Abstract
The mating pathway in yeast Saccharomyces cerevisiae has long been used to reveal new mechanisms of signal transduction. The pathway comprises a pheromone receptor, a heterotrimeric G protein, and intracellular effectors of morphogenesis and transcription. Polarized cell growth, in the direction of a potential mating partner, is accomplished by the G-protein βγ subunits and the small G-protein Cdc42. Transcription induction, needed for cell–cell fusion, is mediated by Gβγ and the mitogen-activated protein kinase (MAPK) scaffold protein Ste5. A potential third pathway is initiated by the G-protein α subunit Gpa1. Gpa1 signaling was shown previously to involve the F-box adaptor protein Dia2 and an endosomal effector protein, the phosphatidylinositol 3-kinase Vps34. Vps34 is also required for proper vacuolar sorting and autophagy. Here, using a panel of reporter assays, we demonstrate that mating pheromone stimulates vacuolar targeting of a cytoplasmic reporter protein and that this process depends on Vps34. Through a systematic analysis of F-box deletion mutants, we show that Dia2 is required to sustain pheromone-induced vacuolar targeting. We also found that other F-box proteins selectively regulate morphogenesis (Ydr306, renamed Pfu1) and transcription (Ucc1). These findings point to the existence of a new and distinct branch of the pheromone-signaling pathway, one that likely leads to vacuolar engulfment of cytoplasmic proteins and recycling of cellular contents in preparation for mating.
Keywords: G protein, mitogen-activated protein kinase (MAPK), yeast, ubiquitin, protein sorting, F-box, fungal mating, pheromone signaling
Introduction
Many extracellular signals are detected by G-protein–coupled receptors (GPCRs).10 In animals, these signals include odors, tastes, light, pH, nucleotides, biogenic amines, peptides, steroids, and phospholipids. In each case, receptor activation results in binding of GTP to a G-protein α subunit and dissociation of Gα from the βγ subunit dimer. Both Gα and Gβγ can then transduce signals through the activation of intracellular enzymes and ion channels. G-protein signaling ends when GTP is hydrolyzed to GDP, a process that is accelerated by members of the regulator of G-protein signaling (RGS) family (1).
GPCRs also play an important role in yeast mating. Genetic analysis in yeast Saccharomyces cerevisiae has led to the identification and characterization of several new signaling proteins, including the first RGS protein Sst2 (2). Sst2 is required for pheromone gradient tracking (3–5), adaptation (6, 7), and noise suppression (8). Other pathway components were identified from mutants that produce an unresponsive sterile (ste) phenotype, including the mating pheromone receptors (Ste2 and Ste3), the G-protein βγ subunits (Ste4 and Ste18), a Gβγ effector (Ste5), downstream protein kinases (Ste20, Ste11, and Ste7), and a transcription factor (Ste12) (2). Further analysis revealed that the G-protein βγ dimer recruits and activates Far1 in complex with Cdc24 (9–13), an exchange factor for the small G-protein Cdc42 (14), as well as Ste5, a scaffold protein required for activation of the MAPKs Fus3 and Kss1 (15–22). Cdc42 promotes cell cycle arrest and morphological changes necessary for mating (23–25). Either Fus3 or Kss1 can sustain the mating transcription program (17, 26–28). Fus3 alone promotes cell cycle arrest and directed expansion toward a pheromone gradient (29–33). Kss1 promotes invasive growth under nutrient-limiting conditions (34–36). Together, these signaling processes lead to the fusion of haploid a and α cells and formation of an a/α diploid.
Given its genetic tractability and the relatively small number of signaling components, the yeast mating pathway has emerged as one of the best-characterized of any signaling system. Moreover, newer genetic screens, combined with biochemical analysis of the affected gene products, have provided evidence for a possible third signaling branch mediated by the Gα subunit Gpa1. In particular, we demonstrated transcriptional activation by a constitutively active (GTPase-deficient) mutant form of Gα, Gpa1Q323L (37–39). A systematic comparison of 4847 gene deletion mutants revealed a transducer role for seven proteins, including Dia2, Vps15, and Vps34 (38). Dia2 is one of an estimated 16 F-box adaptor proteins that recruit substrates for ubiquitination (40, 41). Vps15 and Vps34 are the regulatory and catalytic subunits of the sole phosphatidylinositol (PI) 3-kinase in yeast and are required for proper vacuolar sorting and autophagy (42–44). Thus, the same screen for Gα effectors revealed components of pathways governing ubiquitination and autophagy, representing the two predominant routes for protein degradation in the cell. However, it is not known how these candidate effectors contribute to the mating response.
Ubiquitination is the covalent attachment of a 76-amino acid peptide to specific target proteins. Once a protein is ubiquitinated, ubiquitin can itself be ubiquitinated, resulting in the formation of a polyubiquitin chain. Generally speaking, this process is mediated by three factors: an E1 ubiquitin-activating enzyme; an E2 ubiquitin-conjugating protein; and an E3 ubiquitin ligase (45). The SCF (Skp1/Cullin/F-box) ubiquitin ligase employs an F-box protein to recruit specific substrates for ubiquitination, which are then captured by the proteasome and rapidly degraded (46). Up to 30% of newly synthesized proteins are incorrectly made and degraded in a ubiquitin-dependent manner (47). Other proteins are properly folded and ubiquitinated in response to internal and external cues. For example, ubiquitination of both Ste7 and Sst2 is induced by pheromone, and these modifications are thought to represent feedback loops leading to pheromone desensitization and resensitization, respectively (48–50).
In contrast to ubiquitin-mediated degradation, autophagy is far less specific and also has much greater flexibility in the choice of cargos (51). Whereas ubiquitin targets specific proteins for destruction, autophagy delivers bulk cytoplasmic contents to the vacuole or lysosome. Autophagy can in turn be classified into two broad categories, nonselective and selective, based on the nature of the substrates being consumed. Nonselective autophagy (also macroautophagy) is initiated by nutrient limitation and allows cytoplasmic contents to be broken down and reused. Selective autophagy describes the removal of specific cargos, including organelles such as mitochondria (mitophagy) (52–55). A variant of this pathway, known as the cytoplasm–to–vacuole targeting pathway, is responsible for the proper delivery of vacuolar enzymes (56). As with the mating pathway, genetic screens in yeast revealed dozens of autophagy-related (atg) genes, including components of the PI 3-kinase complex, that mediate nonselective and/or selective autophagy (57–59). Detailed investigations have revealed a remarkable degree of conservation of autophagy components in humans and yeast (60–65).
Having demonstrated a potential link between Gpa1 and both the ubiquitin–proteasome (Dia2) and vacuolar transport (Vps15 and Vps34) systems (38), we sought to investigate the role of these proteins in pheromone signaling. We show here that pheromone promotes transport of a cytoplasmic protein biosensor to the vacuole and that this process requires Vps34 and Vps15. A screen of 14 nonessential F-box deletion mutants reveals that Dia2 alone is needed to sustain pheromone-induced vacuolar targeting. A previously uncharacterized F-box protein (Ydr306, renamed Pfu1) is required to abandon nonproductive mating projections prior to mating. A third family member is required for MAPK induction (Ucc1). On the basis of these findings, we propose that vacuolar targeting represents a third and distinct branch of the mating response pathway, one that is uniquely mediated by the G-protein α subunit, the PI 3-kinase Vps34, and the F-box protein Dia2.
Results
Mating pheromone α-factor promotes vacuolar targeting of a cytoplasmic reporter protein
In the yeast mating pathway, binding of the pheromone α-factor to its receptor leads to dissociation of the G-protein α and βγ subunits. Free Gβγ initiates new gene transcription and morphogenesis in preparation for mating. The Gα protein Gpa1 constrains the levels of free Gβγ but also binds to and activates the PI 3-kinase Vps34 (38). The functional consequences of Gpa1-mediated activation of Vps34 have not been explored. Given that Vps34 is required for autophagy (via Complex I during nutrient limitation) and cytoplasm–to–vacuole transport (via Complex II under basal conditions), we hypothesized that Vps34 might also promote these processes in response to G-protein activation.
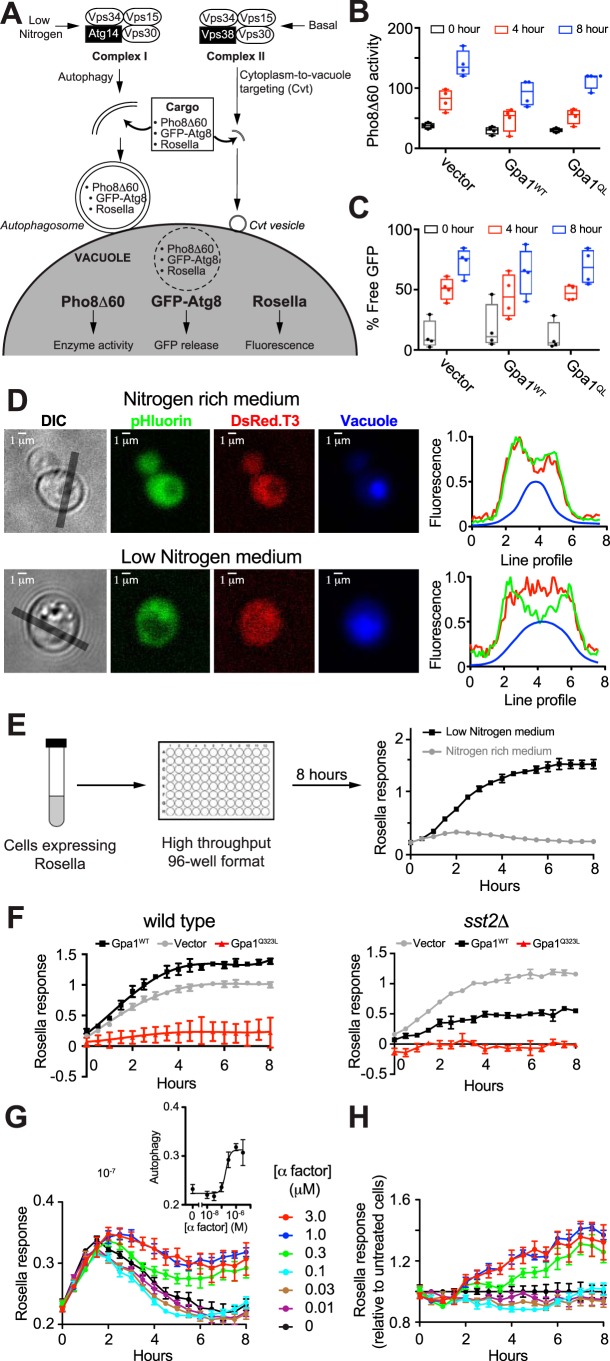
Earlier studies in animal cells revealed that autophagy is inhibited by mutations in Gαi3 that diminish GTP binding or hydrolysis (Q204L) (66–69). To determine whether Gα in yeast promotes autophagy, we transformed WT cells with a single copy plasmid encoding the corresponding GTPase-deficient mutant (Gpa1Q323L), WT Gpa1 (Gpa1WT), or empty vector as a control. As illustrated in Fig. 1A, we considered three independent methods to monitor autophagy in response to Gpa1 activation. The Pho8Δ60 enzymatic assay (70) is based on the alkaline phosphatase Pho8, which is synthesized as an inactive precursor and subsequently activated upon transport to the vacuole via the secretory pathway. Deletion of residues 1–60 of Pho8 (Pho8Δ60) prevents vacuolar transport and maintains the inactive enzyme in the cytosol. Upon induction of bulk autophagy through nitrogen deprivation, cytosolic Pho8Δ60 is engulfed by autophagosomes and targeted to the vacuole for activation. Enzyme activity is measured by spectrophotometry and used as an indicator of bulk autophagy. As shown in Fig. 1B, expression of WT (unactivated) Gpa1 and Gpa1Q323L inhibited autophagy in low-nitrogen conditions relative to a vector control. To determine whether the autophagy machinery is required, we monitored vacuolar processing of GFP–Atg8 (71). Atg8 is a ubiquitin-like protein that is conjugated to phosphatidylethanolamine (72–74), resulting in membrane expansion around portions of the cytoplasm (75, 76) and delivery to the vacuole or lysosome (76–82). The GFP–Atg8 reporter is subsequently cleaved to release GFP. As shown in Fig. 1C, and in contrast to the vacuolar transport assay, Gpa1WT and Gpa1Q323L had no detectable effect on Atg8 processing. We infer from these results that Gpa1 regulates vacuolar transport of cytoplasmic proteins, but it does not appear to regulate Atg8-mediated autophagy directly.
Figure 1.
Mating pheromone α-factor induces transport of cytoplasmic proteins to the vacuole. Three independent methods were used to quantify autophagy in BY4741 yeast. A, PI 3-kinase Vps34 exists as two distinct complexes comprising a regulatory kinase (Vps15), a common effector (Vps30), and unique partners (Atg14 or Vps38). Complex I promotes bulk autophagy in nutrient-limiting conditions such as nitrogen starvation. Complex II mediates transport of vacuolar enzymes in nutrient-rich conditions. Both complexes may contribute to the engulfment of cytoplasmic cargo (including reporters Pho8Δ60, GFP–Atg8, or Rosella) within specialized double-membrane vesicles. Fusion of these vesicles with the vacuolar membrane and subsequent release of cargo is reported by quantitative readouts. B, Pho8Δ60 activity induced by nitrogen deprivation in cells expressing plasmid-borne WT Gpa1, GTPase-deficient Gpa1Q323L, or vector. C, GFP–Atg8 processing into free GFP in low-nitrogen conditions in cells expressing Gpa1WT, Gpa1Q323L, or vector. D, biosensor Rosella, composed of super-ecliptic pHluorin (reporter) and DsRed.T3 (internal control), was used to quantify vacuolar targeting in BY4741 bar1Δ (WT) cells. Presented are representative images and line profiles from individual cells in nitrogen-rich (top) or low-nitrogen (bottom) media showing the partial translocation of Rosella from the cytoplasm (red + green) to the vacuole (red) as indicated by a vacuole-specific dye (blue). Gray lines in differential interference contrast (DIC) images indicate regions selected for line profile analysis. E, cells expressing Rosella were maintained in exponential growth phase for 24 h prior to treatment with a dose range of mating pheromone α-factor or deprived of nitrogen as a positive control, in 96-well microplates. Fluorescence was measured every 30 min for 8 h. Vacuolar targeting of Rosella was quantified as the ratio of red and green fluorescence. Analysis was done using Microsoft Excel and GraphPad Prism. F, time course of vacuolar targeting induced by nitrogen deprivation in WT and sst2Δ cells expressing plasmid-borne WT Gpa1, GTPase-deficient Gpa1Q323L, or vector. G, time course of Rosella response in cells treated with 0–3 μm α-factor. Inset, dose dependence of response at 8 h. H, relative response curves obtained by scaling the time profiles in G relative to untreated cells. Pho8Δ60 and GFP–Atg8 data are from four biological replicates. Boxes extend from the 25th to 75th percentiles. Whiskers range from the minimum to maximum value. Rosella experiments were done at least two times, and data are ± S.D. for four technical replicates.
Although the Pho8Δ60 and GFP–Atg8 assays have revealed core components of the autophagy machinery (83), they lack sensitivity and are not amenable to high-throughput analysis. As an alternative, we adapted a two-color fluorescent biosensor, Rosella, to a high-throughput 96-well microplate format (Fig. 1D) (84). Rosella is a cytoplasmic protein composed of two GFP variants, superecliptic pHluorin (SEP) and DsRed.T3. Whereas SEP is pH-sensitive (85), DsRed.T3 is pH-stable (86). When Rosella is transported from the cytoplasm to the lumen of the vacuole (or lysosome in animal cells), the lower pH quenches SEP fluorescence. In contrast, DsRed.T3 is unaffected and serves as an internal control. Thus, vacuolar delivery of Rosella is reported as the ratio of DsRed.T3 and SEP fluorescence. Rosella has previously been used in end-point assays that include fluorescence microscopy and flow cytometry (84). Here, we used the Rosella biosensor as a model to monitor transport of cytoplasmic proteins to the vacuole over time.
We first validated the method by monitoring Rosella fluorescence in WT BY4741 cells in low-nitrogen (pro-autophagy) medium, as a positive control. Under these conditions, cells exhibited an increased response within 2 h relative to control cells grown in nitrogen-rich medium (Fig. 1E). The response continued to increase before saturating at ∼6 h. We then considered the role of Gpa1. As shown in Fig. 1F (left), expression of Gpa1 weakly stimulated vacuolar targeting, while Gpa1Q323L inhibited the response, as it did in the Pho8Δ60 assay. We note that although the Rosella assay reports vacuolar delivery of the reporter, the Pho8Δ60 assay reports on vacuolar proteolysis. Thus, Gpa1Q323L (like Gαi3Q204L) inhibits vacuolar targeting, despite its documented ability to activate Vps34 catalytic activity. To confirm the inhibitory effect of Gα–GTP we measured the response in cells lacking Sst2, which is required for proper Gpa1–GTP hydrolysis (87). As shown in Fig. 1F (right), deletion of SST2 led to an attenuation of response in otherwise WT (GPA1+) cells expressing an additional, plasmid-borne copy of either Gpa1 or Gpa1Q323L. We infer that constraining Gpa1 to the GTP-bound form results in attenuation of autophagy.
Our data so far suggest a dominant-negative effect of Gpa1–GTP, at least under pro-autophagy growth conditions. These results allow for the possibility that Gpa1–GTP promotes vacuolar targeting of Rosella under normal growth conditions. To test this, we monitored vacuolar targeting in response to receptor activation. For these experiments we used cells lacking the secreted Bar1 protease, so as to minimize pheromone degradation during the extended 8-h time course. Cells were treated with 0–3 μm pheromone, and Rosella fluorescence was monitored every 30 min, for 8 h, in an automated 96-well plate reader (Fig. 1G). For all samples, including the untreated control, we observed an initial increase in vacuolar transport of the Rosella reporter. This transient increase may be attributed to the effects of liquid handling, for example, the transfer of cells from glass culture tubes to polystyrene microplate wells and from constant shaking to periodic shaking. Such changes can be considered a stress situation resulting in a high basal level of response (83). Between 4 and 8 h, the signal returned to baseline at low doses but was comparatively elevated at the higher concentrations of pheromone (0.3 to 3 μm). To correct for the observed changes in basal activity, we calculated the ratio of signal in pheromone-treated and untreated samples (Fig. 1H). These data reveal a peak increase of ∼40% and a half-maximal response of ∼200 nm α-factor (Fig. 1G, inset). The peak response is roughly one-fifth of that induced by nitrogen starvation (Fig. 1E). Based on these findings, we conclude that pheromone stimulation results in a modest but significant increase in vacuolar transport of the cytoplasmic reporter protein.
Pheromone-induced vacuolar targeting is mediated by the PI 3-kinase complex II
Vps34 is the sole PI 3-kinase in yeast and is present in two distinct protein complexes (43, 88–91). Complex I is composed of Vps34, Vps15, Vps30 (Beclin-1 in animal cells), and Atg14. Complex II contains Vps38 (UVRAG) in place of Atg14 (92–94). Whereas Complex I is required for autophagy in nutrient-limiting conditions, Complex II is required for proper delivery of several resident vacuolar enzymes such as carboxypeptidase Y (91) under nutrient-rich conditions (Fig. 1A). Both complexes are located at endosomes. The organization of the two tetrameric complexes has been determined from a cryo-EM structure of mammalian Complex I (95) and a crystal structure of yeast Complex II (96).
To identify which of the two effector complexes is needed for pheromone-induced vacuolar targeting, we analyzed individual gene deletions using the Rosella assay. As anticipated, cells lacking Vps34 and Vps15 failed to exhibit vacuolar targeting in response to pheromone (Fig. 2A and Fig. S1). Whereas the response was unaffected by deletion of Atg14 (Complex I), it was completely abrogated in cells lacking Vps38 (Complex II) (Fig. 2B). Thus, although nutrient-driven autophagy is mediated by Complex I, pheromone-induced vacuolar targeting is mediated by Complex II.
Figure 2.
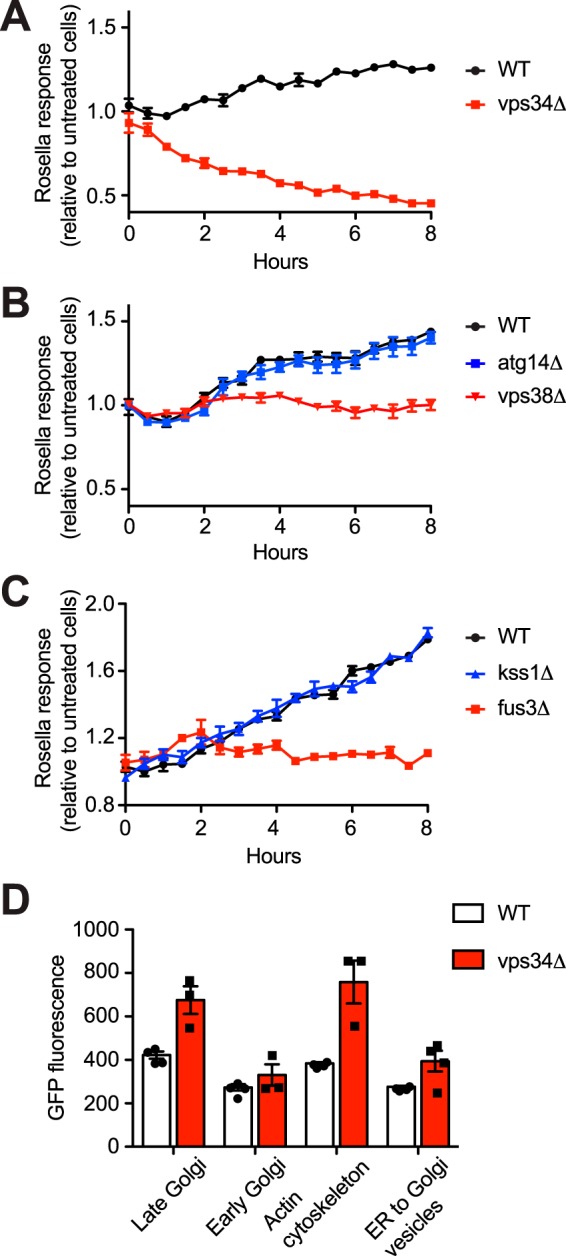
Pheromone-induced vacuolar targeting is mediated by the PI 3-kinase complex II and the MAPK Fus3. Time course of vacuolar targeting induced by 1 μm α-factor in BY4741 bar1Δ cells lacking Vps34 (A), Vps38 or Atg14 (B), and Fus3 or Kss1 (C). D, flow cytometry analysis of organelle abundance in WT and vps34Δ cells. Abundance is reported by fluorescence from GFP fused with Chc1 (late Golgi/clathrin), Cop1 (early Golgi), Sac6 (Actin cytoskeleton), or Sec13 (endoplasmic reticulum-to-Golgi transport vesicles). Rosella experiments were done at least two times, and data are mean ± S.D. from one experiment. Flow cytometry data are ± S.D. for four biological replicates (10,000 cells each).
Pheromone-induced vacuolar targeting is mediated by the MAPK Fus3
Whereas Gβγ recruits and activates Ste5 (97–101), Gpa1/Gα binds to and activates Vps34 (38). Ste5 assembles and activates the protein kinases Ste11, Ste7, and Fus3 but is also required for activation of Kss1 (15–22). In contrast, Vps34 activates Fus3 alone (38). Fus3, but not Kss1, is likewise required for autophagy under nitrogen-limiting conditions (102); under the same conditions Kss1, but not Fus3, promotes invasive/pseudohyphal growth in susceptible strains (34–36). Thus, we anticipated that Fus3 would be needed for pheromone-induced vacuolar targeting of Rosella. As predicted by this model, cells lacking Fus3 failed to respond to pheromone, whereas cells lacking Kss1 responded normally (Fig. 2C). Replacement of phosphorylation sites necessary for MAPK activation (FUS3T180A/Y182F) also eliminated the response (Fig. S2). Mutations in other pathway components, all of which are required for proper Fus3 expression and/or activation, are likewise required for vacuolar targeting. Specifically, we confirmed a requirement for the receptor (Ste2), Gβ (Ste4), scaffold (Ste5), MAPKKK (Ste11), MAPKK (Ste7), and transcription factor (Ste12). Although several of these proteins act downstream of Gpa1, they are necessary for the proper expression of mating pathway component(s) such as the receptor and Fus3; this could also explain why Ste4 is needed for signaling by constitutively active variants of Ste5 and Ste11, despite their well-established functions downstream of the Gβγ subunits (18, 38, 103). Cells lacking GPA1 are permanently arrested in the G1 phase of the cell cycle (104, 105).
Our results indicate that Vps34 is required for pheromone-induced vacuolar transport of the cytoplasmic reporter (Fig. 2A). To determine whether Vps34 also targets specific organelles for vacuolar degradation, we compared the abundance of four proteins with well-characterized and distinct subcellular localizations: Sec13–GFP (marker of endoplasmic reticulum–to–Golgi transport vesicles); Cop1–GFP (early Golgi), Chc1–GFP (late Golgi), and Sac6–GFP (actin cytoskeleton). As shown in Fig. 2D, most of these proteins showed higher abundance in the vps34Δ mutant, suggesting a role for the vacuolar targeting pathway in maintaining the abundance of cellular organelles and the actin cytoskeleton. Below, we examine the fate of these proteins upon mating.
F-box protein Dia2 promotes pheromone-dependent vacuolar targeting
In our previous screen of nearly 5000 gene deletion strains, we determined that Vps34 and Vps15 are required for signal transduction by Gpa1. The same screen revealed a similar requirement for the F-box protein Dia2. Dia2 is one of 16 members of the F-box family, which serves to recruit specific substrates for ubiquitination and for degradation by the proteasome protease complex (41). Although a role in autophagy has not been reported in yeast, F-box proteins are known to regulate autophagy in animal cells. A genome-wide human siRNA screen identified ZBTB16 as an inhibitor of PI 3-phosphate production and starvation-mediated autophagy (106). ZBTB16 mediates proteasome degradation of Atg14L (107). Two other adaptor proteins, FBLXL2 and KLHL20, promote ubiquitination of VPS34 in response to DNA damage (108) and nutrient starvation (109), respectively. Thus, F-box adaptor proteins can serve as important regulators of autophagy-related processes, at least in animal cells. Accordingly, we considered a similar role for the F-box proteins in yeast.
Given the documented role of Dia2 in Gpa1 signaling, we first determined its ability to regulate Rosella targeting. As shown in Fig. 3A, and in support of our model, cells lacking DIA2 exhibit a small impairment of response after 6 h of pheromone treatment. A comparison of pheromone concentrations at the 8-h time point also revealed a more graded pheromone dose-response profile, as compared with WT cells (Fig. 3B). We infer from these results that Dia2 promotes the ubiquitination and degradation of a component in the pheromone-response pathway. Moreover, the target of Dia2 is likely to be long-lived, given the length of time required to observe the attenuation of response.
Figure 3.
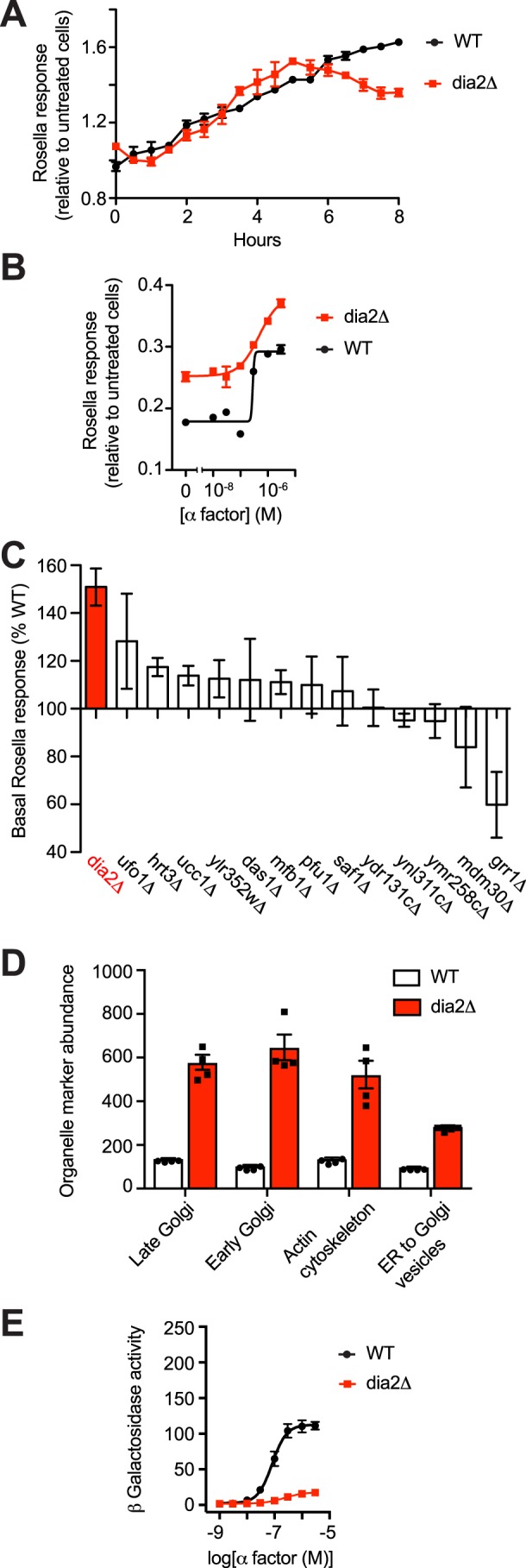
F-box protein Dia2 promotes pheromone-dependent vacuolar targeting. A, time course of Rosella response (relative to untreated cells) induced by 1 μm α-factor in BY4741 bar1Δ cells and cells lacking Dia2. B, dose dependence of response after 8 h of treatment with 0–3 μm α-factor. C, F-box screen. Bars represent percent basal response relative to BY4741 bar1Δ cells. D, flow cytometry analysis of organelle abundance in BY4741 WT and dia2Δ cells, as done above for vps34Δ (Fig. 2D). E, β-gal activity in cells expressing the FUS1-lacZ transcription reporter and treated with 0–3 μm α-factor for 90 min. Rosella experiments were done at least two times with four technical replicates, and data are mean ± S.D. from one experiment. Flow cytometry data are ± S.D. for four biological replicates (10,000 cells each).
To determine whether other F-box adaptor proteins regulate vacuolar targeting, we analyzed individual gene deletions using the Rosella assay. Whereas Dia2 promotes vacuolar targeting of cytoplasmic Rosella in pheromone-treated cells, it had the opposite effect in unstimulated conditions (Fig. 3B). The basal signal was likewise elevated in one other mutant tested (ufo1Δ, Fig. 3C), but that mutant did not exhibit any differences in response to pheromone stimulation.
We then considered whether Dia2 regulates the abundance of organelle marker proteins, in the manner of Vps34. As shown in Fig. 3D, abundance of Sec13–GFP, Cop1–GFP, Chc1–GFP, and Sac6–GFP are likewise elevated in the dia2Δ mutant. The stabilizing effects of the dia2Δ and vps34Δ mutants are in contrast to the observed destabilization of the cytoplasmic protein Rosella. Thus, Dia2 and Vps34 promote pheromone-induced vacuolar targeting of cytoplasmic proteins and also limit the abundance of organelle markers, at least under basal (unstimulated) conditions. We then considered the contribution of Dia2 to maintaining organelle abundance upon mating of haploid cells to produce diploids. To that end, MATa cells expressing the organelle markers were combined with MATα cells and, after selecting for diploids, quantified for GFP by flow cytometry. As shown in Fig. S3, haploid and diploid cells lacking Dia2 exhibited a similar increase in organelle abundance, which was not evident in either WT cells or cells lacking other F-box proteins. We conclude that Dia2 specifically regulates cytoplasmic but not organelle-associated proteins during the mating pathway signaling.
Finally, we considered other aspects of the mating response. In particular, we measured pathway-specific gene transcription with a reporter consisting of the FUS1 promoter fused to the β-gal gene. Compared with WT cells, a vps34Δ mutant has a substantially diminished transcription response (38). As shown in Fig. 3E, deletion of DIA2 likewise results in attenuation of transcription. Thus, Dia2 and Vps34 are both required to sustain pheromone-dependent transcription, and the changes in transcription mirror the changes in pheromone-dependent Rosella targeting reported above. In addition, Dia2, like Vps34, limits abundance of organelle-associated proteins but does so independently of the mating signal.
F-box protein Ucc1 regulates pheromone-dependent MAPK signaling
Taken together, our findings reveal a novel form of vacuolar targeting that is mediated by mating pheromone, the G-protein α subunit Gpa1, the PI 3-kinase Complex II, the MAPK Fus3, and the F-box protein Dia2. Although pheromone-induced vacuolar targeting has several unique features, it nonetheless acts in combination with known components of the pheromone-response pathway. For example, Fus3 is activated downstream of Gα as well as by Gβγ. To further distinguish these processes, we initiated a targeted screen for specific regulators of each output.
Given the specific role of Dia2 in vacuolar targeting, we tested our panel of 14 nonessential F-box deletion strains for any that modulate MAPK- and Cdc42-dependent signaling. We began by measuring activation of Fus3, which requires phosphorylation of two conserved residues, Tyr-180 and Thr-182. Cells were treated with pheromone, resolved by gel electrophoresis and immunoblotting, and probed with antibodies that detect the dual-phosphorylated form of the protein. In this case, the dia2Δ mutant sustained full Fus3 activation, despite the marked decrease in Fus3-mediated transcription and vacuolar targeting (110). Indeed, of the 14 deletion mutants tested, only ucc1Δ resulted in an appreciable change in the phospho-Fus3 signal (Fig. 4A). We conclude that Ucc1, but not Dia2, regulates Fus3 activation.
Figure 4.
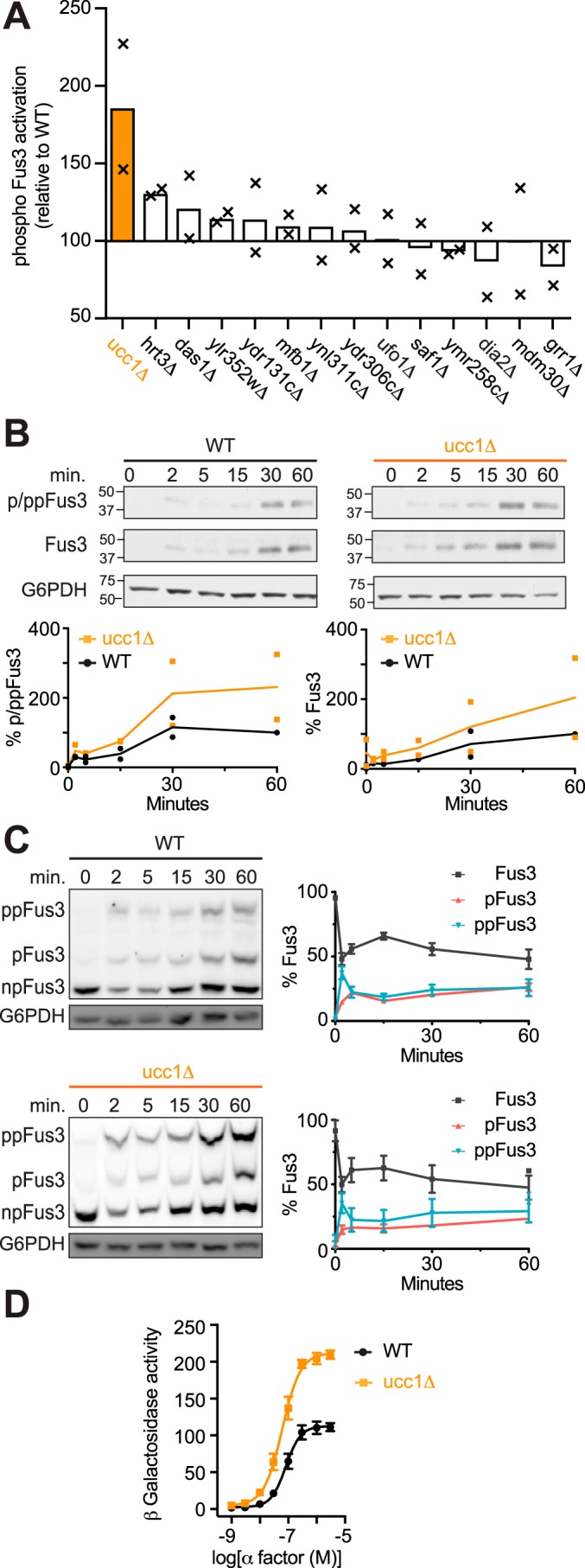
F-box protein Ucc1 promotes MAPK induction. A, F-box screen. Bars represent average phospho-MAPK (Fus3) activation relative to BY4741 (WT). Cells were treated with 3 μm α-factor for 60 min and analyzed by immunoblotting with phospho-p44/42 antibodies to detect activated Fus3. B, time course of Fus3 activation. WT and ucc1Δ cells were treated with 3 μm α-factor and analyzed by immunoblotting with phospho-p44/42 (p/pp-Fus3), Fus3, and G6PDH (load control) antibodies. C, Phos-tag immunoblot analysis of WT and ucc1Δ cells treated with α-factor and analyzed by immunoblotting, as described in B. Protein bands (left) representing dual-phosphorylated (ppFus3), mono-phosphorylated (pFus3), and nonphosphorylated (Fus3) forms of the protein were quantified using ImageJ and presented as a percentage of total Fus3 at 0 min (right). D, β-gal activity in cells expressing the FUS1-lacZ transcription reporter and treated with 0–3 μm α-factor for 90 min. Data represent percentage of maximum activity relative to WT cells. A and B show individual data points (×) and the mean of two biological replicates; all other data are ± S.D.
Ucc1 could work directly on Fus3, a Fus3-binding partner, or on Fus3-mediated transcription. Fus3 is not known to be degraded in a ubiquitin-dependent manner; however, it is well-known to invoke a positive feedback loop that amplifies the signal through increased transcription and induction of Fus3 itself (Fig. 4B). To distinguish between increased phosphorylation and increased abundance of the kinase, we used phos-tag gel electrophoresis to resolve the proportion of phosphorylated and unphosphorylated species (111). As shown in Fig. 4C, and as reported previously, pheromone treatment led to the rapid accumulation of dual-phosphorylated Fus3 and, following a slight delay, accumulation of mono-phosphorylated protein (this mono-phosphorylated pool of kinase does not stimulate but rather inhibits the mating response) (111). As is evident from these data, the proportion of phosphorylated and unphosphorylated protein was unaltered in the ucc1Δ strain. We infer that Ucc1 limits the overall abundance of Fus3 and that the increase in p/ppFus3 abundance (Fig. 4B) reflects an increase in Fus3 expression. In accordance with these findings, deletion of UCC1 led to an ∼2-fold increase in pheromone-induced gene transcription (Fig. 4D). We conclude that Ucc1 is a negative regulator of MAPK signaling, and its primary effect is on MAPK protein expression rather than kinase activation.
F-box protein Ydr306c (Pfu1) regulates pheromone-dependent morphogenesis
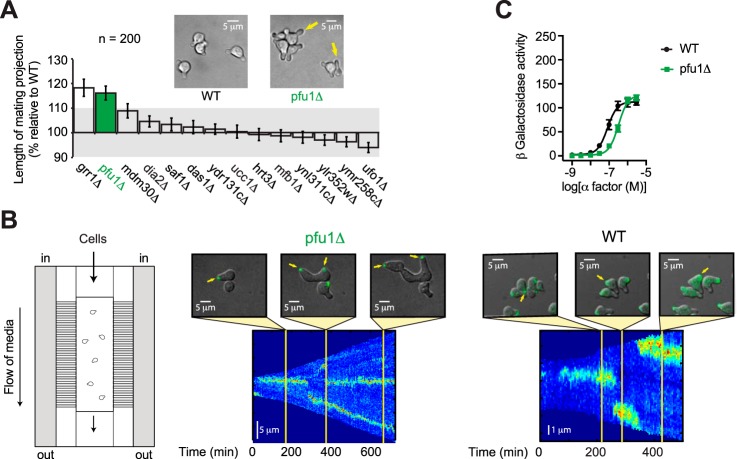
Having identified F-box proteins that regulate pheromone-induced vacuolar targeting (Dia2) and Fus3 induction (Ucc1), we considered the role of F-box proteins in morphogenesis. In preparation for mating, haploid cells form a pear-shaped projection, or “shmoo,” that expands in the direction of the pheromone gradient. Accordingly, we measured the size of the mating projection in WT cells and in cells lacking each of the nonessential F-box proteins. Cells were plated on agar pads supplemented with pheromone for 15 h. We then measured the base–to–tip lengths of mating projections in 200 individual cells for each strain. As shown in Fig. 5A, cells lacking UCC1 or DIA2 displayed little or no change in cell shape as compared with WT cells (Fig. 5A). In contrast, cells lacking GRR1 or YDR306c exhibited mating projections that are longer by 15–20%. Grr1 has a well-characterized role in cell cycle arrest and morphogenesis (112–114). The functional role of YDR306c has not previously been explored.
Figure 5.
F-box protein Pfu1 is required for proper morphogenesis. A, F-box screen. Bars represent average mating projection lengths relative to WT BY4741 (WT) (n = 200 cells). Cells were treated with 30 μm α-factor for 18 h prior to imaging. B, live cell imaging of the polarity marker Bem1–GFP in cells treated with 300 nm α-factor for 18 h. Shown is a schematic of the microfluidics device as well as representative images and kymographs of Bem1–GFP fluorescence. C, β-gal activity in cells expressing the FUS1-lacZ transcription reporter and treated with 0–3 μm α-factor for 90 min. Data represent percentage of maximum activity in WT cells. All data are ± S.D.
YDR306c was of particular interest because it is expressed predominantly in haploid (mating competent) cells, as determined by stable isotope labeling of amino acids and MS analysis (115). Very few proteins are uniquely found in haploid cells, but these include the pheromone receptor, all three G-protein subunits, the scaffold Ste5, the MAPK Fus3 (but not Kss1), and the downstream transcription factor Ste12 (115). All of these proteins are needed to detect and track a pheromone gradient and, with the exception of Fus3, all are essential for mating. Thus, Ydr306 may represent a new member of the mating pathway.
To further characterize the cell polarization function, we monitored oriented growth in cells lacking Ydr306, in response to a spatially uniform field of pheromone, using a custom microfluidics device (Fig. 5B) (30). Cell polarization was scored using the well-characterized polarity marker Bem1–GFP. Bem1 binds directly and selectively to the activated form of Cdc42, which together with its guanine nucleotide–exchange factor Cdc24 comprises the core components of the “polar cap.” The polar cap assembles at the tip of the first mating projection and remains there during active morphogenesis. Failure to find a mating partner causes WT cells to disassemble the cap and form a new polar cap at a different site (5, 116–118).
To quantify changes in the polar cap over time, we plotted the distribution of Bem1–GFP as line scans measured around the cell boundary (kymographs). As shown in Fig. 5B, cells normally expand in one direction for ∼200 min (represented as a single, sharp band of Bem1 fluorescence) before abandoning the cap and expanding in another direction. Establishment of a new polarity site coincides with a break in the kymograph traces, which is followed by a new fluorescent band at a different cell boundary position (y axis). For the first 200 min, ydr306cΔ mutants functioned much like WT cells. Between 200 and 600 min, however, the strains exhibited strikingly different behaviors. Whereas the WT cells abandoned the first projection, the mutant cells formed multiple projections while also maintaining the original site of expansion (Fig. 5B and Movie S1). The exaggerated growth may be due in part to a modest (2-fold) decrease in pheromone sensitivity (Fig. 5C), because WT cells exposed to a weak stimulus also become elongated in this manner (3, 30, 119). Reduced sensitivity is independent of MAPK activation, however, as shown in Fig. 4A (ydr306c). We conclude that YDR306c selectively regulates the morphogenesis branch of the pheromone pathway and, in particular, is required for efficient removal of the polar cap from nonproductive mating fronts. In light of the pheromone-dependent phenotype and its documented role in ubiquitination (41), we named the gene PFU1 (pheromone F-box ubiquitination).
Discussion
Vacuolar recycling of cellular contents has long been known to occur under conditions of nutrient limitation. Here we demonstrate, using the cytoplasmic biosensor Rosella, a parallel process that occurs in response to pheromone stimulation. Pheromone-induced vacuolar targeting is transmitted by the G-protein α subunit, the PI 3-kinase Vps34, and the protein kinase Fus3. The F-box protein Dia2 specifically regulates this event, whereas other F-box proteins selectively regulate MAPK induction and mating morphogenesis (Fig. 6). Whereas Dia2 promotes vacuolar targeting, Ucc1 regulates MAPK abundance, and Pfu1 promotes the disassembly of the polar cap. Pfu1 is of particular interest because it has not previously been characterized, is expressed exclusively in haploids, and limits the lifetime of the mating projection. Collectively, our data support the existence of a new branch of the pheromone-response pathway and reveal new components of the mating apparatus.
Figure 6.
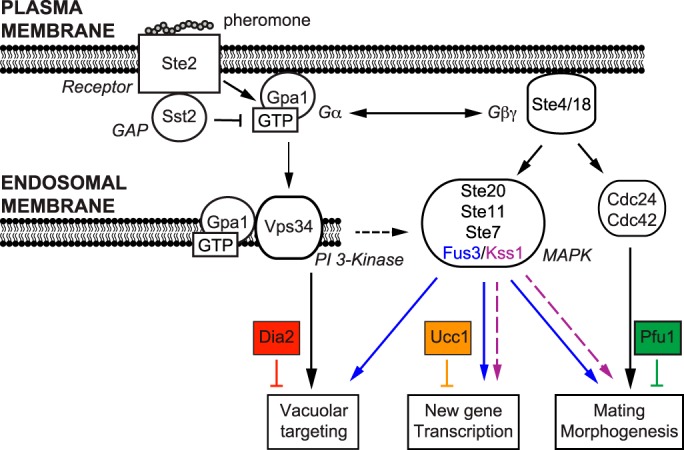
Model of the mating pathway with three distinct branches leading to vacuolar targeting, transcription, and morphogenesis. Binding of mating pheromone to its receptor Ste2 triggers activation of the G protein. Vacuolar targeting is initiated by Gpa1–GTP at endosomal membranes, requires the PI 3-kinase Vps34 and the terminal MAPK Fus3 (but not Kss1), and is positively regulated by the F-box protein Dia2. In contrast, new gene transcription and morphogenesis are initiated by the Gβγ subunits at the plasma membrane, transmitted by either Fus3 or Kss1, and negatively regulated by the F-box protein Ucc1. Proper morphogenesis is initiated by Gβγ, mediated by the small G-protein Cdc42, and is regulated by the F-box protein, Pfu1.
Our findings build on earlier studies linking Gpa1 to Vps34 activation and of Vps34 activation to autophagy. Previous efforts to investigate these processes were hampered, however, by the lack of appropriate genetic tools and sufficiently sensitive assays. Since it was first characterized by EM in the 1950s, a hallmark of autophagy has been the focal degradation of cytoplasmic content within lysosomes. More recently, autophagic activity is reported as the redistribution or degradation of Atg8 (or LC3), increased abundance of autophagosomes, and increased degradation of cytoplasmic proteins. Although these methods have revealed critical pathway components, they are not conducive to large-scale measures of pathway activity (83). Moreover, methods that rely on the appearance or abundance of GFP–Atg8 do not track specific cargo, are difficult to quantify, and can be affected by changes in cell size and morphology (as occurs during mating morphogenesis). Biochemical methods, such as those that rely on Pho8Δ60 alkaline phosphatase activity, are quantitative but require cell lysis, are not amenable to high-throughput analysis, and can be affected by the ubiquitin–proteasome pathway. By comparison, the Rosella reporter constitutes a marker-independent method to measure the sequestration of cytoplasmic cargo in living cells, over time, and under various growth conditions (84). By adapting the Rosella biosensor for use in a microplate reader, we now have a platform for quantitative measurement of Gpa1/Vps34 signaling, in multiple strains, at multiple concentrations of stimulus, and at multiple time points.
Our use of the Rosella bioassay yielded several important new advances related to yeast mating. First and foremost, we have shown that mating pheromone stimulates vacuolar targeting, albeit weakly as compared with the effects of nitrogen starvation. Similar findings have been reported previously for mammalian GPCRs, including the muscarinic cholinergic receptor (120–126) and β-adrenergic receptor (124–126). Other examples are likely to emerge, given that many GPCRs not only regulate but also detect the abundance of nutrients. For instance, there are GPCRs that bind to amino acids (GPRC6A, T1R1, and T1R3), long-chain fatty acids (GPR120 and GPR40), and short-chain fatty acids (GPR41 and GPR43). The T1R1/T1R3 receptors stimulate mechanistic Target of Rapamycin Complex I (mTORC1), which in turn phosphorylates and inhibits the PI 3-kinase needed for autophagosome biogenesis (122, 127).
One unusual aspect of pheromone-induced vacuolar targeting is the positive role for the G-protein α subunit, Gpa1. It has long been recognized that Gpa1 controls the release of Gβγ, which in turn recruits Ste5 and Cdc24, and these processes are sufficient to drive nearly all aspects of the mating response. In contrast, it is Gpa1–GTP that binds to and activates Vps34 (38), and Vps34 activity is absolutely required for autophagy and vacuolar targeting (42, 128). Another unusual aspect of this system is the endomembrane location of the effector protein Vps34. It had long been assumed that G proteins function exclusively at the plasma membrane, in conjunction with cell-surface receptors and extracellular ligands. Our findings indicate that Gpa1 functions at endosomes to promote PI 3-kinase activity. This process is likely to be conserved in mammals, given that VPS34 binds directly to Gαi (129) and is activated by at least one GPCR (130) as well as the nonreceptor exchange factor Ric-8A (129).
Another unusual aspect of pheromone-induced vacuolar targeting is the specific requirement for Fus3. Whereas Fus3 and Kss1 are both activated by pheromone, Fus3 alone is activated by Gpa1. Whereas Fus3 alone promotes cell cycle arrest (29), gradient tracking (30), and vacuolar tracking, either kinase can sustain mating morphogenesis and transcriptional induction (17, 111, 131, 132). However, there is clearly some integration of Gpa1 signaling with other branches of the mating pathway. For example, Gβγ can also promote the activation of Fus3. Thus, our use of F-box deletions is significant because it allowed us to show that the three branches can be functionally separated. An important goal for the future is to identify the relevant substrates for Fus3 and the three F-box proteins. Pfu1 is of particular interest because it is expressed exclusively in haploid cells (115), and it is therefore likely to have a unique and specific role in mating.
In summary, these findings advance our understanding in several important ways. First, we have determined the functional consequences of Vps34 activation by Gpa1. In particular, our results demonstrate that vacuolar targeting is stimulated by mating pheromone and is mediated by Vps34. Although Vps34 is well-known to be required for autophagy and cytoplasm–to–vacuolar targeting, these events had not been investigated in detail in the context of the yeast G-protein–signaling pathway. The fluorescent biosensor Rosella provided the technical breakthrough needed to answer this long-standing question. Moreover, our systematic analysis of F-box deletions provided corroborating evidence for the existence of a third distinct branch of pheromone-signaling pathway. Finally, our studies revealed that one of the F-box proteins Pfu1 acts to limit the lifetime of the mating projection. Pfu1 had not previously been characterized but was of particular interest because it (like the G protein) is expressed exclusively in signaling-competent haploid cells. F-box proteins serve as receptors for the plant hormones auxin and jasmonates (133–135). Thus, homologous proteins could serve as targets for inhibitors that selectively enhance or diminish GPCR signaling and autophagy-related processes in humans.
Experimental procedures
Strains, plasmids, and growth conditions
Yeast strains used in this study were BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ) and BY4741-derived gene deletion mutants obtained from the Yeast Knockout Collection (Invitrogen) for transcription and morphogenesis screens or remade by homologous recombination of PCR-amplified drug resistance genes (136) with flanking homology to the gene of interest (Table S1) for autophagy and Rosella assays. Gpa1Q323L and WT Gpa1 were expressed using plasmid pRS316 (39). Pho8Δ60 was stably integrated into the genome, replacing the native PHO8 gene (gift from Mara Duncan, University of Michigan). The plasmid pRS416–GFP–ATG8 (Addgene 49425) was obtained as a gift from Daniel Klionsky (137), and the GFP–ATG8 construct was subsequently introduced into the pRS415 vector. The autophagy biosensor Rosella was expressed with the pAS1NB–DsRed.T3–SEP plasmid (2μ, ampR, LEU2+) (gift from Mark Prescott and Rodney Devenish, Monash University) (84). Cells were grown in synthetic complete medium supplemented with antibiotics or lacking specific nutrients to maintain plasmid selection and containing 2% w/v dextrose (hereafter, SCD medium or SCD − nutrient). Colonies were inoculated into liquid medium and grown overnight at 30 °C. Saturated cultures were then diluted with fresh medium to A600 = 0.1 and cultured for 6 h, then further diluted to A600 = 0.001, and cultured for 18 h to maintain A600 <1 prior to use. Cells were transferred to low-nitrogen conditions by washing and resuspending in SCD medium lacking ammonium sulfate (SCD − nit).
Autophagy assays
The Pho8Δ60 assay was performed as described previously (70). Cells were grown to absorbance (at 600 nm) ∼1 in SCD medium prior to transfer to SCD − nit medium. Aliquots were collected after 0, 4, and 8 h of starvation, transferred to assay buffer (250 mm Tris-HCl, pH 9.0, 10 mm MgSO4, 10 μm ZnSO4), and lysed with glass beads on an automatic vortex mixer (5 min at 4 °C). The cell lysate was diluted 10-fold and incubated with substrate (5 mm α-naphthyl phosphate, Sigma N7255) at 30 °C for 20 min. Enzyme activity was measured spectrophotometrically (345 nm excitation and 472 nm emission) and presented as emission per amount of protein in the reaction, which was determined via the Bradford method. For the GFP–Atg8 assay (71), cells were similarly propagated in SCD-leucine medium prior to nitrogen starvation. Cell extracts were obtained after 0, 3, and 6 h of starvation. Cell pellets were lysed with TCA buffer, and protein extracts were normalized and resolved by 10% SDS-PAGE. Proteins were detected by immunoblotting with GFP antibodies (sc-999c, clone B-2, Santa Cruz Biotechnology) or glucose-6-phosphate dehydrogenase (G6PDH) antibodies (Sigma). Immunoreactive species were detected with antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch) using ECL-plus reagent (Life Technologies, Inc.) on a Bio-Rad Chemidoc Touch Imaging System. Protein bands were quantified using Image Lab software version 6.0.1 (Bio-Rad).
Rosella assay
Cells lacking the secreted protease Bar1 were exposed to 0–3 μm mating pheromone α-factor in SCD-leucine medium in black clear-bottom 96-well microplates (Greiner 655087 or Corning 3631). As a positive control, a separate culture was starved of nitrogen, as described previously. Background signal was measured using clear medium that did not contain cells. Microplates were sealed to reduce evaporation (adhesive PCR plate seal, Thermo Fisher Scientific AB0558) and placed in a microplate reader (Molecular Devices SpectraMax M5 or i3x) for 8 h at 30 °C. At each time point, samples were shaken, and fluorescence was measured for SEP (488 nm excitation and 530 nm emission) and DsRed.T3 (543 nm excitation and 587 nm emission). Pheromone treatment does not by itself affect cytoplasmic pH (138). All experiments were performed two or more times in quadruplicate. Time-course response profiles were generated by calculating the ratio of background-corrected dsRed.T3 and SEP fluorescence. Dose-response profiles were calculated at the 8-h time point using a variable slope (four parameters) nonlinear regression with least-squares fit (GraphPad Prism). The magnitude of pheromone-induced vacuolar targeting of Rosella was determined at each time point by scaling the pheromone-induced response with basal response in untreated cells.
Organelle culling
Organelle abundance was measured in cells expressing GFP- or RFP-tagged Cop1 (early Golgi), Chc1 (late Golgi/clathrin), Sec13 (endoplasmic reticulum to Golgi), Sac6 (actin cytoskeleton), or Om45 (mitochondria) (139). BY4741 (MATa) cells containing GFP-labeled organelles were combined with BY4742 (MATα) cells expressing RFP-labeled organelles on YPD agar plates overnight. Diploid cells expressing both GFP- and RFP-labeled organelles were selected on SCD − histidine + G418 plates. Colonies were inoculated into SCD medium as described above, and GFP (488 nm excitation and 525/50 nm emission) and RFP (594 nm excitation and 610/20 nm emission) fluorescence were measured on an LSR II flow cytometer (BD Biosciences). BY4741 lacking fluorescent markers was used to measure background signal. The RFP signal was used to confirm that no residual MATa cells were selected in the population of diploid cells. Flow cytometry data were analyzed using FlowJo version 10 and are presented as the average median fluorescence intensity of 10,000 cells from four biological replicates.
Transcription reporter assay
Pathway-specific outputs leading to transcription and morphogenesis were monitored using previously published methods. Transcription was quantified as the activity of the lacZ (β-gal) reporter expressed from the pheromone-inducible FUS1 promoter (140). Cells were grown in SCD − histidine medium to absorbance (at 600 nm) ∼1, exposed to 0–10 μm mating pheromone for 90 min, permeabilized, and incubated with the substrate fluorescein di-β-d-galactopyranoside for another 90 min. β-Gal hydrolyzes the substrate to fluorescein, which is quantified by fluorescence (485 nm excitation and 530 nm emission). A signal is evident after 2 h, similar to the Rosella assay for vacuolar targeting. The assay is stopped after 3 h to avoid depleting the substrate.
Morphogenesis assay
Morphogenesis was measured using brightfield illumination on a Nikon Ti 2000-inverted microscope. Agar pads were prepared by dissolving 2% agar in clear SCD medium and spotting (200 μl) onto a clear glass slide. Another slide was quickly placed on top, and the agarose was allowed to solidify. After removing the top slide, the agar pad was supplemented with 30 μm α-factor in SCD medium. High concentrations of pheromone are needed in WT (BAR1+) cells to allow for the formation of multiple mating projections. Next, 5 μl of cells were mounted on the pad and sealed with a clean glass coverslip as described previously (141). Mating projection lengths were determined using FIJI image analysis software (142). Data are shown as percentage of the average length of mating projections relative to WT cells (n = 200). Morphogenesis-related changes in the localization of the polarity marker protein Bem1 were monitored as described previously (5). Briefly, cells expressing Bem1–GFP were immobilized in a microfluidic chamber and exposed to a constant flow of fresh growth medium containing 300 nm α-factor in both channels. Because the medium is constantly being replenished, low concentrations of pheromone are sufficient to invoke the formation of mating projections. Images were acquired at 5-min intervals for 12 h as a z-series of 1-μm step size, 5 μm around the focal plane, using an Olympus IX83 spinning-disc confocal microscope equipped with an Andor Revolution XD spinning-disk unit. Kymographs were generated using FIJI and MATLAB, as described previously (5).
Phospho-MAPK and Phos-tag immunoblotting
MAPK activation was measured using quantitative immunoblotting as described previously for Hog1 (143). Briefly, cells grown to absorbance (at 600 nm) ∼1 in yeast extract/peptone/2% dextrose (YPD) medium were treated with 3 μm α-factor pheromone, and aliquots were collected at the indicated time points. Cell pellets were lysed with TCA buffer; protein extracts were normalized and resolved by either standard 10 or 12% acrylamide SDS-PAGE or 50 μm Zn2+-Phos-tag in 10% acrylamide SDS-PAGE (Wako Chemicals). Proteins were detected by immunoblotting with phospho-p44/42 antibodies (9101, Cell Signaling Technology), Fus3 antibodies (Santa Cruz Biotechnology), or G6PDH antibodies (Sigma). Immunoreactive species were detected with antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology) using ECL-plus reagent (Life Technologies, Inc.) or with Alexa Fluor 647 (Jackson ImmunoResearch) on a Bio-Rad Chemidoc Touch Imaging System. Protein bands were quantified using the scanning densitometry function within FIJI (142).
Author contributions
N. R., C. L. G., and H. G. D. conceptualization; N. R. and C. L. G. resources; N. R. data curation; N. R. validation; N. R., C. L. G., L. A., S. M. B., J. H. H., J. B. K., J. B. S., S. K. S., N. H. V., and E. Y. investigation; N. R. and C. L. G. methodology; N. R. and H. G. D. writing-original draft; N. R. project administration; N. R. and H. G. D. writing-review and editing; T. C. E., B. E., and H. G. D. supervision; H. G. D. funding acquisition.
Supplementary Material
Acknowledgments
The University of North Carolina Flow Cytometry Core Facility is supported in part by National Institutes of Health P30 CA016086 Cancer Center Core Support Grant to the University of North Carolina Lineberger Comprehensive Cancer Center.
This work was supported by National Institutes of Health Grants R35GM118105 (to H. G. D.) and R01GM114136 (to H. G. D., B. E., and T. C. E.), National Institute of General Medical Sciences, Division of Training, Workforce Development, and Diversity under the Institutional Research and Academic Career Development Award, Grant K12-GM000678 (to C. L. G.), a Barthalmus Undergraduate Research Grant from the State of North Carolina Undergraduate Research and Creativity Symposium (to L. A.), and an individual Summer Undergraduate Research Fellowship from the American Society for Pharmacology and Experimental Therapeutics (to J. B. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3, Table S1, and supporting Movie S1.
- GPCR
- G-protein–coupled receptor
- PI
- phosphatidylinositol
- MAPK
- mitogen-activated protein kinase
- SEP
- super-ecliptic pHluorin
- SCD
- synthetic complete medium with dextrose
- G6PDH
- glucose-6-phosphate dehydrogenase
- RGS
- regulator of G-protein signaling
- RFP
- red fluorescent protein.
References
- 1. Stewart A., and Fisher R. A. (2015) Introduction: G protein-coupled receptors and RGS proteins. Prog. Mol. Biol. Transl. Sci. 133, 1–11 10.1016/bs.pmbts.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 2. Dohlman H. G., and Thorner J. W. (2001) Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70, 703–754 10.1146/annurev.biochem.70.1.703 [DOI] [PubMed] [Google Scholar]
- 3. Segall J. E. (1993) Polarization of yeast cells in spatial gradients of α mating factor. Proc. Natl. Acad. Sci. U.S.A. 90, 8332–8336 10.1073/pnas.90.18.8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorer R., Pryciak P. M., and Hartwell L. H. (1995) Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 131, 845–861 10.1083/jcb.131.4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelley J. B., Dixit G., Sheetz J. B., Venkatapurapu S. P., Elston T. C., and Dohlman H. G. (2015) RGS proteins and septins cooperate to promote chemotropism by regulating polar cap mobility. Curr. Biol. 25, 275–285 10.1016/j.cub.2014.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan R. K., and Otte C. A. (1982) Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2, 21–29 10.1128/MCB.2.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan R. K., and Otte C. A. (1982) Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2, 11–20 10.1128/MCB.2.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixit G., Kelley J. B., Houser J. R., Elston T. C., and Dohlman H. G. (2014) Cellular noise suppression by the regulator of G protein signaling Sst2. Mol. Cell 55, 85–96 10.1016/j.molcel.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Z. S., Leung T., Manser E., and Lim L. (1995) Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol. 15, 5246–5257 10.1128/MCB.15.10.5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valtz N., Peter M., and Herskowitz I. (1995) FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131, 863–873 10.1083/jcb.131.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Butty A. C., Pryciak P. M., Huang L. S., Herskowitz I., and Peter M. (1998) The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282, 1511–1516 10.1126/science.282.5393.1511 [DOI] [PubMed] [Google Scholar]
- 12. Nern A., and Arkowitz R. A. (1999) A Cdc24p–Far1p–Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144, 1187–1202 10.1083/jcb.144.6.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimada Y., Gulli M. P., and Peter M. (2000) Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2, 117–124 10.1038/35000073 [DOI] [PubMed] [Google Scholar]
- 14. Zheng Y., Cerione R., and Bender A. (1994) Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 269, 2369–2372 [PubMed] [Google Scholar]
- 15. Printen J. A., and Sprague G. F. Jr. (1994) Protein–protein interactions in the yeast pheromone-response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi K. Y., Satterberg B., Lyons D. M., and Elion E. A. (1994) Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78, 499–512 10.1016/0092-8674(94)90427-8 [DOI] [PubMed] [Google Scholar]
- 17. Breitkreutz A., Boucher L., and Tyers M. (2001) MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11, 1266–1271 10.1016/S0960-9822(01)00370-0 [DOI] [PubMed] [Google Scholar]
- 18. Andersson J., Simpson D. M., Qi M., Wang Y., and Elion E. A. (2004) Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 23, 2564–2576 10.1038/sj.emboj.7600250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maleri S., Ge Q., Hackett E. A., Wang Y., Dohlman H. G., and Errede B. (2004) Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol. Cell. Biol. 24, 9221–9238 10.1128/MCB.24.20.9221-9238.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kusari A. B., Molina D. M., Sabbagh W. Jr, Lau C. S., and Bardwell L. (2004) A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164, 267–277 10.1083/jcb.200310021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flatauer L. J., Zadeh S. F., and Bardwell L. (2005) Mitogen-activated protein kinases with distinct requirements for Ste5 scaffolding influence signaling specificity in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 1793–1803 10.1128/MCB.25.5.1793-1803.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M., and Lim W. A. (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311, 822–826 10.1126/science.1120941 [DOI] [PubMed] [Google Scholar]
- 23. Simon M. N., De Virgilio C., Souza B., Pringle J. R., Abo A., and Reed S. I. (1995) Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376, 702–705 10.1038/376702a0 [DOI] [PubMed] [Google Scholar]
- 24. Oehlen L. J., and Cross F. R. (1998) The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 8556–8559 10.1074/jbc.273.15.8556 [DOI] [PubMed] [Google Scholar]
- 25. Moskow J. J., Gladfelter A. S., Lamson R. E., Pryciak P. M., and Lew D. J. (2000) Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 7559–7571 10.1128/MCB.20.20.7559-7571.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts C. J., Nelson B., Marton M. J., Stoughton R., Meyer M. R., Bennett H. A., He Y. D., Dai H., Walker W. L., Hughes T. R., Tyers M., Boone C., and Friend S. H. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880 10.1126/science.287.5454.873 [DOI] [PubMed] [Google Scholar]
- 27. Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., and Young R. A. (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404 10.1016/S0092-8674(03)00301-5 [DOI] [PubMed] [Google Scholar]
- 28. Paliwal S., Iglesias P. A., Campbell K., Hilioti Z., Groisman A., and Levchenko A. (2007) MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446, 46–51 10.1038/nature05561 [DOI] [PubMed] [Google Scholar]
- 29. Erdman S., and Snyder M. (2001) A filamentous growth response mediated by the yeast mating pathway. Genetics 159, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hao N., Nayak S., Behar M., Shanks R. H., Nagiec M. J., Errede B., Hasty J., Elston T. C., and Dohlman H. G. (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol. Cell 30, 649–656 10.1016/j.molcel.2008.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Errede B., Vered L., Ford E., Pena M. I., and Elston T. C. (2015) Pheromone-induced morphogenesis and gradient tracking are dependent on the MAPK Fus3 binding to Gα. Mol. Biol. Cell 26, 3343–3358 10.1091/mbc.e15-03-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hegemann B., Unger M., Lee S. S., Stoffel-Studer I., van den Heuvel J., Pelet S., Koeppl H., and Peter M. (2015) A cellular system for spatial signal decoding in chemical gradients. Dev. Cell 35, 458–470 10.1016/j.devcel.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 33. Conlon P., Gelin-Licht R., Ganesan A., Zhang J., and Levchenko A. (2016) Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E5896–E5905 10.1073/pnas.1610081113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Madhani H. D., Styles C. A., and Fink G. R. (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91, 673–684 10.1016/S0092-8674(00)80454-7 [DOI] [PubMed] [Google Scholar]
- 35. Cook J. G., Bardwell L., and Thorner J. (1997) Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390, 85–88 10.1038/36355 [DOI] [PubMed] [Google Scholar]
- 36. Bardwell L., Cook J. G., Voora D., Baggott D. M., Martinez A. R., and Thorner J. (1998) Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12, 2887–2898 10.1101/gad.12.18.2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo M., Aston C., Burchett S. A., Dyke C., Fields S., Rajarao S. J., Uetz P., Wang Y., Young K., and Dohlman H. G. (2003) The yeast G protein a subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol. Cell 12, 517–524 10.1016/S1097-2765(03)00307-1 [DOI] [PubMed] [Google Scholar]
- 38. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., and Dohlman H. G. (2006) Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein α subunit at the endosome. Cell 126, 191–203 10.1016/j.cell.2006.04.045 [DOI] [PubMed] [Google Scholar]
- 39. Apanovitch D. M., Iiri T., Karasawa T., Bourne H. R., and Dohlman H. G. (1998) Second site suppressor mutations of a GTPase-deficient G-protein α subunit. Selective inhibition of βγ-mediated signaling. J. Biol. Chem. 273, 28597–28602 10.1074/jbc.273.44.28597 [DOI] [PubMed] [Google Scholar]
- 40. Palecek S. P., Parikh A. S., and Kron S. J. (2000) Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156, 1005–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kus B. M., Caldon C. E., Andorn-Broza R., and Edwards A. M. (2004) Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54, 455–467 10.1002/prot.10620 [DOI] [PubMed] [Google Scholar]
- 42. Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., and Emr S. D. (1993) Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- 43. Stack J. H., Herman P. K., Schu P. V., and Emr S. D. (1993) A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195–2204 10.1002/j.1460-2075.1993.tb05867.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Backer J. M. (2016) The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 473, 2251–2271 10.1042/BCJ20160170 [DOI] [PubMed] [Google Scholar]
- 45. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 46. Zheng N., and Shabek N. (2017) Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- 47. Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., and Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 10.1038/35008096 [DOI] [PubMed] [Google Scholar]
- 48. Hao N., Yildirim N., Wang Y., Elston T. C., and Dohlman H. G. (2003) Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J. Biol. Chem. 278, 46506–46515 10.1074/jbc.M308432200 [DOI] [PubMed] [Google Scholar]
- 49. Wang Y., and Dohlman H. G. (2002) Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277, 15766–15772 10.1074/jbc.M111733200 [DOI] [PubMed] [Google Scholar]
- 50. Wang Y., Ge Q., Houston D., Thorner J., Errede B., and Dohlman H. G. (2003) Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1–Cullin–F-box complex. J. Biol. Chem. 278, 22284–22289 10.1074/jbc.M301272200 [DOI] [PubMed] [Google Scholar]
- 51. Dikic I. (2017) Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- 52. Kanki T., and Klionsky D. J. (2008) Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393 10.1074/jbc.M802403200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okamoto K., Kondo-Okamoto N., and Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 54. Kanki T., Wang K., Cao Y., Baba M., and Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanki T., and Klionsky D. J. (2010) The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75, 795–800 10.1111/j.1365-2958.2009.07035.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lynch-Day M. A., and Klionsky D. J. (2010) The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359–1366 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsukada M., and Ohsumi Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- 58. Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., and Wolf D. H. (1994) Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280 10.1016/0014-5793(94)00672-5 [DOI] [PubMed] [Google Scholar]
- 59. Harding T. M., Morano K. A., Scott S. V., and Klionsky D. J. (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602 10.1083/jcb.131.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kawamata T., Kamada Y., Suzuki K., Kuboshima N., Akimatsu H., Ota S., Ohsumi M., and Ohsumi Y. (2005) Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 338, 1884–1889 10.1016/j.bbrc.2005.10.163 [DOI] [PubMed] [Google Scholar]
- 61. Stasyk O. V., Stasyk O. G., Mathewson R. D., Farré J. C., Nazarko V. Y., Krasovska O. S., Subramani S., Cregg J. M., and Sibirny A. A. (2006) Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy 2, 30–38 10.4161/auto.2226 [DOI] [PubMed] [Google Scholar]
- 62. Yen W. L., and Klionsky D. J. (2007) Atg27 is a second transmembrane cycling protein. Autophagy 3, 254–256 10.4161/auto.3823 [DOI] [PubMed] [Google Scholar]
- 63. Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- 64. Mizushima N., Yoshimori T., and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 65. Reggiori F., and Klionsky D. J. (2013) Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 10.1534/genetics.112.149013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ogier-Denis E., Couvineau A., Maoret J. J., Houri J. J., Bauvy C., De Stefanis D., Isidoro C., Laburthe M., and Codogno P. (1995) A heterotrimeric Gi3-protein controls autophagic sequestration in the human colon cancer cell line HT-29. J. Biol. Chem. 270, 13–16 10.1074/jbc.270.1.13 [DOI] [PubMed] [Google Scholar]
- 67. Ghidoni R., Houri J. J., Giuliani A., Ogier-Denis E., Parolari E., Botti S., Bauvy C., and Codogno P. (1996) The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur. J. Biochem. 237, 454–459 10.1111/j.1432-1033.1996.0454k.x [DOI] [PubMed] [Google Scholar]
- 68. Ogier-Denis E., Houri J. J., Bauvy C., and Codogno P. (1996) Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J. Biol. Chem. 271, 28593–28600 10.1074/jbc.271.45.28593 [DOI] [PubMed] [Google Scholar]
- 69. Petiot A., Ogier-Denis E., Bauvy C., Cluzeaud F., Vandewalle A., and Codogno P. (1999) Subcellular localization of the Gαi3 protein and Gα interacting protein, two proteins involved in the control of macroautophagy in human colon cancer HT-29 cells. Biochem. J. 337, 289–295 10.1042/0264-6021:3370289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Noda T., and Klionsky D. J. (2008) The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- 71. Shintani T., and Klionsky D. J. (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., and Ohsumi Y. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 73. Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., and Inagaki F. (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9, 611–618 10.1111/j.1356-9597.2004.00750.x [DOI] [PubMed] [Google Scholar]
- 74. Polson H. E., de Lartigue J., Rigden D. J., Reedijk M., Urbé S., Clague M. J., and Tooze S. A. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- 75. Abeliovich H., Dunn W. A. Jr., Kim J., and Klionsky D. J. (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034 10.1083/jcb.151.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xie Z., Nair U., and Klionsky D. J. (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 10.1091/mbc.e07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., and Ohsumi Y. (2007) The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- 78. Fujita N., Itoh T., Omori H., Fukuda M., Noda T., and Yoshimori T. (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 10.1091/mbc.e07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., and Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaufmann A., Beier V., Franquelim H. G., and Wollert T. (2014) Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell 156, 469–481 10.1016/j.cell.2013.12.022 [DOI] [PubMed] [Google Scholar]
- 81. Lee Y. K., and Lee J. A. (2016) Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49, 424–430 10.5483/BMBRep.2016.49.8.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rubinsztein D. C., Shpilka T., and Elazar Z. (2012) Mechanisms of autophagosome biogenesis. Curr. Biol. 22, R29–R34 10.1016/j.cub.2011.11.034 [DOI] [PubMed] [Google Scholar]
- 83. Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C. M., Adams P. D., Adeli K., Adhihetty P. J., Adler S. G., Agam G., Agarwal R., Aghi M. K., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd Ed). Autophagy 12, 1–222 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rosado C. J., Mijaljica D., Hatzinisiriou I., Prescott M., and Devenish R. J. (2008) Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4, 205–213 10.4161/auto.5331 [DOI] [PubMed] [Google Scholar]
- 85. Miesenböck G., De Angelis D. A., and Rothman J. E. (1998) Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- 86. Bevis B. J., and Glick B. S. (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20, 83–87 10.1038/nbt0102-83 [DOI] [PubMed] [Google Scholar]
- 87. Apanovitch D. M., Slep K. C., Sigler P. B., and Dohlman H. G. (1998) Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Gα protein pair in yeast. Biochemistry 37, 4815–4822 10.1021/bi9729965 [DOI] [PubMed] [Google Scholar]
- 88. Herman P. K., Stack J. H., and Emr S. D. (1991) A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. EMBO J. 10, 4049–4060 10.1002/j.1460-2075.1991.tb04981.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stack J. H., DeWald D. B., Takegawa K., and Emr S. D. (1995) Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321–334 10.1083/jcb.129.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kametaka S., Okano T., Ohsumi M., and Ohsumi Y. (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291 10.1074/jbc.273.35.22284 [DOI] [PubMed] [Google Scholar]
- 91. Kihara A., Noda T., Ishihara N., and Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yuan H. X., Russell R. C., and Guan K. L. (2013) Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995 10.4161/auto.26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kim J., Kim Y. C., Fang C., Russell R. C., Kim J. H., Fan W., Liu R., Zhong Q., and Guan K. L. (2013) Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Baskaran S., Carlson L. A., Stjepanovic G., Young L. N., Kim D. J., Grob P., Stanley R. E., Nogales E., and Hurley J. H. (2014) Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife 3, 2014, 10.7554/eLife.05115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J. E., Masson G. R., Johnson C., Steyaert J., Ktistakis N. T., and Williams R. L. (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365 10.1126/science.aac7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Whiteway M. S., Wu C., Leeuw T., Clark K., Fourest-Lieuvin A., Thomas D. Y., and Leberer E. (1995) Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science 269, 1572–1575 10.1126/science.7667635 [DOI] [PubMed] [Google Scholar]
- 98. Feng Y., Song L. Y., Kincaid E., Mahanty S. K., and Elion E. A. (1998) Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8, 267–278 10.1016/S0960-9822(98)70108-3 [DOI] [PubMed] [Google Scholar]
- 99. Pryciak P. M., and Huntress F. A. (1998) Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone-response pathway. Genes Dev. 12, 2684–2697 10.1101/gad.12.17.2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sette C., Inouye C. J., Stroschein S. L., Iaquinta P. J., and Thorner J. (2000) Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11, 4033–4049 10.1091/mbc.11.11.4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang Y., Chen W., Simpson D. M., and Elion E. A. (2005) Cdc24 regulates nuclear shuttling and recruitment of the Ste5 scaffold to a heterotrimeric G protein in Saccharomyces cerevisiae. J. Biol. Chem. 280, 13084–13096 10.1074/jbc.M410461200 [DOI] [PubMed] [Google Scholar]
- 102. Umekawa M., and Klionsky D. J. (2012) Ksp1 kinase regulates autophagy via the target of rapamycin complex 1 (TORC1) pathway. J. Biol. Chem. 287, 16300–16310 10.1074/jbc.M112.344952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Inouye C., Dhillon N., and Thorner J. (1997) Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278, 103–106 10.1126/science.278.5335.103 [DOI] [PubMed] [Google Scholar]
- 104. Dietzel C., and Kurjan J. (1987) The yeast SCG1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell 50, 1001–1010 10.1016/0092-8674(87)90166-8 [DOI] [PubMed] [Google Scholar]
- 105. Miyajima I., Nakafuku M., Nakayama N., Brenner C., Miyajima A., Kaibuchi K., Arai K., Kaziro Y., and Matsumoto K. (1987) GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 50, 1011–1019 10.1016/0092-8674(87)90167-X [DOI] [PubMed] [Google Scholar]
- 106. Lipinski M. M., Hoffman G., Ng A., Zhou W., Py B. F., Hsu E., Liu X., Eisenberg J., Liu J., Blenis J., Xavier R. J., and Yuan J. (2010) A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell 18, 1041–1052 10.1016/j.devcel.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang T., Dong K., Liang W., Xu D., Xia H., Geng J., Najafov A., Liu M., Li Y., Han X., Xiao J., Jin Z., Peng T., Gao Y., Cai Y., et al. (2015) G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. Elife 4, e06734 10.7554/eLife.06734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xiao J., Zhang T., Xu D., Wang H., Cai Y., Jin T., Liu M., Jin M., Wu K., and Yuan J. (2015) FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 29, 184–196 10.1101/gad.252528.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liu C. C., Lin Y. C., Chen Y. H., Chen C. M., Pang L. Y., Chen H. A., Wu P. R., Lin M. Y., Jiang S. T., Tsai T. F., and Chen R. H. (2016) Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol. Cell 61, 84–97 10.1016/j.molcel.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 110. Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R. 3rd., Madhani H. D. (2004) Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119, 991–1000 10.1016/j.cell.2004.11.052 [DOI] [PubMed] [Google Scholar]
- 111. Nagiec M. J., McCarter P. C., Kelley J. B., Dixit G., Elston T. C., and Dohlman H. G. (2015) Signal inhibition by a dynamically regulated pool of monophosphorylated MAPK. Mol. Biol. Cell 26, 3359–3371 10.1091/mbc.e15-01-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Barral Y., Jentsch S., and Mann C. (1995) G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9, 399–409 10.1101/gad.9.4.399 [DOI] [PubMed] [Google Scholar]
- 113. Skowyra D., Koepp D. M., Kamura T., Conrad M. N., Conaway R. C., Conaway J. W., Elledge S. J., and Harper J. W. (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284, 662–665 10.1126/science.284.5414.662 [DOI] [PubMed] [Google Scholar]
- 114. Schweitzer K., Cocklin R., Garrett L., Desai F., and Goebl M. (2005) The ubiquitin ligase SCFGrr1 is necessary for pheromone sensitivity in Saccharomyces cerevisiae. Yeast 22, 553–564 10.1002/yea.1234 [DOI] [PubMed] [Google Scholar]
- 115. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., and Mann M. (2008) Comprehensive mass-spectrometry–based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 10.1038/nature07341 [DOI] [PubMed] [Google Scholar]
- 116. Moore T. I., Tanaka H., Kim H. J., Jeon N. L., and Yi T. M. (2013) Yeast G-proteins mediate directional sensing and polarization behaviors in response to changes in pheromone gradient direction. Mol. Biol. Cell 24, 521–534 10.1091/mbc.e12-10-0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dyer J. M., Savage N. S., Jin M., Zyla T. R., Elston T. C., and Lew D. J. (2013) Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23, 32–41 10.1016/j.cub.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. McClure A. W., Minakova M., Dyer J. M., Zyla T. R., Elston T. C., and Lew D. J. (2015) Role of polarized G protein signaling in tracking pheromone gradients. Dev. Cell 35, 471–482 10.1016/j.devcel.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Erdman S., Lin L., Malczynski M., and Snyder M. (1998) Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140, 461–483 10.1083/jcb.140.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. You Y. J., Kim J., Cobb M., and Avery L. (2006) Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3, 237–245 10.1016/j.cmet.2006.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kang C., You Y. J., and Avery L. (2007) Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 21, 2161–2171 10.1101/gad.1573107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kang C., and Avery L. (2009) Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 23, 12–17 10.1101/gad.1723409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Oh D. Y., Talukdar S., Bae E. J., Imamura T., Morinaga H., Fan W., Li P., Lu W. J., Watkins S. M., and Olefsky J. M. (2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 10.1016/j.cell.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Aránguiz-Urroz P., Canales J., Copaja M., Troncoso R., Vicencio J. M., Carrillo C., Lara H., Lavandero S., and Díaz-Araya G. (2011) β(2)-Adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim. Biophys. Acta 1812, 23–31 10.1016/j.bbadis.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 125. Lizaso A., Tan K. T., and Lee Y. H. (2013) β-Adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243 10.4161/auto.24893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang L., Lu K., Hao H., Li X., Wang J., Wang K., Wang J., Yan Z., Zhang S., Du Y., and Liu H. (2013) Decreased autophagy in rat heart induced by anti-β1-adrenergic receptor autoantibodies contributes to the decline in mitochondrial membrane potential. PLoS ONE 8, e81296 10.1371/journal.pone.0081296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wauson E. M., Zaganjor E., Lee A. Y., Guerra M. L., Ghosh A. B., Bookout A. L., Chambers C. P., Jivan A., McGlynn K., Hutchison M. R., Deberardinis R. J., and Cobb M. H. (2012) The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol. Cell 47, 851–862 10.1016/j.molcel.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wenk M. R., Lucast L., Di Paolo G., Romanelli A. J., Suchy S. F., Nussbaum R. L., Cline G. W., Shulman G. I., McMurray W., and De Camilli P. (2003) Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat. Biotechnol. 21, 813–817 10.1038/nbt837 [DOI] [PubMed] [Google Scholar]
- 129. Boularan C., Kamenyeva O., Cho H., and Kehrl J. H. (2014) Resistance to inhibitors of cholinesterase (Ric)-8A and Gαi contribute to cytokinesis abscission by controlling vacuolar protein-sorting (Vps)34 activity. PLoS ONE 9, e86680 10.1371/journal.pone.0086680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Windmiller D. A., and Backer J. M. (2003) Distinct phosphoinositide 3-kinases mediate mast cell degranulation in response to G-protein-coupled versus FcϵRI receptors. J. Biol. Chem. 278, 11874–11878 10.1074/jbc.M211787200 [DOI] [PubMed] [Google Scholar]
- 131. Farley F. W., Satterberg B., Goldsmith E. J., and Elion E. A. (1999) Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone-response pathway on the MAP kinase Fus3p. Genetics 151, 1425–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sabbagh W. Jr, Flatauer L. J., Bardwell A. J., and Bardwell L. (2001) Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8, 683–691 10.1016/S1097-2765(01)00322-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Dharmasiri N., Dharmasiri S., and Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- 134. Kepinski S., and Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- 135. Katsir L., Schilmiller A. L., Staswick P. E., He S. Y., and Howe G. A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105 10.1073/pnas.0802332105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Longtine M. S., McKenzie A. 3rd., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 137. Strømhaug P. E., Bevan A., and Dunn W. A. Jr. (2001) GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 276, 42422–42435 10.1074/jbc.M104087200 [DOI] [PubMed] [Google Scholar]
- 138. Isom D. G., Sridharan V., Baker R., Clement S. T., Smalley D. M., and Dohlman H. G. (2013) Protons as second messenger regulators of G protein signaling. Mol. Cell 51, 531–538 10.1016/j.molcel.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 140. Hoffman G. A., Garrison T. R., and Dohlman H. G. (2002) Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 344, 617–631 10.1016/S0076-6879(02)44744-1 [DOI] [PubMed] [Google Scholar]
- 141. Tanaka K., Kitamura E., and Tanaka T. U. (2010) Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods 51, 206–213 10.1016/j.ymeth.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. English J. G., Shellhammer J. P., Malahe M., McCarter P. C., Elston T. C., and Dohlman H. G. (2015) MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast. Sci. Signal 8, ra5 10.1126/scisignal.2005774 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.