction with glycemia in providing protection might be relevant to other molecules implicated in diabetic complications.
In keeping with the collagen pathway, the synonymous exonic variant rs118124843, which reached genome-wide significance for the “Micro” phenotype, is located near DDR1, the gene encoding the discoidin domain-containing receptor 1. On the basis of chromatin conformation interaction data from Capture HiC Plotter,65 the rs118124843 containing fragment interacts with six gene promoter regions, including DDR1, suggesting that the variant regulates DDR1 expression across multiple tissues (Supplemental Table 12). DDR1 is a collagen receptor66 shown to bind type IV collagen,67 and is highly expressed in kidneys, particularly upon renal injury.68 Upon renal injury, Ddr1-deficient mice display lower levels of collagen,69 decreased proteinuria, and an increased survival rate compared with wild-type controls,70 with Ddr1/Col4a3 double-knockout mice displaying protection from progressive renal fibrosis and prolonged lifespan compared with Col4a3 knockout mice alone.69 Thus, through its role in collagen binding, DDR1 has been suggested as a possible therapeutic target for kidney disease.69
The association of rs12615970, an intronic variant on chromosome 2 near the COLEC11 gene, met genome-wide significance for the CKD phenotype, as well as nominal significance for multiple albuminuria-based traits. The rs12615970 containing fragment was found to interact with COLEC11, ALLC, and ADI1 transcription start sites in chromatin conformation data on GM12878 cell line (Supplemental Table 12).65,71 Collectin-11 is an innate immune factor synthesized by multiple cell types, including renal epithelial cells with a role in pattern recognition and host defense against invasive pathogens, through binding to fructose and mannose sugar moieties.72,73 Mice with kidney-specific deficiency of COLEC11 are protected against ischemia-induced tubule injury because of their reduced complement deposition,74 and mutations in COLEC11 have been identified in families with 3MC syndrome, a series of rare autosomal recessive disorders resulting in birth defects and abnormal development, including kidney abnormalities.75
The intronic variant rs144434404, associated at study-wide significance with the microalbuminuria phenotype, resides within the bone morphogenetic protein 7 (BMP7) gene. BMP7 encodes a secreted ligand of the TGF-β superfamily of proteins. Developmental processes are regulated by the BMP family of glycosylated extracellular matrix molecules via serine/threonine kinase receptors and canonical Smad pathway signaling. Coordinated regulation of both BMP and BMP-antagonist expression is essential for developing tissues, and changes in the levels of either BMP or BMP-antagonists can contribute to disease progression such as fibrosis and cancer.76 BMP7 is required for renal morphogenesis, and Bmp7 knockout mice die soon after birth, because of reduced ureteric bud branching.77–79 Maintenance of Bmp7 expression in glomerular podocytes and proximal tubules of diabetic mice prevents podocyte loss and reduces overall diabetic renal injury.38 More recently, we have identified a mechanism through which BMP7 orchestrates renal protection through Akt inhibition, and highlights Akt inhibitors as potential antifibrotic therapeutics.80 It is also noteworthy that the BMP7 antagonist grem-1 is implicated in DKD,81–83 and gremlin has been implicated as a biomarker of kidney disease.84
Strengths of this analysis include the large sample size, triple that of the previous largest GWAS; the uniform genotyping and QC procedures; standardized imputation for all studies (1000 Genomes reference panel); the inclusion of exome array content; the exploration of multiple standardized phenotype definitions of DKD; and supportive data from various sources of human kidney samples. Several of the loci identified have known correlations with kidney biology, suggesting that these are likely true associations with DKD. However, we acknowledge a number of limitations. First, nine variants have low MAF and were driven by only two cohorts, indicating that further validation will be required to increase confidence in these associations. Second, seven variants were significantly associated with microalbuminuria only, a trait shown to be less heritable in previous studies. We included these loci to maximize comprehensiveness in reporting novel DKD associations. Replication in independent samples and functional confirmation is required to validate all of these loci. Although the gene-level, gene set, and pathway analyses had limited power, these analyses identified several additional potential DKD loci and pathways, some with relevance to kidney biology, that require further follow-up. Finally, although we included only controls with a minimum diabetes duration of 15 years, we cannot fully rule out that some of the controls would progress to DKD in the future, as the improvements in diabetes treatment in the past decades have postponed the onset of complications. We also excluded cases with short diabetes duration to avoid renal complications that might be due to other causes. These phenotypic definitions were meant to overcome the limitation that in clinical practice kidney biopsy specimens are rarely taken from individuals with diabetes to verify the diagnosis. As for any late-onset disease, these challenges in phenotypic definition may have reduced our power to detect additional associations. We note, however, that this relatively small degree of contamination would lead to loss of power and increased type 2 error rather than false positive findings; therefore, it does not undermine the robustness of the associations reported here.
Diabetic complications are unquestionably driven by hyperglycemia and partially prevented by improved glycemic control in both T1D and T2D, but there has been doubt over what contribution, if any, inherited factors contribute to disease risk. In line with previous genetic studies, this study with a markedly expanded sample size identified several loci strongly associated with DKD risk. These findings suggest that larger studies, aided by novel analyses and including T2D, will continue to enhance our understanding of the complex pathogenesis of DKD, paving the way for molecularly targeted preventive or therapeutic interventions.
Disclosures
We acknowledge the following conflicts of interest: Groop has received investigator-initiated research grants from Eli Lilly and Roche, is an advisory board member for AbbVie, AstraZeneca, Boehringer Ingelheim, Cebix, Eli Lilly, Janssen, Medscape, Merck Sharp & Dohme, Novartis, Novo Nordisk and Sanofi, and has received lecture fees from AstraZeneca, Boehringer Ingelheim, Eli Lilly, Elo Water, Genzyme, Merck Sharp & Dohme, Medscape, Novo Nordisk and Sanofi. McCarthy is a Wellcome Investigator and an NIHR Senior Investigator. He serves on advisory panels for Pfizer, NovoNordisk, and Zoe Global, has received honoraria from Merck, Pfizer, NovoNordisk and Eli Lilly, has stock options in Zoe Global, and has received research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. Rossing has received consultancy and/or speaking fees (to his institution) from AbbVie, Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly, MSD, Novo Nordisk and Sanofi Aventis and research grants from AstraZeneca and Novo Nordisk, and shares in Novo Nordisk.
Dr. Onengut-Gumuscu reports grants from JDRF, during the conduct of the study. Dr. Caramori reports grants from Bayer Pharmaceuticals, personal fees from Elcelix, outside the submitted work. Dr. de Boer reports personal fees from Boehringer-Ingelheim, personal fees from Ironwood, non-financial support from Medtronic, non-financial support from Abbott, outside the submitted work. Dr. Snell-Bergeon reports grants from NIH, grants from JDRF, during the conduct of the study; other from GlaxoSmithKline, outside the submitted work. Dr. Weitgasser reports personal fees from Abbott, personal fees from Astra Zeneca, personal fees from Boehringer-Ingelheim, grants and personal fees from Eli Lilly, personal fees from MSD, grants and personal fees from NovoNordisk, personal fees from Dexcom, personal fees from Roche, grants and personal fees from Sanofi, personal fees from Servier, personal fees from Takeda, personal fees from Spar, outside the submitted work. Dr. McCarthy reports grants, personal fees and other from Pfizer, grants, personal fees and other from Merck, grants, personal fees and other from Novo Nordisk, grants from Takeda, grants from Servier, grants from Sanofi Aventis, grants from Boehringer Ingelheim, grants from Astra Zeneca, grants from Janssen, grants, personal fees and other from Eli Lilly, grants from Abbvie, grants from Roche, during the conduct of the study; grants, personal fees and other from Pfizer, grants, personal fees and other from Eli Lilly, grants, personal fees and other from Merck, other from Zoe Global, other from Genentech (wef June 2019), outside the submitted work. Dr. Costacou reports grants from National Institutes of Health (NIH), during the conduct of the study. Dr. McKnight reports grants from Northern Ireland Public Health Agency (Research and Development Division) and Medical Research Council as part of the USA-Ireland-Northern Ireland research partnership and Department for the Economy NI 15/IA/3152 during the conduct of the study. Dr. Kretzler reports grants from NIH, non-financial support from University of Michigan, during the conduct of the study; grants from JDRF, grants from Astra-Zeneca, grants from NovoNordisc, grants from Eli Lilly, grants from Gilead, grants from Goldfinch Bio, grants from Merck, grants from Janssen, grants from Boehringer-Ingelheim, grants from Elpidera, grants from European Union Innovative Medicine Innitiative, outside the submitted work; In addition, Dr. Kretzler has a patent Biomarkers for CKD progression issued. Dr. Susztak reports grants from GSK, Regeneron, Boehringer, Gilead, Bayer, Lilly, Merck, outside the submitted work. Dr. Colhoun reports grants from AStraZeneca LP, other from Bayer, grants and other from Eli Lilly and Company, other from Novartis Pharmaceuticals, grants and other from Regeneron, grants from Pfizer Inc., other from Roche Pharmaceuticals, grants and other from Sanofi Aventis, grants and personal fees from Novo Nordisk, outside the submitted work. Dr. Groop reports personal fees from AbbVie, personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, personal fees from Eli Lilly, personal fees from Elo Water, personal fees from Janssen, personal fees from Medscape, personal fees from Mundipharma, personal fees from MSD, personal fees from Novartis, personal fees from Novo Nordisk, personal fees from Sanofi, outside the submitted work. Dr. Marre reports personal fees from Novo-Nordisk, personal fees from Servier, personal fees from Merck, outside the submitted work; and Marre is president of the Fondation Francophone pour la Recherche sur le Diabète, a French, non for profit institution. This institution is supported financially by the following companies: Novo-Nordisk, Merck, Sanofi, Eli Lilly, Astra-Zeneca, Roche, Abbott. Dr. Hirschhorn reports other from Camp4 Therapeutics, outside the submitted work. Dr. Florez reports personal fees from Janssen, outside the submitted work. Dr. Hiraki reports grants from Juvenile Diabetes Research Foundation (JDRF), during the conduct of the study. Dr. Rossing reports grants and other from Astra Zeneca, other from Boehringer Ingelheim, other from Bayer, grants and other from Novo Nordisk, other from MSD, other from Eli Lilly, other from astellas, other from Abbvie, other from Mundi Pharma, other from Sanofi, other from Gilead, outside the submitted work.
Funding
This study was supported by a grant from the JDRF (17-2013-7) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01 DK081923 and R01 DK105154. Salem was supported in part by JDRF grant 3-APF-2014-111-A-N, and National Heart, Lung and Blood Institute grant R00 HL122515. Todd was supported by NIDDK grant K12-DK094721. Sandholm received funds from the European Foundation for the Study of Diabetes (EFSD) Young Investigator Research Award funds and Academy of Finland (299200). Godson, Andrews, Brennan, Martin, Hughes, and Doyle are supported by Science Foundation Ireland - Health Research Board (SFI-HRB) US Ireland Research Partnership SFI15/US/B3130. Nelson was supported in part by the Intramural Research Program of the NIDDK. The FinnDiane study was funded by JDRF (17-2013-7), Folkhälsan Research Foundation, the Wilhelm and Else Stockmann Foundation, the Liv och Hälsa Foundation, Helsinki University Central Hospital Research Funds (EVO), the Novo Nordisk Foundation (NNF OC0013659), and Academy of Finland (275614 and 316664). The Pittsburgh Epidemiology of Diabetes Complications Study (EDC) study was supported by NIDDK grant DK34818 and by the Rossi Memorial Fund. The Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR) study was funded by National Eye Institute grant EY016379. The Sweden study was supported by Family Erling-Persson (Brismar) and Stig and Gunborg Westman foundations (Gu). SUMMIT Consortium: this work was supported by Innovative Medicines Initiative (IMI) (SUMMIT 115006); Wellcome Trust grants 098381, 090532, and 106310; National Institutes of Health grant R01-MH101814; and JDRF grant 2-SRA-2014-276-Q-R; and other grants from Swedish Research Council, European Research Council Advanced (ERC-Adv) research grant 269045-GENE TARGET T2D, Academy of Finland grants 263401 and 267882, and Sigrid JUselius Foundation. The George M. O’Brien Michigan Kidney Translational Core Center, funded by NIH/NIDDK grant 2P30-DK-081943 for the bioinformatics support. The Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection (GoDARTS) was funded by the Wellcome Trust (072960/Z/03/Z, 084726/Z/08/Z, 084727/Z/08/Z, 085475/Z/08/Z, 085475/B/08/Z) and as part of the European Union’s IMI-SUMMIT program.
Supplementary Material
Acknowledgments
Conceptualization: Godson, Maxwell, Groop, Hirschhorn, and Florez; Formal analysis: Salem, Cole, Sandholm, Valo, Di Liao, Cao, Pezzolesi, Smiles, Qiu, Nair, Park, Liu, Menon, Kang, R. Klein, B.E. Klein, Canty, Paterson, Chen, van Zuydam, and Onengut-Gumuscu; Investigation: Pezzolesi, Smiles, Skupien Qiu, Nair, Haukka, McKnight, Snell-Bergeon, Maahs, Maxwell, Paterson, Forsblom, Ahlqvist, Ahluwalia, Lajer, Boustany-Kari, Kang, Maeastroni, Tregouet, Gyorgy, Bull, Palmer, Stechemesser, Paulweber, Weitgasser, Rovīte, Pīrāgs, Prakapiene, Radzeviciene, Verkauskiene, Panduru, Groop, McCarthy, Gu, Möllsten, Falhammar, Brismar, Rossing, Costacou, Zerbini, Marre, Hadjadj, Forsblom, Chen, and Onengut-Gumuscu; Resources: Smiles, Harjutsalo, McKnight, Nelson, Caramori, Mauer, Gao, Snell-Bergeon, Maahs, Guo, Miller, Maxwell, Paterson, Chen, and Onengut-Gumuscu; Data curation: Hiraki, Di Liao, Cao, Smiles, Harjutsalo, Skupien, McKnight, R. Klein, B.E. Klein, Lee, Gao, Mauer, Caramori, Snell-Bergeon, Maahs, Guo, Miller, Maxwell, Chen, and Onengut-Gumuscu; Writing, original draft: Salem, Todd, Sandholm, Cole, Brennan, Andrews, Doyle, Hughes, Hiraki, Paterson, and Godson; Writing, review and editing: Salem, Todd, Sandholm, Cole, Brennan, Andrews, Doyle, Hughes, Hiraki, Paterson, Godson, Qiu, Park, Skupien, Mauer, Caramori, de Boer, Martin, McKnight, McKay, Maxwell, Susztak, McCarthy, Groop, Rich, Forsblom, Hirschhorn, and Florez; Visualization: Salem, Todd, Sandholm, Cole, Pezzolesi, Qiu., Di Liao, Cao, Park, Skupien, R. Klein, B.E. Klein, Lee, McKnight, McKay, Maxwell, and Paterson; Project administration: Todd; Supervision: Paterson, Krolewski, Groop, Godson, Maxwell, Colhoun, Rich, Kretzler, Susztak, Hirschhorn, and Florez; Funding acquisition: Paterson, Krolewski, Florez, and Hirschhorn. All authors approved the final version of the manuscript.
AusDiane acknowledges Dr. Bernhard Baumgartner from Department of Medicine, Diakonissen-Wehrle Hospital, Salzburg, Austria. LatDiane acknowledges the following doctors and researchers: A. Bogdanova, D. Grikmane, I. Care, A. Fjodorova, R. Graudiņa, A. Valtere, D. Teterovska, I. Dzīvīte-Krišāne, I. Kirilova, U. Lauga-Tūriņa, K. Geldnere, D. Seisuma, N. Fokina, S. Šteina, L. Jaunozola, I. Balcere, U. Gailiša, E. Menise, S. Broka, L. Akmene, N. Kapļa, J. Nagaiceva, A. Pētersons, A. Lejnieks, I. Konrāde, I. Salna, A. Dekante, A. Grāmatniece, A. Saliņa, A. Silda, V. Mešečko, V. Mihejeva, K. Kudrjavceva, I. Marksa, M. Cirse, D. Zeme, S. Kalva-Vaivode, J. Kloviņš, L. Nikitina-Zaķe, S. Skrebinska, Z. Dzērve, and R. Mallons. LitDiane acknowledges Dr. Jurate Lasiene, Prof. Dzilda Velickiene, Dr. Vladimiras Petrenko and nurses from the Department of Endocrinology, Hospital of Lithuanian University of Health Sciences. RomDiane acknowledges the following doctors and researchers: M. Anghel, DM Cheta, BA Cimpoca, D. Cimponeriu, CN Cozma, N. Dandu, D. Dobrin, I Duta, AM Frentescu, C. Ionescu-Tirgoviste, R. Ionica, R. Lichiardopol, AL Oprea, NM Panduru, A. Pop, G. Pop, R. Radescu, S. Radu, M. Robu, C. Serafinceanu, M. Stavarachi, O Tudose from the N.C Paulescu National Institute for Diabetes Nutrition and Metabolic Diseases from Bucharest; and M. Bacu, D. Clenciu, C. Graunteanu, M. Ioana, E. Mota, and M. Mota from Craiova Emergency County Hospital. SDR: Thanks to Maria Sterner and Malin Neptin for GWAS genotyping in the Scannia Diabetes Registry. The FinnDiane study thanks M. Parkkonen, A. Sandelin, A-R. Salonen, T. Soppela, and J. Tuomikangas for skillful laboratory assistance in the FinnDiane study. We also thank all the participants of the FinnDiane study and gratefully acknowledge all the physicians and nurses at each centre involved in the recruitment of participants (Supplemental Table 14). GoDARTS: We are grateful to all the participants in this study, the general practitioners, the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The study complies with the Declaration of Helsinki. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymised data and NHS Tayside, the original data owner. For a full list of SUMMIT consortium members, see Supplemental Table 15.
The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Data and Software Availability
GWAS summary statistics for all ten DKD phenotypes and two adjustment models are available for download at the AMP-Type 2 Diabetes Knowledge Portal (http://www.type2diabetesgenetics.org/informational/data), under “JDRF Diabetic Nephropathy Collaborative Research Initiative GWAS” datasets. Individual-level genotype data cannot be shared for all cohorts because of restrictions set by study consents and by European Union and national regulations regarding individual genotype data.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Biologic Underpinnings of Type 1 Diabetic Kidney Disease,” on pages 1782–1783.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019030218/-/DCSupplemental.
Supplemental Table 1. Cohorts contributing to analyses.
Supplemental Table 2. Pseudo-R2 of all SNPs across all GWAS as calculated by the McKelvey and Zavoina method.
Supplemental Table 3. Characteristics of RASS participants. Categorical variables display counts and percentage. Continuous values are mean±SD.
Supplemental Table 4. Multivariate analysis of association between rs55703767 and GBM width.
Supplemental Table 5. Look-up of the lead loci in GWAS on eGFR in the general population (Gorski et al.31).
Supplemental Table 6. Look-up of the lead loci in GWAS in the SUMMIT consortium (van Zuydam et al.13).
Supplemental Table 7. Association at lead loci stratified by HbA1c<7.5%.
Supplemental Table 8. Association of rs55703767 with DN in DCCT/EDIC subgroups.
Supplemental Table 9. Significant (P<0.05/18,222 genes tested =2.74×10–6) gene level associations with DKD in MAGMA.
Supplemental Table 10. Top nominally significant gene-level associations (P<1.0×10–5) with DKD in PASCAL.
Supplemental Table 11. Significant gene set and pathway analysis results.
Supplemental Table 12. eQTL associations and chromatin conformation interactions for the lead SNPs.
Supplemental Table 13. TWAS results with P<1×10–4.
Supplemental Table 14. Physicians and nurses at health care centers participating in the collection of FinnDiane patients.
Supplemental Table 15. Members of the SUMMIT consortium.
Supplemental Figure 1. Manhattan and QQ plots for each case-control definition and covariate model (minimal and full).
Supplemental Figure 2. Regional chromosomal location plots and forest plots by cohort of newly discovered DKD associations.
Supplemental Figure 3 (S2). Correlation of expression of COL4A3 with degree of fibrosis and eGFR in microdissected kidney samples.
Supplemental Figure 4 (S3). Genotype–phenotype associations at the lead loci when stratified by mean HbA1c <7.5% in the FinnDiane study.
Supplemental Figure 5 (S4): Genotype–phenotype associations at the lead rs55703767 (COL4A3) locus when stratified by mean HbA1c <7.5% in up to 3226 individuals with T2D from the GoDARTS.
Supplemental Figure 6 (S6). Fishplots comparing significance and directionality between minimal and fully adjusted models for each of the ten phenotype definitions.
Supplemental Figure 7 (S5). Association at previously reported loci (P<5×10–8) for renal complications in individuals with diabetes.
Supplemental Figure 8 (S7). Forest plots of the associations at the previously reported lead loci from the GENIE consortium with largely overlapping studies.
Supplemental Figure 9 (S7). Meta-analysis results for the loci that have previously been associated with DKD, or with eGFR or AER in the general population.
Supplemental Figure 10 (S8). eQTL analysis in microdissected tubule samples.
Supplemental Figure 11 (S9). Functional annotation of TAMM41.
Supplemental Appendix 1. Cohort descriptions and references.
References
- 1.Centers for Disease Control and Prevention : National Diabetes Statistics Report, 2017 Estimates of Diabetes and its Burden in the United States Background, Atlanta, GA, Centers for Disease Control and Prevention, US Department of Health and Human Services, 2017 [Google Scholar]
- 2.Tuttle KR, Bakris GL, Bilous RW, Chiang JL, de Boer IH, Goldstein-Fuchs J, et al.: Diabetic kidney disease: A report from an ADA Consensus conference. Am J Kidney Dis 64: 510–533, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Krolewski M, Eggers PW, Warram JH: Magnitude of end-stage renal disease in IDDM: A 35 year follow-up study. Kidney Int 50: 2041–2046, 1996 [DOI] [PubMed] [Google Scholar]
- 4.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J: Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53: 2449–2454, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Quinn M, Angelico MC, Warram JH, Krolewski AS: Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 39: 940–945, 1996 [DOI] [PubMed] [Google Scholar]
- 6.Seaquist ER, Goetz FC, Rich S, Barbosa J: Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 320: 1161–1165, 1989 [DOI] [PubMed] [Google Scholar]
- 7.Sandholm N, Van Zuydam N, Ahlqvist E, Juliusdottir T, Deshmukh HA, Rayner NW, et al.: The FinnDiane Study Group; The DCCT/EDIC Study Group; GENIE Consortium; SUMMIT Consortium : The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol 28: 557–574, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyengar SK, Sedor JR, Freedman BI, Kao WH, Kretzler M, Keller BJ, et al.: Family Investigation of Nephropathy and Diabetes (FIND) : Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family investigation of nephropathy and diabetes (FIND). PLoS Genet 11: e1005352, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, et al.: ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium : Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandholm N, Forsblom C, Mäkinen VP, McKnight AJ, Osterholm AM, He B, et al.: SUMMIT Consortium; FinnDiane Study Group : Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia 57: 1143–1153, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, et al.: FIND Consortium; FinnDiane Study Group and the GENIE Consortium : Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol 24: 1537–1543, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, et al.: DCCT/EDIC Research Group : New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet 8: e1002921, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Zuydam NR, Ahlqvist E, Sandholm N, Deshmukh H, Rayner NW, Abdalla M, et al.: Finnish Diabetic Nephropathy Study (FinnDiane); Hong Kong Diabetes Registry Theme-based Research Scheme Project Group; Warren 3 and Genetics of Kidneys in Diabetes (GoKinD) Study Group; GENIE (GEnetics of Nephropathy an International Effort) Consortium; Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) Research Group; SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) Consortium : A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 67: 1414–1427, 2018. 29703844 [Google Scholar]
- 14.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR: Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44: 955–959, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchsberger C, Abecasis GR, Hinds DA: minimac2: Faster genotype imputation. Bioinformatics 31: 782–784, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Ji L: Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95: 221–227, 2005 [DOI] [PubMed] [Google Scholar]
- 17.McKelvey RD, Zavoina W: A statistical model for the analysis of ordinal level dependent variables. J Math Sociol 4: 103–120, 1975 [Google Scholar]
- 18.Mauer M, Zinman B, Gardiner R, Suissa S, Sinaiko A, Strand T, et al.: Renal and retinal effects of enalapril and losartan in type 1 diabetes. N Engl J Med 361: 40–51, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen CD, Frach K, Schlöndorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133–140, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, et al.: Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes 58: 469–477, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, et al.: European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Ko YA, Yi H, Qiu C, Huang S, Park J, Ledo N, et al.: Genetic-variation-driven gene-expression changes highlight genes with important functions for kidney disease. Am J Hum Genet 100: 940–953, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu C, Huang S, Park J, Park Y, Ko YA, Seasock MJ, et al.: Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 24: 1721–1731, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al.: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al.: The NIH Roadmap Epigenomics Mapping Consortium : The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 28: 1045–1048, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al.: GTEx Consortium : Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 9: 1825, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yagame M, Kim Y, Zhu D, Suzuki D, Eguchi K, Nomoto Y, et al.: Differential distribution of type IV collagen chains in patients with diabetic nephropathy in non-insulin-dependent diabetes mellitus. Nephron 70: 42–48, 1995 [DOI] [PubMed] [Google Scholar]
- 28.Caramori ML, Parks A, Mauer M: Renal lesions predict progression of diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol 24: 1175–1181, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fufaa GD, Weil EJ, Lemley KV, Knowler WC, Brosius FC 3rd, Yee B, et al.: Structural predictors of loss of renal function in American Indians with type 2 diabetes. Clin J Am Soc Nephrol 11: 254–261, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauer M, Caramori ML, Fioretto P, Najafian B: Glomerular structural-functional relationship models of diabetic nephropathy are robust in type 1 diabetic patients. Nephrol Dial Transplant 30: 918–923, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, et al.: 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7: 45040, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorn LM, Forsblom C, Fagerudd J, Thomas MC, Pettersson-Fernholm K, Saraheimo M, et al.: FinnDiane Study Group : Metabolic syndrome in type 1 diabetes: Association with diabetic nephropathy and glycemic control (the FinnDiane study). Diabetes Care 28: 2019–2024, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Morris AD, Boyle DI, MacAlpine R, Emslie-Smith A, Jung RT, Newton RW, et al.: DARTS/MEMO Collaboration : The diabetes audit and research in Tayside Scotland (DARTS) study: Electronic record linkage to create a diabetes register. BMJ 315: 524–528, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan DM; DCCT/EDIC Research Group : The diabetes control and complications trial/epidemiology of diabetes interventions and complications study at 30 years: Overview. Diabetes Care 37: 9–16, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Diabetes Control and Complications Trial Research Group : Implementation of treatment protocols in the diabetes control and complications trial. Diabetes Care 18: 361–376, 1995 [DOI] [PubMed] [Google Scholar]
- 36.Blunsom NJ, Gomez-Espinosa E, Ashlin TG, Cockcroft S: Mitochondrial CDP-diacylglycerol synthase activity is due to the peripheral protein, TAMM41 and not due to the integral membrane protein, CDP-diacylglycerol synthase 1. Biochim Biophys Acta Mol Cell Biol Lipids 1863: 284–298, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, et al.: Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab 17: 709–718, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, de Caestecker M, Kopp J, Mitu G, Lapage J, Hirschberg R: Renal bone morphogenetic protein-7 protects against diabetic nephropathy. J Am Soc Nephrol 17: 2504–2512, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Sandholm N, Haukka JK, Toppila I, Valo E, Harjutsalo V, Forsblom C, et al.: Confirmation of GLRA3 as a susceptibility locus for albuminuria in Finnish patients with type 1 diabetes. Sci Rep 8: 12408, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S: Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLOS Comput Biol 12: e1004714, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Leeuw CA, Mooij JM, Heskes T, Posthuma D: MAGMA: Generalized gene-set analysis of GWAS data. PLOS Comput Biol 11: e1004219, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA: Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics 3: 33, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al.: DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 6: 20–28, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Loo YM, Gale M Jr.: Immune signaling by RIG-I-like receptors. Immunity 34: 680–692, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mollen KP, Anand RJ, Tsung A, Prince JM, Levy RM, Billiar TR: Emerging paradigm: Toll-like receptor 4-sentinel for the detection of tissue damage. Shock 26: 430–437, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Mora C, Navarro JF: Inflammation and diabetic nephropathy. Curr Diab Rep 6: 463–468, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Wada J, Makino H: Inflammation and the pathogenesis of diabetic nephropathy. Clin Sci (Lond) 124: 139–152, 2013 [DOI] [PubMed] [Google Scholar]
- 48.Lin M, Yiu WH, Wu HJ, Chan LY, Leung JC, Au WS, et al.: Toll-like receptor 4 promotes tubular inflammation in diabetic nephropathy. J Am Soc Nephrol 23: 86–102, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J, Chadban SJ, Zhao CY, Chen X, Kwan T, Panchapakesan U, et al.: TLR4 activation promotes podocyte injury and interstitial fibrosis in diabetic nephropathy. PLoS One 9: e97985, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colhoun HM, Marcovecchio ML: Biomarkers of diabetic kidney disease. Diabetologia 61: 996–1011, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKnight AJ, Patterson CC, Sandholm N, Kilner J, Buckham TA, Parkkonen M, et al.: Warren 3/UK GoKinD Study Group : Genetic polymorphisms in nitric oxide synthase 3 gene and implications for kidney disease: A meta-analysis. Am J Nephrol 32: 476–481, 2010 [DOI] [PubMed] [Google Scholar]
- 52.Prabhakar SS: Role of nitric oxide in diabetic nephropathy. Semin Nephrol 24: 333–344, 2004 [DOI] [PubMed] [Google Scholar]
- 53.Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Böger CA, Endlich K, et al.: CARDIoGRAM Consortium; ICBP Consortium; CARe Consortium; WTCCC2 : Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function. Hum Mol Genet 21: 5329–5343, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swan EJ, Maxwell AP, McKnight AJ: Distinct methylation patterns in genes that affect mitochondrial function are associated with kidney disease in blood-derived DNA from individuals with Type 1 diabetes. Diabet Med 32: 1110–1115, 2015 [DOI] [PubMed] [Google Scholar]
- 55.Müller-Deile J, Dannenberg J, Schroder P, Lin MH, Miner JH, Chen R, et al.: Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int 92: 836–849, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodgin JB, Nair V, Zhang H, Randolph A, Harris RC, Nelson RG, et al.: Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes 62: 299–308, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanetti D, Rao A, Gustafsson S, Assimes TL, Montgomery SB, Ingelsson E: Identification of 22 novel loci associated with urinary biomarkers of albumin, sodium, and potassium excretion. Kidney Int 95: 1197–1208, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holme A, Hurcombe JA, Straatman-Iwanowska A, Inward CI, Gissen P, Coward RJ: Glomerular involvement in the arthrogryposis, renal dysfunction and cholestasis syndrome. Clin Kidney J 6: 183–188, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleppel MM, Fan W, Cheong HI, Michael AF: Evidence for separate networks of classical and novel basement membrane collagen. Characterization of alpha 3(IV)-alport antigen heterodimer. J Biol Chem 267: 4137–4142, 1992 [PubMed] [Google Scholar]
- 60.Khoshnoodi J, Pedchenko V, Hudson BG: Mammalian collagen IV. Microsc Res Tech 71: 357–370, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kashtan CE, Ding J, Garosi G, Heidet L, Massella L, Nakanishi K, et al.: Alport syndrome: A unified classification of genetic disorders of collagen IV α345: A position paper of the Alport Syndrome Classification Working Group. Kidney Int 93: 1045–1051, 2018 [DOI] [PubMed] [Google Scholar]
- 62.Xie J, Wu X, Ren H, Wang W, Wang Z, Pan X, et al.: COL4A3 mutations cause focal segmental glomerulosclerosis. J Mol Cell Biol 6: 498–505, 2014 [DOI] [PubMed] [Google Scholar]
- 63.Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, Savige J: Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat 32: 127–143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guan M, Ma J, Keaton JM, Dimitrov L, Mudgal P, Stromberg M, et al.: Association of kidney structure-related gene variants with type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum Genet 135: 1251–1262, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schofield EC, Carver T, Achuthan P, Freire-Pritchett P, Spivakov M, Todd JA, et al.: CHiCP: A web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics 32: 2511–2513, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alves F, Vogel W, Mossie K, Millauer B, Höfler H, Ullrich A: Distinct structural characteristics of discoidin I subfamily receptor tyrosine kinases and complementary expression in human cancer. Oncogene 10: 609–618, 1995 [PubMed] [Google Scholar]
- 67.Vogel W, Gish GD, Alves F, Pawson T: The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell 1: 13–23, 1997 [DOI] [PubMed] [Google Scholar]
- 68.Dorison A, Chantziantoniou C: DDR1: A major player in renal diseases. Cell Adhes Migr 12: 299–304, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gross O, Girgert R, Beirowski B, Kretzler M, Kang HG, Kruegel J, et al.: Loss of collagen-receptor DDR1 delays renal fibrosis in hereditary type IV collagen disease. Matrix Biol 29: 346–356, 2010 [DOI] [PubMed] [Google Scholar]
- 70.Kerroch M, Guerrot D, Vandermeersch S, Placier S, Mesnard L, Jouanneau C, et al.: Genetic inhibition of discoidin domain receptor 1 protects mice against crescentic glomerulonephritis. FASEB J 26: 4079–4091, 2012 [DOI] [PubMed] [Google Scholar]
- 71.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al.: Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47: 598–606, 2015 [DOI] [PubMed] [Google Scholar]
- 72.Selman L, Hansen S: Structure and function of collectin liver 1 (CL-L1) and collectin 11 (CL-11, CL-K1). Immunobiology 217: 851–863, 2012 [DOI] [PubMed] [Google Scholar]
- 73.Hansen S, Selman L, Palaniyar N, Ziegler K, Brandt J, Kliem A, et al.: Collectin 11 (CL-11, CL-K1) is a MASP-1/3-associated plasma collectin with microbial-binding activity. J Immunol 185: 6096–6104, 2010 [DOI] [PubMed] [Google Scholar]
- 74.Farrar CA, Tran D, Li K, Wu W, Peng Q, Schwaeble W, et al.: Collectin-11 detects stress-induced L-fucose pattern to trigger renal epithelial injury. J Clin Invest 126: 1911–1925, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rooryck C, Diaz-Font A, Osborn DP, Chabchoub E, Hernandez-Hernandez V, Shamseldin H, et al.: Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat Genet 43: 197–203, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walsh DW, Godson C, Brazil DP, Martin F: Extracellular BMP-antagonist regulation in development and disease: Tied up in knots. Trends Cell Biol 20: 244–256, 2010 [DOI] [PubMed] [Google Scholar]
- 77.Zeisberg M, Shah AA, Kalluri R: Bone morphogenic protein-7 induces mesenchymal to epithelial transition in adult renal fibroblasts and facilitates regeneration of injured kidney. J Biol Chem 280: 8094–8100, 2005 [DOI] [PubMed] [Google Scholar]
- 78.Vukicevic S, Kopp JB, Luyten FP, Sampath TK: Induction of nephrogenic mesenchyme by osteogenic protein 1 (bone morphogenetic protein 7). Proc Natl Acad Sci U S A 93: 9021–9026, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo G, Hofmann C, Bronckers AL, Sohocki M, Bradley A, Karsenty G: BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev 9: 2808–2820, 1995 [DOI] [PubMed] [Google Scholar]
- 80.Higgins DF, Ewart LM, Masterson E, Tennant S, Grebnev G, Prunotto M, et al.: BMP7-induced-Pten inhibits Akt and prevents renal fibrosis. Biochim Biophys Acta Mol Basis Dis 1863: 3095–3104, 2017 [DOI] [PubMed] [Google Scholar]
- 81.Roxburgh SA, Kattla JJ, Curran SP, O’Meara YM, Pollock CA, Goldschmeding R, et al.: Allelic depletion of grem1 attenuates diabetic kidney disease. Diabetes 58: 1641–1650, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dolan V, Murphy M, Sadlier D, Lappin D, Doran P, Godson C, et al.: Expression of gremlin, a bone morphogenetic protein antagonist, in human diabetic nephropathy. Am J Kidney Dis 45: 1034–1039, 2005 [DOI] [PubMed] [Google Scholar]
- 83.McMahon R, Murphy M, Clarkson M, Taal M, Mackenzie HS, Godson C, et al.: IHG-2, a mesangial cell gene induced by high glucose, is human gremlin. Regulation by extracellular glucose concentration, cyclic mechanical strain, and transforming growth factor-beta1. J Biol Chem 275: 9901–9904, 2000 [DOI] [PubMed] [Google Scholar]
- 84.Afkarian M, Hirsch IB, Tuttle KR, Greenbaum C, Himmelfarb J, de Boer IH: Urinary excretion of RAS, BMP, and WNT pathway components in diabetic kidney disease. Physiol Rep 2: e12010, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.