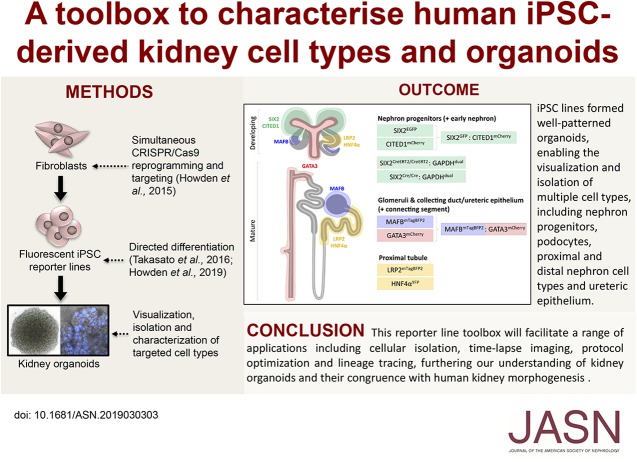
Significance Statement
Kidney organoids generated from human induced pluripotent stem cells (iPSCs) show great potential for modeling kidney diseases and studying disease pathogenesis. However, the relative accuracy with which kidney organoids model normal morphogenesis, as well as the maturity and identity of the renal cell types they comprise, remain to be fully investigated. The authors describe the generation and validation of ten fluorescent CRISPR/Cas9 gene-edited iPSC reporter lines specifically designed for the visualization, isolation, and characterization of cell types and states within kidney organoids, and demonstrate the use of these lines for cellular isolation, time-lapse imaging, protocol optimization, and lineage-tracing applications. These tools offer promise for better understanding this model system and its congruence with human kidney morphogenesis.
Keywords: stem cell, kidney development, molecular biology
Visual Abstract
Abstract
Background
The generation of reporter lines for cell identity, lineage, and physiologic state has provided a powerful tool in advancing the dissection of mouse kidney morphogenesis at a molecular level. Although use of this approach is not an option for studying human development in vivo, its application in human induced pluripotent stem cells (iPSCs) is now feasible.
Methods
We used CRISPR/Cas9 gene editing to generate ten fluorescence reporter iPSC lines designed to identify nephron progenitors, podocytes, proximal and distal nephron, and ureteric epithelium. Directed differentiation to kidney organoids was performed according to published protocols. Using immunofluorescence and live confocal microscopy, flow cytometry, and cell sorting techniques, we investigated organoid patterning and reporter expression characteristics.
Results
Each iPSC reporter line formed well patterned kidney organoids. All reporter lines showed congruence of endogenous gene and protein expression, enabling isolation and characterization of kidney cell types of interest. We also demonstrated successful application of reporter lines for time-lapse imaging and mouse transplantation experiments.
Conclusions
We generated, validated, and applied a suite of fluorescence iPSC reporter lines for the study of morphogenesis within human kidney organoids. This fluorescent iPSC reporter toolbox enables the visualization and isolation of key populations in forming kidney organoids, facilitating a range of applications, including cellular isolation, time-lapse imaging, protocol optimization, and lineage-tracing approaches. These tools offer promise for enhancing our understanding of this model system and its correspondence with human kidney morphogenesis.
Continued progress in the field of organoid culture is rapidly bridging the gap between traditional two-dimensional cell culture and animal models, better recapitulating the in vivo environment through enabling three-dimensional (3D) cellular organization and interaction. Although 3D culture systems for primary tissues have been in use for decades, recent advances in the field of embryonic stem cell– and induced pluripotent stem cell (iPSC)–derived organoids have been a significant development in the context of human research, bypassing the limited manipulation that is afforded by animal models and the many ethical and sample availability issues faced by human research. As such, organoids have the capacity to revolutionize our understanding of human development, regeneration, disease pathogenesis, and drug responses, with potentially far-reaching therapeutic applications in the future.
Organoids derived from iPSCs are now being exploited as a model system for a variety of organs, ranging from tissues of the gastrointestinal system through to the brain (reviewed in Fatehullah et al.1). Stemming from our knowledge of critical growth factors and signaling molecules during development, we and others have recently devised stepwise differentiation protocols that mimic kidney organogenesis in vivo to generate kidney progenitors from human iPSCs.2–6 Our own protocol generates complex 3D organoids of multiple maturing renal cell types with the capacity to self-organize into the expected renal subcompartments, such as nephrons, stroma, endothelial, and perivascular structures.5,7 Despite this exciting progress, organoids do not yet accurately recapitulate the human kidney in situ. One barrier is the complexity and cellular diversity of this organ, consisting of at least 26 different cell types8 that comprise the millions of epithelial, stromal, and vascular cells within the organ. An additional complicating factor is that much of our knowledge of kidney development has arisen from transgenic rodent models, such as gene knockout or lineage-tracing models.9,10 As a result, limited viability in culture, cellular immaturity, and atypical compartmental organization are hurdles faced by all groups in the kidney organoid field.
Akin to the leap in our understanding of mammalian kidney biology after the increased use of transgenic animal models, fluorescent reporter line generation represents a powerful tool in the organoid space, enabling real-time interrogation of individual cells as well as their isolation and characterization. Traditionally relying on viral or homologous recombination-based targeting methods (reviewed in Den Hartogh et al.11), recent advances in gene editing approaches, including the sequence-specific nuclease CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system12–16 and simultaneous reprogramming and gene targeting techniques,17 have revolutionized the field with respect to the specificity, efficiency, and flexibility of reporter line generation.
Here, we report the generation of a toolbox of iPSC reporter lines engineered specifically to mark an array of kidney cells types that encompass various renal subcompartments, including progenitors, proximal and late distal nephron, and ureteric epithelium. Not only do these lines allow the isolation and validation of individual cell types, they also enable the live assessment of differentiation and organoid development as it progresses. Such tools are likely to prove invaluable to the field of kidney organoid generation in the future through establishing improved and individualized protocols for differentiation and maturation, as well as far-reaching applications in the field of kidney research and medicine.
Methods
Reporter Line Access
All reporter lines will be available for distribution from the Washington University Kidney Translational Research Center (KTRC), St. Louis, MO. Complete descriptions including validations of each iPSC reporter line and a request form to access individual lines are available on the ReBuilding a Kidney (RKB) Consortium data repository (https://www.rebuildingakidney.org) and are fully accessible at https://doi.org/10.25548/16-DYNR.18 In brief, the request form will guide interested users regarding access to appropriate Material Transfer Agreements and how to contact the KTRC for the process of receiving lines.
Vector Design
Plasmids containing sgRNA specific to the gene of interest were generated by annealing 24-mer oligodeoxyribonucleotides (ODNs) encoding the desired protospacer sequence and appropriate overhangs, followed by ligation into the BbsI sites of the pSMART-sgRNA vector, as previously described.19 See Table 1 for CRISPR/Cas9 target sites (protospacers) used in this study. Gene targeting vectors encoding templates for knock-in experiments comprised the desired reporter gene (EGFP, mCherry, mTagBFP2, or YFP) flanked by sequences (typically approximately 0.5 kb) corresponding to sequence immediately upstream and downstream of the desired target site. Gene targeting vectors were generated using two gBlocks (Integrated DNA Technologies) encoding the targeting cassette, which were inserted into the AatII and EcoRI sites of the pDNR-Dual (Clontech) plasmid vector. The generation of gene targeting constructs encoding templates for the generation of SIX2EGFP, SIX2CreERT2, SIX2Cre, GAPDHdual, and MAFBmTagBFP2 iPSCs has been described previously.19,20 All plasmids were propagated in DH5α Escherichia coli (Bioline, New South Wales, Australia) and prepared for transfection using a Plasmid Maxi kit (QIAGEN, Hilden, Germany).
Table 1.
CRISPR/Cas9 target sites (protospacers) used for reporter line generation
| Gene | Sequence (5′-3′) |
|---|---|
| CITED1 | TGCCAAGTGTCCCTAAAGA |
| SIX2 | GTCAGCCAACCTCGTGGACC |
| GAPDH | CTTCCTCTTGTGCTCTTGCT |
| MAFB | CTCGCTTCAGCGATGGCCG |
| LRP2 | GGAGTTGGGTCTCTTCTCGA |
| HNF4A | TTCCCCACTGTGCCGCTTT |
| GATA3 | GAGGCCATGGAGGTGACGG |
Generation of Knock-In iPSCs
All iPSCs containing a knock-in of a single fluorescent reporter were derived from human foreskin fibroblasts (American Type Culture Collection numbers CRL-2429 or PCS-201–010) using a previously described protocol that combines reprogramming and gene editing in one step.19 Briefly, episomal reprogramming plasmids (pEP4E02SET2K, pEP4E02SEN2L, pEP4E02SEM2K, and pSimple-miR302/367), in vitro transcribed mRNA encoding the SpCas9-Gem variant,21 and corresponding sgRNA and gene targeting plasmids were introduced into fibroblasts using the Neon Transfection System as described below. In vitro transcribed mRNA encoding a truncated version of the EBNA1 protein was also included to enhance nuclear uptake of the reprogramming plasmids.22 The generation of dual reporter iPSCs (containing two different reporter genes) required a subsequent round of gene targeting in the corresponding iPSCs (Table 2). In vitro transcribed mRNA encoding the SpCas9-Gem variant,21 and corresponding sgRNA and gene targeting plasmids, were introduced into iPSCs using the Neon Transfection System as described below. To identify correctly targeted iPSCs, genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN) and PCR analysis was performed using primers that flank the ODNs flanking the 5′ and 3′ recombination junction. Heterozygous and homozygous clones were distinguished using ODNs that flank the intended target site. A summary of the reporter lines generated in this study is shown in Table 2.
Table 2.
iPSC reporter line details and localizations
| Line Name | Gene Targeted | Reporter/Insert and Zygosity | Targeting Approach | Parental Line | Reporter Expression/Line Purpose | CHIR Exposure during Diff. |
|---|---|---|---|---|---|---|
| Single reporter lines | ||||||
| SIX2EGFP | SIX2 | EGFP (homozygous) | 3′ with T2A | CRL-2429 (ATTC) a,b | Identifies NPs, early nephron, stromal populations | 3 d, 8 µM |
| CITED1mCherry | CITED1 | mCherry (hemizygous) | 3′ with T2A | CRL-2429a | Identifies early intermediate mesoderm, NPs, early nephron | 3 d, 8 µM |
| GATA3mCherry | GATA3 | mCherry (heterozygous) | 5′ with T2A | CRL-2429a | Identifies connecting segment (SLC12A1−), collecting duct, GATA3+ interstitial population | 4 d, 6 µM |
| MAFBmTagBFP2 | MAFB | mTAGBFP2 (heterozygous) | 5′with polyA signal | CRL-2429a,b | Identifies podocytes | 4 d, 6 µM |
| LRP2mTagBFP2 | LRP2 | mTAGBFP2 (heterozygous) | 3′ with T2A | CRL-2429a | Identifies proximal tubule | 4 d, 6 µM |
| HNF4αYFP | HNF4α | YFP (homozygous) | 3′with T2A | PCS-201–010 (ATTC)a | Identifies proximal tubule | 4 d, 6 µM |
| Dual reporter lines | ||||||
| SIX2EGFP:CITED1mCherry | SIX2/CITED1 | EGFP/mCherry (hom.:hem.) | 3′ with T2A | SIX2EGFP iPSCs | Identifies NPs, early nephron, stromal populations | 3 d, 8 µM |
| MAFBBFP:GATA3mCherry | MAFB/GATA3 | mTAGBFP2/mCherry (het.:het.) | 5′with polyA signal: 5′ with T2A | MAFBmTagBFP2 iPSCs | Identified podocytes (MAFB+), collecting duct/connecting segment (GATA3+), GATA3+ interstitial population | 4 d, 6 µM |
| Lineage-tracing lines | ||||||
| SIX2Cre/Cre | SIX2 | Cre (homozygous) | 3′with T2A | CRL-2429a,b | SIX2 lineage tracing (see SIX2Cre/Cre:GAPDHdual line below) | N/A |
| SIX2CreERT2 | SIX2 | Cre/ERT2 (homozygous) | 3′with T2A | CRL-2429a,b | Inducible SIX2 lineage tracing (see SIX2CreERT2/CreERT2: GAPDHdual line below) | N/A |
| SIX2Cre/Cre:GAPDHdual | SIX2/GAPDH | Cre/loxP-flanked EGFP-mCherry (hom.:het.) | 3′with T2A:3′ with T2A | SIX2Cre/Cre above (Cre expressed from endogenous SIX2 locus)b | SIX2 lineage tracing (endogenous SIX2 activation results in permanent switching of EGFP to mCherry) | 4 d, 7 µM |
| SIX2CreERT2/CreERT2: GAPDHdual | SIX2/GAPDH | CreERT2/loxP-flanked EGFP-mCherry (hom.:het.) | 3′with T2A:3′ with T2A | SIX2CreERT2/CreERT2 above (CreERT2 expressed from endogenous SIX2 locus)b | Inducible SIX2 lineage tracing (endogenous SIX2 activation combined with tamoxifen-induced Cre results in permanent switching of EGFP to mCherry) | 4 d, 7 µM |
Indicates gene targeting and reprogramming performed simultaneously.
Indicates lines that have been reported previously. CRL-2429 and PCS-201–010 are foreskin fibroblast lines.
Cell Transfection
Transfections were performed using the Neon Transfection System (Thermo Fisher Scientific). Human fibroblasts or iPSCs were harvested with TrypLE (Thermo Fisher Scientific) 2 days after passaging and resuspended in Buffer R at a final concentration of 1×107 cells/ml. Electroporation was performed in a 100 μl tip using 1400 V for 20 ms and two pulses for human fibroblasts, or 1100 V for 30 ms and one pulse for human iPSCs. After electroporation, fibroblasts were transferred to six-well Matrigel-coated plates containing DMEM+15% FBS and switched to reprogramming medium (TeSR-E7+100 μM sodium butyrate) after 3 days, with medium changes every other day. Electroporated human iPSCs were plated on six-well Matrigel-coated plates containing Essential 8 medium with 5 μM Y-27632 (Tocris).
Cell Culture, Differentiation, and Organoid Generation
Human foreskin fibroblasts were cultured in DMEM (Thermo Fisher Scientific) supplemented with 15% FBS (Hyclone) and 1× MEM Non-Essential Amino Acids Solution (Thermo Fisher Scientific) at 37°C, 5% CO2, and 5% O2. All iPSC lines were maintained and expanded at 37°C, 5% CO2, and 5% O2 in Essential 8 medium (Thermo Fisher Scientific) on Matrigel-coated plates with daily media changes, and passaged every 3–4 days with EDTA in 1× PBS as previously described.23 The genomic integrity of each iPSC line was confirmed by karyotyping using SNP arrays (Illumina Infinium CoreExome-24 v1.1, performed by VCGS, Melbourne, Australia) and G-banding (GTW banding method, performed by the Cytogenetics Research Core, Washington University), as well as immunofluorescence of pluripotency markers. Reporter iPSC lines were differentiated as described previously,20 with minor variations in the initial seeding density for differentiation in six-well plates (8×104 cells per well) and CHIR99021 exposure duration (4–8 µM for 3–5 days; refer to Supplemental Figure 1A, Table 2). Organoids were generated either by bioprinting20,24 or using a manual technique7 as described previously, and cultured for 14–18 days before harvest.
Flow Cytometry and FACS
Organoids for flow cytometry or FACS were transferred into a cell culture plate and manually dissociated using a scalpel for 1 minute before the addition of warmed (37°C) Accutase solution (STEMCELL Technologies, Vancouver, Canada), using approximately 1 ml per six organoids. Enzymatic dissociation was continued at 37°C in the incubator, retrieving the cells to briefly pipette-mix every 3 minutes, for a total of 15 minutes. Once dissociated, Accutase was inactivated by adding an equal volume of DMEM/F12 (Life Technologies) with 1% FCS and placing the plate on ice for 1 minute. The cells were then passed through 40 and 70 μm cell strainers with an additional 1 ml of DMEM/F12+1% FCS, centrifuged (1500 rpm for 3 minutes) and resuspended in 300–500 µl of FACS wash (PBS with 1% FCS). Flow cytometry and FACS was performed using the LSRFortessa Cell Analyzer and FACSARIA Fusion cell sorter (BD Biosciences), respectively. Sorted populations were collected into TeSR-E6 media (STEMCELL Technologies). Data acquisition and analysis was performed using FACsDiva (BD Biosciences) and FlowLogic software (Inivai Technologies). Gating was performed on live cells on the basis of forward- and side-scatter analysis.
Gene Expression Analyses
After FACS, sorted cells were centrifuged at 1500 rpm for 3 minutes. After aspiration of the supernatant, cell pellets were lysed. Then RNA extraction, cDNA synthesis, and quantitative RT-PCR were performed using the Bioline Isolate II Mini/Micro RNA Extraction Kit, SensiFAST cDNA Synthesis Kit, and the SensiFAST SYBR Lo-ROX Kit (Bioline), respectively, as per manufacturer’s instructions. Each quantitative RT-PCR reaction was performed in triplicate using the primer pairs detailed in Table 3. Data were graphed and analyzed in Prism 7 (GraphPad). Single-cell RNA-sequencing, used for analysis of the GATA3 interstitial population, was performed according to Howden et al.20
Table 3.
Forward and reverse primers used for quantitative RT-PCR
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| GAPDH | CTCTCTGCTCCTCCTGTTCGAC | TGAGCGATGTGGCTCGGCT |
| CITED1 | AACTTCTGCCAAGGCTCTGA | CACTGCTTTGCGATCTTTCA |
| CUBN | AACTTCCTAATCCCCAGCGG | GTCCACCTCCTCAGTTCCTG |
| GATA3 | GCCCCTCATTAAGCCCAA | TTGTGGTGGTCTGACAGTTCG |
| HNF4α | ACCCTCGTCGACATGGACA | GCCTTCTGATGGGGACGTG |
| LRP2 | GGTGGGGCCTTCTATGAACC | AGAGCTGTCCCATCATTGGC |
| MAFB | GCCAAACCGCATAGAGAACG | TGATGCAAAATGCCCGGAAC |
| SIX2 | TCCTGGTCCCTCCGTATGTA | TAGGGGCAGATAGACCACCA |
Immunofluorescence, Microscopy, and Live Imaging
For immunofluorescence, organoids were cut off Transwell filters using a scalpel, transferred to a 48-well plate, and fixed in ice cold 2% paraformaldehyde (Sigma-Aldrich) for 20 minutes, followed by 15 minutes washing in three changes of PBS. All blocking and staining incubations were performed at 4°C overnight on a rocking platform. Organoids were blocked in 10% donkey serum with 0.3% Triton-X 100 (Sigma-Aldrich). Antibodies were diluted in 0.1% Triton-X 100/PBS and are detailed in Table 4. Primary antibodies were detected with Alexa Fluor-conjugated fluorescent secondary antibodies (Life Technologies), diluted at 1:500. Nuclei were detected with DAPI diluted at 1:2000 in PBS included with the secondary antibody solution. After incubations in primary and secondary antibody solutions, organoids were washed in three changes of PBS for 3 hours (1 hour per wash). Imaging of fixed organoids was performed on the ZEISS LSM 780 (Carl Zeiss, Oberkochen, Germany) or Dragonfly Spinning Disk (Andor Technology, Belfast, United Kingdom) confocal microscopes, as well as the ZEISS Apotome.2 fluorescence microscope (Carl Zeiss). Live imaging of MAFBmTagBFP2:GATA3mCherry organoids was performed on the Dragonfly Spinning Disk confocal within a humidified incubation chamber set at 37°C and 5% O2.
Table 4.
Antibodies and lectins used for immunofluorescence
| Specificity | Host Species | Dilution Range | Manufacturer and Identifier |
|---|---|---|---|
| NANOG | Mouse | 1:50 | DSHB (PCRP-NANOGP1–2D8) |
| CITED1 | Rabbit | 1:300 | Thermo Fisher Scientific (RB-9219-P1) |
| GATA3 | Goat | 1:300 | R&D Systems (AF2605) |
| GFP | Chicken | 1:300 | Sapphire Biosciences (Ab13970) |
| ECADHERIN | Mouse | 1:300 | BD Biosciences (610181) |
| EpCAM (Alexa488 conjugate) | Mouse | 1:300 | BioLegend (324210) |
| LAMININ | Rabbit | 1:400–1:500 | Sigma-Aldrich (L9393) |
| mCherry (RFP) | Rabbit | 1:300–1:400 | MBL Medical & Biologic Laboratories Co. Ltd. (PM005) |
| MECA-32 | Rat | 1:100 | Novus Biologic (NB100–77668) |
| mTagBFP2 | Rabbit | 1:500 | Evrogen (AB233) |
| NEPHRIN | Sheep | 1:300 | R&D Systems (AF4269) |
| PECAM-1 | Goat | 1:100 | Santa Cruz Biotechnology (SC-1506) |
| Proximal tubule brush border membrane | Lotus tetragonolobus lectin (LTL) | 1:300–1:500 | Vector Laboratories (B-1325) |
| SIX2 | Rabbit | 1:300 | Proteintech Group (11562–1-AP) |
| SLC12A1 | Rabbit | 1:300 | Proteintech (18970–1-AP) |
Organoid Transplantation
Transplantation of D7+12 (7 days of monolayer differentiation followed by 12 days of organoid culture) LRP2mTagBFP2 organoids was performed with approved animal ethics protocols (MCRI AEC A873 and A888) using immunocompromised 10-week-old female NSG mice (Jackson Laboratories) and D7+12 organoids as described previously for subcapsular transplants.25 For omental transplants, 8-week-old female NSG mice were anesthetized with isoflurane and a midline incision made to expose the abdominal fat. D7+9 organoids were removed from the Transwell culture plate using a scalpel blade to cut the surrounding filter, leaving enough margin to pick up the filter-attached organoid using tweezers. Organoids were placed on the exposed abdominal fat, secured with TISSEEL fibrin sealant (Baxter, Alabama), and the incision closed. For both subcapsular and omental transplants, mice were euthanized via cervical dislocation to retrieve the transplanted organoids 21 days after surgery. Harvested organoids were processed for analysis using the fixation and staining protocols outlined above.
Results and Discussion
Generation and Validation of Reporter Lines
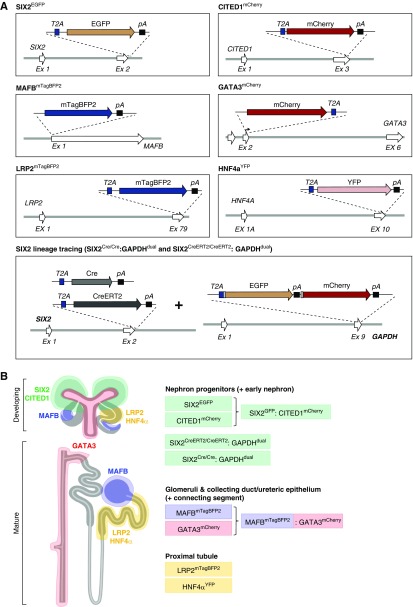
Generation of all single reporter lines was performed via simultaneous reprogramming and targeting of primary human fibroblasts, as previously described19,21 (refer to Methods and Table 2). For single and dual reporter lines, cassettes encoding fluorescent reporter proteins were introduced into gene loci known to mark particular cell types or stages of development within the developing kidney (Figure 1, A and B). SIX2 lineage-tracing lines were generated as described previously.20 Successfully targeted clones were verified by PCR analysis and the untargeted allele in heterozygous clones were assessed for CRISPR/Cas9-induced insertion/deletion mutations. All iPSCs carrying heterozygous knock-in of a reporter gene did not contain evidence of indel formation on the untargeted allele except for the LRP2mTagBFP reporter, which was found to contain a 5-bp deletion at the extreme 3′ end of the LRP2 (megalin) coding region and is not expected to affect function. For each reporter line, we confirmed expression of pluripotency factors (such as OCT4 and NANOG) and genomic integrity was confirmed by both molecular karyotyping and G-banding (individual reports available on the RKB Consortium data repository [https://www.rebuildingakidney.org]; examples provided for the SIX2EGFP and SIX2EGFP:CITED1mCherry iPSC lines in Supplemental Figure 1B). Reporter lines were all validated with respect to their capacity to pattern into nephron-containing kidney organoids using a standard panel of four antibodies/lectins selected to show the presence of podocytes within glomeruli (NPHS1 [nephrin]), proximal tubules (LTL [lotus tetragonolobus lectin]), distal nephron epithelial segments (including loop of Henle and distal convoluted tubule; LTL-negative/ECADHERIN or EPCAM-positive) and presumptive ureteric epithelium and connecting segment (GATA3/ECADHERIN-positive) (Figures 2A and 3, Ai and Bi), as well as additional segmentation markers such as SLC12A1 (NKCC2, thick ascending limb of the loop of Henle) and HNF4α (proximal tubule), as demonstrated in MAFBmTagBFP2:GATA3mCherry organoids (Figure 3Bi). As expected, in cells expressing each of the various fluorescent reporters, we could confirm expression of the endogenous protein from the corresponding target locus (Figures 2 and 3).
Figure 1.
Fluorescent reporter line constructs and expected distribution of reporter signals in developing and mature kidney structures. (A) Schematic diagrams of the targeting strategies used to generate single, dual, and lineage-tracing iPSC reporter lines. SIX2EGFP:CITED1mCherry and MAFBmTagBFP2:GATA3mCherry dual reporter lines were generated through retargeting of the single SIX2EGFP and MAFBmTagBFP2 iPSC lines, respectively. (B) Simplified schematic highlighting the key most important developing and mature kidney cell types and segments that users may wish to investigate using each iPSC line.
Figure 2.
Validation and characterization of iPSC reporter lines for metanephric mesenchyme and NPs. (A) Immunofluorescence of a representative D7+12 SIX2EGFP:CITED1mCherry organoid demonstrating expression of nephron segment-specific markers for proximal tubules (LTL+; blue), distal tubules, (ECADHERIN [ECAD]+/LTL−; green), ureteric epithelium/connecting segment (ECAD+/GATA3+; green and red), and podocytes of the glomeruli (NPHS1+; gray). (B) Detection of endogenous EGFP (green) and mCherry (red) reporter expression via confocal microscopy (top panels) and flow cytometry (bottom panels; see Supplemental Figure 1B for control) in live D7+5 to D7+7 organoids derived from SIX2EGFP, CITED1mCherry and SIX2EGFP:CITED1mCherry lines. (C) Confirmation of congruence between EGFP reporter expression and endogenous SIX2/SIX2. Immunofluorescence (Ci) in D7+12 SIX2EGFP organoids depicts colocalization of EGFP (green) and SIX2 (red) staining. EGFP+ cells isolated via FACS and subjected to quantitative RT-PCR (qRT-PCR) (Cii) showing enrichment of SIX2 expression (green bar) compared with control (whole D7+11 organoids, gray bar). (D) Confirmation of congruence between mCherry reporter expression and endogenous CITED1/CITED1. Immunofluorescence (Di) in D7 CITED1mCherry differentiated iPSCs depicting co-localization of mCherry (red) and CITED1 (green) staining. FACS-isolated mCherry+ cells subjected to qRT-PCR (Dii) confirms enrichment of CITED1 expression (red bar) compared with control (whole D7+11 organoid, gray bar). In qRT-PCR for (Cii) and (Dii), error bars represent SEM and significance was determined using a t test on normalized (∆Ct) values (**P≤0.01; ***P≤0.001). Scale bars in (A–D) represent 50 µm.
Figure 3.
Validation and characterization of iPSC reporter lines for proximal/distal nephron and ureteric epithelium. (A) Characterization of proximal nephron iPSC lines, LRP2mTagBFP2 and HNF4αYFP. Immunofluorescence of D7+12 organoids (Ai) confirmed expression of markers for proximal tubules (LTL+; blue), distal tubules, (ECADHERIN [ECAD]+/LTL−; green), ureteric epithelium/connecting segment (ECAD+/GATA3+; green and red), and podocytes of the glomeruli (NPHS1+; gray). YFP and mTagBFP2 reporter fluorescence was detectable both by live confocal imaging (Aii) and flow cytometry (Aiii) of LRP2mTagBFP2 and HNF4αYFP organoids. FACS-isolated mTagBFP2+ and YFP+ cells (Aiv; blue and yellow bars) from LRP2mTagBFP2 and HNF4αYFP organoids showed enriched endogenous expression of their targeted genes, LRP2 and HNF4α, compared with the reporter-negative control populations (gray bars) via quantitative RT-PCR (qRT-PCR). Sorted reporter-positive populations also showed enrichment for additional proximal tubule markers such as CUBN. Error bars depict SEM and significance was determined using a t test on normalized (∆Ct) values (*P≤0.05; ***P≤0.001; ****P<0.001). Transplanted D7+9 LRP2mTagBFP2 organoids (Avii) retrieved from subcapsular (left panel) and omental (right panels) transplants 21 days after surgery (D7+9+21) showed endogenous mTagBFP2 expression and human and mouse derived vasculature surrounding mTagBFP2+ tubules (cyan) via immunofluorescence (vasculature marker; PECAM-1 [red], mouse-specific cellular antigen; MECA-32 [green]). (B) Characterization of distal nephron/ureteric epithelium iPSC lines, GATA3mCherry and MAFBmTagBFP2:GATA3mCherry. Immunofluorescence of representative D7+12 MAFBmTagBFP2:GATA3mCherry organoids (Bi) demonstrates expression of nephron segment-specific markers for proximal tubules (LTL+; top panel [blue] and HNF4α; bottom panel [green]), distal tubules, (ECAD+/LTL−; top panel [green/blue]), thick ascending limb of the loop of Henle (SLC12A1+; bottom panel [red]), ureteric epithelium/connecting segment (ECAD+/GATA3+; top panel [green/red] and bottom panel [GATA3 only, gray]), and podocytes of the glomeruli (NPHS1+; top panel [gray]). Endogenous mCherry (red) and mTagBFP2 (blue) reporter expression GATA3mCherry and MAFBmTagBFP2:GATA3mCherry organoids (Bii) was detected via live confocal imaging and flow cytometry. Immunofluorescence of GATA3mCherry organoids (Biii) confirmed colocalization of mCherry (red) with GATA3 (green) protein and qRT-PCR of FACS-isolated mCherry+ cells from these organoids showed enriched GATA3 gene expression (red bar) compared with the mCherry−/EpCam+ epithelial control cell population (gray bar). Error bars represent SEM and significance was determined using a t test on normalized (∆Ct) values (****P<0.001). (C) Representative still images from confocal time-lapse imaging of a D7+9 MAFBmTagBFP2:GATA3mCherry organoid across a 35-hour period demonstrates endogenous mCherry (red) and mTagBFP2 (blue) expression. Scale bars in (A and B) represent 50 µm. Scale bar in (C) represents 500 µm.
Reporters for Characterization of Metanephric Mesenchyme and Nephron Progenitors
During normal kidney organogenesis, two main progenitor pools, the ureteric bud (UB) and the mesenchymal nephron progenitors (NPs), undergo a series of reciprocal inductive interactions that are critical to both maintain these populations and drive differentiation.26 Branching morphogenesis of the UB gives rise to an intricate collecting duct system for drainage of urine, whereas NPs condense around the branching UB tips, undergoing successive rounds of induction, mesenchymal-to-epithelial transition, and nephron formation (reviewed in Saxén and Sariola27). Although it is clear that kidney organoids contain a NP population able to give rise to segmented nephrons, a significant hurdle faced by all studies in the field is the lack of defined self-renewing NP niches,20 a feature that will be critical for organoid growth and maturation. Furthermore, despite recent studies of the maintenance of mouse NPs and human endogenous/pluripotent stem cell-derived NPs,28–31 our knowledge of how to maintain these self-renewing progenitors in organoids while retaining a nephron induction capacity remains limited.
Extensive studies in rodents have shown that Six2 and Cited1 specifically mark the NPs within a capping mesenchyme around the tips of the branching ureteric tree and have confirmed the critical requirement of Six2 for NP self-renewal, and thus continued nephrogenesis.32,33 In order to further characterize, visualize, and isolate this presumptive NP population in organoids, three iPSC lines were generated: a SIX2EGFP reporter line (previously reported in Howden et al.20), a CITED1mCherry reporter line, and a dual SIX2EGFP:CITED1mCherry iPSC reporter line (Figures 1 and 2). SIX2EGFP, CITED1mCherry, and SIX2EGFP:CITED1mCherry iPSC lines were able to form kidney organoids (Figure 2, A and B) and expression of EGFP and/or mCherry fluorescent reporters was visible for all three lines, determined by live imaging and flow cytometry (Figure 2B). Furthermore, EGFP (Figure 2Ci) and mCherry (Figure 2Di) reporter proteins were confirmed within SIX2- and CITED1-positive cells, respectively, and were enriched for these gene transcripts after cell isolation using FACS (Figure 2, Cii and Dii).
CITED1/mCherry expression was apparent from day 2 of differentiation, consistent with expression of CITED1 in the posterior primitive streak34–36 before becoming restricted to NPs and early developing epithelial structures.37 We consistently observed CITED1/mCherry expression in the vast majority of cells at day 7 of differentiation (Supplemental Figure 2, A and B) and a reduction in the fraction of CITED1/mCherry-expressing cells post-organoid formation (Figure 2B). A time course of SIX2EGFP expression revealed EGFP detection by approximately D7+3 (Supplemental Figure 2C). These distinct expression characteristics highlight the fact that caution is required when assuming NP identity with any one marker alone. Within organoids, both SIX2/EGFP and CITED1/mCherry expression were observed in early epithelial structures (Figure 2, B and C), with approximately 6% of the population double positive at D7+5 using the SIX2EGFP:CITED1mCherry dual reporter line (Figure 2B). This persistence of expression in early nephron is supported by recent data in fetal human kidneys.37 Together, these three lines will be valuable resources for further optimizing this critical population within organoids.
More recently, we have described the first application of lineage tracing within organoids, utilizing a fluorescent reporter line to trace SIX2-expressing populations throughout differentiation and organoid maturation.20 In this instance, Cre recombinase or CreERT2 cassettes were introduced into the endogenous SIX2 locus. A dual fluorescence cassette (mCherry gene downstream of a loxP-flanked EGFP gene) was then inserted into the endogenous GAPDH locus to generate tamoxifen-inducible (SIX2CreERT2/CreERT2:GAPDHdual) and noninducible (SIX2Cre/Cre:GAPDHdual) SIX2 lineage-tracing iPSCs.20 Induction of endogenous SIX2 expression, or SIX2 expression combined with tamoxifen exposure, results in a permanent switching of EGFP to mCherry reporter gene expression (Supplemental Figure 2Di). Using these tools, we confirmed that nephron epithelia within kidney organoids are derived from a SIX2-expressing progenitor (Supplemental Figure 2Dii). However, the use of the SIX2CreERT2/CreERT2:GAPDHdual line and tamoxifen induction at different times throughout kidney organoid differentiation revealed that SIX2-expressing progenitors lose their capacity to contribute to such structures over time.20 The isolation of the SIX2-expressing cells using the SIX2EGFP and SIX2EGFP:CITED1mCherry reporter lines described here will enable a dissection of the changes occurring in this population during organoid culture.
Reporter Lines for Monitoring Proximal Nephron Maturation
There is considerable interest in the application of organoids for in vitro nephrotoxicity and drug efficacy studies.38 To date, the majority of cell-based nephrotoxicity studies have focused on the use of two-dimensional proximal tubule cultures. However, such approaches face limitations owing to reduced drug transporter expression.39,40 The 3D composition of kidney organoids suggests that these have the potential to better recapitulate the in vivo response. Recent studies have highlighted progress toward more mature iPSC-derived podocytes41,42 and proximal tubules, showing evidence of substrate uptake in some cases.3,5,25 However, these segments are still immature, with whole kidney organoid transcriptional profiles resembling those of first trimester human kidney.5,43
To enable further improvement of the proximal nephron and its segmentation, we have developed three reporter lines to identify podocytes (MAFBmTagBFP; previously reported in Hale et al.41; Figure 1, Supplemental Figure 3) and proximal tubules (LRP2mTagBFP2 and HNF4αYFP, Figures 1 and 3), well accepted markers of these segments in developing kidney.44,45 All three reporter lines were confirmed to form correctly patterned organoids (Figure 3Ai, Supplemental Figure 3Ai), with endogenous reporter expression detectable by both live confocal imaging and flow cytometry (Figure 3Aii and iii, Supplemental Figure 3Aii and iii). Robust reporter gene expression was evident by approximately D7+7 to D7+10 for MAFBmTagBFP2 and HNF4αYFP, and D7+14 for LRP2mTagBFP2. FACS isolation from organoids using mTagBFP2 and YFP reporters enabled specific enrichment of MAFB-, LRP2-, and HNF4α-expressing cells, as well as additional proximal tubule markers such as CUBN in the LRP2mTagBFP2 and HNF4αYFP lines (Figure 3Aiv, Supplemental Figure 3Aiv), and correct localization of reporter expression within cells expressing the corresponding endogenous protein was also established (Figure 3Av and vi, Supplemental Figure 3Av). Using the MAFBmTagBFP2 line, we investigated how closely reporter gene expression correlates with the onset of endogenous MAFB protein. Although a low level of reporter expression was detected in a small number of cells by flow cytometry in D7+5 organoids, endogenous MAFB protein, detected using immunofluorescence of fixed organoids, could not be detected until the D7+7 time point. However, a positive correlation between reporter gene expression and the ability to detect endogenous MAFB protein was observed across the time course.
We have previously reported the use of the MAFBmTAGBFP2 reporter line for the temporal characterization of the developing glomerular epithelial compartment across organoid development.41 This confirmed the initial expression of MAFB in the early proximal S-shaped body and eventual restriction to a NPHS1+/NPHS2+ podocyte population. We have also demonstrated the utility of this line in vivo after transplantation of organoids into immunocompromised mice,25 where patent glomerular capillaries were shown to arise within MAFBmTagBFP2-postive glomeruli of transplanted organoids. Here we provide similar evidence of tubular fluorescence after subcapsular and omentum transplantation of LRP2mTagBFP2 organoids into immunocompromised mice (Figure 3Avii). This will ultimately facilitate live multiphoton imaging and the recovery and transcriptional analysis of perfused organoids to investigate the impact of flow on tubular maturation.
Single and Double Reporter Lines for the Optimization of Late Distal Nephron and Ureteric Epithelium
The generation of a ureteric tip environment is known to be necessary for NP survival and self-renewal, whereas the recreation of a contiguous ureteric epithelium to which distal nephrons can connect is likely to be critical for the ultimate functionality of a stem cell-derived kidney tissue. To allow further characterization of the late distal and ureteric epithelium within kidney organoids, we generated an iPSC reporter line that harbors mCherry within the endogenous GATA3 locus. Although GATA3 is detected to some extent in the late distal nephron, it is a common marker of the collecting duct system in both human and mouse kidneys46–49 and was previously confirmed in kidney organoids at levels appropriate for use as a reporter of this segment.7 A second dual fluorescent reporter line was engineered via the retargeting of the GATA3mCherry line with MAFBmTagBFP2.
Both the single GATA3mCherry and dual MAFBmTagBFP2:GATA3mCherry lines showed a capacity to form segmented organoids (example of immunofluorescence shown for the dual line, Figure 3Bi). Reporter fluorescence was detectable via both live imaging and flow cytometry (Figure 3Bii), correlating with endogenous proteins (Figure 3Biii, top panel) and facilitating specific enrichment of cells expressing the targeted gene (Figure 3Biii, bottom panel). In addition to its epithelial expression, GATA3/mCherry was found to mark a subset of interstitial, nonepithelial cells, frequently associated with NPHS1+ podocytes of the glomeruli (Supplemental Figure 3B, left panel). This was also supported by analysis of existing single-cell RNA-sequencing datasets,20 showing ECADHERIN expression in just a subset of GATA3+ cells (Supplemental Figure 3B, right panel), and supports previous reports of GATA3 gene expression within the glomerular mesangium and adjacent endocapillary cells of human and mouse kidney.47
Debate in the field continues around whether both human iPSC-derived NPs and UB progenitors can be simultaneously generated in a single differentiation, owing to differences in the specification of both lineages during mesodermal differentiation in the mouse.4,50 We have previously demonstrated that a shorter exposure to the canonical WNT agonist, CHIR, during the first stage of differentiation results in patterning to a more anterior intermediate mesoderm such that resulting organoids contain a larger population of GATA3+ECADHERIN+ epithelium coincident with forming nephrons.5,7 To explore this further, the dual reporter line (MAFBmTagBFP2:GATA3mCherry) was used to monitor patterning and segmentation in real-time using live confocal microscopy. Organoids generated from the MAFBmTagBFP2:GATA3mCherry line were imaged using time-lapse microscopy from D7+9 onwards for 35 hours (Figure 3C, Video 1). GATA3/mCherry expression was detected in epithelial structures by D7+5–D7+6 (Supplemental Figure 3C, Supplemental Video 1), before the commencement of live imaging. Subsequent MAFB/mTagBFP2 expression became identifiable by D7+9 (+5 hours) and progressively intensified within each forming nephron across the experiment time frame (Figure 3C, Supplemental Video 1, Video 1), demonstrating both evidence for coincidence of these two populations within a given organoid, a dichotomy in onset of gene expression and the value of such dual reporters for the real-time monitoring and characterization of nephrogenesis in kidney organoids.
In summary, we describe here the development of a toolbox of CRISPR/Cas9 gene edited human iPSC reporter lines specifically developed for the analysis of kidney organoids. Previous studies have described selected iPSC-derived reporter lines for similar purposes, including a SIX2GFP, NPHS1GFP and a SIX2GFP:NPHS1mKate line.51,52 This extended suite of reporter lines, including multiple markers for renal subcompartments as well as dual reporter lines and lineage-tracing lines, will enable a more thorough analysis of morphogenesis within different organoid protocols and represent an invaluable resource for rapidly improving existing differentiation protocols with respect to cell type, maturation, and minimization of off-target populations.
Disclosures
Prof. Little reports grants from National Institutes of Health, grants from National Health and Medical Research Council of Australia, during the conduct of the study. In addition, Prof. Little has a patent AU2014277667 licensed to Organovo Inc.
Funding
This work was supported by the National Institutes of Health (grants DK107344-01 to Prof. Little and DK107350 to Assoc. Prof. Jain) and the National Health and Medical Research Council, Australia (grant GNT1100970). Prof. Little is a Senior Principal Research Fellow of the National Health and Medical Research Council (grant GNT1136085). The Murdoch Children’s Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. The MCRI gene editing facility is supported by the Stafford Fox Foundation.
Supplementary Material
Acknowledgments
Conceptualization: Prof. Little, Dr. Howden, Methodology, Dr. Howden, Validation, Dr. Vanslambrouck, Dr. Howden, Mr. Wilson, Ms. Tan, and Ms. Soo; investigation: Dr. Vanslambrouck, Dr. Howden, Mr. Wilson, Ms. Tan, Ms. Soo, and Dr. Spijker; writing, original draft: Dr. Vanslambrouck; writing, review and editing: Dr. Vanslambrouck, Dr. Howden, and Dr. Little; resources, conceiving and planning iPSC line distribution: Assoc. Prof. Jain; resources, iPSC line validation and distribution: Assoc. Prof. Jain, Ms. Neilson, and Asst. Prof. Cui; supervision: Dr. Vanslambrouck, Dr. Howden, and Prof. Little; funding acquisition: Dr. Howden and Prof. Little.
These tools were generated as part of the ReBuilding a Kidney consortium, funded by the National Institutes of health USA (DK107344 and DK107350). We thank Ms. Bendi Gong at the Washington University St. Louis for performing pluripotency marker immunostaining experiments. We acknowledge Ms. Cristina Williams, Dr. Todd Valerius, Ms. Laura Pearlman and Ms. Hongsuda Tangmunarunkit from the reBuilding a Kidney consortium for establishing the online iPSC reporter line request process.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019030303/-/DCSupplemental.
Supplemental Figure 1. Organoid generation protocol and validation of iPSC lines.
Supplemental Figure 2. Reporter expression dynamics.
Supplemental Figure 3. Characterization and validation of MAFBmTagBFP2 organoids and GATA3-expressing populations within single and dual GATA3mCherry reporter lines.
Supplemental Video 1. Time-lapse imaging of the D7+9 MAFBmTagBFP2:GATA3mCherry organoid.
References
- 1.Fatehullah A, Tan SH, Barker N: Organoids as an in vitro model of human development and disease. Nat Cell Biol 18: 246–254, 2016 [DOI] [PubMed] [Google Scholar]
- 2.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, et al.: Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6: 8715, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV: Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 33: 1193–1200, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, et al.: Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14: 53–67, 2014 [DOI] [PubMed] [Google Scholar]
- 5.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, et al.: Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526: 564–568, 2015 [DOI] [PubMed] [Google Scholar]
- 6.Toyohara T, Mae S, Sueta S, Inoue T, Yamagishi Y, Kawamoto T, et al.: Cell therapy using human induced pluripotent stem cell-derived renal progenitors ameliorates acute kidney injury in mice. Stem Cells Transl Med 4: 980–992, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takasato M, Er PX, Chiu HS, Little MH: Generation of kidney organoids from human pluripotent stem cells. Nat Protoc 11: 1681–1692, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Awqati Q, Oliver JA: Stem cells in the kidney. Kidney Int 61: 387–395, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Abe T, Fujimori T: Reporter mouse lines for fluorescence imaging. Dev Growth Differ 55: 390–405, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Humphreys BD, DiRocco DP: Lineage-tracing methods and the kidney. Kidney Int 86: 481–488, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Den Hartogh SC, Passier R: Concise review: Fluorescent reporters in human pluripotent stem cells: Contributions to cardiac differentiation and their applications in cardiac disease and toxicity. Stem Cells 34: 13–26, 2016 [DOI] [PubMed] [Google Scholar]
- 12.Cho SW, Kim S, Kim JM, Kim JS: Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232, 2013 [DOI] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al.: Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E: A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J: RNA-programmed genome editing in human cells. Elife 2: e00471, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al.: RNA-guided human genome engineering via Cas9. Science 339: 823–826, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howden SE, Maufort JP, Duffin BM, Elefanty AG, Stanley EG, Thomson JA: Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Reports 5: 1109–1118, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little MH: (Re)Building a Kidney Consortium. 2019. Available at: 10.25548/16-DYNR. Accessed June 28, 2019 [DOI] [Google Scholar]
- 19.Howden SE, Thomson JA, Little MH: Simultaneous reprogramming and gene editing of human fibroblasts. Nat Protoc 13: 875–898, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howden SE, Vanslambrouck JM, Wilson SB, Tan KS, Little MH: Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation. EMBO Rep 20: e47483, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howden SE, McColl B, Glaser A, Vadolas J, Petrou S, Little MH, et al.: A Cas9 variant for efficient generation of indel-free knockin or gene-corrected human pluripotent stem cells. Stem Cell Reports 7: 508–517, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howden SE, Wardan H, Voullaire L, McLenachan S, Williamson R, Ioannou P, et al.: Chromatin-binding regions of EBNA1 protein facilitate the enhanced transfection of Epstein-Barr virus-based vectors. Hum Gene Ther 17: 833–844, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al.: Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8: 424–429, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JW, Chambon A, Bishard K, Hartung A, Arndt D, Brugnano J, et al. : Bioprinted pluripotent stem cell-derived kidney organoids provide opportunities for high content screening. bioRxiv, 2018 [Google Scholar]
- 25.van den Berg CW, Ritsma L, Avramut MC, Wiersma LE, van den Berg BM, Leuning DG, et al.: Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation In vivo. Stem Cell Reports 10: 751–765, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little MH, Mcmahon AP: Mammalian kidney development: principles, progress, and projections. Cold Spring Harb Perspect Biol 4: pii: a008300, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxén L, Sariola H: Early organogenesis of the kidney. Pediatr Nephrol 1: 385–392, 1987 [DOI] [PubMed] [Google Scholar]
- 28.Brown AC, Muthukrishnan SD, Oxburgh L: A synthetic niche for nephron progenitor cells. Dev Cell 34: 229–241, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Araoka T, Wu J, Liao HK, Li M, Lazo M, et al.: 3D culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell 19: 516–529, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pode-Shakked N, Gershon R, Tam G, Omer D, Gnatek Y, Kanter I, et al.: Evidence of In vitro preservation of human nephrogenesis at the single-cell level. Stem Cell Reports 9: 279–291, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanigawa S, Taguchi A, Sharma N, Perantoni AO, Nishinakamura R: Selective In vitro propagation of nephron progenitors derived from embryos and pluripotent stem cells. Cell Reports 15: 801–813, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle S, Shioda T, Perantoni AO, de Caestecker M: Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn 236: 2321–2330, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, et al.: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beddington RS, Robertson EJ: Axis development and early asymmetry in mammals. Cell 96: 195–209, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Dunwoodie SL, Rodriguez TA, Beddington RS: Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech Dev 72: 27–40, 1998 [DOI] [PubMed] [Google Scholar]
- 36.Schlange T, Andrée B, Arnold H, Brand T: Expression analysis of the chicken homologue of CITED2 during early stages of embryonic development. Mech Dev 98: 157–160, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Lindström NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, et al.: Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J Am Soc Nephrol 29: 806–824, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soo JY, Jansen J, Masereeuw R, Little MH: Advances in predictive in vitro models of drug-induced nephrotoxicity. Nat Rev Nephrol 14: 378–393, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisel P, Renner O, Nies AT, Schwab M, Schaeffeler E: Solute carrier transporter and drug-related nephrotoxicity: The impact of proximal tubule cell models for preclinical research. Expert Opin Drug Metab Toxicol 10: 395–408, 2014 [DOI] [PubMed] [Google Scholar]
- 40.Qi W, Johnson DW, Vesey DA, Pollock CA, Chen X: Isolation, propagation and characterization of primary tubule cell culture from human kidney. Nephrology (Carlton) 12: 155–159, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Hale L, Howden SE, Phipson B, Lonsdale A, Er PX, Ghobrial I, et al. : Human kidney organoid glomeruli provide an improved approach to interrogate podocyte biology and model podocytopathy at scale. Nat Commun 9: 5167, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, et al.: Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 1: 0069, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD: Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23: 869–881.e8, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindström NO, Tran T, Guo J, Rutledge E, Parvez RK, Thornton ME, et al.: Conserved and divergent molecular and anatomic features of human and mouse nephron patterning. J Am Soc Nephrol 29: 825–840, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiagarajan RD, Georgas KM, Rumballe BA, Lesieur E, Chiu HS, Taylor D, et al.: Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS One 6: e17286, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Combes AN, Phipson B, Lawlor KT, Dorison A, Patrick R, Zappia L, et al. : Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand-receptor crosstalk. Development 146: dev178673, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Labastie MC, Catala M, Gregoire JM, Peault B: The GATA-3 gene is expressed during human kidney embryogenesis. Kidney Int 47: 1597–1603, 1995 [DOI] [PubMed] [Google Scholar]
- 48.Menon R, Otto EA, Kokoruda A, Zhou J, Zhang Z, Yoon E, et al.: Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development 145: dev164038, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oosterwegel M, Timmerman J, Leiden J, Clevers H: Expression of GATA-3 during lymphocyte differentiation and mouse embryogenesis. Dev Immunol 3: 1–11, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takasato M, Little MH: The origin of the mammalian kidney: Implications for recreating the kidney in vitro. Development 142: 1937–1947, 2015 [DOI] [PubMed] [Google Scholar]
- 51.Boreström C, Jonebring A, Guo J, Palmgren H, Cederblad L, Forslöw A, et al.: A CRISP(e)R view on kidney organoids allows generation of an induced pluripotent stem cell-derived kidney model for drug discovery. Kidney Int 94: 1099–1110, 2018 [DOI] [PubMed] [Google Scholar]
- 52.Sharmin S, Taguchi A, Kaku Y, Yoshimura Y, Ohmori T, Sakuma T, et al.: Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J Am Soc Nephrol 27: 1778–1791, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.