Abstract
Liquid–liquid phase separation (LLPS) facilitates the formation of condensed biological assemblies with well-delineated physical boundaries, but without lipid membrane barriers. LLPS is increasingly recognized as a common mechanism for cells to organize and maintain different cellular compartments in addition to classical membrane-delimited organelles. Membraneless condensates have many distinct features that are not present in membrane-delimited organelles and that are likely indispensable for the viability and function of living cells. Malformation of membraneless condensates is increasingly linked to human diseases. In this review, we summarize commonly used methods to investigate various forms of LLPS occurring both in 3D aqueous solution and on 2D membrane bilayers, such as LLPS condensates arising from intrinsically disordered proteins or structured modular protein domains. We then discuss, in the context of comparisons with membrane-delimited organelles, the potential functional implications of membraneless condensate formation in cells. We close by highlighting some challenges in the field devoted to studying LLPS-mediated membraneless condensate formation.
Keywords: protein/protein interaction, cell signaling, cell biology, synapse, cellular regulation, biological condensates, Phase separation
Introduction
In eukaryotic cells, reaction components are spatiotemporally compartmentalized so that materials are concentrated and activities are localized and protected from damaging activities, such as proteolysis, changes in pH, and undesired covalent modifications. Classical organelles are membrane-enclosed where the lipid bilayer provides a physical barrier to separate their interior contents from the exterior environment. Examples include Golgi apparatus, mitochondria, and endoplasmic reticulum (ER).3 However, many organelles are not membrane-enclosed (often referred to as membraneless compartments in the literature), and such organelles include but are not limited to germ granules, stress granules, nucleoli, centrosomes, and synapses in neurons. In these membraneless compartments, due to the lack of physical separation, molecules can freely exchange with their counterparts in the surrounding bulk solution. Sharp concentration gradients are maintained between the proteinaceous (and sometimes protein and nucleic acid mixtures) interior and the much more diluted exterior. Reaction machineries can reversibly assemble and disassemble within a short time window, as fast as a few seconds. Reaction constituents can be integrated or removed to control specific activities. While recognized for many years, the mechanisms governing the formation of membraneless organelles have remained unclear until about 10 years ago. The first direct experimental evidence came from the study of P granules in germ cells of Caenorhabditis elegans (1). P granule is a collection of RNA and RNA-binding proteins (RBPs) localized at the posterior cortex of a dividing embryo. P granules appear as spherical droplets with liquid-like properties, and they fuse with one another, deform under shear stress, and flow off the surface of the nucleus. Fluorescence recovery after photobleaching (FRAP) analysis demonstrated rapid turnover rates of constituent proteins, which is indicative of fast molecular rearrangements. These observations together suggested that P granules form through liquid–liquid phase separation (LLPS), distinct from canonical macromolecular assemblies. Since then, the list of membraneless organelles that are organized by LLPS has been ever-growing. Nevertheless, early concerns had been raised over the specificity of phase-separated condensates observed in vitro and their biological significance in vivo. Comprehensive studies were followed to show that the concept of phase separation can help to explain the formation and organization of non-membrane-bound biomolecular compartments as well as their physical and material properties that cannot be understood with the classical physical chemistry theories for dilute solutions. It now comes to the realization that LLPS might be a general mechanism to drive compartmentalization in the absence of lipid bilayers (2–7), and this has greatly motivated the field to re-investigate mechanisms underlying formation and functional implications of membraneless organelles from new perspectives. However, caution should be exercised that not all condensed phase properties observed in vitro can be extrapolated to living cells. Rigorous characterizations, both in vitro and in vivo, are required to demonstrate the existence of LLPS of a particular biological system under physiological conditions. Here, we first review our understanding about the molecular codes that contribute to LLPS formation. We discuss some of the common techniques for characterization of LLPS. We then discuss the functional implications of LLPS-driven organelle assemblies. Finally, we propose a few new potential research directions inspired by current works on LLPS.
What is phase separation?
By definition, phase separation refers to the immiscibility of two solutions whereby they separate into two states. In biological systems, this often leads to a large volume of dilute liquid phase and a small volume of concentrated liquid phase. In biology, phase separation is not unheard of. In protein crystallization, when a crystallization reagent is added to the protein solution oil-like droplets can be observed to separate from the bulk solution. LLPS in this case is indicative of a metastable transition state from which crystals may grow by changing temperature, precipitant concentration, protein concentration, etc. Molecules are miscible in solution until reaching their solubility limit. Phase separation happens when the macromolecule/macromolecule or solute/solute interactions are energetically favored over the macromolecule/solute interactions, and the gain in free energies is favored over its loss in entropic tendency toward a homogeneous solution state (6, 8, 9). A free energy minimum is then reached, but the two phases with different solute concentrations are at the same Gibbs free energy (4). For each molecular system, a phase diagram can be constructed by systematically screening through conditions such as temperature, salt concentration, pH, or macromolecular concentration. Phase diagram helps one to identify conditions that promote phase separation and to determine the likelihood of phase separation happening under physiological conditions (Fig. 1A). Phase boundary, which is defined by the binodal line, indicates the boundary that two distinct phases stably co-exist in solution. Outside the binodal curve, molecules stay as homogeneous mixtures. Between the binodal and spinodal curves lies a metastable region where liquid demixes via a nucleation process. Within the spinodal zone, spinodal decomposition occurs. In the other words, spontaneous phase separation takes place in the spinodal zone where molecules rapidly transit from a less stable region to a more stable phase-separated region bypassing the metastable nucleation zone.
Figure 1.
Types of multivalent interactions driven by intrinsically disordered elements in LLPS systems. A, phase diagram constructed by varying protein concentration and storage conditions such as buffer reagents and temperature. Solid line depicts the boundary at which molecules reach their solubility limit and become immiscible with the surrounding solution. Gray box highlights the confocal image showing the homogeneous solution state of the NR2B C-terminal tail (labeled with Alexa Cy3) in the absence of PSD scaffold proteins. Conditions within the spinodal curve (indicated as dashed line) are where spinodal decomposition occurs. Example of the fluorescence image, highlighted by the green box, shows that the membrane-tethered NR2B tail (labeled with Alexa Cy5) formed clusters on supported lipid bilayers upon the addition of major PSD scaffold proteins. Phase separation is only observed in the presence of a nucleation process when conditions lie in between the binodal (indicated as solid line) and spinodal curves. Representative image, highlighted by the yellow box, shows the clustered state of the NR2B tail (labeled with Alexa Cy3) in 3D solution in the presence of major PSD scaffold proteins (adapted from Ref. 45). Scale bar, 10 μm. B, aromatic residues in intrinsic disorder containing proteins are involved in π–π or cation–π interactions with positively charged residues such as Arg and Lys. RGG repeats are frequently found in LCRs. C, patterned charge distributions to facilitate electrostatic interactions between oppositely charged residues. D, secondary structural elements are involved in multivalent intermolecular interactions, such as the kinked cross–β-sheets formed by a segment of FUS LCR (PDB code 6BWZ).
Multivalency is a key determining factor underlying LLPS in biological systems (10). Molecules can undergo inter- or intramolecular interactions to assemble into oligomers or polymers that tend to have a lowered solubility limit and thus are more likely to demix with the surrounding solution. In a folded domain protein, multiple binding sites, either for itself or for its binding partners, promote phase separation (Fig. 2). In proteins with a higher content of intrinsic disorder, multivalent weakly self-attracting interactions collectively drive phase separation (Fig. 1, B–D). Aggregations of misfolded cytosolic or nuclear proteins have been associated with a broad range of neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis (ALS) (11–13). Solid fibrils formed by disordered proteins represent another form of phase separation–solution to solid transition. Pathological aggregation and its close link to brain diseases are discussed in several recent reviews (14–19). For the scope of this review, we focus on recent works on LLPS. Below, we discuss the sequence properties coding for LLPS, the biological functions of condensate formation, and the technical developments for in vitro characterization of phase separation systems.
Figure 2.
Types of multivalent interactions driven by modular domains in LLPS systems. A, interaction network of N-WASP, nephrin, and NCK. B, schematic representations showing the network of multivalent interactions involving major PSD proteins. Solid line indicates direct modular domain interactions. Dashed line indicates indirect recruitment of actin filaments via Shank3 and Homer proteins. C, schematic interaction network of presynaptic active-zone proteins RIM and RIM-binding protein together with the cytoplasmic tail of the N-type voltage-gated Ca2+ channel (NCav).
Codes for LLPS
Tremendous progress has been made over the past decade in trying to understand the molecular features in common that drive phase separation. In this section, we discuss examples of phase separation promoted by intrinsically disordered sequences or more specifically folded domain/target interactions.
Multivalency driven by intrinsic disordered sequences
Intrinsically-disordered proteins represent an abundant class of proteins involved in phase separation. Low-complexity regions (LCRs) show biased amino acid preferences including Gly, Ser, Asn, Glu, Phe, and Tyr. These amino acids often appear in repeats, such as RG, FG, and YG, that are important for forming ribonucleoprotein (RNP) granules in P bodies, P granules, stress granules in cytoplasm or nucleoli, and paraspeckles in the nucleus (20–25). The lack of a defined three-dimensional (3D) structure in LCRs favors weakly adhesive interactions that drive phase separation. A good example that was studied in detail is Fused in Sarcoma (FUS) proteins. Full-length FUS proteins have been shown to undergo LLPS at close to physiological concentrations in the presence of crowding reagent or when cooled to 4 °C (26, 27). A gel-like state is observed when FUS protein concentration reaches hundreds of micromolars, well-above its physiological concentration. FUS LCR alone can assemble into hydrogels at sub-millimolar concentrations (28), and the hydrogels trap and retain the LC domains of other RBPs such as hnRNPA1, Sup35, TIA1, and TDP43, although to different levels. The liquid droplets of FUS protein can further mature into fibrous aggregates resembling the pathological protein fibers found in ALS patients. Mutations in the prion-like domain, which induce the early onset of ALS, further promote the solution-solid transition. Tremendous progress has been made in recent years in revealing the emergent sequence determinants in LCR that promote phase separation. Noticeably, the types of interactions critical for phase separation are commonly known to drive protein folding or interactions. We discuss below how these sequence features can drive molecular interactions in new ways.
Intrinsically-disordered proteins rich in aromatic residues are favored to form π–π stacking interactions that can drive phase separation (Fig. 1B) (25, 28–30). Mutation of these aromatic residues to serine can strongly decrease the amount of protein enrichment into a condensed phase. Apart from side-chain π–π interactions, small residues with relatively exposed backbone peptide bonds can also form the so-named planar π interactions (31). Gly, Ser, Thr, and Pro residues are indeed frequently found in LCRs of RBPs. RG/RGG repeats are also found in multiple LCR-containing proteins such as in the nuage protein DEAD-box helicase 4 (Ddx4) (25), the P-granule protein LAF-1 (22), and the neuronal granule protein Fragile X mental retardation protein (FMRP) (32, 33). Arginine can form cation–π interactions with aromatic residues, either intramolecularly or intermolecularly (Fig. 1B). An increase in the number of cation–π interactions by arginine substitutions in FUS protein, for example, can significantly promote its ability to phase separate and lower the threshold concentration of the solution-gel transition (30, 34). Conversely, substitution of arginine and tyrosine or phenylalanine with alanine disrupts cation–π interactions and consequently the ability to phase separate. Similarly, arginine methylation in Ddx4 (25), hnRNPA2 (35), and FMRP (33) reduces or abolishes their phase separation likely because of the weakened intermolecular cation–π interactions. Charged residues also contribute to droplet formation both in vitro and in vivo (Fig. 1C). Ddx4, for example, contains blocks of net negative or positive charges, typically 8–10 residues in length with 3–8 charged residues (25). Interestingly, these charge blocks appear in clusters with alternating positive and negative charge distributions. Removal of such opposite charge patterning inhibited Ddx4 phase separation. Apart from amino acid side-chain interactions, phase separation can be driven by secondary structural elements. Examples include a short, evolutionarily-conserved helical segment in TDP-43 C-terminal domain involved in intermolecular helical interactions (36) and a 57-residue segment in FUS LCR involved in the formation of cross–β-sheets stabilized by hydrogen bonding and π–π stacking interactions (37). Recent crystallographic studies of LCRs in FUS, hnRNPA1, and nup98 revealed another type of interaction between secondary structural elements that drive phase separation. These regions are highly abundant in aromatic residues, which are involved in inter- and intra-sheet stabilizations. In addition, the LCRs form kinked β-sheets to allow close encountering of the backbones for hydrogen bonding or van der Waals interactions and subsequently to stabilize the packing of neighboring β-sheets. Such regions are therefore referred to as low-complexity aromatic-rich kinked segments (Fig. 1D) (38).
Based on current knowledge of the relationship between emerging sequence features and phase separation, a spectrum of predictive tools has been developed to enable researchers to identify regions in intrinsically disordered proteins that might be involved in LLPS and to understand the molecular mechanisms behind solution-solution/gel transitions. This has been extensively discussed in a review written by Alberti et al. (3)
Multivalent interactions driven by defined modular protein domains
Experiments to manipulate the valency of a folded protein have proven an inverse correlation between the number of binding domains or motifs and the saturation concentration above which the system undergoes phase separation. For instance, repeats of SH3 domain bind Pro-rich motifs (PRMs) and phase separate into condensed droplets upon mixture at high concentration (10). The phase boundary (i.e. the threshold concentration for LLPS) is lowered when the number of binding modules increases suggesting that LLPS is strongly dependent on the valency of interactions. Similarly, multivalent nucleic acid/protein interaction systems are known to undergo LLPS both in vitro and in cells when certain critical numbers of valency are reached (39, 40). There are now many examples of phase separation systems driven by modular domain interactions. One example is the multivalent protein network involving the transmembrane protein nephrin, the adaptor protein NCK and its ligand N-WASP that regulate actin assembly in podocytes of kidney (Fig. 2A) (10, 41). NCK contains three SH3 domains, each of which can bind to the six PRMs in N-WASP; two proteins assemble into higher-order oligomers that phase separate. This process is accelerated by nephrin addition where phosphotyrosine (pTyr) residues in nephrin bind to SH2 domains in NCK. The assembled droplets can further recruit Arp2/3 complexes for actin polymerization. An analogous system is observed in T-cell receptor signaling (42). LAT, a transmembrane protein for T-cell activation, is phosphorylated at multiple Tyr sites that are required for T-cell signaling. Addition of Grb2, an adaptor protein, and Sos1, a guanine nucleotide exchange factor for Ras GTPase, caused phase separation of pLAT through interaction between pTyr in LAT and the SH2 domain in Grb2 and between SH3 domains in Grb2 and PRMs in Sos1. The number of pTyr residues affects binding valency in the system and consequently the efficiency of receptor clustering on the supported lipid bilayer. In both systems, properties of reconstituted condensates in vitro strongly correlate with the observations in living cells.
In neurons, synapses assemble between axons and dendrites. Beneath the postsynaptic membranes, there lies an electron-dense layer of material known as the postsynaptic density (PSD). Addition of SynGAP, a negative PSD activity regulator, to the major PSD scaffold protein PSD-95 caused droplet formation (43). Phase separation was completely abrogated if the interaction interface or SynGAP trimer interface was impaired. PSD-95 also clustered with SAPAP, Shank, and Homer, which are major PSD scaffold proteins, but with much higher efficiency compared with SynGAP (Fig. 2B) (44). This increased propensity to form droplets is likely because of increasing valency provided by multivalent interaction interfaces among PSD constituents. Importantly, all of the interactions involved in forming the PSD protein network are highly specific and with strong affinities, and these interactions involve well-folded protein-binding domains (Fig. 2B). Strikingly, the assembly of PSD droplets was dispersed when Homer1a, a monomeric splice variant of Homer1, was added to the pre-assembled condensates (44). This suggests Homer1 oligomerization plays a crucial role in promoting LLPS. Reconstituted PSD condensates can further cluster the cytoplasmic tail of N-methyl-d-aspartic acid receptor subunit and nucleate actin polymerization both in solution and on the supported lipid bilayer.
The presynaptic active zone is also organized by phase separation (45). As viewed under an electron microscope, the active zone comprises densely-packed proteins, which organize into a layer of electron-dense projection below the presynaptic membranes. RIM and RIM-binding protein, two major active-zone scaffold proteins, formed liquid droplets upon mixing. RIM-binding protein contains three SH3 domains, each of which binds to Pro-rich motifs in RIM (Fig. 2C). The cytoplasmic tail of voltage-gated Ca2+ channel (NCav) is also enriched into RIM/RIM-binding protein condensates, and this co-clustering significantly lowers the threshold concentration to undergo LLPS. When the NCav C-terminal tail is attached to membrane, RIM, RIM-binding protein, and NCav co-cluster on supported lipid bilayer, providing a mechanistic explanation to the tight coupling of Ca2+ influx and neurotransmitter release in presynaptic termini.
In addition to these systems, aggregation of Rubisco by the protein CcmM (46) and interaction between the tetravalent RNA-binding protein PTB and an RNA oligonucleotide (10, 47) are also shown to phase separate through multivalent folded domain interactions. In RBPs, phase separation can be driven by modular domain interactions apart from those weak, self-adhesive interactions. In particular, RGG repeats and RNA recognition motifs (RRMs) are involved in RNA binding. Although binding between individual repeats and RNA is relatively weak, multiple RGG repeats together generate a high-affinity interaction and cross-link proteins and RNAs into higher-order oligomers (33, 36, 48–50). Repeats of RRMs on RBPs and multiple RRM-binding sites on RNA provide a further degree of multivalency to promote phase separation. Addition of RNA to hnRNPA1 caused formation of liquid droplets (24). In other cases, RNA is readily recruited to the pre-assembled liquid droplets or hydrogels via charge/charge interactions (33, 51–54). It appears that intrinsically disordered regions and high-affinity domain interactions can both contribute to phase separation. In reality, it is likely that both types of multivalency are coupled to promote droplet formation in many systems.
Methods for observation of phase separation
When it comes to work with phase separation in vitro, multiple approaches are generally combined to describe the phenomenon of phase separation, to distinguish it from nonphysiologically relevant aggregations or simple binding-induced molecular assemblies, and probably most importantly, to link to its biological functions in cells.
3D solution system
The most intuitive observation of phase separation in a test tube may be the turbidity and opalescence of a solution when components are mixed under certain conditions (protein concentration, stoichiometry, salt concentration, pH, temperature, etc.). Such simple sedimentation assay can be used to quantitatively evaluate the fractional distribution of proteins in each phase (see Ref. 43 for an example). A transparent “pellet” observed after centrifugation implies a liquid phase rather than aggregates or precipitates. Alternatively, direct measurement of the turbidity (46, 55, 56) is also helpful for estimating the extent of phase separation. Practically, due to the non-negligible gravity of the condensed phase droplets, researchers should be cautious about the heterogeneity within a final solution, and two experimental setups must be considered as follows: 1) a vertical, instead of horizontal, light beam is recommended for absorbance measurement to gain a more reliable result; 2) a sample solution needs to be vortexed immediately before the measurement to ensure a homogeneous suspension.
Imaging assays are necessary to confirm the liquid-like properties of the condensed phase. Differential interference contrast (DIC) imaging is the most straightforward method to depict the coexistence of two (or more) distinctive phases (Fig. 3A). The spherical morphology, fusion upon contact, and droplet fission, as well as the deformation of droplets under shear forces, together demonstrate the liquid-like properties of the condensed phase (Fig. 3, B–D) (1, 4, 6). Combining with fluorophore labeling, either co-localization or the coexistence of sub-compartments can be visualized (Fig. 3A) (57). However, we should always be alert for potential imaging artifacts and should not merely rely on imaging assays for the following reasons. 1) Cross-talk between multiple channels is not easy to be completely blocked, and it will always give an image when high laser power and long exposure times are applied. Thus, we recommend using single fluorophore labeling, if possible, in a given system except for co-localization or other necessary conditions. 2) The conjugation of a fluorophore (or genetically-encoded fluorescent tag) may affect the properties of labeled protein, and relatively subtle impact may be augmented in a much more concentrated phase. In addition, overexposure of high percentage labeled protein may cause photobleaching of the fluorophore under continuous laser power; therefore, we recommend researchers to dilute the labeled protein with an unlabeled one to achieve sparse labeling (usually, ∼1% is sufficient). It is helpful to use a sedimentation assay with labeled protein to check the potential effect of labeling on its ability to phase separate. 3) It is hard to accurately judge whether a protein of interest is specifically retained in the condensed phase, especially when the fluorescence contrast to the surrounding environment is not high enough. Because the matrix pore size (i.e. the void space between the protein network mesh) of the condensed phase is deemed to be large enough to accommodate normal protein, as indicated by our recent observation that the dodecameric CaMKII (∼550 kDa) can penetrate the reconstituted condensed PSD phase in vitro (44), it is not surprising that even some irrelevant proteins can go through but do not prefer the condensed phase. Nonetheless, fluorescent imaging can provide valuable information with rigorous controls. Recently, we developed an absolute concentration estimation method based on measured fluorescence intensity (Fig. 3E) (44). A standard curve is first constructed by plotting the fluorescence of protein measured at known concentrations. Based on this curve, we can then back-calculate the exact concentration of components in a condensed phase, even though the concentration in the surrounding dilute phase cannot be precisely estimated due to the very-low signal-to-noise ratio. Protein concentration in the dilute phase may be determined taking into account its fractional distribution observed in sedimentation assay. Concentration ratio and therefore volume ratio between condensed and dilute phases can ultimately be estimated (45).
Figure 3.
Techniques for characterizing the condensed phase formed in 3D solution. A, DIC (left image) coupled with fluorescence imaging (middle and right images) of the phase droplets and multiple labeling of different components demonstrate their co-localization. B–D, dynamic properties of the condensed phase. E, fluorescence intensity–based absolute concentration estimation (adapted from Refs. 44, 45). A standard curve of fluorescence intensity to dye concentration is initially generated for calibration. Z direction scanning is performed to determine the proper focal plane for concentration estimation. At each Z stack, the fluorescence intensity distribution is plotted. Within the Z dimension of selected droplets, average fluorescence intensities are then compared across different layers. In a given system, the fluorescence intensity is constant regardless of the droplet size, and therefore the absolute protein concentration within a condensed phase can be calculated from the standard curve.
FRAP analysis is increasingly adopted to demonstrate the mobility and dynamics of molecules within liquid droplets (Fig. 3C). Molecule exchange within the condensed phase (half-bleach) or exchange between the condensed and diluted phases (half-/whole-bleach) can be faithfully captured by FRAP experiments. However, we should be cautious in assessing the fitting of fluorescence recovery curves because it can always give us a result regardless of whether the model is appropriate or not. Fluorescence recovery depends on the movement rates of molecules, but this “movement” consists of diffusion (in the dilute phase, condensed phase, and the interface) and interaction (dissociation koff and association kon). It is therefore difficult to derive an exact value of either characteristic diffusion rate constant (τ) or diffusion coefficient (D) before figuring out a theoretical model, even though the apparent τ and D provide certain referential meanings. Besides, a less mobile or immobile fraction may occur, and a second bleach immediately after the plateau of the first bleach is suggested to confirm this. The immobile fraction can be generated from a systematic background, which is an intrinsic property or is produced during imaging time by rapid hardening/aging. In some systems, the phase droplets are initially fluid, but their viscoelasticity increases over the time, and molecules eventually cannot exchange with their counterparts in the surrounding solution. This process is known as hardening/aging, although the mechanism behind it is currently unknown.
Apart from direct visualization, several techniques have recently been brought into the phase separation field to describe the material properties of biological condensates. The isolated droplets make it possible to monitor the material properties of individual phase via atomic force microscopy (AFM), from which the stiffness, viscosity, elasticity, pore size, and other soft material parameters can be quantitatively extracted. This information will provide useful insights into the behavior of biological condensates in cells. For example, AFM measurements of the mechanical properties of PSD droplets indicate that the six-component PSD condensate is more gel-like compared with the two-component PSD condensates reconstituted in vitro (44). It is thus reasonable to speculate that under physiological conditions PSD may be a more gel-like structure due to the more complex valences and more crowded environment, which fits well to the previous electron microscopy (EM) observations (58). Measured material properties also provide explanations toward the observed protein dynamics in vitro. The time-dependent hardening indicated by elastic modulus values suggested an aging process of reconstituted PSD condensates, consistent with the observation that PSD constituents demonstrated a time-dependent decreasing of the signal recovery in FRAP analysis (44).
How are molecules organized within the condensed phase? Researchers show great interest toward understanding the atomic details of the condensed phase. The intrinsic heterogeneity, highly-dynamic properties, and the numerous transient interactions existing in the liquid-like phase make it extremely difficult to obtain structural information (59). Nevertheless, protein concentrations within the condensed phase remain the same in a given condition (pH, T, salt, etc.) (3, 44), bringing hope to acquire some configuration rules from structural studies. A recent study combing cryo-EM and cryo-electron tomography (cryo-ET) to solve the structure of the Rubisco–CcmM complex under LLPS conditions may give us some inspiration (46). The cryo-ET analysis of clusters of Rubisco complexes revealed that the median nearest-neighbor distance is around 150 Å, and the linker region sequesters two complex modules, which makes it possible to solve the complex structure within the condensed phase by single particle cryo-EM.
2D membrane system
Signal transduction between cells cannot skip over membranes. Supported lipid bilayer has been a popular working model to mimic cell membrane in vitro for years (60). People have noticed a large number of membrane proteins such as adhesion molecules, receptors, and channels that are required to be assembled/enriched/clustered together to transmit signals, but conventional protein/protein interactions can hardly elucidate the coupling principle until phase separation came into sight. Reconstitution of transmembrane protein clustering on the supported lipid bilayer is important for studying the mechanism of formation and the functional consequences of these microclusters (Fig. 4A). A recent review by Case et al. (61) has summarized the significance of LLPS in transmembrane signaling. Here, we discuss some applications and their caveats when dealing with the supported lipid bilayer in phase separation systems.
Figure 4.
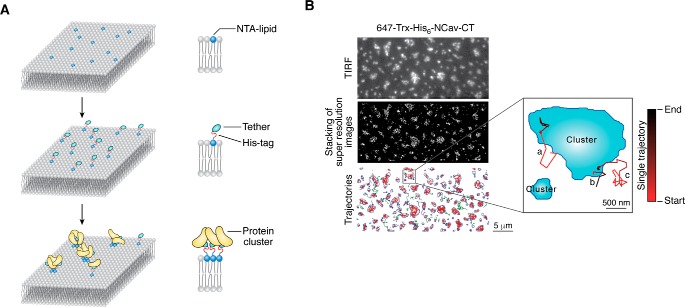
Condensed phase formed on 2D supported lipid bilayer. A, schematic diagram of microdomain formation on 2D supported lipid bilayer. Membrane proteins homogeneously distribute on the supported lipid bilayer via tethering of the His-tag to Ni2+-NTA-decorated lipids. Protein clusters are observed on lipid bilayers after the addition of other components to drive phase separation. B, STORM analysis of membrane proteins, the cytoplasmic tail of NCav as an example, on supported lipid bilayer (adapted from Ref. 45). Image captured under TIRF microscopy mode first sketches the contours of the condensed phase, which turns out to perfectly overlap with the image reconstructed from STORM analysis. Trajectories of individual molecules are followed by single molecular tracking assay, both inside and outside the condensed phase. Direction of movement is marked by gradient color from black to red.
To visualize the clustering of membrane proteins on the supported lipid bilayer, either total internal reflection fluorescence (TIRF) or confocal microscopy can be performed considering that the membrane thickness is much below the optical microscopy resolution. Traditional imaging methods like co-localization, fusion, dispersion, and FRAP can also be conducted to describe the ensemble behaviors of proteins on the supported lipid bilayer. In addition, all the materials are restricted to a single membrane sheet which theoretically accounts for all the signal sources, making it more convenient for quantification. First, fluorescence intensity-based quantification methods as described for analysis of droplets in 3D solution is applicable. The same fluorophore can be conjugated to either lipid or protein, and the absolute number of lipids can be calculated by the known coating surface area and lipid headgroup size. A standard curve of fluorescence intensity with respect to the number of fluorophores can then be plotted. Assuming the illumination property of a fluorophore remains the same no matter if conjugated to lipid or to protein, protein density can thus be converted by referring to the standard curve generated from the labeled lipid (41, 45). An alternative approach is to take advantage of the bilayer membrane to perform single molecule counting. The protein of interest is labeled with two different fluorophores, where one is sparse enough for single molecule counting and the other for experiments. One can then calculate the molecular number from the known concentration ratio of the sparsely-labeled fraction before coating (42, 62).
Furthermore, the confinement of membrane protein on the supported lipid bilayer allows one to trace proteins at the single molecular level (Fig. 4B). STORM provides a method to delineate behaviors of membrane-localized protein microclusters (45). The distribution of molecules is directly counted (i.e. bypassing the fluorescence intensity conversion in bulk imaging experiments), and the concentration ratio can be extracted. The dynamics of individual molecules is directly evaluated by tracking their individual movement trajectories instead of depending on the overall fluorescence recovery as in FRAP analysis. Trajectories of single molecules over time can be categorized, and it has been shown that molecules within the condensed phase move significantly slower than their counterparts in the dilute phase (Fig. 4B). Super-resolution imaging is a powerful technique for illustrating features of individual molecules, but deliberations need to be taken for the compatibility of the two systems. For example, the imaging buffer for STORM experiments contains a thiol compound to enable photoswitching. However, when a His-tagged protein attaches to the membrane via interaction with DGS/NTA-Ni2+ lipid to mimic its membrane localization, the reducing reagent can interfere with membrane attachment. Fortunately, this system is tolerant to a sub-dosage of 2-mercaptoethanol to some extent, although a sufficient amount of photoswitching can still be achieved (45). Another concern is the requirement of the oxygen scavenger system that consists of glucose, glucose oxidase, and catalase. High concentration of glucose (∼10%) may affect the ability of the molecular components to phase separate, although the influence is case–by–case, and rigorous controls are required. In addition, the intermediate product, hydrogen peroxide, will oxidize and destroy the lipid membrane if it failed to be eliminated by catalase. Therefore, the relative stoichiometry of imaging buffer components and the duration of imaging time are crucial to maintain a reduced environment at all times.
Open questions
The application of a 2D supported lipid bilayer system for characterization of LLPS is still at the initial stage and in the ascendant. Many important questions remain to be considered to promote the development of this system and, in return, to facilitate the thorough comprehension of this field. In the current systems, the coating of membrane proteins mainly relies on NTA-Ni2+/His interaction for simplicity. Clustering of membrane proteins, however, will drag synchronized movement and clustering of NTA-lipids, which may affect the overall membrane properties. Besides, the protein-coating efficiency depends on the proportion of NTA-lipid (DGS-NTA), but the percentage of DGS-NTA itself will affect membrane fluidity as indicated by FRAP analysis (63). Continuous efforts are thus demanded to overcome this problem. For example, transmembrane proteins might be inserted into supported lipid bilayer independent of NTA-Ni2+/His interaction. The lipid components we are looking at in this study are way too simplified compared with those in natural conditions. Importantly, lipid compositions can change over time and in response to cellular activities, and protein/lipid interactions are often involved in signal transduction and regulation. In addition, lipid itself can undergo phase separation, which represents another fascinating research field (64). What happens if protein phase separation comes across lipid phase separation? Membrane bilayers constituting more close–to–physiology lipid compositions should certainly be taken into consideration over the long haul. The concept of the “membraneless compartment” is gradually becoming a consensus, and studies performed with supported lipid bilayer increasingly uncover the relationship between protein condensates in solution and protein clusters on lipid membranes. Early evidence has already shown a direct connection between synaptic vesicle pool and its buffering surrounding synapsin phase separation in presynaptic termini (56). In the future, it will be interesting to study how membraneless and membrane-bound compartments are coupled using a combination of 3D solution and 2D membrane systems.
Functional implications
We have so far discussed the molecular mechanisms that drive phase separation and how to characterize LLPS in solution and on lipid bilayers in vitro. Studies on in vitro reconstitution systems shed light on the biological significance of phase separation. In this section, we propose a few potential functional implications of having non-membrane-enclosed biological condensates in cells.
Compartmentalization without physical barriers
Membrane-mediated molecular confinements guarantee specific proteins/nuclei acids subcellular localization thus allowing distinct functions of each organelle. However, membrane-bound organelles with limited types are insufficient to support diverse cellular processes with multiple functions. Cytoplasm should be further segregated to control each unique chemical reaction without potential disturbances. Functional proteins are often found to have their preferential sites in a cell with a sharp concentration gradient to the neighboring environment. Phase separation among different biomacromolecules facilitates spontaneous formation of different subcellular compartments without the help of lipid membranes. Because phase separation is driven by intrinsic properties of an exact protein and its binding partners, such compartmentalization can be highly specific to its inner components. The transition is achieved in a membrane-independent manner; therefore, cells can simultaneously condensate different materials into various compartments, each with defined constitution and function (Fig. 5). Moreover, forming membraneless organelles would be “energy friendly” to cells because lipid biogenesis and membrane identity maintenance consume a huge amount of energy. Phase separation by molecules under physiological conditions is a natural process with no demands for extra energy; thus, cells do not need to actively deliver materials toward each condensate against a huge concentration gradient. The membraneless and liquid-like properties allow a newly-formed condensate to fuse with another to enlarge its size, a process that could also be energy-costly if every condensate is enclosed by membranes (Fig. 5).
Figure 5.
Biological functions of LLPS-mediated membraneless compartments.
Achieve high local concentrations for molecular interactions and rapid chemical reactions
Self-condensation is one of the key features and probably also the most important function of phase separation. Compared with macromolecular complexes formed by traditional interaction mode, a demixed phase enables molecules to be massively enriched into a restricted subcellular region and subsequently to increase their local concentrations by hundreds of folds. This massive increase in concentration brings at least two non-negligible changes toward materials inside the condensates. First, for scaffolding proteins involved in assembly of the entire architecture, weak interactions between molecules, which usually is almost undetectable in aqueous solution, can be dramatically amplified and contribute to the properties of biological condensates (65, 66). That could explain why sometimes a single amino acid substitution on a given protein, which hardly changes its behavior in homogeneous solution, might severely influence its ability to phase separate. Those previously identified weak interactions should also be re-evaluated and taken into the consideration of phase separation, because they may no longer be nonspecific or without any functional implications. Second, higher local concentration of enzymes enriched in condensates might show altered activities or kinetics during chemical reactions. If a given enzyme gets concentrated into the condensed phase with an open conformation, the active recruitment or exclusion of its substrate determines whether a chemical reaction would be promoted or inhibited (Fig. 5). CaMKII is the most abundant enzyme in synapses, co-localizes with its numerous substrates in PSD, and exhibits neuronal activity-dependent translocation into synapse from the dendritic shaft (67). Upon kinase activation, one might foresee enhanced phosphorylation of CaMKII substrates to activate downstream signaling pathways. Actin polymerization provides another example of how phase separation can promote reaction kinetics. In nephrin and LAT systems, the amount of actin assembly is dramatically up-regulated when the signaling components undergo phase separation (discussed above). It has recently been demonstrated that the increased membrane dwell time of N-WASP, in the condensed phase, promotes its association with the Arp2/3 complex and subsequently the actin polymerization rate compared with the homogeneous solution state (10, 41, 68, 69). The formation of astral microtubules from the centrosome is also promoted when tubulin monomers become massively enriched into a reconstituted centrosome condensate by SPD5 (66). Stress granules provide a contrasting example where protein translation is sequestered by actively “squeezing” mRNAs and some of the translational machineries from cytoplasm into densely-packed condensates (69). Future experiments should be focused on both accurately measuring the enzyme kinetics and systemically proposing reaction theories in condensed phases.
Allow fast changes of molecules upon signaling
The membrane-enclosed structure shows reduced molecular dynamics because its inner materials are completely segregated by lipid bilayers. Condensates formed by phase separation can confine molecules to a given region but meanwhile allow them to freely exchange with their counterparts in the surrounding solution. This type of molecular dynamics provides a unique feature of phase separation-mediated condensation to rapidly rearrange its interior constituents in response to different stimuli. Compositional reorganization within a particular compartment can be accomplished by selectively altering the behavior of a given protein with covalent modifications that favor or disfavor its local environment. For example, synapsin undergoes phase separation by itself and further cluster synaptic vesicles (56). This condensation can be dissolved upon synapsin phosphorylation by CaMKII. Arginine methylation on FUS protein did not affect its phase separation ability, but it dramatically decreased the hardness of FUS droplets, indicating that post-translational modification could also modulate the material properties of a given condensate (34). Phase separation might also undergo an overall weakening when the key organizer is depleted or competed off by other regulatory molecules. The dispersion of reconstituted PSD phase droplets by an alternatively spliced form of Homer1 provides another good example of biological condensate regulation (44). Because multivalent intermolecular interactions (both strong and weak) are the driving force of phase separation, one could imagine that the condensation process can be extremely sensitive to changes in the outside environment, including salt concentration, pH, temperature, redox conditions, etc. Tuning biomolecular interactions might bring huge influences on a condensed phase, making each condensed system a perfect biosensor that enables the cell to recognize various signals and make rapid responses to them.
Sub-segregation via phase–in–phase, phase–to–phase, or surface coating
Organelles with multiple membrane layers are not commonly used in living cells. Mitochondria and chloroplasts are the only two known systems with double layers of lipid membrane that allow their inner materials to be further segregated to facilitate multistep reactions during respiration and photosynthesis. Sub-segregations within an organelle can provide new isolated regions with distinct functions but meanwhile allow each segregated part to communicate with each other. This smart design might also be adopted by membraneless condensates to support themselves with multiple functions. Sub-segregation can happen when multiple proteins co-cluster into the same condensates with one of them forming a smaller droplet and being totally embedded among the other, a phenomenon termed as phase–in–phase (Fig. 5). For example, three sub-compartments (NPM1, FIB1, and POLR1E) of nucleoli in Xenopus laevis form distinct and immiscible liquid phases where FIB1 and POLR1E condensed into smaller-sized puncta inside a single NPM1 condensate (57). A completely buried phase is isolated by outer layer proteins thus preventing potential dynamic exchange. At the same time, protein concentrations would further increase as the total volume of a droplet gets smaller, which might vastly speed up reactions inside the condensate. When the sizes of two sub-segregations become similar, a layer–to–layer structure could form with two droplets sharing a common interface but with each exposed to the outside environment. A condensation organized in a phase–to–phase pattern can generate multiple functional interfaces for biomolecular interactions and signaling transductions with specific orientations (Fig. 5). During germline development in C. elegans, two P-granule proteins, ZNFX-1 and WAGO-4, become phase separated from the P granule to form an independent liquid phase. This newly-formed phase further assembles into tri-condensation with the P granule and Mutator foci in a phase–to–phase manner to spatiotemporally regulate epigenetic inheritance during development (70). The detailed molecular mechanisms for phase–in/to–phase is still not well-understood, and it is believed that both the interaction among inner materials and the surface tension of individual droplets may govern the sub-segregation process. Surface coating is another unique type of sub-segregation when some molecules only localize to the surface of a transitioned phase (Fig. 5). Recruitment of particular molecules to the droplet surface might change the surface properties of a given condensate and offer it with new functions. In addition, biomolecules, even without the help of transmembrane or membrane-binding domains, can be confined into a 2D system, which dramatically alters their activities. This might be achieved when a given protein has two featured surfaces, one of which favors the inner environment of a condensed phase but the other disfavors and gets excluded. Thus, surface coating is regarded as an equilibrium between protein attraction and exclusion from materials inside the condensate. Surface coating to a selected phase droplet may also affect its material properties. Molecules on the condensate surface can regulate dynamic exchange of its inner materials, influence free diffusion of small molecules, and even alter fusion process of droplets. A recent study reported that EPG-2, a scaffold protein in the C. elegans P granule, can specifically decorate the surface of SEPA/PGL-1/-3 droplets and modulate the condensate properties (71).
Direct communications between membraneless and membrane-bound organelles
Membraneless organelles formed by phase separation could communicate with membrane-bound organelles via direct interactions (Fig. 5). Such communication might help to specify the localization of membrane-bound organelles, to reorganize protein distributions on membrane surface, and to introduce new functions to organelles. TIS11B, an RNA-binding protein, for example, forms membraneless granules that directly attaches to the ER (72). TIS granules specifically retain certain mRNAs and exclude others to enable accurate protein translation in the ER. RNA granule is also observed to associate with late endosomes residing close to mitochondria in neuronal axons to regulate local synthesis of axonal proteins (73).
Phase separation and evolution
Previous subsections discuss the functional implications of phase separation in terms of offering a living cell with multiple functions. Here, we postulate on the possible biological importance of phase separation from the angle of life evolution. It is hard for one to imagine that the earliest form of life directly starts with membrane-bound organelles as lipids are not typical information carriers like nuclei acids. In addition, the biogenesis of lipids depends on other molecules with catalytic activities, such as protein or RNA. The origin of life is believed to depend on RNA because it carries the genetic information and possesses enzymatic activity that may allow them to self-reproduce. RNA is so unique as it forms a long chain with multiple binding sites for other RNAs or proteins, which might also explain why it could always easily undergo LLPS with different RBPs or even by itself. Compartmentalization mediated by phase separation may reveal how proteins and nucleic acids assemble into condensed bioreactors in the ocean before the emergence of lipid membranes. Indeed, membraneless condensation is observed not only in higher animals but also in ancient cyanobacteria, indicating a common and conserved biological process that could be shared by all the living creatures on earth (46). We refer readers to several recent reviews (6, 74, 75) that also discussed the emergence of the LLPS process during evolution.
Conclusions and perspective
In cells, biomaterials can be organized into membrane-bound or membraneless compartments. Significant progress has been made over the past decade in understanding the mechanisms underlying the formation and organization of non-membrane-enclosed organelles. Many of these systems are driven by LLPS via which collections of molecules demixed from the bulk solution/cytoplasm to form biological condensates. Modular domain proteins and intrinsic disorder containing proteins exhibit multivalent intermolecular interactions either via specific, high-affinity interactions or weak adhesions that drive phase separation. Analysis of features of molecules involved in LLPS has started to reveal sequence determinants in intrinsically disordered regions that promote phase separation. Although our understanding is still rudimentary, it is clear that certain sequence patterns are heavily involved and can determine the material properties of a condensate. A large collection of methods has been developed to study the dynamics, composition, and physical properties of condensed droplets in vitro. We still need more quantitative assays, measurements, and descriptions for future phase separation studies. For instance, theoretical work is required for understanding the underlying physical and chemical principles of LLPS. Bioengineering tools may be designed to precisely control phase separations in vitro or in vivo. Another appealing, yet very challenging, idea is to reveal the atomic details of the condensed droplets. In particular, could there be a single structure that might be “solved”? Cryo-EM studies have been conducted on some condensates trying to answer this question, although little success has been achieved so far. This is somewhat expected because many of the phase separations are contributed by intrinsically disordered elements. It is difficult to imagine how the multivalent interactions might be restricted to oligomers of homogeneously distributed sizes. Nevertheless, we cannot rule out the possibility that a core structure of defined stoichiometry and conformation might exist among other flexible and heterogeneous structures, especially where the condensate formation is driven by modular domain interactions. To “solve” the atomic structure of phase condensates might be too optimistic at current stages, but it is possible that we might reveal the molecular organizations within the condensates using cryo-ET and cryo-EM techniques. Is there a layered organization within the phase droplets? How do molecules assemble into supramolecular complexes that phase separate from the bulk solution? Results from in vitro characterizations of phase droplets would provide insights into how the macroscopic properties of condensates might contribute to their biological functions in cells. There remains much to answer about the biological condensates. How are the enzyme kinetics regulated in the condensed phase? Recent studies using nephrin and LAT systems have shown that increased dwell times of enzymes in the condensed phase lead to faster reaction rates (62, 68). Is this a general mechanism for other molecular systems? Compared with membrane-bound organelles, LLPS-mediated membraneless structures have their own advantages; but it also brings many potential problems. For example, how to achieve specificity in organization? How to prevent unwanted fusion or mixing without a membrane barrier? How does sub-segregation happen inside an organelle and how to maintain this specific pattern? Is this physiologically relevant and functionally regulated? Will the concept of phase separation help us to re-examine many diseases that are hard to be explained by physical chemistry principles of dilute solution systems? For instance, alteration in the material properties of many neuronal protein condensates might contribute to neurodegenerative diseases. The concentration dependence of phase separation might help explain the dosage sensitivity of SynGAP, a negative regulatory protein of PSD assembly, in psychiatric diseases. Answers to these questions will provide us with in-depth insights into mechanisms underlying the formation and regulation of biological condensates in cells and to understand how nature evolves this type of compartmentalization in life. In the end, although the list of non-membrane-bound organelles formed by LLPS continues to expand, researchers should always ask themselves the following: is the LLPS-driven protein condensation observed in vitro biologically relevant? If so, what is its contribution to cellular functions? Can the in vivo observations be explained by mechanisms other than liquid phase separation (76)? Tailored experiments need to be designed to distinguish between these possibilities. Nonetheless, we can be assured that LLPS-mediated biological condensate formation is an emerging life science research field with numerous exciting opportunities.
This work was supported in part by Grants AoE-M09-12 and C6004-17G from RGC of Hong Kong and Grant 510178 from The Simons Foundation for Autism Research. The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- RBP
- RNA-binding protein
- FMRP
- Fragile X mental retardation protein
- LLPS
- liquid–liquid phase separation
- PSD
- postsynaptic density
- FUS
- Fused in Sarcoma
- RNP
- ribonucleoprotein
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- ALS
- amyotrophic lateral sclerosis
- LCR
- low-complexity region
- SH
- Src homology
- PRM
- Pro-rich motif
- FRAP
- fluorescence recovery after photobleaching
- RRM
- RNA recognition motif
- AFM
- atomic force microscopy
- cryo-ET
- cryo-electron tomography
- TIRF
- total internal reflection fluorescence
- DIC
- differential interference contrast
- STORM
- stochastic optical reconstruction microscopy
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- DGS
- 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl.
References
- 1. Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A. A. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 2. Alberti S. (2017) Phase separation in biology. Curr. Biol. 27, R1097–R1102 10.1016/j.cub.2017.08.069 [DOI] [PubMed] [Google Scholar]
- 3. Alberti S., Gladfelter A., and Mittag T. (2019) Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell 176, 419–434 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banani S. F., Lee H. O., Hyman A. A., and Rosen M. K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., and Fuxreiter M. (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 7. Shin Y., and Brangwynne C. P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 8. Flory P. J. (1942) Thermodynamics of high polymer solutions. J. Chem. Phys. 10, 51–61 10.1063/1.1723621 [DOI] [Google Scholar]
- 9. Lin Y. H., Forman-Kay J. D., and Chan H. S. (2018) Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry 57, 2499–2508 10.1021/acs.biochem.8b00058 [DOI] [PubMed] [Google Scholar]
- 10. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiti F., and Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- 12. Knowles T. P., Vendruscolo M., and Dobson C. M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 10.1038/nrm3810 [DOI] [PubMed] [Google Scholar]
- 13. Taylor J. P., Hardy J., and Fischbeck K. H. (2002) Toxic proteins in neurodegenerative disease. Science 296, 1991–1995 10.1126/science.1067122 [DOI] [PubMed] [Google Scholar]
- 14. Elbaum-Garfinkle S. (2019) Matter over mind: Liquid phase separation and neurodegeneration. J. Biol. Chem. 294, 7160–7168 10.1074/jbc.REV118.001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gan L., Cookson M. R., Petrucelli L., and La Spada A. R. (2018) Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 21, 1300–1309 10.1038/s41593-018-0237-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jucker M., and Walker L. C. (2018) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat. Neurosci. 21, 1341–1349 10.1038/s41593-018-0238-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nedelsky N. B., and Taylor J. P. (2019) Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol. 15, 272–286 10.1038/s41582-019-0157-5 [DOI] [PubMed] [Google Scholar]
- 18. Soto C., and Pritzkow S. (2018) Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 21, 1332–1340 10.1038/s41593-018-0235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor J. P., Brown R. H. Jr., and Cleveland D. W. (2016) Decoding ALS: from genes to mechanism. Nature 539, 197–206 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chong P. A., Vernon R. M., and Forman-Kay J. D. (2018) RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 10.1016/j.jmb.2018.06.014 [DOI] [PubMed] [Google Scholar]
- 21. Decker C. J., Teixeira D., and Parker R. (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frey S., Richter R. P., and Görlich D. (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- 24. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 27. Murakami T., Qamar S., Lin J. Q., Schierle G. S., Rees E., Miyashita A., Costa A. R., Dodd R. B., Chan F. T., Michel C. H., Kronenberg-Versteeg D., Li Y., Yang S. P., Wakutani Y., Meadows W., et al. (2015) ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin Y., Currie S. L., and Rosen M. K. (2017) Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 10.1074/jbc.M117.800466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., and Hyman A. A. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA-binding proteins. Cell 174, 688–699.e16 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vernon R. M., Chong P. A., Tsang B., Kim T. H., Bah A., Farber P., Lin H., and Forman-Kay J. D. (2018) π-π contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486 10.7554/eLife.31486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V. H., Janke A. M., Baatsen P., Vercruysse T., Kolaitis R. M., Daelemans D., et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsang B., Arsenault J., Vernon R. M., Lin H., Sonenberg N., Wang L. Y., Bah A., and Forman-Kay J. D. (2019) Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. U.S.A. 2019, 201814385 10.1073/pnas.1814385116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qamar S., Wang G., Randle S. J., Ruggeri F. S., Varela J. A., Lin J. Q., Phillips E. C., Miyashita A., Williams D., Ströhl F., Meadows W., Ferry R., Dardov V. J., Tartaglia G. G., Farrer L. A., et al. (2018) FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734.e15 10.1016/j.cell.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryan V. H., Dignon G. L., Zerze G. H., Chabata C. V., Silva R., Conicella A. E., Amaya J., Burke K. A., Mittal J., and Fawzi N. L. (2018) Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7 10.1016/j.molcel.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conicella A. E., Zerze G. H., Mittal J., and Fawzi N. L. (2016) ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murray D. T., Kato M., Lin Y., Thurber K. R., Hung I., McKnight S. L., and Tycko R. (2017) Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hughes M. P., Sawaya M. R., Boyer D. R., Goldschmidt L., Rodriguez J. A., Cascio D., Chong L., Gonen T., and Eisenberg D. S. (2018) Atomic structures of low-complexity protein segments reveal kinked β-sheets that assemble networks. Science 359, 698–701 10.1126/science.aan6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain A., and Vale R. D. (2017) RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Du M., and Chen Z. J. (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banjade S., and Rosen M. K. (2014) Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife 3, 10.7554/eLife.04123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su X., Ditlev J. A., Hui E., Xing W., Banjade S., Okrut J., King D. S., Taunton J., Rosen M. K., and Vale R. D. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng M., Shang Y., Araki Y., Guo T., Huganir R. L., and Zhang M. (2016) Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 166, 1163–1175.e12 10.1016/j.cell.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng M., Chen X., Guan D., Xu J., Wu H., Tong P., and Zhang M. (2018) Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell 174, 1172–1187.e16 10.1016/j.cell.2018.06.047 [DOI] [PubMed] [Google Scholar]
- 45. Wu X., Cai Q., Shen Z., Chen X., Zeng M., Du S., and Zhang M. (2019) RIM and RIM-BP form presynaptic active-zone-like condensates via phase separation. Mol. Cell 73, 971–984.e5 10.1016/j.molcel.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 46. Wang H., Yan X., Aigner H., Bracher A., Nguyen N. D., Hee W. Y., Long B. M., Price G. D., Hartl F. U., and Hayer-Hartl M. (2019) Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566, 131–135 10.1038/s41586-019-0880-5 [DOI] [PubMed] [Google Scholar]
- 47. Lin Y., Protter D. S., Rosen M. K., and Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saha S., Weber C. A., Nousch M., Adame-Arana O., Hoege C., Hein M. Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., Pozniakovski A., Eckmann C. R., Jülicher F., and Hyman A. A. (2016) Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 10.1016/j.cell.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schwartz J. C., Wang X., Podell E. R., and Cech T. R. (2013) RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918–925 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., and Gladfelter A. S. (2015) RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burke K. A., Janke A. M., Rhine C. L., and Fawzi N. L. (2015) Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 53. Mitrea D. M., Cika J. A., Guy C. S., Ban D., Banerjee P. R., Stanley C. B., Nourse A., Deniz A. A., and Kriwacki R. W. (2016) Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, e13571 10.7554/eLife.13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Monahan Z., Ryan V. H., Janke A. M., Burke K. A., Rhoads S. N., Zerze G. H., O'Meally R., Dignon G. L., Conicella A. E., Zheng W., Best R. B., Cole R. N., Mittal J., Shewmaker F., and Fawzi N. L. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M. (2017) Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Milovanovic D., Wu Y., Bian X., and De Camilli P. (2018) A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 10.1126/science.aat5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., and Brangwynne C. P. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Petersen J. D., Chen X., Vinade L., Dosemeci A., Lisman J. E., and Reese T. S. (2003) Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J. Neurosci. 23, 11270–11278 10.1523/JNEUROSCI.23-35-11270.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu H., and Fuxreiter M. (2016) The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 165, 1055–1066 10.1016/j.cell.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Richter R. P., Bérat R., and Brisson A. R. (2006) Formation of solid-supported lipid bilayers: an integrated view. Langmuir 22, 3497–3505 10.1021/la052687c [DOI] [PubMed] [Google Scholar]
- 61. Case L. B., Ditlev J. A., and Rosen M. K. (2019) Regulation of transmembrane signaling by phase separation. Annu. Rev. Biophys. 48, 465–494 10.1146/annurev-biophys-052118-115534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang W. Y. C., Alvarez S., Kondo Y., Lee Y. K., Chung J. K., Lam H. Y. M., Biswas K. H., Kuriyan J., and Groves J. T. (2019) A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 10.1126/science.aau5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Su X., Ditlev J. A., Rosen M. K., and Vale R. D. (2017) Reconstitution of TCR signaling using supported lipid bilayers. Methods Mol. Biol. 1584, 65–76 10.1007/978-1-4939-6881-7_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heberle F. A., and Feigenson G. W. (2011) Phase separation in lipid membranes. Cold Spring Harb. Perspect. Biol. 3, a004630 10.1101/cshperspect.a004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Woodruff J. B., Wueseke O., Viscardi V., Mahamid J., Ochoa S. D., Bunkenborg J., Widlund P. O., Pozniakovsky A., Zanin E., Bahmanyar S., Zinke A., Hong S. H., Decker M., Baumeister W., Andersen J. S., et al. (2015) Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science 348, 808–812 10.1126/science.aaa3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Woodruff J. B., Ferreira Gomes B., Widlund P. O., Mahamid J., Honigmann A., and Hyman A. A. (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 67. Lisman J., Yasuda R., and Raghavachari S. (2012) Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182 10.1038/nrn3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Case L. B., Zhang X., Ditlev J. A., and Rosen M. K. (2019) Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 10.1126/science.aau6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Decker C. J., and Parker R. (2012) P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4, a012286 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wan G., Fields B. D., Spracklin G., Shukla A., Phillips C. M., and Kennedy S. (2018) Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683 10.1038/s41586-018-0132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang G., Wang Z., Du Z., and Zhang H. (2018) mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506.e22 10.1016/j.cell.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 72. Ma W., and Mayr C. (2018) A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein/protein interactions. Cell 175, 1492–1506.e19 10.1016/j.cell.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cioni J. M., Lin J. Q., Holtermann A. V., Koppers M., Jakobs M. A. H., Azizi A., Turner-Bridger B., Shigeoka T., Franze K., Harris W. A., and Holt C. E. (2019) Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell 176, 56–72.e15 10.1016/j.cell.2018.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Poudyal R. R., Pir Cakmak F., Keating C. D., and Bevilacqua P. C. (2018) Physical principles and extant biology reveal roles for RNA-containing membraneless compartments in origins of life chemistry. Biochemistry 57, 2509–2519 10.1021/acs.biochem.8b00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cinar H., Fetahaj Z., Cinar S., Vernon R. M., Chan H. S., and Winter R. H. A. (2019) Temperature, hydrostatic pressure, and osmolyte effects on liquid–liquid phase separation in protein condensates: physical chemistry and biological implications. Chemistry 2019, 10.1002/chem.201902210 [DOI] [PubMed] [Google Scholar]
- 76. Raff J. W. (2019) Phase separation and the centrosome: a fait accompli? Trends Cell Biol. 29, 612–622 10.1016/j.tcb.2019.04.001 [DOI] [PubMed] [Google Scholar]