SUMMARY
Movement of ribosomes on mRNA is often interrupted by secondary structures that present mechanical barriers and play a central role in translation regulation. We investigate how ribosomes couple their internal conformational changes with the activity of translocation factor EF-G to unwind mRNA secondary structures using high-resolution optical tweezers with single-molecule fluorescence capability. We find that hairpin opening occurs during EF-G catalyzed translocation and is driven by the forward rotation of the small subunit head. Surprisingly, modulating the magnitude of the hairpin barrier by force shows that ribosomes respond to strong barriers by shifting their operation to an alternative 7-fold slower kinetic pathway prior to translocation. Shifting into a slow gear results from an allosteric switch in the ribosome that may allow it to exploit thermal fluctuations to overcome mechanical barriers. Finally, we observe that ribosomes occasionally open the hairpin in two successive sub-codon steps, revealing a previously unobserved translocation intermediate.
Keywords: Ribosome, translocation mechanism, mRNA secondary structures, translation regulation, allosteric switch, single-molecule fluorescence, optical tweezers
Graphical Abstract
In Brief / eTOC Blurb
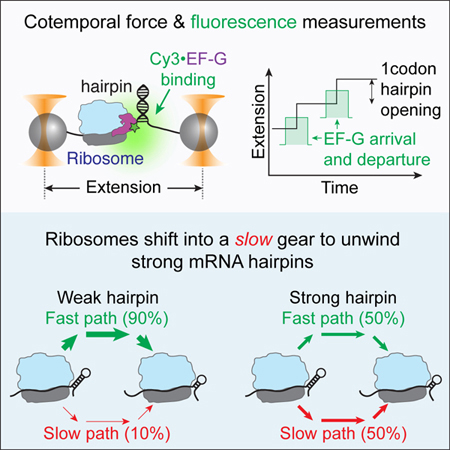
Desai et al. used optical tweezers with single-molecule fluorescence detection to show that mRNA hairpin opening and translocation by the ribosome occur concurrently. Moreover, they unravel an allosteric gear-shift mechanism of translation regulation where ribosomes switch into an alternative slower kinetic pathway in response to strong mRNA hairpins.
INTRODUCTION
Ribosomes are macromolecular machines responsible for the synthesis of proteins in all cells. During translation initiation, ribosomes assemble on a messenger RNA (mRNA) and subsequently enter the elongation phase (Schmeing and Ramakrishnan, 2009). Here, they move along the mRNA in steps of one codon (3 nucleotides) to decode its message and correspondingly add an amino acid to the growing polypeptide chain (Schmeing and Ramakrishnan, 2009). This mechanical movement of the ribosome on the mRNA is called translocation and is catalyzed by the GTPase activity of a trans-acting elongation factor, EF-G (Rodnina et al., 1997). Translocation is also accompanied by large-scale conformational changes within the ribosome. After peptide bond formation, the small (30S) and the large (50S) ribosomal subunits rotate with respect to each other spontaneously, resulting in the translocation of the acceptor ends of the tRNAs in the 50S subunit (Cornish et al., 2008; Frank and Agrawal, 2000; Moazed and Noller, 1989b; Sharma et al., 2016; Zhang et al., 2009). During this process, however, the codon of the mRNA and the anti-codon of the tRNAs that reside in a cleft between the head and the body domains of the 30S subunit have not yet moved. Several lines of evidence indicate that subsequent forward and/or reverse rotation of the 30S head completes this movement, resulting in the net translocation of the mRNA relative the ribosome by one codon (Guo and Noller, 2012; Ratje et al., 2010; Zhou et al., 2014).
During translocation, ribosomes often encounter mechanical barriers due to structures such as hairpins and pseudoknots adopted locally by the mRNA (Ding et al., 2014; Katz and Burge, 2003; Kertesz et al., 2010; Li et al., 2012; Mustoe et al., 2018). Crystal structures have revealed that the entry tunnel of the ribosome can only accommodate single-stranded RNA (Yusupova et al., 2001). Biochemical studies have shown that while ribosomes can intrinsically unwind mRNA secondary structures during translocation (Takyar et al., 2005), the rate of translation is reduced in front of such barriers (Qu et al., 2011; Wen et al., 2008); as such, these barriers are thought to play a key regulatory role in cotranslational protein folding (Chaney and Morris, 1978; Guisez et al., 1993; Watts et al., 2009), mRNA localization (Chartrand et al., 2002; Young and Andrews, 1996), protein abundance (Duan et al., 2003; Nackley et al., 2016), and, in combination with additional cis elements, can induce ribosomal frame-shifting leading to the synthesis of alternative protein products (Jacks et al., 1988; Tsuchihashi, 1991). Despite a strong effect of mRNA secondary structures on translation, a number of questions remain as to how ribosomes couple their helicase activity with their translocation. Specifically, when are mRNA secondary structures opened during a given translation cycle? How is the activity of EF-G coupled to strand opening and translocation? And, how are the internal conformational changes of the ribosome involved in barrier crossing?
Positively charged amino acid residues located at the mRNA entry tunnel are known to play a crucial role in the destabilization of secondary structures prior to translocation (Qu et al., 2011). Mutating these residues do not affect translocation along a single-stranded mRNA but prevents progress on a double-stranded RNA template, indicating that unwinding of secondary structures at the entry tunnel may occur prior to translocation (Takyar et al., 2005). Alternatively, unwinding could occur concomitantly with EF-G binding, whereby the free energy gained upon binding is used to destabilize the hairpin. Finally, unwinding could occur after EF-G binding and concomitantly with mRNA translocation, by the forces generated during the forward or the reverse rotation of the 30S head domain (Liu et al., 2014).
Moreover, the mechanism by which translation rate is modified by these barriers is debated. Previous single molecule optical tweezers studies postulate that mRNA hairpins should directly and selectively reduce the rate of EF-G catalyzed translocation (Qu et al., 2011). Whereas, single molecule fluorescence studies have proposed that mRNA secondary structures allosterically modify the ribosome to delay E-site tRNA release, thereby affecting parts of translation cycle not related to translocation (Chen et al., 2013a). A limitation of these measurements is that they only monitor either the mechanical or the fluorescence coordinate, and therefore cannot directly correlate motions within the ribosome and/or the activity of trans-acting factors such as EF-G to hairpin opening. Such questions are also broadly applicable to many molecular motors that couple the activity of protein or chemical factors to their movement along a substrate (Bustamante et al., 2011).
Therefore, in this study, we use a recently developed high-resolution optical tweezers with single molecule fluorescence capability, a.k.a ‘fleezers’ (Comstock et al., 2011), to directly measure hairpin opening while simultaneously visualizing the binding and release of EF-G. We find that a downstream hairpin, irrespective of its stability, is always only opened after EF-G binding, unequivocally establishing that translocation and hairpin opening are temporally coupled. Moreover, using antibiotic perturbation experiments, we show that the unwinding of secondary structures results from the force generated upon forward 30S head rotation. Then, we increase the stability of the downstream barrier by decreasing the force applied to the hairpin and, surprisingly discover that strong hairpins only marginally reduce the rate of translocation; instead, they act as allosteric switches that globally slow down the translation cycle, biasing ribosomes into a kinetically altered 7-fold slower pathway. Finally, the use of two orthogonal — mechanical and fluorescence — channels to monitor translation, enabled us to unravel a novel unwinding intermediate where each hairpin opening event occurs via two successive sub-codon steps while EF-G remains bound. These findings uncover fundamental insights on the mechanism of mRNA unwinding and translocation by the ribosome.
RESULTS
Ribosomes Open mRNA Hairpins During Translocation
To test whether unwinding occurs prior to, concomitantly with or after EF-G binding, we perform an optical tweezers assay wherein we simultaneously measure mRNA hairpin opening in parallel with EF-G arrival and release, using high-resolution optical tweezers with single-molecule fluorescence capability (Comstock et al., 2011; Wen et al., 2008) (Figures 1A and 1B). This assay uses an mRNA hairpin with repeating valine codons that is tethered by its 5’ and 3’ ends to two polystyrene beads, each held in an optical trap (Figures 1A and S1). A ribosome is stalled at the base of the hairpin by omission of Val-tRNAVal. After capturing the stalled ribosome complex on the beads, translation is restarted by supplying a mixture containing tRNAVal, valyl-tRNA synthetase, valine, Cy3-labeled EF-G, EF-Tu, ATP and GTP.
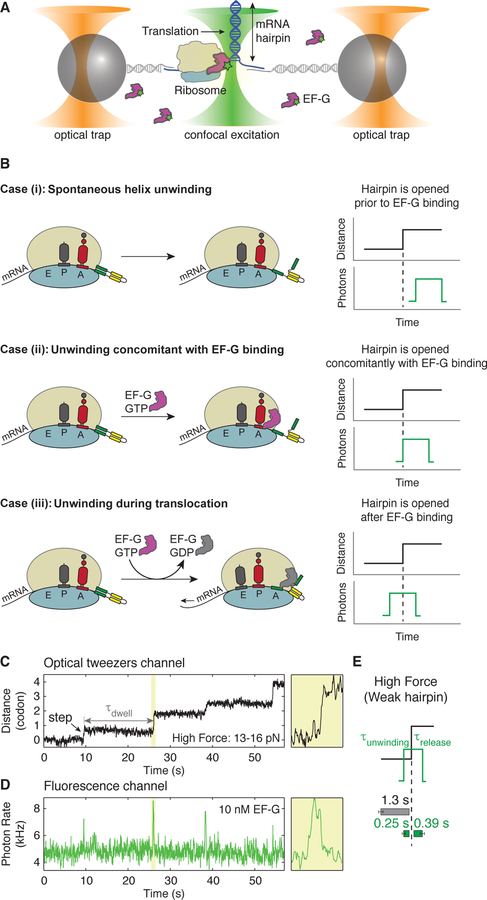
Figure 1. Ribosome opens an mRNA hairpin after EF-G binding.
(A) Experimental setup for simultaneous measurement of hairpin opening and EF-G binding using a time-shared optical trap with single-molecule confocal imaging, ‘fleezers’. Time resolution of the optical tweezers channel is 7.5 ms and that of fluorescence channel is 10 ms. Assembly of ribosome-stalled complexes is outlined in Figure S1.
(B) Possible scenarios of when hairpin unwinding occurs with respect to EF-G binding.
Case (i): the hairpin is spontaneously opened prior to EF-G binding due to the destabilization energy contributed by the ribosome (Qu et al., 2011).
Case (ii): the hairpin is opened concomitant with EF-G binding. Here, either EF-G binding itself induces a conformational change (for example, the forward or reverse movement of the 30S head) that results in the opening of the hairpin, or the 30S head rotates back and forth as a Brownian ratchet, and only the binding of EF-G effectively rectifies the position of the 30S head leading to hairpin opening.
Case (iii): the hairpin is opened a variable amount of time after EF-G binding. Opening could result from events such as EF-G•GTP tight binding, GTP hydrolysis, Pi release, EF-G•GDP release or internal conformational changes of the ribosome.
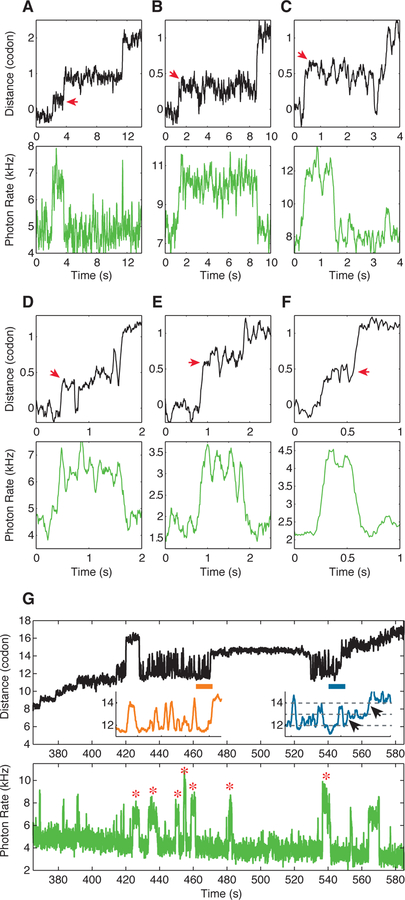
(C, D) An example of a single-ribosome ‘fleezers’ trajectory along an mRNA hairpin held under high external force of 13–16 pN with 10 nM Cy3-labeled-EF-G. (C) Optical tweezers channel, ribosome opens the hairpin in 1 codon steps (= 6 nts of hairpin opened) separated by dwells, τdwell. Data was recorded at 133 Hz and displayed at 13 Hz. Yellow box shows a zoomed-in event. (D) Fluorescence channel, each spike in fluorescence corresponds to the binding of an EF-G. Yellow box shows a zoomed-in event. Data is recorded at 100 Hz and displayed at 10 Hz. Additional zoomed-in events shown in Figure S2.
(E) Summary of average τdwell (grey) and average EF-G residence times before unwinding (τunwinding, green) and after unwinding (τrelease, green) for a weak hairpin held under external force of 13–16 pN (n = 55 events, 9 molecules). Error bars represent SEM.
This scheme enables us to measure translocation in real time through two channels: (i) the optical tweezers channel to follow the step-wise opening of the hairpin (Figure 1C) and (ii) the fluorescence channel to monitor the binding of fluorescently-labeled EF-G (Figure 1D). In this experiment, each extension step corresponds to the unwinding of exactly one codon (Figure 1C). The waiting time between two consecutive codon steps is termed a ‘dwell’, during which the mRNA is stationary on the ribosome and a number of chemical events occur, including the addition of one amino acid to the nascent polypeptide chain. Likewise, each spike in the fluorescence channel corresponds to the binding and dissociation of a single EF-G molecule (Figure 1D).
Strikingly, at each codon, we observe a strict order of events: hairpin unwinding occurs always after EF-G arrival; similarly, release of EF-G occurs always after the unwinding event (yellow box in Figures 1C and 1D, Figure S2). The total time that EF-G is bound to the ribosome is the sum of the time between EF-G binding and the unwinding of the hairpin, τunwinding, and the time between unwinding and EF-G release, τrelease (Figure 1E). On average, τunwinding is 0.25 ± 0.07 s and τrelease is 0.39 ± 0.1 s when the hairpin is held at high force (> 13 pN) (Figures 1E and S2, distributions of τunwinding and τrelease are shown in Figures 4D and S6A respectively). Therefore, unwinding occurs only after EF-G binding and not before by the independent helicase action at the mRNA entry site, nor does it occur simultaneously with EF-G arrival via the free energy gained upon binding. As the mRNA entry tunnel can only accommodate ssRNA (Yusupova et al., 2001), hairpin opening must then occur after EF-G binding and concomitantly with mRNA translocation.
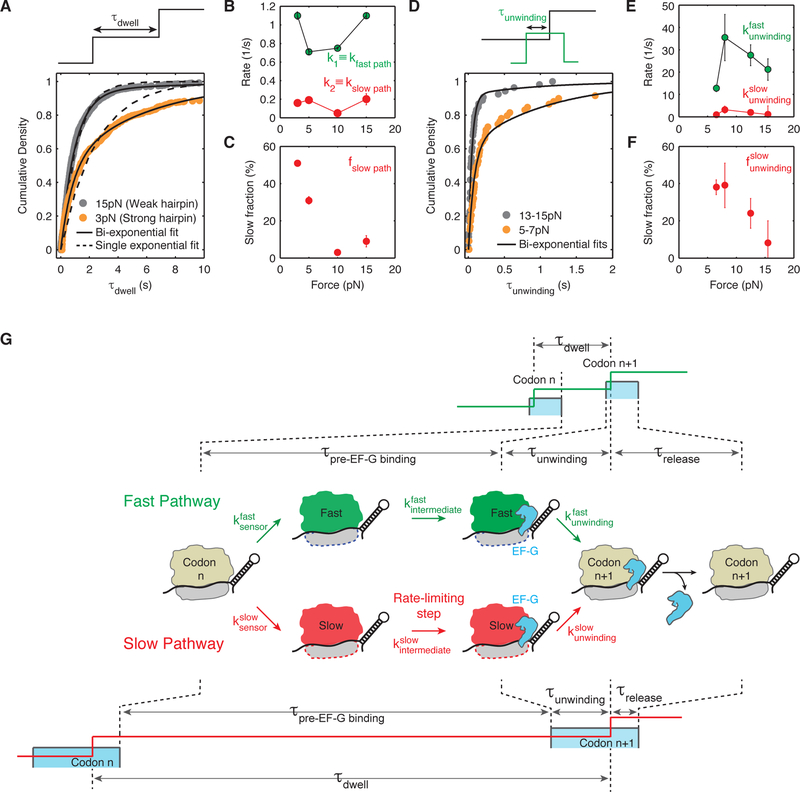
Figure 4. Ribosome translates through a hairpin via two parallel pathways that bifurcate before EF-G binding and converge after hairpin opening.
(A) Cumulative density of τdwell, the time ribosome spends at each codon, is best fit by a mixture of two exponentials given by: .
(B) Summary of fast and slow pathway rates, kfast path and kslow path obtained from fits to the cumulative distribution of τdwell at forces 3 pN (n = 551 events, 19 molecules), 5 pN (n = 256 events, 8 molecules), 10 pN (n = 776 events, 19 molecules) and 15 pN (n = 889 events, 24 molecules). Error bars represent 95% confidence intervals. Concentration of EF-G is 10 µM. kfast path and kslow path, remain constant at ~0.92 codon/s and ~0.15 codon/s respectively at various forces.
(C) Fraction of events going through the slow pathway, ƒslow path, as obtained from fitting shown in (A). ƒslow path increases from ~10 % at high force (> 10 pN) to ~30–50 % at low force (< 7 pN). Error bars represent 95% confidence intervals.
(D) Cumulative density of τunwinding, the time between EF-G binding and hairpin opening, is best fit by a mixture of two exponentials given by: .
(E) Summary of bi-exponential rates and obtained from fits to the cumulative distribution of τunwinding at forces 5–8 pN (n = 62 events, 14 molecules), 7–9 pN (n = 46 events, 10 molecules), 12–13 pN (n = 80 events, 20 molecules) and 14–17 pN (n = 53 events, 22 molecules). These experiments were performed in passive mode, i.e., the force is maintained in a particular regime rather than being held constant. Concentration of EF-G is 10 nM. Error bars represent 95% confidence intervals.
(F) Fraction of events going through the slow unwinding pathway, , increase from ~15% at high force (> 12 pN) to ~40% at low force (< 9 pN). Error bars represent 95% confidence intervals.
(G) Proposed kinetic scheme: first, the ribosome ‘senses’ the hairpin barrier and irreversibly switches into either a ‘fast’ state (shown in green) or a ‘slow’ state (shown in red) via rates or respectively. The ratio, , fraction of translation events that go through either pathway as shown in (C) and (F). Then, the ribosome in either ‘fast’ or ‘slow’ state must undergo an intermediate transition ( or ) that becomes rate limiting at low force and determines the overall rates shown in (B). It is possible that this intermediate transition is the rate of EF-G binding (see Discussion). Then, the ribosome unwinds the hairpin via rates and shown in (E). Finally, EF-G is released in a similar fashion in either pathway suggesting that the bifurcated pathways converged upon unwinding (Figure S6). A cartoon of the fleezers trajectory for both the fast and slow pathway is shown above and below the kinetic scheme. Notice that while τunwinding increases in the slow pathway, it does not increase enough to account for the total increase in τdwell.
Recall that a translocation cycle begins after peptide bond formation when the 30S and 50S subunits rotate with respect to each other resulting in the movement of the acceptor ends of the tRNAs in the 50S subunit by one codon. Then, after EF-G binding, two coordinated conformational changes occur within the ribosome that result in the movement of the tRNAs and the mRNA in the 30S subunit (Guo and Noller, 2012; Mohan et al., 2014; Ratje et al., 2010; Zhou et al., 2014). First, the 30S head rotates forward, moving the tRNAs and the mRNA relative to the 30S body by one codon. Second, the 30S head detaches from the tRNAs and the mRNA and moves back into the non-rotated position. As a result, the entire ribosome moves itself relative to the mRNA by one codon and completes a translocation cycle.
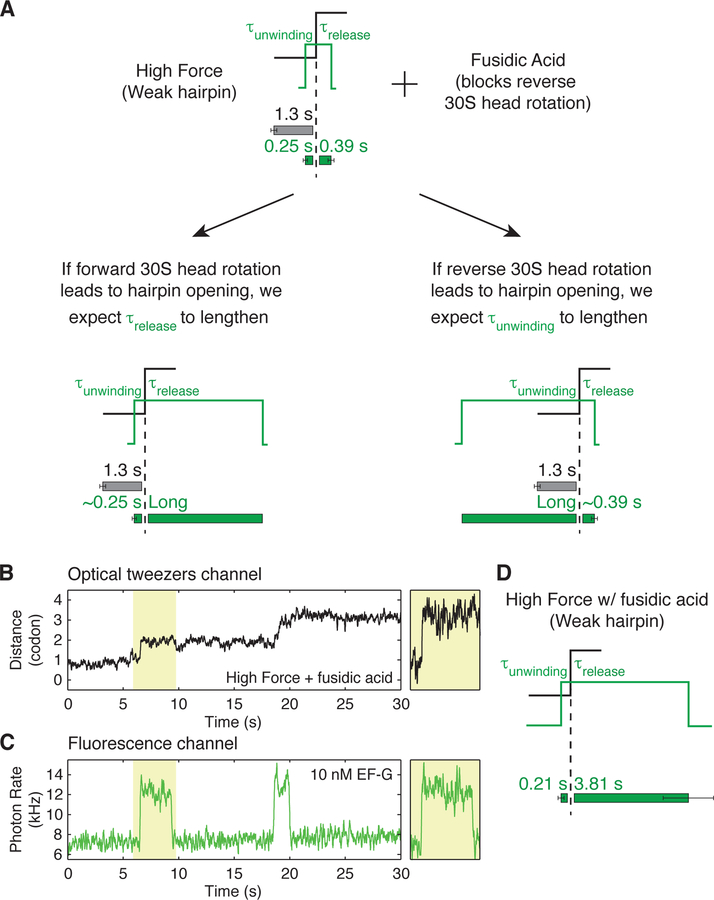
In order to distinguish whether forward or reverse 30S head rotation results in hairpin opening, we performed experiments with antibiotic fusidic acid (Figure 2). Previous studies have shown that while fusidic acid binds to an early translocation intermediate, its kinetic effect on forward 30S head rotation and subsequent tRNA-mRNA movement is small. Conversely, fusidic acid greatly reduces the rate of late translocation events such as reverse 30S head rotation, E-site tRNA release and EF-G dissociation (Belardinelli and Rodnina, 2017; Borg et al., 2015; Ramrath et al., 2013; Wasserman et al., 2016). Therefore, by measuring whether τunwinding or τrelease increases in the presence of fusidic acid, we can determine the relative timing of 30S head rotation and hairpin opening (Figure 2A). Interestingly, we find that τunwinding is unchanged (0.21 ± 0.1 s), but τrelease increases dramatically (~10-fold) to 3.81 ± 0.84 s in the presence of fusidic acid (Figures 2B–D). These results establish that hairpin opening occurs before 30S head reverse rotation, and that the helicase and translocase activities of the ribosome are tightly coupled and occur simultaneously.
Figure 2. Hairpin opening occurs during forward 30S head rotation.
(A) Diagram of predicted EF-G residence times in presence of antibiotic fusidic acid for unwinding resulting from either forward or reverse 30S head rotation.
(B, C) An example of a single-ribosome ‘fleezers’ trajectory in presence of 50–200 µM fusidic acid and 10 nM Cy3-EF-G. Hairpin is held under high external force of 13–16 pN. (C) Optical tweezers channel, data was recorded at 133 Hz and displayed at 13 Hz.
(D) Fluorescence channel, data is recorded at 100 Hz and displayed at 10 Hz. Additional zoomed-in events shown in Figure S3. (D) Summary of average EF-G residence times before and after unwinding show that τrelease increases dramatically in the presence of fusidic acid (n = 25 events, 13 molecules). Error bars represent SEM.
Strong mRNA Hairpins Reduce Translation Rate
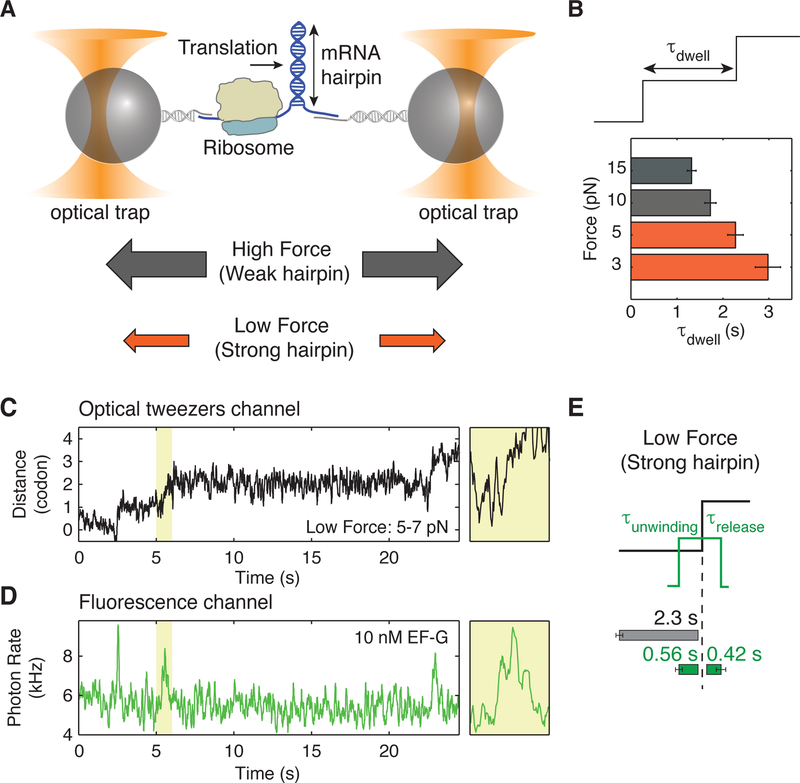
Next, we ask, how does the ribosome respond to the presence of a stronger mRNA hairpin? Does the rate of hairpin unwinding decrease upon increased strength of the hairpin? Or, are other parts of the translation cycle modified by the downstream hairpin? In our assay, we can modulate the strength of the barrier by applying force to destabilize the junction (Bustamante et al., 2004; Qu et al., 2011; Tinoco and Bustamante, 2002) (Figure 3A). A key advantage of using force rather than GC content to alter the hairpin stability is that the mRNA coding sequence and identity of the tRNAs remain invariant; so that steps in the translation cycle unrelated to mechanical movement, such as decoding, remain unaffected.
Figure 3. Force-dependence of translation rates.
(A) Schematic showing the effect of force on the stability of the hairpin junction. High applied force results in a weaker hairpin junction.
(B) Average time per codon increases at low forces as the hairpin presents a stronger barrier (n = 889 events from 24 molecules at 15 pN, n = 776 events from 19 molecules at 10 pN, n = 256 events from 8 molecules at 5 pN and n = 551 events from 19 molecules at 3 pN). The concentration of EF-G is 10 µM.
(C, D) An example of a single-ribosome ‘fleezers’ trajectory with 10 nM Cy3-EF-G and hairpin is held under low external force (5–7 pN). (C) Optical tweezers channel, data was recorded at 133 Hz and displayed at 13 Hz. (D) Fluorescence channel, data is recorded at 100 Hz and displayed at 10 Hz. Additional zoomed-in events shown in Figure S4.
(E) Summary of average τdwell (grey) and average EF-G residence times before unwinding (τunwinding, green) and after unwinding (τrelease, green) for a strong hairpin held under low external force of 5–7 pN (n = 62 events, 14 molecules). Error bars represent SEM. When compared to data collected at high force (Figure 1E), τdwell increases by ~1 s at low force, but τunwinding only increases by ~0.3 s and τrelease remains statistically invariant.
We measure the average residence time of the ribosome at each codon (τdwell) as a function of force applied to destabilize the hairpin junction (Figure 3B). At saturating concentrations of EF-G (10 µM), we find that ribosomes spend ~1.3 s/codon in front of a weak barrier (high force > 13 pN) and ~2.5 s/codon in front of a strong barrier (low force < 7pN, Figure 3B). Hence, the rate of translation decreases 2-fold from ~0.8 codon/s to ~0.4 codon/s as the strength of the downstream barrier increases. The translation rate measured at high force in our single-molecule experiments is slightly slower than the 3–10 codon/s reported in in-vitro translation assays performed in bulk, possibly due to temperature effects as our measurements are made at room temperature as opposed to 37°C (Holtkamp et al., 2015; Pavlov and Ehrenberg, 1996).
The Effect of a Strong Hairpin on Translation Rate is Not Restricted to the Unwinding Step
What change in the single-ribosome trajectories accounts for this two-fold decrease in translation rate observed with increased strength of the barrier? We find earlier that the dwell duration per codon (τdwell) increases on average by ~1 s at low force when the junction is more stable (Figure 3B). Intuitively we would expect that because EF-G catalyzed translocation is the force-sensitive step, its lengthening should account for the full dwell time increase observed in front of a barrier. Therefore, since the hairpin is opened only after EF-G binding, we expect that the residence time of EF-G before unwinding (τunwinding) would correspondingly increase by ~1 s/codon at low force in our assay, possibly due to a slower rate of forward head rotation (Figures 3C, 3D, and S4).
Surprisingly, application of low force only increases τunwinding by ~0.35 s on average, while τrelease remains statistically invariant (Figure 3E). Hence, our results directly indicate that an additional kinetic event either prior to EF-G binding or after EF-G release becomes rate limiting and accounts for the lengthening of the total dwell time by the increased strength of the junction barrier. This is unlikely to be A-site tRNA binding or peptide bond formation, which have been shown to be insensitive to downstream mRNA secondary structures (Chen et al., 2013a; 2014; Kim et al., 2014). And we can exclude photobleaching artifacts of the bound EF-G since we determine the average bleaching times from fusidic acid measurements to be at least 5 s, which are 5-fold longer than the total residence time of EF-G on the ribosome (Figure 2D).
Translation Occurs via Two Parallel Pathways
Further analysis of the single ribosome trajectories showed that fitting of the distribution of total dwell time (τdwell) requires a minimum of two exponentials (Figures 4A and S5) according to,
where P(t) is the probability of observing a dwell of duration t; k1 and k2 are the respective rates of the two exponential distributions; ƒ1 is the fraction of events that a given ribosome translates with rate k1; and ƒ2 is the fraction of events that it translates with rate k2. The need of two exponentials to describe the distribution of dwell times indicates that the kinetic mechanism of the ribosome elongation cycle is not a simple linear scheme but that it bifurcates into two alternative pathways with rates k1 and k2.
Significantly, the stability of the hairpin junction determines the fraction of time that the ribosome chooses one or the other path. At high force of 15 pN, during which the mRNA hairpin is highly destabilized, we observe that ~90% of the translation events occur at the rate k1 (~1.1 codon/s) while the remaining events occur through a “slow” pathway with the rate k2 (~0.2 codon/s) (Figure 4B – rates k1 and k2, and Figure 4C – fraction of events with rate k2). Notice that the rate k2 is ~6-fold slower than k1 and hence we define k1 ≡ kfast path and k2 ≡ kslow path. As the force applied to the hairpin is reduced and its stability increases, the values of kfast path and kslow path remain relatively constant (Figure 4B); however, the fraction of translation events that enter the slow pathway increases to 30–50% as the force decreases below 7 pN (Figure 4C). These slow events do not arise from a subset of impaired ribosomes (static dispersion) since the biphasic translation rate is observed for any individual ribosome trajectory, indicating that the same ribosome can switch between the two pathways (dynamic dispersion) (Figure S5). Thus, the presence of a strong barrier biases the ribosomes into the slower pathway with minimal alteration of the pathway rates, and accounts for the 2-fold decrease in the average translation rate observed in these conditions.
Pathway Identity is Maintained Through EF-G Catalyzed Hairpin Opening
Next, we ask whether the translation kinetic pathway bifurcation occurs prior to EF-G binding through hairpin unwinding or after unwinding during EF-G release and resetting of the ribosome (see Data S1). Interestingly, the distribution of lifetimes of τunwinding (the residence time of EF-G before unwinding), is also best fit by a double exponential (Figure 4D) with fast and slow rates that are also not significantly force-sensitive (Figure 4E). At high and low force, is ~24 /s and is ~2 /s (Figure 4E). Significantly, however, the fraction of unwinding events displaying slow kinetics increases from ~15% at high force (above 12 pN) to ~40% at low force (below 9 pN, Figure 4F).
Notice that both kdwell (Figure 4B) and kunwinding (Figure 4E) are described by a mixture of two exponentials, but more importantly that the fraction of events going through the slow branch increase in approximately the same proportions for both the distributions (kdwell and kunwinding) as a function of force (Figures 4C and 4F). These results indicate that the translation pathway bifurcates prior to EF-G binding and the ribosome remains in the fast and slow branches through EF-G binding and subsequent hairpin unwinding. Consistently, analysis of τrelease reveals that the distribution of this parameter is insensitive to the strength of the hairpin (Figure S6), indicating that the two pathways have converged during the unwinding events.
However, it is important to note that the rates of unwinding ( and , Figure 4E) are much faster than the overall rates of the fast and slow pathways (kfast path ~0.9 codons/s; kslow path ~0.15 codons/s, Figure 4B). This observation implies that there must exist an earlier kinetic transition (denoted below as kintermediate) in both the fast and slow pathways that is rate-limiting for each of the respective branches.
Kinetic Scheme for Translation Through mRNA Hairpins
From the above results, we conclude that the translation pathway bifurcates before EF-G binding and converges after EF-G-catalyzed hairpin unwinding (Figure 4G and Data S1). Our data require three sequential steps to describe the kinetics of translation through mRNA secondary structures. First, there is a “hairpin sensor” step that occurs rapidly and irreversibly, that gives rise to the bifurcation of the translation pathway into a “fast” and a “slow” branch with rates and , respectively. The ratio of these rates is force-sensitive and is equal to the fraction of events going through each pathway, i.e., . Second, an intermediate kinetic step occurs in both pathways ( and ). This step is insensitive to force and determines the rates of translation through the two paths (Figure 4B). These two sequential ‘sensor’ and ‘intermediate’ steps are necessary to explain the force-dependent population shift and the force-independent rates of the two pathways, respectively. In the third step, the ribosome opens the hairpin in the presence of EF-G via rates and , neither of which is rate-limiting in the respective pathways. The two pathways converge after the unwinding event, EF-G is released, and the ribosome is reset for another round of elongation. Taken together, these results indicate that the presence of a strong barrier at the RNA junction allosterically switches the ribosome before EF-G binding from a fast to a slow overall translation speed. We also tested other possible kinetic schemes particularly one where the ribosome could be stuck in an inactive conformation prior to EF-G binding and found that this scheme does not satisfy all the data obtained here (see Data S1 for details).
Detection of Translocation Intermediates
In the presence of a strong barrier, we occasionally observe that unwinding, instead of taking place in a single transition, occurs in two — smaller than one codon — steps, a process accompanied by unusually long EF-G binding durations (Figures 5A–F). The first sub-step covers on average 42 ± 17 % of the total step size and occurs soon after EF-G binding. In the majority of events, the dwell time in the intermediate state lasts up to a few seconds, and EF-G release is observed only after the second step of unwinding has occurred (Figures 5A, 5B and 5D–F). However, in a few events (such as the one occurring at ~0.5 s in Figure 5C) the first half-step is reversible, and the full step occurs only at ~3.5 s via the intermediate. In this particular case, there is a potential EF-G binding event at ~3.5 s when the full step occurs, but due to low intensity of the signal, we cannot conclusively determine the presence of EF-G. It is important to note that the use of labeled EF-G in these experiments and the lengthening of its dwell time on the ribosome clearly indicate that the sub-steps are not merely a manifestation of hairpin dynamics.
Figure 5. Novel ribosome-dependent hairpin dynamics.
(A-F) Examples of single-ribosome ‘fleezers’ trajectories in presence of 10 nM Cy3-labeled-EF-G where the unwinding step is split into two sub-steps that add up to one codon. Notice that EF-G remains bound until the entire step is completed. For comparison, a second unwinding step that occurs in a single transition is shown in A. The sub-steps are highlighted with red arrows.
(G) An example of a single-ribosome ‘fleezers’ trajectory in presence of 10 nM Cy3-labeled-EF-G showing a region of hairpin hopping where several EF-G molecules unproductively bind for long time scales (shown with stars).
In contrast, we sometimes observe large-scale excursions (~2 codons) of the ribosome along the mRNA (Figure 5G). These movements closely resemble those previously described during frame shifting along a slippery sequence (Yan et al., 2015). Interestingly, during these excursions, several EF-G molecules are seen to bind and unbind with prolonged dwell times on the ribosome (see EF-G events marked by a star in Figure 5G). Thus, in the presence of a strong mRNA junction, the ribosome appears to switch into a diffusive mode over the extent of two codons during which multiple EF-Gs are seen to bind and unbind.
Mechanism of Translocation Through mRNA Secondary Structures
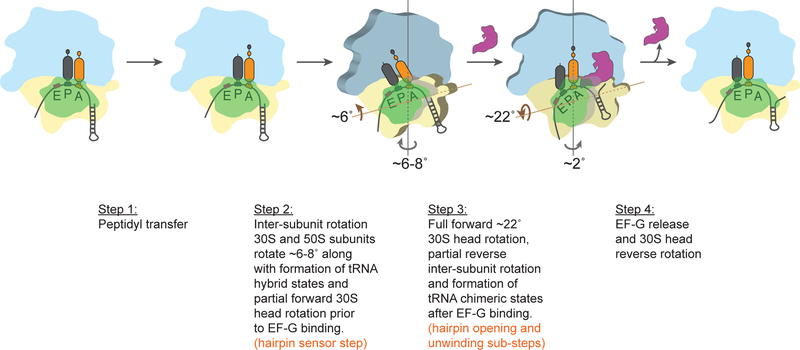
All together, these results lead us to propose a mechanism where the ribosome can operate in either of two distinct kinetic modes or ‘gears’, both of which are competent to bind EF-G and translocate. After peptide bond formation, two major conformational changes are known to occur in the ribosome prior to translocation: (1) inter-subunit rotation and (2) partial forward 30S head rotation (Figure 6, Step 2). In this state, the 30S and 50S subunits spontaneously rotate ~6–8° with respect to each other, concomitant with formation of tRNA hybrid states (Frank and Agrawal, 2000; Moazed and Noller, 1989b; Zhang et al., 2009). Structural evidence shows that this state is accompanied by an intermediate degree of rotation (5–7°) of the 30S head domain with respect to the 30S body, along an axis orthogonal to that of the inter-subunit rotation, prior to EF-G binding (Mohan et al., 2014). In fact, deconvolution of FRET measurement data from donor and acceptor positions at S13 and L33 or at S6 and at L9, respectively, reveal a similar mobility in the 30S head domain prior to EF-G arrival (Belardinelli et al., 2016). Importantly, this partial head rotation is distinct from the full ~22° forward head rotation that occurs after EF-G binding that is accompanied by a partial reverse rotation of the 30S body (Guo and Noller, 2012; Ratje et al., 2010; Zhou et al., 2014). As shown here, the full forward head rotation results in the opening of the hairpin junction at the entry port (Step 3 in Figure 6).
Figure 6. Model for tight coupling between helicase and translocase activity of the ribosome.
At the beginning of each elongation cycle, the ribosome decodes the mRNA codon in the A-site and transfers the peptide chain from the P-site tRNA to the A-site tRNA (Step 1). Then, two major conformational changes occur in the ribosome prior to EF-G binding: inter-subunit rotation and partial 30S head rotation (Step 2). We propose that the ribosome could ‘sense’ the presence of an mRNA hairpin during partial head rotation (Step 2) to shift into a ‘slow’ gear. After EF-G binding, the head further rotates forward to a full 22° swivel as the body domain rotates back along the inter-subunit rotation axis (Step 3). Our results demonstrate that downstream mRNA hairpins are opened during this step. Finally, EF-G is released, and the ribosome is reset for another round of elongation (Step 4). The 50S subunit is shown in blue, the 30S body domain is shown in yellow and the 30S head domain is shown in green. Inter-subunit rotation axis is shown in grey and head-rotation axis is shown in brown. The magnitude of rotation is represented by the size of the arrow.
Consequently, as the ribosome senses the hairpin prior to EF-G binding, we propose that the intermediate head rotation that occurs during inter-subunit rotation could operate as the sensor that switches the ribosome between the fast vs slow gears and that leads to the subsequent rate-limiting step of each pathway. Even though the detailed structural intermediates involved during the slow gear path remain unknown, since the hairpin junction abuts the interface of the 30S head and the 30S body, we favor a model where two distinct head rotation modes result in the ‘fast’ and ‘slow’ gears of ribosome operation. We propose that the 30S head “senses” the downstream junction during its initial rotation (Step 2, Figure 6), and switches into an altered conformation in response to a strong barrier. Indeed, several structures of the ribosome trapped using antibiotics or non-hydrolyzable analogs of EF-G•GTP that block translocation, show this partial degree of head rotation (~6°) hinting at the possibility that such conformational change gives rise to the ‘low’ gear (Brilot et al., 2013; Chen et al., 2013c; Mohan et al., 2014; Ratje et al., 2010; Svidritskiy et al., 2014; Tourigny et al., 2013; Zhang et al., 2016).
DISCUSSION
Many cellular processes are carried out by sophisticated molecular machines that move in a precise manner to produce forces, torques and displacements. These machines operate through cycles that couple many chemical events such as internal conformational changes, nucleotide binding and hydrolysis, release of catalysis products, and binding/unbinding of trans-acting factors to their mechanical task (Bustamante et al., 2011). How are these various chemical and mechanical steps coordinated in the operational cycle of these machines remains a central question in biophysics. Although the advent of single molecule methods has greatly improved the ability to extract mechanistic details from the individual molecular trajectories, the challenge is compounded by the fact that, more often than not, these measurements are one-dimensional, tracking a single event of the cycle.
Indeed, optical tweezers experiments, in which forces and displacements can be detected with millisecond resolution, have provided unprecedented details about the mechanical operation of such machines (Moffitt et al., 2008). In this way, the stepping of nucleic acid motors, such as polymerases, helicases and translocases, has been resolved at the single base-pair level (Chemla, 2010). Likewise, torque generation by topoisomerases, DNA translocases, and flagellar motors has been measured directly and provided significant mechanistic insights into their operation (Moffitt et al., 2006; Ryu et al., 2000). However, these mechanical measurements are blind to the concomitant chemical events of the machines’ operational cycles. Alternatively, the use of single molecule fluorescence methods have enabled researchers to follow, for example, in real time, some of the conformational changes of these machines during their operation, but at the expense of disregarding their progress along the mechanical coordinate (Joo et al., 2008).
The recent development of a new generation of optical tweezers instruments, endowed with single-molecule fluorescence capability (referred to here as ‘fleezers’ for fluorescence optical tweezers), has opened the possibility of monitoring molecular machine trajectories along two (and in principle several) orthogonal reaction coordinates in a co-temporal manner (Comstock et al., 2011; Hohng et al., 2007; Lang et al., 2004; van Mameren et al., 2008). Hence, these instruments make it possible to simultaneously record, correlate, and causally relate chemical events (binding of ligands, conformational changes, etc.) with the corresponding mechanical events (force or torque generation and displacement). Here we have used a fleezers instrument (Comstock et al., 2011; Whitley et al., 2017) to investigate the coupling of ribosome translocation to the binding and activity of elongation factor EF-G, and to uncover how downstream secondary structures in the mRNA template regulate this process.
Mechanism of helicase and translocase activities of the ribosome
During protein synthesis, ribosomes translocate along the mRNA in steps of one codon. This movement is often impeded by structural elements in the downstream mRNA such as hairpins and pseudoknots that are situated at the entry pore on the small 30S subunit. Previous optical tweezers studies have proposed that ribosomes unwind mRNA secondary structures via two modes of active helicase activity: one in which the positively charged amino acid residues at the surface of the mRNA entry tunnel destabilize helical regions prior to EF-G binding, and the other in which the forces generated during EF-G catalyzed translocation actively open the secondary structures (Qu et al., 2011). Here, by cotemporally monitoring both the mechanical and the fluorescence coordinates of a ribosome translating on an mRNA hairpin, we find that junction opening always occurs a variable length of time after EF-G binding, unequivocally establishing that the helicase and translocase activities of the ribosome occur simultaneously. This observation therefore indicates that the surface of the mRNA entry tunnel is not responsible for the opening of secondary structure prior to translocation (Qu et al., 2011), but it is still possible and likely that these positively charged amino acids contribute to the destabilization of downstream hairpins.
Direct measurements of the one-codon displacement of the mRNA on the ribosome have shown that ribosomes are capable of generating forces that can unwind downstream secondary structures during translocation (Liu et al., 2014). Such forces could be exerted during either the forward or the reverse rotation of the 30S head domain that results in the coordinated movement of the mRNA codons and the tRNA anticodons in the 30S subunit. By using an antibiotic to stall the reverse 30S head rotation, we can identify the forward 30S head rotation, that ensues after EF-G binding, as the likely candidate for the force generation step of the ribosome that results in hairpin opening. Consistent with this finding, a previous single-molecule study has reported a small rotation of EF-G with respect to the ribosome after GTP hydrolysis that could exert force to “unlock” the ribosome and drive translocation (Chen et al., 2016). In this case, unlocking corresponds to a ribosome intermediate where the 30S head has fully rotated forward, whereas the 30S body has moved back towards its non-rotated state. Furthermore, structures of the ribosome captured in intermediate states of translocation also support that hairpin opening must occur prior to or during forward 30S head rotation as the mRNA entry tunnel cannot accommodate dsRNA in the 30S head rotated state (Zhou et al., 2014). Overall, our results show a tight temporal coupling between EF-G binding, internal dynamics of the ribosome, and mRNA translocation/hairpin opening.
This coupling is maintained even in cases where the presence of a mechanical barrier of an mRNA hairpin junction forces the ribosome to move in sub-codon steps; in these cases, we find that the EF-G residence time is prolonged until the full codon translocation has been completed. We speculate that these sub-codon steps could correspond to the head trapped in intermediate states of rotation as have been observed structurally in vacant ribosomes or in the presence of EF-G analogs (Mohan et al., 2014; Pulk and Cate, 2013). We suggest that such a strict coordination between EF-G and the mechanical movement of the ribosome could be necessary to ensure that the reading frame of the mRNA is always maintained.
Along the same lines, the ~2 codon diffusive movement of the ribosome at low force is accompanied with several futile EF-G binding events. As the mRNA hairpin used in these experiments consists of repeating valine codons, such large-scale movement of the ribosome along the mRNA could arise from the re-pairing of peptidyl tRNA to the neighboring valine codons. This hypothesis is consistent with a previous observation that similar slips in the reading frame occur during frame shifting only when the slippery sequence is positioned at the decoding site of the ribosome (Yan et al., 2015). Moreover, EF-G binding does not seem to be the rate-limiting event to exit this diffusive state of the ribosome, as several EF-G molecules are seen to bind and unbind. Rather, given that we do not provide amino-acyl tRNAs for alternative reading frame codons, it is possible that the arrival of EF-G must coincide with the ribosome re-registering in the correct reading frame to allow the ribosome to exit this state and continue translation.
Translation regulation by secondary structures
The stability of downstream mRNA secondary structures has been shown to modulate the rate of translation (Chen et al., 2013a; 2014; Qu et al., 2011; Wen et al., 2008). Here, we have used force to modulate the strength of the mRNA barrier in front of the ribosome, to investigate how the magnitude of this barrier affects the translation cycle. Until now, the naïve expectation has been that the resulting reduced translation rate is due to the slowing down of the mRNA unwinding mechanical step before a strong barrier (Qu et al., 2011). Surprisingly, however, we find that the hairpin opening step of the ribosome catalyzed by EF-G is only marginally affected in the presence of stronger hairpins. Instead, the ribosome responds to stronger barriers by shifting its operation ~50% of the time into an alternative kinetic pathway, i.e., a slower ‘gear’.
We postulate that this shift into an alternative slower pathway in front of strong mechanical barriers occurs prior to EF-G binding. A previous study has shown that a frameshifting stimulating stem loop destabilizes the inter-subunit rotated/hybrid state of the ribosome and biases the ribosome towards the non-rotated/classical state (Kim et al., 2014). EF-G binding to such a destabilized hybrid state could be impaired and possibly result in the slower pathway here. Consistent with this interpretation, in another study where a frameshifting stimulating pseudoknot is present at the mRNA entry port, a moderate inhibitory effect on EF-G binding was observed (Caliskan et al., 2014).
However, the presence of downstream barriers in the context of frameshifting also significantly slowed down events after EF-G binding such as unlocking of the ribosome (Kim et al., 2014), and late translocation events such as 30S head reverse rotation, E-site tRNA release and EF-G dissociation (Caliskan et al., 2014; Chen et al., 2013a; Kim et al, 2014). While we do observe that in the presence of an mRNA hairpin, the ribosome can take an alternative slower pathway to unwind the hairpin after EF-G binding ( in Figures 4D–F) thereby slowing down the unlocking of the ribosome, we do not observe any delay in EF-G release as a function of the strength of the barrier (Figure S6). The latter result is in contrast with previous studies but could be rationalized because these late stage translocation transitions are likely specific to the presence of a slippery sequence during frame-shifting, which is not present in our study. In fact, several studies have now shown that the slipping of the tRNA-mRNA complex occurs during the reverse 30S head rotation (Caliskan et al., 2014, Chen et al., 2013a; 2014; Kim et al, 2014). Consistently, structural data suggests that the paddles that hold the mRNA reading frame in position disengage only during reverse 30S head rotation (Zhou et al., 2013), allowing for the uncoupled movement of the reading frame and subsequent frame-shifting.
Moreover, the identity of the secondary structure could also play an important role in determining which parts of the translation cycle are slowed down. For example, pseudoknots are mechanically stronger barriers than mRNA hairpins and have been shown to result in a tilted conformation of the 30S head domain during reverse 30S head rotation (Ramrath et al., 2012). It is possible that either such a tilted conformation favors frameshifting or that the ribosome must frame-shift to relieve this tilted state and proceed translation normally. It is also important to note that in frame-shifting conditions, the ribosome is not translating through an mRNA structural barrier and only encounters the junction upon approaching the slippery sequence and therefore many frameshifting studies might not have captured the bifurcated pathway that we observe prior to EF-G binding. In the future, it would be very interesting to pursue the current experiments in frameshifting conditions and we predict that a strong effect on τrelease and downstream resetting of the ribosome would be observed.
Although the structural details of the ribosome conformation resulting in the slow gear are yet to be established, we speculate that the 30S head domain plays an important role in shifting between the two distinct gears in presence of an mRNA structural barrier (Figure 6). Interestingly, several studies have seen that ribosomes take alternate translation pathways in response to a wide variety of challenging conditions such as in the absence of GTP hydrolysis (on EF-G, Belardinelli et al., 2016), starvation of amino-acyl tRNAs during frameshifting (Caliskan et al., 2017) and in the presence of antibiotics (Peske et al., 2004). Whether the mechanism of the switch into the slower pathway is the same for all these conditions is yet to be determined.
Finally, shifting into a slower gear to unwind secondary structures could be thermodynamically more favorable as the ribosome could potentially take advantage of the thermal fluctuations of the junction, thus minimizing energy dissipation. Indeed, a recent study shows that the amount of energy dissipated in a non-equilibrium process can be significantly reduced if the process slows down in those areas of the potential energy surface where there is greater friction (Sivak and Crooks, 2016). This prediction has been experimentally confirmed in a recent single-molecule study that uses coincidentally a DNA hairpin (Tafoya et al., 2018); it was speculated that the high thermodynamic efficiency displayed by molecular machines could be explained if these have evolved to switch into a slower ‘gear’ in regions of their potential energy landscape associated with high dissipation, such as mRNA hairpins.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Carlos Bustamante (carlosb@berkeley.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Source organism
Plasmid for mRNA hairpin was cloned in SURE 2 Supercompetent cells. All other plasmids were cloned in Escherichia coli DH5α cells. All proteins were purified from Escherichia coli BL21 (DE3) cells. Ribosomes were harvested from Escherichia coli MRE600 cells.
METHODS DETAILS
Construction of mRNA hairpin plasmid
To generate the DNA sequence containing the complete mRNA used in the experiments two gBlocks (IDT), each containing one arm of the long hairpin structure, were ordered. gBlock1 (covering the 5’ side of the hairpin sequence) was digested with KpnI and BsaI, whereas gBlock2 (covering the 3’ side of the hairpin sequence) was digested with BsaI and EcoRI. BsaI digestion generated the UUUU tetraloop that caps the long hairpin structure. The resulting DNA pieces were inserted into the KpnI and EcoRI sites of pBluescript SK+. This generated the following sequence (sequence contributed by pBluescript SK+ including the T7 promoter as well as KpnI and EcoRI sites is underlined): TAATACGACTCACTATAGGGCGAATTGGGTACCAGTGGCTGAGGCTTAACTAGTTCTAGAAA TAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGACTACAAGGATGACGATGACAAGA AGGTGGTCGTGGTAGTAGTGGTTGTTGTGGTAGTCGTTGTTGTGGTGGTCGTAGTCGTGGTC GTGGTGGTTGTTGTAGTTGTTGTTGTTGTTGTAGTGGTTGTGGTAGTGGTAGTCGTGGTGGTT GTTGTAGTGGTTGTTGTAGTGGTGGTGGGCCACGCGGCGAAAGCCTCATTGGTCTCGTTTTC GAGACCAATGAGGCTTTCGCCGCGTGGCCCACCACCACTACAACAACCACTACAACAACCA CCACGACTACCACTACCACAACCACTACAACAACAACAACAACTACAACAACCACCACGAC CACGACTACGACCACCACAACAACGACTACCACAACAACCACTACTACCACGACCACCGTG TACAGAACGCAATGAATTC
The resulting plasmid was then linearized by digestion with EcoRI to generate the template for in vitro transcription.
mRNA hairpin synthesis
mRNA hairpin was made by in vitro transcription of the plasmid linearized with EcoRI using MEGAscript T7 Transcription Kit. 0.5 µg of template was used per 20 µL of reaction and transcription was carried out at 37°C for 4 hours. mRNA was then purified via a phenol-chloroform extraction followed by ethanol precipitation and a MicroSpin G25 column. mRNA was stored at −20°C.
dsDNA handle synthesis
The mRNA hairpin was tethered between two beads via a 5’ handle and 3’ handle as described in Figure S1. Both the handles are ~2.5 kb in length and were generated by PCR using modified primers. The 5’handle forward and the 3’handle reverse primers were labeled with biotin at their respective 5’ ends through which these handles are attached to streptavidin coated polystyrene beads. The 5’handle reverse primer has an inverted base that efficiently terminates Phusion DNA polymerase to generate a 21 nts overhang that anneals to the 5’ end of the mRNA hairpin. The 3’handle forward primer has 18 inverted bases at its 5’end that efficiently terminates either Phusion or Taq polymerase and results in an 18 nts overhang that anneals to the 3’ end of the mRNA hairpin.
Protein purification and labeling
The ribosomes were harvested and purified from Escherichia coli MRE600 cells (Moazed and Noller, 1989a) and S-100 enzymes were purified as described previously (Traub et al., 1981). Initiation factors (Lancaster and Noller, 2005), wild-type EF-G (Wilson and Noller, 1998) and EF-Tu (Boon et al., 1992) were purified via the engineered His-tag using a HisTrap column followed by anion exchange chromatography with a HiTrap Q column. Plasmid for Val-RS was obtained from Prof. Susan Marqusee and purified via an engineered His-tag. Plasmid for S73C EF-G was obtained from Prof. Joseph Puglisi. To fluorescently label EF-G (Chen et al., 2013b), S73C EF-G was dialyzed into a labeling buffer (20 mM Tris•HCl pH 7.5, 100 mM NaCl and 0.1 mM TCEP) and 6x molar excess of Cy3-monomaleimide was added. Protein + dye solution was rotated gently in a nutator at room temperature for 2 hours and then at 4°C over night. Free Cy3 was removed by passing the labeled protein through three 10DG desalting columns and then through a sephacryl S300 size exclusion column. Labeled EF-G was separated from unlabeled EF-G through hydrophobic interactions chromatography with a TSKgel Phenyl 5-PW column. All proteins were stored in 25 mM Tris•HCl pH 7.5, 60 mM NH4Cl, 10 mM MgCl2 and 5 mM β-Me unless stated otherwise. All proteins were flash frozen in liquid nitrogen and stored in −80°C.
Translation Buffer
The in vitro translation buffer is composed of 40 mM HEPES•KOH pH 7.5, 60 mM NH4Cl, 10 mM Mg(OAc)2, and 6 mM 2-mercaptoethanol. Additionally, either 10 mM sodium azide (for non-fluorescence experiments) or 12.5 mM ascorbic acid (for fluorescence experiments) was added to the translation buffer to prolong tether lifetimes in optical tweezers.
Preparation of Ribosome Initiation Complexes
Initiation complexes were assembled in bulk by mixing 2 µM of 70S ribosomes, 0.1 µM mRNA, 1.75 µM charged f-Met tRNA, 1 mM GTP and 1 µM of each initiation factor (IF1, 2 and 3) in translation buffer and incubating at 37°C for 15 min. They were flash frozen in 2 µL aliquots; each aliquot contains 0.2 pmol of mRNA and 4 pmol of 70S ribosomes.
Deacylation of Total tRNAs
Total tRNAs were dissolved in ultra-pure water to a concentration of ~ 1 U/µL and stored in 50 µL aliquots at −80°C. To deacylate and unfold tRNAs, 300 µL of 45 mM Tris•HCl pH 8.0 was added to a 50 µL aliquot of total tRNA and incubated at 85°C for 2 min and then room temperature for 10 min. 6.25 µL of 1 M MgCl2 was added and the reaction was incubated at 37°C for 30 min to allow the tRNAs to refold. Then, 30uL of 3M KOAc pH 5.3 was added and the reaction was placed on ice till cold. Finally, the deacylated tRNAs were purified via two phenol extractions and one chloroform extraction followed by ethanol precipitation. The deacylated tRNAs were resuspended in 10mM KOAc pH 5.3 and stored in −80°C.
Charging of DYK tRNAs
25 U of deacylated total tRNAs were charged using 20 µL of DEAE purified S100 extract in a total volume of 200 µL with a buffer composed of 50 mM HEPES•KOH pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, 250 µM of each amino acid (D, Y, and K) and 4 mM ATP. This reaction was incubated at 37°C for 20 min. Then, 20 µL of 3M KOAc was added and the reaction was placed on ice till cold. Finally, two phenol extractions and one chloroform extraction were performed followed by ethanol precipitation. The charged DYK tRNAs were resuspended in 10mM KOAc pH 5.3 and stored in −80°C.
Preparation of Stalling Mix
A 50 µL reaction of 1 mM GTP, 1 mM ATP, 24 µM EF-Tu, 4 µM EF-G and 1 µL of Superase•In was made in translation buffer and incubated at 37°C for 15 min. 6.4 U of charged DYK tRNAs were added to the reaction and further incubated at 37°C for 5 min. The reaction was then placed on ice until cold and diluted with 350 µL of translation buffer containing 1 mM ATP and 1mM GTP. This stalling mix was flash frozen in 10 µL aliquots and stored in −80°C.
Preparation of Ribosome Stalled Complexes
One 10 µL aliquot of stalling mix was added to one 2 µL aliquot of ribosome initiation complexes and the resulting sample was incubated at 37°C for 7 min. 1 µL of 250 nM 5’handle was added to the reaction and further incubated at 37°C for 2 min and then at room temperature for 5 min. The sample was kept on ice for the rest of the day. A fresh sample of stalled complexes was made every day.
Assembly of Ribosome Stalled Complexes on Streptavidin Beads
20 µL of 0.1% solution of streptavidin coated polystyrene beads was made in translation buffer and vortexed at high speed for 30 min. To make the ‘sample beads’, 2 µl of the ribosome stalled complexes were deposited on 0.75 µl of 0.1 % streptavidin coated polystyrene beads (0.84 µm diameter) and incubated at room temperature for 5 min. ‘Handle beads’ were made by mixing 1 µl of 0.1 % streptavidin coated polystyrene beads with 1 µl of 50 nM 3’ handle. To form a tether, a ‘sample bead’ with ribosome-mRNA-5’handle complex was held in an optical trap and brought close to a ‘handle bead’ with the 3’ handle held in the other optical trap. Upon the hybridization of the 3’ overhang of the 3’ handle bead with the 3’ side of the mRNA, a tether was formed (Figure S1A). We then unfold the partially translated mRNA hairpin to verify that the ribosome is accurately stalled with the first valine codon in the P-site (Figure S1B). Then, the tethered stalled ribosome is held at either a constant force with force feedback (for experiments without fluorescence) or a semi-passive constant force with a range of 13–16 pN for high force or 5–7 pN for low force (experiments with fluorescence), and translation is restarted by the addition of a factor mix. The factor mix, composed of 1 mM ATP, 1 mM GTP, 400 µM valine amino acid, 2 µM Val-RS, 2 µM valine tRNA, 4 µM EF-Tu, and variable amount of EF-G (dependent on the experimental condition), is incubated at 37°C for 20 min to pre-charge valine tRNA. When, Cy3-EF-G was used, a cocktail composed of 4 mM Trolox, 1 mM 1,3,5,7-cyclooctatetraene, 1 mM p-nitrobenzyl alcohol, 0.8% (w/v) glucose, 300 µg/ml glucose oxidase, 40 µg/ml catalase and 12.5 mM ascorbic acid was added to the factor mix to prolong fluorophore and tether lifetimes. Catalase was pre-incubated with RNase-Out to suppress RNase activity. Factor mix was made fresh every 2 hours due to pH change caused by the activity glucose oxidase and catalase.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hidden Markov Model step finding analysis (non-fluorescence ribosome trajectories)
An automated algorithm was developed to detect one-codon steps while being robust against the main source of spurious noise, i.e. non-cooperative hairpin unzipping (such as the one seen in Figure S5A, middle trace around time = 20 s). We model the stepping process as follows: at any given time t, the ribosome may be at any codon position c in the hairpin (1 ≤ c ≤ cmax = 50); and some more base pairs (n ≥ 0) may additionally be open at the base of the hairpin. For practicality, we assume n < nmax = 10 (this assumption is discussed below). Assuming that the initial tether extension is d and the size of one base ssRNA is s, the tether extension at any given time is d + 2s × (3c + n) where, (3c + n) gives the total number of basepairs open. For now, we will assume that d and s are known, fixed values. The measurement noise can be approximated as normally distributed with a standard deviation σ, which is added to the actual tether extension.
At any given time point, one of four transitions may occur:
the ribosome advances by one codon (c → c + 1); this is associated by a decrease of n by 3 – if at least that many bases were opened – (n → max(n − 3,0)),
the ribosome does not advance (c → c), but the hairpin opens by one base (n → n + 1),
the ribosome does not advance (c → c), but the hairpin closes by one base – if at least one base is open (n → max(n − 1,0)),
neither c nor n change.
Let pfwd, popen, pclose, and 1 − pfwd − popen − pclose be the respective probabilities of the four events.
Overall, this setup fully describes a hidden Markov model with cmax × nmax states (each indexed by a pair (c, n)), with the structure of the transition matrix implied by the transitions listed above. The parameters pfwd, popen, pclose, and σ can fitted using the standard Baum-Welch algorithm (Rabiner, 1989). Note that the transition matrix, of size (cmaxnmax, cmaxnmax), is actually sparse —there are only 4cmaxnmax nonzero entries—; this important observation allows us to speed up the summations in our custom Cython (Behnel et al., 2011) Baum-Welch implementation, both in the forward-backward step and the update step, by skipping most summands.
In practice, it is difficult to a priori obtain d or s with high accuracy. Instead, we start with approximate ranges of estimates for them (d from the approximate position at the start of the trace, s from a freely-jointed chain estimate). We then repeat the above HMM fit with various values of d and s sampled in their respective ranges, and pick the (d, s) pair for which the HMM fit yields the highest final likelihood.
The output of the procedure is thus a (d, s) pair and a HMM fit, which includes in particular a γt(c, n) matrix (in the notation used by (Rabiner, 1989)) indicating the probability, at any time point, that the system is at a state (c, n). In practice, we do not care about the number of additional open base pairs; we thus marginalize γt over n (i.e., for each value of c, sum over all values of n) to obtain the probability γt(c) that the ribosome is at codon c at time t. We then select for each time t the codon c with the maximum probability.
However, there remain two issues with our approach. First, the HMM as described above yields, in practice, a clearly unsatisfactory fit, where the value of the codon position is always underestimated, and all fluctuations in the signal are assigned to fluctuations in the number of additional open base pairs. This arises because changes in n have better resolution than changes in c and can go both forward and backward; it is thus easier for the algorithm to adjust n than c). To avoid this effect, we force popen/pclose (at every iteration of the Baum-Welch algorithm) to a fixed ratio, estimated from the thermodynamic stability of the hairpin as computed by mfold (adjusting for the destabilization from the tension and the additional ribosome destabilization) (Qu et al., 2011; Zuker, 2003). In practice, this ratio is small (~0.1) and suitably penalizes excessive fluctuations in n.
Second, note that our estimate of c may not be equally good at every time point. In particular, when the number of additional open base pairs is much more than one codon, any reasonable knowledge of the actual ribosome position is essentially lost (this is the reason why we could assume n < nmax: there is little information in the time intervals where n ≥ 10 anyways). Such time intervals are characterized by the quantity being relatively small; in other words, there is a large probability that there is more than one codon worth’s of additional open base pairs. We chose to set a cutoff on this value: we excluded any region where γt(c, n < 3) < 1⁄2. Additionally, if additional basepairs are opened, we discard downstream steps depending on the number of basepairs opened. If n ≤ 3, we discard the current and the following step. If 4 ≤ n ≤ 6, we discard the current and the following two steps. If 7 ≤ n ≤ 9, we discard the current and the following three steps. Finally, if n > 9, we discard the current and the next four steps.
All high force data (force ≥ 10 pN) was downsampled to 133 Hz and all low force data (force ≤ 5 pN) was downsampled to 66 Hz, except data collected at 5 pN with 30 nM EF-G, where data had to be downsampled to 44 Hz for convergence of the fit. At high force, at least one to two points were required to establish a dwell and at low force five to seven points were required due to a decrease in signal to noise at low force.
Identification of transitions for fluorescence ribosome trajectories
Fluorescence and tweezers data was truncated to ± 5 seconds around the event of interest. Transitions in the data were fitted using the following two algorithms:
A classic two-state hidden Markov model (using hmmlearn, https://hmmlearn.readthedocs.io/en/0.2.0/).
The Pruned Exact Linear Time (PELT) segmentation method (Killick et al., 2012) (we used a custom Cython (Behnel et al., 2011) implementation of the algorithm), which efficiently finds, for a given time series, the best least-square fit to it by a stepwise constant function with a given number of steps. More accurately, PELT finds the best fit for a given per-step penalty; we used a dichotomic search to find the penalty that yielded a given number of steps.
Bi-exponential distribution fitting
All fits were made to cumulative distributions using Matlab’s inbuilt fit function. Bayesian information criterion was used to establish when a bi-exponential fit was better than a single exponential fit. Several initialization conditions were tested to ensure convergence of fits. All conditions converged to a single result despite different initialization condition except data collected at 5 pN with 10 µM EF-G. We constrain this fit by using the same kslow path as that obtained from 5 pN data at 1 µM EF-G since we know that kslow path is independent of EF-G concentration (Table S1). Fits were also performed by an orthogonal method, Maximum likelihood estimation, and the resulting fits were within error of current fits and no trends were changed. For 10 µM EF-G, we obtained n = 889 events from 24 molecules at 15 pN, n = 776 events from 19 molecules at 10 pN, n = 256 events from 8 molecules at 5 pN and n = 551 events from 19 molecules at 3 pN. For 1 µM EF-G, we obtained n = 747 events from 23 molecules at 15 pN and n = 403 events from 13 molecules at 5 pN. For 100 nM EF-G, we obtained n = 732 events from 21 molecules at 15 pN and n = 509 events from 18 molecules at 5 pN. For 30 nM EF-G, we obtained n = 333 events from 9 molecules at 15 pN and n = 304 events from 11 molecules at 5 pN. For fluorescence measurements, we obtained n = 55 events from 9 molecules at 13–16 pN and n = 62 events from 14 molecules at 5–7 pN.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SURE 2 Supercompetent Cells | Agilent Technologies | Cat # 200152 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ascorbic Acid | Sigma-Aldrich | Cat # A92902 |
| ATP solution | GE Healthcare | Cat # 45-001-34 |
| BsaI-HF | NEB | Cat # R3535 |
| Catalase | EMD Millipore | CAS # 9001-05-2 |
| Chloroform | Sigma-Aldrich | Cat # C2432 |
| Cy3 Maleimide Mono-Reactive | GE Lifesciences | Cat # PA23031 |
| Cyclooctatetraene | Sigma-Aldrich | Cat # 138924 |
| EcoR1-HF | NEB | Cat # R3101S |
| Glucose Oxidase from Aspergillus Niger | Sigma-Aldrich | Cat # G2133 |
| GTP solution | GE Healthcare | Cat # 45-001-345 |
| KpnI-HF | NEB | Cat # R3142S |
| L-Ascorbic Acid | Sigma-Aldrich | Cat # A92902 |
| Phenol solution | Sigma-Aldrich | Cat # P4557 |
| Phenol:Chloroform:Isoamyl Alcohol | Sigma-Aldrich | Cat # P2069 |
| p-Nitrobenzyl alcohol | Sigma-Aldrich | Cat # N6251 |
| RNaseOUT | Invitrogen | Cat # 10777019 |
| Superase•In | Invitrogen | Cat # AM2694 |
| Trolox | Sigma-Aldrich | Cat # 238813 |
| Valine tRNA | Sigma-Aldrich | Cat # R2645 |
| Critical Commercial Assays | ||
| MEGAscript® T7 Kit | Invitrogen | Cat # AM1333 |
| Oligonucleotides | ||
| 5’handle forward primer: /5Biosg/GGGCGACACGGAAATGTTGAATAC |
IDT | N/A |
| 5’handle reverse primer: 5’-GTCCATATGTATATCTCCTTC(revT or /iSpC3/)GAGCGCTTGTTTCGGCGTGG-3’ anneals to: 5’- GAAGGAGAUAUACAUAUGGAC −3’ |
IDT | N/A |
| 3’handle forward primer: 3’GCACATGTCTTGCGTTACTTAA-5’−5’- GAACGCAATGCGTCTGGGCGC-3’ anneals to: 5’- CGUGUACAGAACGCAAUGAAUU-3’ |
IDT | N/A |
| 3’handle reverse primer: /5Biosg/TTTTTCTAAATACATTCAAATATGTATCCG |
IDT | N/A |
| gBlock1: TAATACGACTCACTATAGGGCGAATTGGGTACCAGT GGCTGAGGCTTAACTAGTTCTAGAAATAATTTTGTTT AACTTTAAGAAGGAGATATACATATGGACTACAAGG ATGACGATGACAAGAAGGTGGTCGTGGTAGTAGTG GTTGTTGTGGTAGTCGTTGTTGTGGTGGTCGTAGTC GTGGTCGTGGTGGTTGTTGTAGTTGTTGTTGTTGTT GTAGTGGTTGTGGTAGTGGTAGTCGTGGTGGTTGTT GTAGTGGTTGTTGTAGTGGTGGTGGGCCACGCGGC GAAAGCCTCATTGGTCTCGTTTTGCGCGC |
IDT | N/A |
| gBlock2: TATATATTTTCGAGACCAATGAGGCTTTCGCCGCGT GGCCCACCACCACTACAACAACCACTACAACAACCA CCACGACTACCACTACCACAACCACTACAACAACAA CAACAACTACAACAACCACCACGACCACGACTACGA CCACCACAACAACGACTACCACAACAACCACTACTA CCACGACCACCGTGTACAGAACGCAATGAATTCGC GCGC |
IDT | N/A |
| Recombinant DNA | ||
| pBluescript SK+ | Stratagene | N/A |
| Software and Algorithms | ||
| Cython | (Behnel et al., 2011) | N/A |
| Matlab | Mathworks | N/A |
| Other | ||
| Econo-pac 10DG Desalting Column | Bio-rad | Cat # 7322010 |
| HiPrep 16/60 Sephacryl S-300 HR Column | GE Healthcare | Cat # 17-1167-01 |
| HiTrap Q HP Column | GE Healthcare | Cat # 17115401 |
| HisTrap HP Column | GE Healthcare | Cat # 17524801 |
| illustra MicroSpin G-25 Column | GE Healthcare | Cat # 27-5325-01 |
| Streptavidin Coated Beads (0.84 µm diameter) | Spherotech | Cat # SVP-08-10 |
| TSKgel Phenyl-5PW Column, Glass | Tosoh Bioscience GmbH | P/N # 0008804 |
Highlights.
mRNA hairpin is opened after EF-G binding, and prior to 30S head reverse rotation
EF-G catalyzed translocation is not affected by mRNA secondary structure stability
Ribosomes operate in two alternative (fast and slow) gears during translation
Increased hairpin stability increases the flux through the slow gear
ACKNOWLEDGEMENTS
We thank Thy Minh Thy Pham and William John van Patten for their help in preparation of reagents; Charles E. Wickersham, Prof. Matthew Comstock, and Prof. Taepjik Ha for their help in building the instrument; Lisa Alexander, Liang Meng Wee, Maya Sen, and Shannon Yan for useful discussions. This research was supported by the Howard Hughes Medical Institute (instrumentation and ribosome biochemistry); by the National Institutes of Health grants R01GM071552 and R01GM032543 (fluorescence experiments); and the Nanomachine program (KC1203) funded by the Office of Basic Energy Sciences of the U.S. Department of Energy (DOE) contract no. DE-AC02-05CH11231 (data analysis algorithms). F.F. acknowledges support from EMBO postdoctoral fellowship; M.R. acknowledges support from the Damon Runyon Cancer Research Foundation (DRG-2096-11).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The published article includes all relevant data generated or analyzed during this study. Additional data and codes are available from the corresponding author on request.
REFERENCES
- Behnel S, Bradshaw R, Citro C, Dalcin L, Seljebotn DS, and Smith K (2011). Cython: The Best of Both Worlds. Comput. Sci. Eng 13, 31–39. [Google Scholar]
- Belardinelli R, and Rodnina MV (2017). Effect of Fusidic Acid on the Kinetics of Molecular Motions During EF-G-Induced Translocation on the Ribosome. Sci. Rep 7(1):10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, and Rodnina MV (2016). Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol 23, 342–348. [DOI] [PubMed] [Google Scholar]
- Boon K, Vijgenboom E, Madsen LV, Talens A, Kraal B, and Bosch L (1992). Isolation and functional analysis of histidine◻tagged elongation factor Tu. Eur. J. Biochem 210, 177–183. [DOI] [PubMed] [Google Scholar]
- Borg A, Holm M, Shiroyama I, Hauryliuk V, Pavlov M, Sanyal S, and Ehrenberg M (2015). Fusidic Acid Targets Elongation Factor G in Several Stages of Translocation on the Bacterial Ribosome. J. Biol. Chem 290, 3440–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, and Grigorieff N (2013). Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. USA 110, 20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Cheng W, and Mejia YX (2011). Revisiting the Central Dogma One Molecule at a Time. Cell 145, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N, Katunin VI, Belardinelli R, Peske F, and Rodnina MV (2014). Programmed –1 Frameshifting by Kinetic Partitioning during Impeded Translocation. Cell 157, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N, Wohlgemuth I, Korniy N, Pearson M, Peske F, and Rodnina MV (2017). Conditional Switch between Frameshifting Regimes upon Translation of dnaX mRNA. Molecular Cell 66, 558–567.e4. [DOI] [PubMed] [Google Scholar]
- Chaney WG, and Morris AJ (1978). Nonuniform size distribution of nascent peptides: The role of messenger RNA. Arch. Biochem. Biophys 191, 734–741. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Huttelmaier S, Donato D, and Singer RH (2002). Asymmetric Sorting of Ash1p in Yeast Results from Inhibition of Translation by Localization Elements in the mRNA. Mol. Cell 10, 1319–1330. [DOI] [PubMed] [Google Scholar]
- Chemla YR (2010). Revealing the base pair stepping dynamics of nucleic acid motor proteins with optical traps. Phys. Chem. Chem. Phys 12, 3080–3095. [DOI] [PubMed] [Google Scholar]
- Chen C, Cui X, Beausang JF, Zhang H, Farrell I, Cooperman BS, and Goldman YE (2016). Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. USA 113, 7515–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS, and Goldman YE (2013a). Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol 20, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE, and Puglisi JD (2014). Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O’Leary SE, and Puglisi JD (2013b). Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol 20, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Feng S, Kumar V, Ero R, and Gao Y-G (2013c). Structure of EF-G-ribosome complex in a pretranslocation state. Nat. Struct. Mol. Biol 20, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Comstock MJ, Ha T, and Chemla YR (2011). Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods 8, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, and Ha T (2008). Spontaneous Intersubunit Rotation in Single Ribosomes. Mol. Cell 30, 578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, and Assmann SM (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, and Gejman PV (2003). Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet 12, 205–216. [DOI] [PubMed] [Google Scholar]
- Ermolenko DN, and Noller HF (2011). mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol 18, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, and Agrawal RK (2000). A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322. [DOI] [PubMed] [Google Scholar]
- Guisez Y, Robbens J, Remaut E, and Fiers W (1993). Folding of the MS2 Coat Protein in Escherichia coli is Modulated by Translational Pauses Resulting from mRNA Secondary Structure and Codon Usage: A Hypothesis. J. Theor. Biol 162, 243–252. [DOI] [PubMed] [Google Scholar]
- Guo Z, and Noller HF (2012). Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA 109, 20391–20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DMJ, and Ha T (2007). Fluorescence-Force Spectroscopy Maps Two-Dimensional Reaction Landscape of the Holliday Junction. Science 318, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W, Kokic G, Jager M, Mittelstaet J, Komar AA, and Rodnina MV (2015). Cotranslational protein folding on the ribosome monitored in real time. Science 350, 1104–1107. [DOI] [PubMed] [Google Scholar]
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, and Varmus HE (1988). Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331, 280–283. [DOI] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, and Ha T (2008). Advances in Single-Molecule Fluorescence Methods for Molecular Biology. Annu. Rev. Biochem 77, 51–76. [DOI] [PubMed] [Google Scholar]
- Katz L, and Burge CB (2003). Widespread Selection for Local RNA Secondary Structure in Coding Regions of Bacterial Genes. Genome Res. 13, 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, and Segal E (2010). Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R, Fearnhead P, and Eckley IA (2012). Optimal Detection of Changepoints With a Linear Computational Cost. J. Am. Stat. Assoc 107, 1590–1598. [Google Scholar]
- Kim H-K, Liu F, Fei J, Bustamante C, Gonzalez RL, and Tinoco I (2014). A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. USA 111, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, and Noller HF (2005). Involvement of 16S rRNA Nucleotides G1338 and A1339 in Discrimination of Initiator tRNA. Mol. Cell 20, 623–632. [DOI] [PubMed] [Google Scholar]
- Lang MJ, Fordyce PM, Engh AM, Neuman KC, and Block SM (2004). Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods 1, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. (2012). Global Analysis of RNA Secondary Structure in Two Metazoans. Cell Rep 1, 69–82. [DOI] [PubMed] [Google Scholar]
- Liu T, Kaplan A, Alexander L, Yan S, Wen J-D, Lancaster L, Wickersham CE, Fredrick K, Noller H, Tinoco IJ, et al. (2014). Direct measurement of the mechanical work during translocation by the ribosome. eLife 3, e04563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, and Noller HF (1989a). Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57, 585–597. [DOI] [PubMed] [Google Scholar]
- Moazed D, and Noller HF (1989b). Intermediate States in the Movement of Transfer-RNA in the Ribosome. Nature 342, 142–148. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, and Bustamante C (2006). Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA 103, 9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Smith SB, and Bustamante C (2008). Recent Advances in Optical Tweezers. Annu. Rev. Biochem 77, 205–228. [DOI] [PubMed] [Google Scholar]
- Mohan S, Donohue JP, and Noller HF (2014). Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. USA 111, 13325–13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, and Weeks KM (2018). Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 173, 181–195.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, and Diatchenko L (2016). Human Catechol- O-Methyltransferase Haplotypes Modulate Protein Expression by Altering mRNA Secondary Structure. Science 314, 1930–1933. [DOI] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Zhou J, and Mohan S (2017). The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol 24, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, and Ehrenberg M (1996). Rate of Translation of Natural mRNAs in an Optimized in Vitro System. Arch. Biochem. Biophys 328, 9–16. [DOI] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, and Wintermeyer W (2004). Conformational Changes of the Small Ribosomal Subunit During Elongation Factor G-dependent tRNA–mRNA Translocation. Journal of Molecular Biology 343, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Pulk A, and Cate JHD (2013). Control of Ribosomal Subunit Rotation by Elongation Factor G. Science 340, 1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wen J-D, Lancaster L, Noller HF, Bustamante C, and Tinoco I (2011). The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner LR (1989). A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77, 257–286. [Google Scholar]
- Ramrath DJF, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, and Spahn CMT (2013). Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. USA 110, 20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJF, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, et al. (2012). The complex of tmRNA–SmpB and EF-G on translocating ribosomes. Nature 485, 526–529. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, et al. (2010). Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, and Wintermeyer W (1997). Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37. [DOI] [PubMed] [Google Scholar]
- Ryu WS, Berry RM, and Berg HC (2000). Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, and Ramakrishnan V (2009). What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242. [DOI] [PubMed] [Google Scholar]
- Sharma H, Adio S, Senyushkina T, Belardinelli R, Peske F, and Rodnina MV (2016). Kinetics of Spontaneous and EF-G-Accelerated Rotation of Ribosomal Subunits. Cell Rep 16, 2187–2196. [DOI] [PubMed] [Google Scholar]
- Sivak DA, and Crooks GE (2016). Thermodynamic geometry of minimum-dissipation driven barrier crossing. Phys. Rev. E 94, 052106. [DOI] [PubMed] [Google Scholar]
- Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, and Korostelev AA (2014). Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations. Structure 22, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafoya S, Large S, Liu S, Bustamante C, and Sivak D (2018). Using a System’s Equilibrium Behavior to Reduce Its Energy Dissipation in Non-Equilibrium Processes. bioRxiv 291989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, and Noller HF (2005). mRNA Helicase Activity of the Ribosome. Cell 120, 49–58. [DOI] [PubMed] [Google Scholar]
- Tinoco I Jr & Bustamante C (2002). The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem 101–102, 513–533. [DOI] [PubMed] [Google Scholar]
- Tourigny DS, Fernández IS, Kelley AC, and Ramakrishnan V (2013). Elongation Factor G Bound to the Ribosome in an Intermediate State of Translocation. Science 340, 1235490–1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Mizushima S, Lowry CV, and Nomura M (1981). In RNA and Protein Synthesis, Moldave K, ed. (New York: Academic Press-Elsevier; ), pp.521–538. [Google Scholar]
- Tsuchihashi Z (1991). Translational frame shifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res 19, 2457–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mameren J, Peterman EJG, and Wuite GJL (2008). See me, feel me: methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Res 36, 4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman MR, Alejo JL, Altman RB, and Blanchard SC (2016). Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol 23, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, and Weeks KM (2009). Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, Noller HF, Bustamante C, and Tinoco I (2008). Following translation by single ribosomes one codon at a time. Nature 452, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley KD, Comstock MJ, and Chemla YR (2017). High-Resolution “Fleezers”: Dual-Trap Optical Tweezers Combined with Single-Molecule Fluorescence Detection In Optical Tweezers, Gennerich A, ed. (New York: Humana Press-Springer Science+Business Media; ), pp. 183–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KS, and Noller HF (1998). Mapping the Position of Translational Elongation Factor EF-G in the Ribosome by Directed Hydroxyl Radical Probing. Cell 92, 131–139. [DOI] [PubMed] [Google Scholar]
- Yan S, Wen J-D, Bustamante C, and Tinoco I (2015). Ribosome Excursions during mRNA Translocation Mediate Broad Branching of Frameshift Pathways. Cell 160, 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, and Andrews DW (1996). The signal recognition particle receptor alpha subunit assembles co◻translationally on the endoplasmic reticulum membrane during an mRNA◻ encoded translation pause in vitro. EMBO J 15, 172–181. [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JHD, and Noller HF (2001). The Path of Messenger RNA through the Ribosome. Cell 106, 233–241. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yan K, Liu G, Song G, Luo J, Shi Y, Cheng E, Wu S, Jiang T, Lou J, et al. (2016). EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome. Nat. Struct. Mol. Biol 23, 125–131. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, and Cate JHD (2009). Structures of the Ribosome in Intermediate States of Ratcheting. Science 325, 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, and Noller HF (2013). Crystal Structures of EF-G–Ribosome Complexes Trapped in Intermediate States of Translocation. Science 340, 1236086–1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, and Noller HF (2014). How the ribosome hands the A-site tRNA to the P site during EF-G–catalyzed translocation. Science 345, 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.