Abstract
Human Schlafen 11 (SLFN11) is an interferon-stimulated gene (ISG) that we previously have demonstrated to ablate translation of HIV proteins based on the virus's distinct codon preference. Additionally, lack of SLFN11 expression has been linked to the resistance of cancer cells to DNA-damaging agents (DDAs). We recently resolved the underlying mechanism, finding that it involves SLFN11-mediated cleavage of select tRNAs predominantly employed in the translation of the ATR and ATM Ser/Thr kinases, thereby establishing SLFN11 as a novel tRNA endonuclease. Even though SLFN11 is thus involved in two of the most prominent diseases of our time, cancer and HIV infection, its regulation remained thus far unresolved. Using MS and bioinformatics-based approaches combined with site-directed mutagenesis, we show here that SLFN11 is phosphorylated at three different sites, which requires dephosphorylation for SLFN11 to become fully functionally active. Furthermore, we identified protein phosphatase 1 catalytic subunit γ (PPP1CC) as the upstream enzyme whose activity is required for SLFN11 to cleave tRNAs and thereby act as a selective translational inhibitor. In summary, our work has identified both the mechanism of SLFN11 activation and PPP1CC as the enzyme responsible for its activation. Our findings open up future studies of the PPP1CC subunit(s) involved in SLFN11 activation and the putative kinase(s) that inactivates SLFN11.
Keywords: transfer RNA (tRNA), cancer, cancer therapy, DNA repair, drug resistance
Introduction
SLFN113 is part of the SLFN family of genes, which are exclusively found in mammals with the notable exception of poxviruses (1). Six human and nine murine SLFN isoforms differing in length ranging from 337 to 910 amino acids (referred to as short, medium, and long SLFNs) have been identified, with humans lacking all short family members (1). Remarkably, SLFNs share no significant sequence similarity with other proteins besides a weakly conserved NH2-terminal putative ATPase associated with diverse cellular activities (AAA) domain. AAA proteins typically form hexamers, utilize ATP hydrolysis for enzymatic function, and are reportedly involved in many diverse cellular processes such as DNA replication and repair, protein degradation and unfolding, and cell motility (2). The long SLFNs, such as SLFN11, contain a sequence in their COOH-terminal region distantly reminiscent of DNA/RNA helicase domains found in prokaryotes (1).
The first Schlafen gene, the short murine Slfn1, was discovered in 1998 in primary lymphoid tissues (3), noting that the Slfn1 gene is differentially regulated during thymocyte development, particularly in the transition from CD4CD8-double negative to -double positive transition. As it is now clear that SLFNs are interferon-induced genes, it is likely that their up-regulation at this point is a consequence of the constitutive interferon β expression we discovered in the thymic medulla (4). Intriguingly, Slfn1-knockout mice displayed no apparent phenotype, whereas in striking contrast, forced transgenic expression of Slfn1 in T cells greatly reduced T-cell growth and development, with a reduction in the number of thymocytes to 1–30% compared with normal mice (3).
In the following decade, very few reports on SLFNs appeared, and no cellular function or molecular mechanism would be assigned to this novel protein family. In 2012, we identified SLFN11 as a potent inhibitor of retrovirus replication (5). The foundation of our discovery was the observation that HEK293 cells produced significantly fewer HIV particles than HEK293T when transfected with a proviral vector (pNL4–3.Luc.R+E−). One notable distinction between these cell lines is the fact that HEK293T cells do not express any SLFN proteins. Ectopic expression of the individual SLFNs in HEK293T cells exposed SLFN11 as the family member that acted as a powerful inhibitor of HIV production, and its ablated expression in HEK293 cells resulted in a significant boost in HIV synthesis. Intriguingly, only viral proteins, but not HIV-associated RNAs, were suppressed by SLFN11, suggesting that SLFN11 functions as a translational inhibitor (5). Furthermore, no effect of SLFN11 on global translation was apparent. Investigation into the molecular mechanism revealed that SLFN11 selectively affected HIV protein synthesis by exploiting the distinct codon bias of the virus compared with the mammalian genome. We noted a change in tRNA expression profiles in HIV-infected cells expressing SLFN11 and found that SLFN11 binds tRNA in vitro.
Concurrent with our study, two independent laboratories reported that loss of SLFN11 expression results in the resistance of cancer cells to DNA-damaging agents (DDAs), the single largest class of chemotherapeutic drugs (6, 7). In contrast, SLFN11 expression was irrelevant to cellular responses to other chemotherapeutics such as kinase inhibitors and tubulin inhibitors. Importantly, SLFN11 expression could predict the survival of a group of ovarian cancer patients, implying a potential use of SLFN11 as a biomarker for sensitivity to DDAs (6). Relating to our findings on inhibition of HIV production, we analyzed the codon usage of components of the DNA damage response. Indeed, ataxia telangiectasia and Rad3-related protein (ATR) and ataxia telangiectasia mutated (ATM) exhibit a similar codon usage bias as HIV, and both exhibited reduced protein expression despite unaffected or even increased mRNA levels after DDA treatment but solely in cells expressing SLFN11 (8). We showed that SLFN11 inhibits ATR protein synthesis via cleavage of type II tRNAs representing codons disproportionally found in the ATR coding sequence. Type II tRNAs, which represent nearly all leucines and serines, harbor an additional variable stem loop compared with their type I counterparts that are not affected by SLFN11 (see also Fig. 2a). The attenuation of ATR translation by SLFN11 is central to DDA-induced cell death, as the cytotoxic response to DDAs can be restored in SLFN11-deficient cells by knockdown or pharmacological inhibition of ATR (8). Similarly, DDA sensitivity of SLFN11-deficient cells can be reestablished through gapmer-mediated reduction of a specific type II tRNA (tRNA-Leu-TAA), reinforcing the link between SLFN11 and tRNA alteration. Complementing our findings, the recent elucidation of the crystal structure of the NH2-terminal region of rat SLFN13 characterized SLFNs as a novel class of tRNA endonucleases (9).
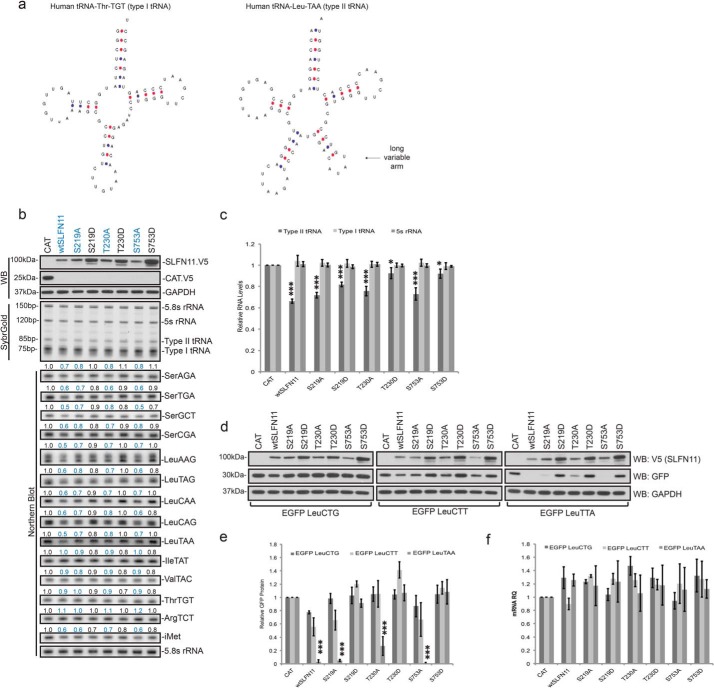
Figure 2.
Effect of SLFN11 phosphorylation site mutants on type II tRNA levels and codon-biased translation. a, depiction of type I versus type II tRNA structure. Source: gtrnadb.ucsc.edu.4 b, WT SLFN11 and the indicated mutants were expressed in HEK293T cells for 48 h. Protein quantities were analyzed by Western blotting (WB), and RNA levels were visualized by SYBR Gold staining and Northern blotting. Numbers indicate quantified Northern blotting signals normalized to 5.8S rRNA c, SYBR Gold–stained tRNA bands were quantified and normalized to 5.8S rRNA (biological replicates, mean ± S.D. (error bars), n = 3; *, p < 0.05; ***, p < 0.001). d, protein expression of EGFP encoded by constructs using only the indicated codon for all leucine residues in HEK293T cells in the absence or presence of the indicated SLFN11 mutants as determined by immunoblotting. e, quantified EGFP protein expression levels from samples as in d with ImageJ software (biological replicates, mean ± S.D. (error bars), n = 3; ***, p < 0.001). f, relative EGFP mRNA levels corresponding to protein samples shown in e (biological replicates, mean ± S.D. (error bars), n = 3). RQ, relative quantification.
Considering the prominent role SLFN11 fulfills in facilitating both an antiretroviral response and an appropriate cell fate decision after DNA damage has occurred, it is naturally desirable to understand how its activation is regulated. To this end, we employed MS to identify sites of possible post-translational modification. Through this approach, we identified three phosphorylation sites in SLFN11 that modulate its ability to cleave type II tRNA and consequently inhibit translation of HIV (or other Leu(TTA) codon–harboring) genes. Surprisingly, our subsequent studies illustrate that dephosphorylated SLFN11 represents the active form, and we reveal protein phosphatase 1 catalytic subunit γ (PPP1CC) as a regulator of SLFN11 function. These results bring us one step closer to understanding the function and regulation of this important component of the antiviral and DNA damage responses.
Results
Identification of SLFN11 phosphorylation sites
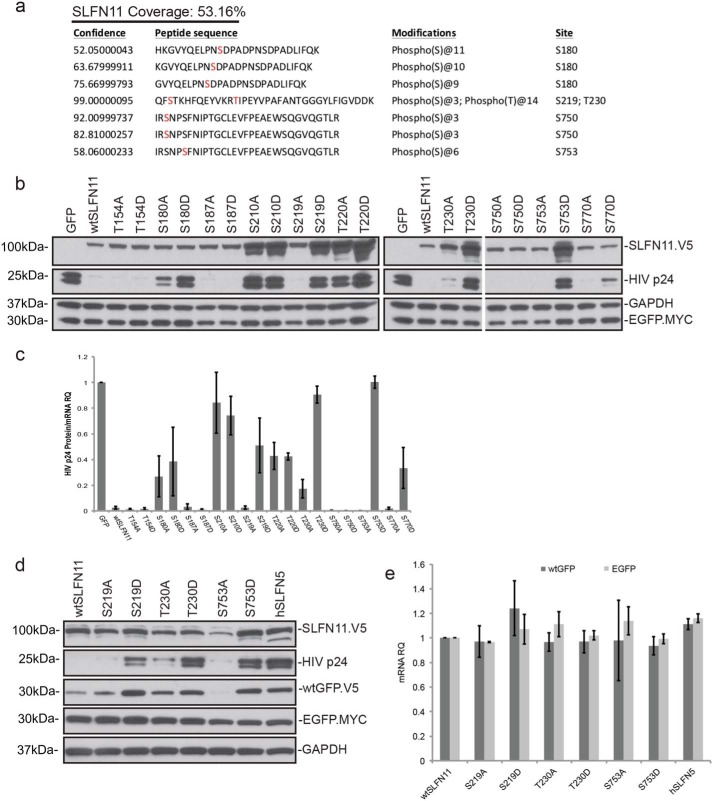
To investigate whether SLFN11 contains any phosphorylation (or other post-translational modification) sites, we performed MS of immunoprecipitated SLFN11, which revealed five putative Ser/Thr phosphorylation sites, namely Ser-180, Ser-219, Thr-230, Ser-750, and Ser-753 (Fig. 1a). To test whether these sites fulfill any regulatory function, we employed mutagenic studies on those residues. Additionally, we also mutated Thr-154 because it is a conserved residue among the SLFN family of proteins (10), Ser-187 and Thr-220 because they are located on the same peptide as the indicated phosphorylated residue, and finally Ser-210 and Ser-770 because they are part of conserved putative phosphorylation motifs found in database searches. Each site was mutated to either Ala, which does not contain a hydroxyl group and is therefore not able to be phosphorylated, or Asp, which contains a negative charge that mimics phosphorylation. To assess the impact of these mutations on SLFN11 function, WT or mutant SLFN11 constructs were cotransfected into HEK293T cells (which do not express endogenous SLFN11 (5)) along with the HIV proviral construct pNL4–3.Luc.R+E− and with EGFP-encoding plasmids, which serve as a transfection control as described previously (5). Monitoring levels of the HIV p24 capsid protein derived from the gag gene revealed that changing Ser-210 or Thr-220 to either Ala or Asp yielded SLFN11 mutants that failed to suppress p24 protein synthesis, hinting that these amino acid changes likely interfere with structural elements of SLFN11. In striking contrast, although Ser-219, Thr-230, or Ser-753 to Asp conversion caused a complete loss of function of SLFN11 as measured by suppression of p24 translation, the translational inhibitory potential upon this viral protein was fully retained when these residues were converted to Ala (Fig. 1b). Importantly, these residues represent the three sites we found earlier to be phosphorylated by mass spectrometric analysis in latent SLFN11 (Fig. 1a). It is noteworthy that the SLFN11 mutants that failed to limit p24 synthesis were actually expressed at substantially higher levels than functional SLFN11 proteins (Fig. 1b), although the variability among the different SLFN11 mRNA levels is insignificant (Fig. S1a), suggesting that SLFN11 translation itself is to some extent self-regulated. Importantly, expression of EGFP from a codon-optimized, cotransfected vector was not affected by any of the SLFN11 proteins, thus serving as a transfection, specificity, and toxicity control. However, as we had already noted in earlier studies that elevated p24 protein levels can alter p24 mRNA levels, possibly through a positive feedback loop (Fig. S1b), we also enumerated p24 protein levels after normalization to the corresponding p24 mRNA. As shown in Fig. 1c, even after compensation for pretranslational events, the differences among the SLFN11 phosphorylation site mutants remain significant.
Figure 1.
Effect of SLFN11 phosphorylation site mutations on its translation-inhibitory properties. a, V5-tagged SLFN11 was expressed in HEK293T cells and immunoprecipitated. The indicated post-translational modifications were identified by mass spectrometric analysis. b, the indicated SLFN11 mutants were expressed in HEK293T, and p24 protein derived from cotransfected pNL4–3.Luc.R+E− was determined by anti-p24 Western blotting. c, HIV p24 protein levels from b were quantified and normalized over HIV p24 mRNA (biological replicates, mean ± S.D. (error bars), n = 3). d, HEK293T cells were cotransfected with plasmids encoding the indicated SLFN11 mutants, pNL4–3.Luc.R+E−, V5-tagged wtGFP, and Myc-tagged EGFP, and resulting protein levels were visualized by Western blotting. e, wtGFP and EGFP mRNA levels of d quantitated by qPCR (biological replicates, mean ± S.D. (error bars), n = 3). RQ, relative quantification.
To unequivocally demonstrate that mutation of Ser-219, Thr-230, and Ser-753 specifically affected the inhibitory properties of SLFN11 on codon usage–dependent translation, we employed a previously reported strategy utilizing the distinct impact of SLFN11 expression on wtGFP compared with codon-optimized EGFP (5, 8). wtGFP harbors a similar codon usage bias as HIV and is susceptible to translational suppression by SLFN11 (5), whereas translation of codon-optimized EGFP is refractory to the effects of SLFN11. As illustrated in Fig. 1d, the impact of the individual SLFN11 mutants on wtGFP expression parallels their effect on p24 protein expression, notwithstanding the fact that neither wtGFP nor EGFP mRNAs were altered by any of the SLFN11 mutants (Fig. 1e).
The fact that mutation of Ser-219, Thr-230, or Ser-753 to the phosphomimetic amino acid Asp rendered such SLFN11 mutants inactive, whereas conversion of these residues to Ala yielded fully functional SLFN11 proteins raised the possibility that SLFN11 is activated by dephosphorylation on these residues. To explore this concept, HEK293 cells were transfected with the HIV proviral vector pNL4–3.Luc.R+E− prior to treatment with okadaic acid, a potent inhibitor of protein phosphatase 1. Indeed, with increasing concentrations of okadaic acid, the suppressive effect of SLFN11 on p24 expression vanished (Fig. S1c), supporting the notion that Ser/Thr phosphatase activity is a prerequisite for SLFN11 function.
Effect of SLFN11 phosphorylation site mutations on tRNA cleavage
In a recent report, we elaborated on the role of SLFN11 as a putative endonuclease that mediates the cleavage of tRNAs during the DNA damage response initiated by chemotherapeutic agents and the consequential translational suppression of mRNAs derived from genes such as ATM and ATR, which disproportionally utilize distinct codons (8). All tRNAs are subdivided into two categories: type I tRNAs are ∼72–74 bases long, whereas type II tRNAs contain an extra variable stem loop, elongating the molecule by around 10 bases (Fig. 2a and Ref. 11). The two classes of tRNA can be distinguished by their discrete mobilities during electrophoresis on Tris borate-EDTA-urea gels (Fig. S2). Our investigations revealed that SLFN11 displays a high specificity for type II tRNAs as substrates and demonstrated that ectopic expression of SLFN11 in HEK293T cells was sufficient to down-regulate type II, but not type I, tRNAs by up to 50% (8).
Based on our results that SLFN11 (Ser-219, Thr-230, or Ser-753 → Asp) fail to abrogate p24 or wtGFP translation, we reasoned that these mutants might lack the ability to facilitate type II tRNA cleavage. To test this hypothesis, we performed analysis of tRNAs derived from HEK293T cells ectopically expressing the various SLFN11 phosphorylation site mutants. When total RNA from these cells was resolved on Tris borate-EDTA-urea gels and stained with SYBR Gold, it became readily apparent that the “active” SLFN11 Ser/Thr → Ala mutants were equally capable of reducing overall type II tRNA as was WT SLFN11, whereas the Ser/Thr → Asp mutants failed to do so (Fig. 2, b, top, and c). Importantly, the SLFN11 Ser/Thr → Ala mutants retained their specificity for type II tRNA, as type I tRNA and 5S rRNA remained unchanged between samples (Fig. 2, b, top, and c). Further analysis of the samples by Northern blotting, probing for all type II tRNAs and selected type I tRNAs, corroborated the SYBR Gold staining conclusions. Quantification of the Northern blots revealed that all type II tRNAs were down-regulated up to 40% by the respective Ser/Thr → Ala mutants, whereas type I tRNAs remained unaffected (Figs. 2b and S3). It should be noted that the transfection efficiency in these studies averaged ∼50% (not shown), leaving an equal number of cells lacking any form of SLFN11. Consequently, the predictably unaltered type II tRNA levels in these cells will mask the full impact of SLFN11 in transfected cells to some degree, and thus the true reduction in type II tRNAs likely exceeds the observable diminution in the combined cell populations.
Impact of SLFN11 phosphorylation site mutations on translation-inhibitory capability
The cleavage of type II tRNAs via SLFN11 correlates directly with its impact on the synthesis of HIV proteins or wtGFP, suggesting that type II tRNA down-regulation is central to selective translation inhibition by SLFN11. Strikingly, however, as we reported in Li et al. (8), we discovered that even though SLFN11 promotes the cleavage of all type II tRNAs, only mRNAs relying on frequent use of two select codons, Leu(TTA) and Leu(CTT), are exquisitely susceptible to translational inhibition by SLFN11. This was illustrated by assessing the translation of fully codon-optimized, SLFN11-“resistant” EGFP and derivatives thereof in which every Leu or Ser codon was replaced with one distinct codon. For example, expression of EGFP from a vector in which all Leu are represented by the codon Leu(CTG) was unaltered by the presence of WT SLFN11 (8). In striking contrast, when all leucines in EGFP were coded via Leu(CTT) or Leu(TTA), SLFN11 exerted a pronounced inhibitory effect on the translation of these proteins (Fig. 2d and Ref. 8). Central to the present study, we uncovered that the SLFN11 phosphorylation site mutants faithfully replicated the impact they had on both p24 and wtGFP translation as well as on the synthesis of EGFP proteins whose leucines were encoded by either Leu(CTT) or, even more pronounced, by Leu(TTA) (Fig. 2d, center and right panels). As anticipated, neither WT SLFN11 nor any Ser/Thr → Ala mutants had any effect on EGFP when leucines were encoded via Leu(CTG). In striking opposition, however, WT SLFN11 as well as all three Ser/Thr → Ala mutants caused massive translational inhibition of EGFPs with leucines solely encoded by TTA or CTT codons. Crucially, all three Ser/Thr → Asp SLFN11 mutants completely lost the ability to inhibit the synthesis of EGFP Leu(CTT) or Leu(TTA) (Fig. 2, d and e). Quantitation by qPCR assured that the respective mRNAs were not affected by any of the SLFN11 proteins (Fig. 2f), corroborating the model that SLFN11 acts strictly via translational repression of mRNAs that harbor Leu(TTA) or Leu(CTT) codons.
DNA-damaging agents trigger a reduction in SLFN11 phosphorylation
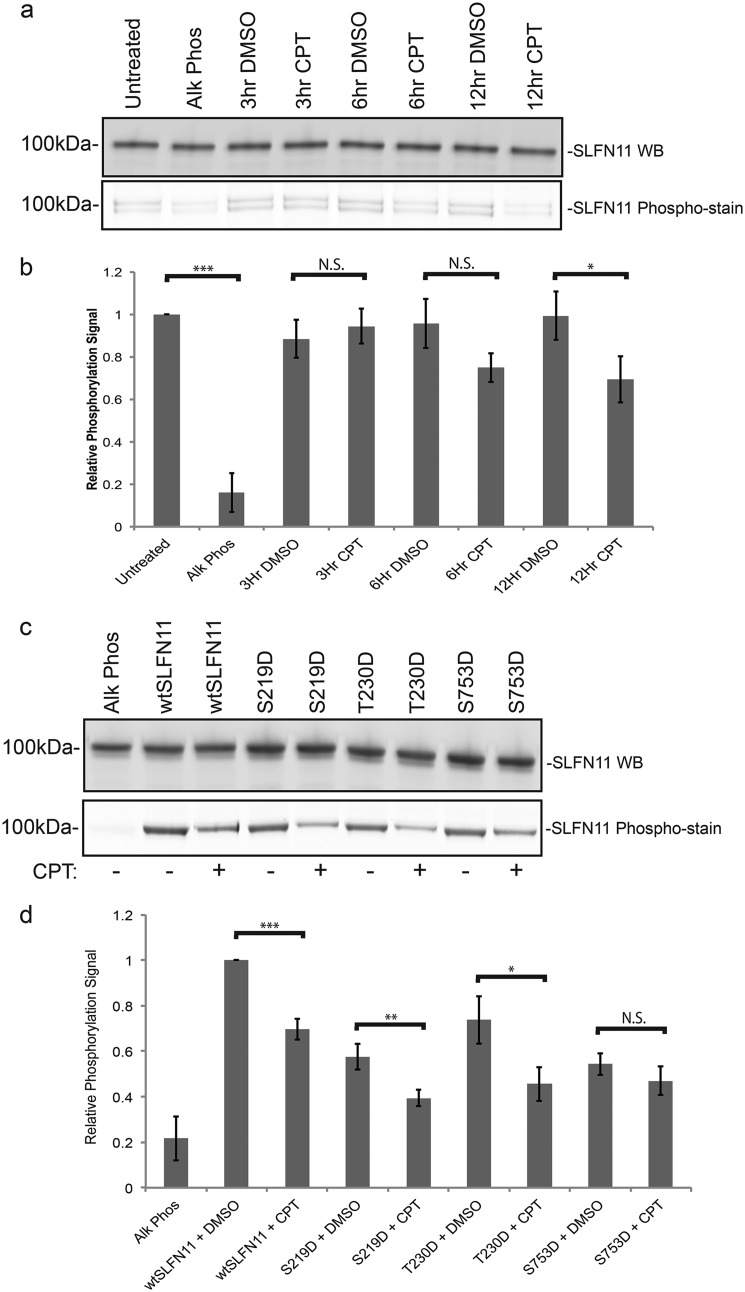
As our results thus far strongly indicated that Ser/Thr dephosphorylation of SLFN11 promotes its activity against type II tRNAs and associated translational events that facilitate cellular responses to HIV infections or DNA damage, it was thus of logical interest to determine whether SLFN11 was indeed dephosphorylated in response to exposure of cells to DDAs. As no phosphospecific antibodies against SLFN11 are currently available, we employed a novel Phospho-TagTM phosphoprotein gel stain that allows quantification of the extent of phosphorylation of the protein under investigation, albeit not in a site-specific manner. When immunoprecipitated SLFN11 from cells exposed to the topoisomerase inhibitor camptothecin (CPT) for 3, 6, or 12 h was analyzed by means of Phospho-Tag phosphoprotein gel stain analysis, we detected a clear decrease in SLFN11 phosphorylation in a time-dependent manner (Fig. 3a, bottom panel). Normalization of the phosphostain band intensity over the corresponding SLFN11 protein amount determined by Western blotting revealed a clear progressive dephosphorylation of SLFN11 over time (Fig. 3b).
Figure 3.
Effect of camptothecin treatment on SLFN11 phosphorylation. a, exogenously expressed SLFN11 was immunoprecipitated with anti-V5 antibody–conjugated beads from HEK293T cells treated with DMSO or 200 nm CPT as indicated and then visualized by Western blotting (WB) or Phospho-Tag phosphoprotein gel staining. b, phosphostain signals were quantified with ImageQuant software after normalization to Western blotting band intensity as shown in a (biological replicates, mean ± S.D. (error bars), n = 3; *, p < 0.05; ***, p < 0.001; N.S., not significant). c, as in a except SLFN11 phosphorylation site mutants from cells treated with DMSO or 200 nm CPT for 12 h were analyzed. d, phosphostain signals were quantified with ImageQuant software after normalization to Western blotting band intensity as shown in c (biological replicates, mean ± S.D. (error bars), n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S., not significant). Alk Phos, alkaline phosphatase.
We next set out to test the effect of CPT on the three SLFN11 (S219D, T230D, and S753D) mutants (Fig. 3c). Even though replacement of Ser/Thr residues with the amino acid Asp frequently produces a phosphomimetic effect that can alter protein function, Asp residues do not react with the Phospho-Tag stain, and in agreement with that notion, all three SLFN11 Ser/Thr → Asp mutants yielded a slightly reduced baseline band intensity on Phospho-Tag–stained gels when compared with WT SLFN11 (Fig. 3, c and d). Upon CPT treatment, the SLFN11 S219D and T230D mutants show a decrease in phosphorylation similar in extent to WT SLFN11. In contrast, S753D mutation of SLFN11 apparently precludes dephosphorylation on other residues as CPT treatment did not produce a significant change in overall SLFN11 phosphorylation, suggesting that dephosphorylation of Ser-753 might be the primary step in SLFN11 activation in the context of DNA damage.
PPP1CC binds SLFN11 and controls its activity
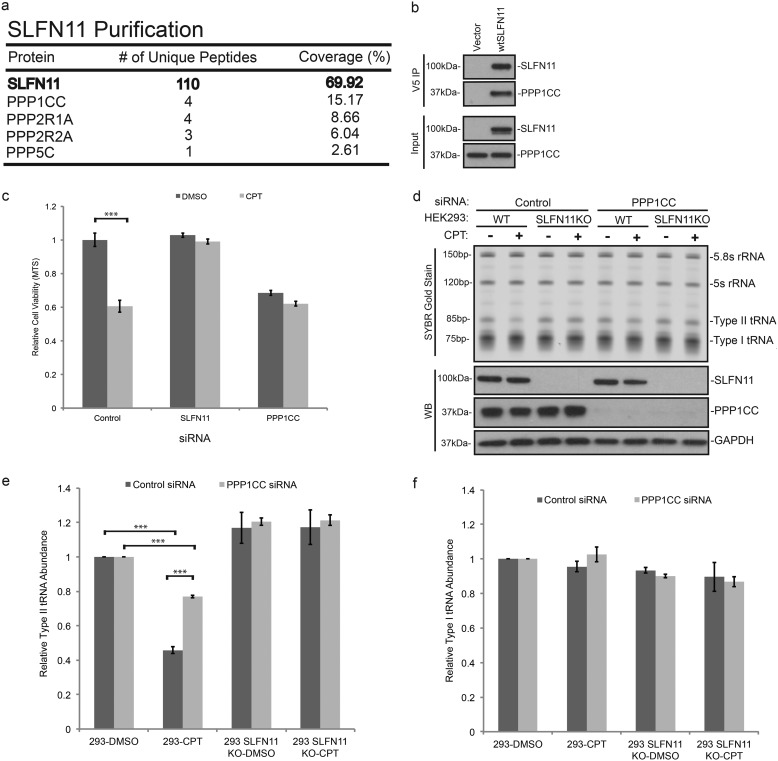
Our mass spectrometry examination and mutational analysis of SLFN11, as well as the results of the okadaic acid experiment (Fig. S1c), all support the hypothesis that SLFN11 is activated by dephosphorylation, implying the involvement of a Ser/Thr phosphatase in the upstream activation pathway. In an effort to identify the enzyme, we employed mass spectrometric analysis of SLFN11 immunoprecipitated under low-stringency conditions to detect interacting proteins. Among the proteins we were able to reproducibly coisolate with SLFN11 was PPP1CC (Fig. 4a). Subsequent coimmunoprecipitation experiments analyzed by Western blotting confirmed that PPP1CC strongly associates with SLFN11 (Fig. 4b), whereas probing for the presence of other protein phosphatases such as PPP1A, PPP1B, PPP2AC, PPP2BC, PPP4C, and PPP5C yielded no evidence of SLFN11 binding (data not shown). To explore whether PPP1CC plays any role in the response to DDAs similar to SLFN11, we tested the effect of PPP1CC knockdown via siRNA on the susceptibility of cells to CPT-induced cell death. As we had already demonstrated extensively in previous studies, the siRNA-mediated ablation of SLFN11 expression renders HEK293 cells resistant to DNA damage–mediated cell death (Fig. 4c, middle bars). Very similarly, elimination of PPP1CC expression also led to an apparent loss of any CPT effect (Fig. 4c, right). Although these results are consistent with our model, we noted that the depletion of PPP1CC per se already impacted cell survival to some extent. As these survival studies extend over several days, we deemed it likely that the loss of PPP1CC alters cellular processes beyond SLFN11 function. To circumvent this issue, we tested the impact of the loss of PPP1CC on DNA damage–induced type II tRNA cleavage, which not only occurs after much shorter exposure to CPT but is also a much more direct and specific readout of SLFN11 function.
Figure 4.
PPP1CC regulates SLFN11 activity. a, V5-tagged SLFN11 was expressed in HEK293T cells and immunoprecipitated. The indicated peptides corresponding to associated proteins were identified by mass spectrometric analysis. b, total cell lysate was subject to immunoprecipitation as in a with anti-V5 IgG, and the resulting immunoprecipitate was probed for the presence of PPP1CC. c, relative viability of HEK293 cells transfected with control, SLFN11-, or PPP1CC-targeting siRNAs was measured by MTS assay after 48 h of 200 nm CPT or DMSO treatment (biological replicates, mean ± S.D. (error bars), n = 3; ***, p < 0.001). d, HEK293 cells and HEK293/SLFN11KO cells were transfected with either control or PPP1CC-targeting siRNA for 48 h prior to treatment with either DMSO or 200 nm CPT for 12 h. Total RNA was visualized by SYBR Gold staining, and the presence of respective proteins was determined by immunoblotting. e and f, relative type II and type I tRNA levels, respectively, from samples shown in d were quantified and normalized over 5.8S rRNA (biological replicates, mean ± S.D. (error bars), n = 3; ***, p < 0.001). WB, Western blotting.
As shown in Fig. 4d, CPT treatment of HEK293 cells yielded a striking reduction in type II tRNAs, which was substantially prevented by PPP1CC knockdown (Fig. 4, d and e). Most importantly, when the same experiment was conducted in HEK293 cells where SLFN11 was permanently ablated via CRISPR, neither CPT treatment nor PPP1CC knockdown had any effect on type II tRNA abundance. Furthermore, type I tRNA levels remained unchanged in every scenario (Fig. 4f), thus excluding nonspecific, cytotoxic effects of the investigational conditions within the time frame of the experiment.
SLFN11 mutant–expressing stable cell lines are differentially sensitized upon DNA damage
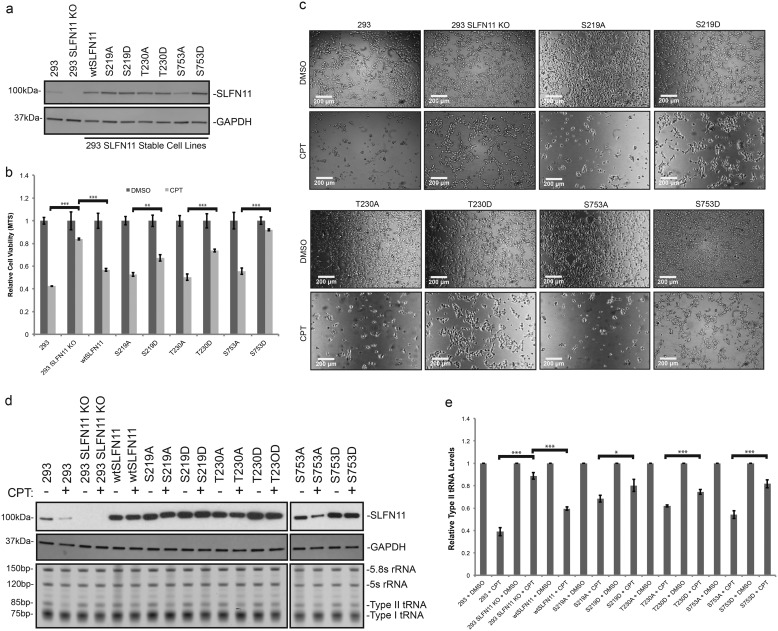
To evaluate the biological impact of mutations of the phosphorylation sites, we created cell lines permanently expressing the respective SLFN11 phosphorylation site mutants in HEK293 SLFN11-CRISPRKO cells (Fig. 5a). CPT treatment of these cell lines revealed greater sensitivity of the Ser → Ala mutants compared with their Ser → Asp counterparts in MTS assays (Fig. 5b). Microscopic visualization of cells after CPT treatment reinforced the notion of increased changes in cell morphology and in cell death when SLFN11 Ser/Thr residues were replaced by alanines compared with aspartic acid mutants (Fig. 5c). In accordance, SLFN11 Ser/Thr → Ala mutants displayed greater down-regulation of type II tRNA upon CPT treatment when compared with their Ser/Thr → Asp counterparts (Fig. 5, d and e). It should be noted that not only did SLFN11 S753A yield the lowest expression level compared with other mutants, it also produced the greatest difference in both cell killing and type II tRNA down-regulation compared with SLFN11 S753D, suggesting Ser-753 as the major regulatory site controlling SLFN11 activity.
Figure 5.
SLFN11 mutant–expressing stable cell lines are differentially sensitized upon DNA damage. a, stable expression of SLFN11 mutants in HEK293/SLFN11KO cell lines as created by lentiviral infection. b, relative viability of stable SLFN11 mutant–expressing HEK293 cell lines was measured by MTS assay after 48 h of 200 nm CPT or DMSO treatment (biological replicates, mean ± S.D. (error bars), n = 3; **, p < 0.01; ***, p < 0.001). c, microscopic images at 10× magnification of stable SLFN11 mutant–expressing HEK293 cells after 48 h of 200 nm CPT or DMSO treatment. d, total RNA from stable SLFN11 mutant–expressing HEK293 cell lines treated for 48 h with 200 nm CPT or DMSO was visualized by SYBR Gold staining, and the presence of the respective proteins was determined by immunoblotting. e, relative type II tRNA levels from samples shown in d were quantified and normalized to 5.8S rRNA (biological replicates, mean ± S.D. (error bars), n = 3; *, p < 0.05; ***, p < 0.001).
Discussion
Our original mechanistic studies revealed that SLFN11 inhibits the synthesis of HIV proteins without global translational suppression based on a distinct preference of synonymous codons between host cell and viral proteins. It appears that SLFN11 evolved as a viral restriction factor, as not only do HIV elite controllers display elevated SLFN11 protein levels (12), but SLFN11 homologs derived from various primates exhibit striking efficacy against HIV despite a high degree of sequence conservation (10).
Contemporaneous with our findings of SLFN11 as a viral antagonist, two independent studies demonstrated a striking correlation between SLFN11 expression and the susceptibility of tumor cells to DDAs (6, 7). Notably, the efficacy of chemotherapeutic agents that do not rely on the DNA damage response is unaltered by the absence of SLFN11 (6–8). We hypothesized that the molecular basis for SLFN11's contribution to DDA sensitivity might similarly involve a selective translational interference based on codon usage bias, and we were indeed able to demonstrate that two key players in DNA damage response, ATR and ATM, are subject to translational suppression by SLFN11 (8). Significantly, their codon usage diverges from that of abundantly expressed cellular proteins whose synthesis is unaffected by SLFN11. In further mechanistic studies, we demonstrated the SLFN11-dependent cleavage of type II tRNAs, thus predisposing ATR or ATM to translational inhibition by SLFN11. The greatest impact was found in the reduction of tRNA-Leu-TAA, which is disproportionally frequently employed by ATR and ATM. Indeed, of the 352 leucines in ATR, 73 use the codon TTA (21%), and of 389 leucine residues in ATM, 91 are encoded by TTA (23%). In striking contrast, only a single TTA codon can be found among the 19 leucine residues of GAPDH (5%) (8).
Our examination of SLFN11 function in both the antiviral and the DNA damage responses exposed that the protein's NH2-terminal half contains the functional domain responsible for the degradation of type II tRNAs, whereas the COOH-terminal domain is responsible for the regulation of SLFN11 activation (8, 13, 14). The NH2-terminal region is highly conserved among SLFN family members but does not bear any resemblance to other proteins. The recent elucidation of the crystallographic structure of the NH2-terminal domain of rat SLFN13 and associated function studies divulged an intrinsic endoribonuclease activity in vitro, with tRNAs as the preferential substrate (9). In concert, these results establish SLFN family members as a novel class of tRNA-specific endoribonucleases with the enzymatic activity residing in the NH2-terminal region of the protein and with an apparent specificity for type II tRNAs.
In light of the critical role of SLFN11 in two major pathologies, it was obvious that identification of the regulatory processes that govern its function is of utmost importance. Mass spectrometric analysis as well as motif searches indicated numerous potential phosphorylation sites. Systematic mutational analysis of these candidate sites allowed us to identify three residues, Ser-219, Thr-230, and Ser-753, whose mutation impacted the inhibitory properties of SLFN11 on HIV p24 protein synthesis. Surprisingly, however, substitution of these amino acids with Ala retained SLFN11 function, whereas replacement with the phosphomimic Asp rendered the resulting proteins inactive. This observation led us to the theory that SLFN11 is activated by dephosphorylation, a notion supported by the fact that the Ser/Thr phosphatase inhibitor okadaic acid was able to abolish the inhibitory effect of SLFN11 on p24 translation. As we had noted previously, SLFN11 can trigger changes in HIV p24 RNA levels as well. We believe this is likely due to p24 being subject to positive feedback regulation via HIV Tat, which enhances transcription of viral genes (15). Crucially, normalization of p24 protein to its corresponding RNA confirmed that SLFN11 clearly predominantly affected translation rather than transcription. Lastly, the effects of the SLFN11 mutations were also apparent when we determined their impact on the expression of wtGFP versus codon-optimized EGFP, the latter of which is refractory to suppression by SLFN11 (Fig. 1, d and e).
Type II tRNAs, which correspond to all serine and leucine codons, are subject to cleavage by SLFN11, presumably being recognized as substrates by the presence of a fourth stem loop that is not present in type I tRNAs (Fig. 2a). Paralleling the pattern SLFN11 phosphorylation site mutations elicited on p24 or wtGFP translation, Asp substitutions of Ser-219, Thr-230, or Ser-753 rendered SLFN11 incapable of reducing type II tRNA levels, whereas conversions to Ala were of no consequence to SLFN11 activity. As described in Li et al. (8), even though all type II tRNAs are reduced by SLFN11, only the synthesis of proteins engaging the use of Leu(TTA) and to a lesser extent Leu(CTT) is attenuated by SLFN11. This was demonstrated through the use of individual EGFP-encoding vectors in which all leucine or serine residues were represented by a single codon, with the original EGFP construct as the control. EGFP protein derived from most constructs exhibited marginal to no alteration on account of the presence of SLFN11, but remarkably, SLFN11 completely abolished the expression of EGFP_Leu(TTA) and significantly lowered the levels of EGFP_Leu(CTT) (8). Again, SLFN11 Ser-219, Thr-230, or Ser-753 to Ala mutants exhibited the same activity as WT SLFN11, whereas all three Ser/Thr → Asp mutations produced SLFN11 proteins that appeared functionally inactive (Fig. 2, d and e).
The mutation studies and the impact of the Ser/Thr phosphatase inhibitor okadaic acid on the inhibitory effect of SLFN11 on p24 translation strongly support the notion that SLFN11 activation requires dephosphorylation on at least three Ser/Thr residues. Unfortunately, no SLFN11 phosphospecific antibodies are currently available; however, evaluation of the overall extent of phosphorylation of SLFN11 by means of Phospho-Tag stain clearly illustrated a decrease in SLFN11 phosphorylation in response to CPT. Notably, no such decline was evident in the SLFN11 Ser-753 → Asp mutant. Ser-753 is the most COOH-terminal of the three phosphorylation sites and is absent in the SLFN11N truncation mutant (residues 1–579) that is sufficient for the degradation of type II tRNAs (5, 8). Thus, a likely scenario is that dephosphorylation of SLFN11 occurs in a sequential manner, with Ser-753 dephosphorylation representing the initial step.
The model that SLFN11 is activated by dephosphorylation naturally implies that a Ser/Thr or dual-specificity phosphatase acts upstream. Indeed, mass spectrometric and Western blotting identification of SLFN11-associated proteins revealed the presence of PPP1CC. The enzymatic activity of this catalytic subunit can be influenced by over 200 regulatory subunits that contribute to the formation of highly substrate-specific holoenzymes (16). Consistent with PPP1CC acting in the SLFN11 activation process, its ablated expression results in the loss of SLFN11 function with respect to type II tRNA reduction and efficacy of DDAs (Fig. 4). It is noteworthy that PPP1CC has already been implicated in the DNA damage response (17, 18). Importantly, abolished PPP1CC expression does not produce similar outcomes in cells lacking SLFN11, substantiating the concept of both proteins acting in the same pathway.
In summary, our studies into the regulation of SLFN11 identified both the activation mechanism and the responsible enzyme. Considering the profound influence SLFN11 exerts in two prominent pathologies, future investigations aimed at the identification of the PPP1CC regulatory subunit(s) and the presumably inactivating kinase are unquestionably warranted.
Materials and methods
Cell lines, plasmids, antibodies, and chemicals
All cell lines were maintained at 37 °C, 5% CO2 in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, 1× minimum essential medium nonessential amino acids, 1 mm sodium pyruvate, and 50 μm 2-mercaptoethanol. The HEK293 (CRL-1573) and HEK293T (CRL-3216) cell lines were acquired from ATCC. MTS cell viability assays, construction of pcDNA6/CAT/V5-His, pcDNA6/SLFN11/V5-His, pcDNA6.2/wtGFP/V5, pcDNA6.2/EGFP/Myc, and EGFP expression constructs with synonymous Leu codons were described previously (5, 8) as was the proviral HIV-1 vector pNL4–3.Luc.R+E−. Point mutations in pcDNA6/SLFN11/V5-His expression vectors were created with the GeneTailorTM Site-Directed Mutagenesis System (Invitrogen). To obtain HEK293 derivative cell lines in which SLFN11 expression was obliterated using CRISPR-Cas9, cells were transfected with pSpCas9(BB)-2A-Puro (PX459) all-in-one CRISPR-Cas9 construct and selected for puromycin resistance (SLFN11 CRISPR-Cas9 guide RNA 4, GCAGCCTGACAACCGAGAAA, obtained from GenScript). Surviving cells were cloned by limiting dilution and screened for SLFN11 expression by immunoblotting. To generate HEK293 SLFN11 mutant–expressing stable cell lines, we followed the experimental strategy described by Murai et al. (13). Briefly, plasmids encoding SLFN11 mutants were amplified using 5′-ATCGGATCCGCGGCCAACATGGAGGCAAATCAGTGC-3′ and 5′-ATTGTCGACGCGGCCCTACTTATCGTCGTCATCCTTGTAATCATGGCCACCCCACGGAA-3′ as forward and reverse primers, respectively, further including the sequence for the FLAG tag, and cloned into the pCDH-EF1-MCS-PGK-copGFP lentiviral expression vector (System Biosciences) by an In-Fusion HD cloning kit (Clontech). The SLFN11-encoding lentiviral vectors were cotransfected into HEK293T cells together with psPAX2 (Addgene, catalog number 12260) and pMD2.g (Addgene, catalog number 12259) packaging plasmids. Viral particles were collected to infect HEK293 SLFN11 CRISPRKO cells whose generation was described above. SLFN11-positive cells were isolated using fluorescence-activated cell sorting (FACS) based on coexpressed GFP (GFP expression levels were gated to similar fluorescence intensity levels). Rabbit polyclonal anti-V5 tag (G-14)-R (catalog number sc-83849-R), murine monoclonal anti-Myc tag (9E10) (catalog number sc-40), and anti-SLFN11 (E-4) (catalog number sc-374339) antibodies and murine isotype control IgG (catalog number sc-2025) were purchased from Santa Cruz Biotechnology, and the murine anti-HIV-1 p24 (catalog number MA171515) was obtained from Thermo Scientific Pierce. Antibodies against GFP (D5.1) (catalog number 2956S) and GAPDH (14C10) (catalog number 2118S) were purchased from Cell Signaling Technology, and leporine polyclonal anti-PPP1CC antibody was purchased from ProteinTech (catalog number 11082-1-AP).
Mass spectrometry
The samples for MS analysis of SLFN11 peptides in Fig. 1a were prepared by transfecting pcDNA6/SLFN11/V5-His into HEK293T cells for 48 h, lysing the cells with 1× Cell Lysis Buffer from Cell Signaling Technology (catalog number 9803) supplemented with phosphatase and protease inhibitor mixtures from Millipore (catalog numbers 524625, 524625, and 539134) as well as 1 mm phenylmethylsulfonyl fluoride. Approximately 10 mg of protein was subjected to immunoprecipitation with anti-V5 antibody. For Fig. 4a, pcDNA6/SLFN11/V5-His was transfected into HEK293T cells, which were subsequently washed with ice-cold PBS, lysed with 0.5% IGEPAL, 50 mm Tris-HCl, pH 7.4, 50 mm NaCl, protease inhibitor mixtures, and 1 mm phenylmethylsulfonyl fluoride. A total of 10 mg of protein was subjected to immunoprecipitation with anti-V5 antibody. Mass spectrometric analysis was performed at the University of California, San Diego proteomics facility for LC-MS/MS analysis on a Lumos hybrid mass spectrometer (Thermo) interfaced with nanoscale reversed-phase UPLC (Thermo Dionex UltiMateTM 3000 RSLC Nano System). Data analysis was carried out using ByonicTM software (Protein Metrics Inc.).
Cell transfection, lysis, immunoprecipitations, and immunoblotting
Reverse transfection of cells was performed using ON-TARGETplus SMARTpool siRNAs (GE Dharmacon) and RNAiMAX reagent (Invitrogen, catalog number 13778075). The siRNAs used were SLFN11 (catalog number L-016764-01-0005), PPP1CC (catalog number L-006827-00-0005), and nontargeting (catalog number D-001810-10-05). Cells were exposed to siRNAs for 48 h prior to treatment with 200 nm CPT and DMSO for 12 h. Cells were lysed in 1× NuPAGE LDS Sample Buffer (Invitrogen) containing 2.5% 2-mercaptoethanol, heated at 90 °C for 5 min, and homogenized using a QIAshredder (Qiagen). For coimmunoprecipitations, HEK293T cells expressing exogenous SLFN11 for 48 h were washed in ice-cold PBS and lysed in buffer containing 0.5% IGEPAL CA630, 5 mm EDTA, 50 mm HEPES, 50 mm NaCl, and 10% glycerol. Lysates were adjusted to 2 mg of total protein per sample and subjected to immunoprecipitation with anti-V5 tag antibody–conjugated magnetic beads (MBL International Corp., catalog number M167-11) for 2 h, sedimented, and washed three times with lysis buffer. Where indicated, the immunoprecipitate was treated with alkaline phosphatase (Invitrogen, catalog number EF0651) for 45 min at 37 °C. Beads were resuspended in 1× NuPAGE LDS Sample Buffer containing 2.5% 2-mercaptoethanol and heated at 90 °C for 5 min. All samples were resolved by 4–12% SDS-PAGE (Invitrogen) prior to transfer onto polyvinylidene difluoride membranes. After blocking and sequential incubation with target-specific primary and horseradish peroxidase–conjugated secondary antibodies, Western Lightning ECL Pro (PerkinElmer Life Sciences) was used to visualize proteins on X-ray film, and signal intensity was quantified with ImageJ64.
Phosphoprotein gel staining
Samples were resolved on 4–12% NuPAGE Bis-Tris gels (Invitrogen), which were subsequently fixed with 50% methanol and 10% acetic acid for 2 h. Gels were washed three times for 10 min, incubated in Phospho-Tag phosphoprotein gel stain (GeneCopoeia, catalog number P005A) for 1.5 h and subsequently destained in 3 × 30-min steps with Diamond Pro-Q phosphoprotein gel destaining solution. After rinsing twice with ultrapure water for 5 min, gels were scanned on a Typhoon imager at 532 nm with a 555 BP R6G laser. Bands were quantified with ImageQuant software.
Total RNA preparation and mRNA qPCR
Total cellular RNA was isolated with TRIzol (Invitrogen) and cleaned with the TURBO DNA-free kit (Ambion). Reverse transcription was performed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). The qPCRs were carried out on an Applied Biosystems StepOne Plus Real-Time PCR System using iTaq Universal SYBR Green Supermix (Bio-Rad) following the manufacturers' protocols. Relative levels of mRNAs of interest were calculated based on ΔCt values and subsequent normalization to GAPDH mRNA levels. The following qPCR primers were used in these assays: GAPDH: forward, 5′-TCCACTGGCGTCTTCACC-3′; reverse, 5′-GGCAGAGATGATGACCCTTTT-3′; EGFP.MYC: forward, 5′-CGCCGACCCAGCTTTCTTGTA-3′; reverse, 5′-TGATCAGCTTCTGCTCGCCG-3′; wtGFP.V5: forward, 5′-CTGGAGTTGTCCCAATTCTTG-3′; reverse, 5′-TCACCCTCTCCACTGACAGA-3′; SLFN11: forward, 5′-AAGGCCTGGAACATAAAAAGG-3′; reverse, 5′-GGAGTATATCGCAAATATCCTGGT-3′; HIV-1 p24: forward, 5′-TGCATGGGTAAAAGTAGTAGAAGAGA-3′; reverse, 5′-TGATAATGCTGAAAACATGGGTA-3′.
tRNA analysis
SYBR Gold stain and Northern blotting of tRNAs was described previously (8).
Statistical analysis
For all statistical analyses, two-sample equal variance (homoscedastic) Student's t tests (two-tailed) were performed using Microsoft Excel. All biological replicates and technical replicates are specified accordingly. Experimental sample sizes were chosen according to commonly accepted ranges for in vitro studies in this field and to achieve statistical significance. For all experiments without statistical analyses, one representative result of at least three independent experiments is shown.
Data availability
All data generated or analyzed during this study are included in this published article (and its supporting information files) or are available from the corresponding author upon reasonable request.
Author contributions
D. M., M. L., and M. D. conceptualization; D. M., M. L., and M. D. data curation; D. M. and M. L. formal analysis; D. M., M. L., and M. D. validation; D. M., R. M. L., M. L., and M. D. investigation; D. M. and M. L. visualization; D. M., R. M. L., and M. L. methodology; D. M. and M. L. writing-original draft; M. D. resources; M. D. supervision; M. D. funding acquisition; M. D. project administration; M. L. and M. D. writing-review and editing.
Supplementary Material
This work was supported by National Institutes of Health Grants R01-GM101982 and R21-AI124199 (to M. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SLFN
- Schlafen
- ISG
- interferon-stimulated gene
- DDA
- DNA-damaging agent
- PPP1CC
- protein phosphatase 1 catalytic subunit γ
- ATR
- ataxia telangiectasia and Rad3-related protein
- ATM
- ataxia telangiectasia mutated
- AAA
- ATPase associated with diverse cellular activities
- HEK
- human embryonic kidney
- EGFP
- enhanced GFP
- qPCR
- quantitative PCR
- CPT
- camptothecin
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- LDS
- lithium dodecyl sulfate
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- KO
- knockout
- MS
- mass spectrometry.
References
- 1. Bustos O., Naik S., Ayers G., Casola C., Perez-Lamigueiro M. A., Chippindale P. T., Pritham E. J., and de la Casa-Esperón E. (2009) Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 447, 1–11 10.1016/j.gene.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Snider J., Thibault G., and Houry W. A. (2008) The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9, 216 10.1186/gb-2008-9-4-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwarz D. A., Katayama C. D., and Hedrick S. M. (1998) Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9, 657–668 10.1016/S1074-7613(00)80663-9 [DOI] [PubMed] [Google Scholar]
- 4. Otero D. C., Baker D. P., and David M. (2013) IRF7-dependent IFN-β production in response to RANKL promotes medullary thymic epithelial cell development. J. Immunol. 190, 3289–3298 10.4049/jimmunol.1203086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li M., Kao E., Gao X., Sandig H., Limmer K., Pavon-Eternod M., Jones T. E., Landry S., Pan T., Weitzman M. D., and David M. (2012) Codon-usage-based inhibition of HIV protein synthesis by human Schlafen 11. Nature 491, 125–128 10.1038/nature11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zoppoli G., Regairaz M., Leo E., Reinhold W. C., Varma S., Ballestrero A., Doroshow J. H., and Pommier Y. (2012) Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. U.S.A. 109, 15030–15035 10.1073/pnas.1205943109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li M., Kao E., Malone D., Gao X., Wang J. Y. J., and David M. (2018) DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 25, 1047–1058 10.1038/s41594-018-0142-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang J. Y., Deng X. Y., Li Y. S., Ma X. C., Feng J. X., Yu B., Chen Y., Luo Y. L., Wang X., Chen M. L., Fang Z. X., Zheng F. X., Li Y. P., Zhong Q., Kang T. B., et al. (2018) Structure of Schlafen13 reveals a new class of tRNA/rRNA-targeting RNase engaged in translational control. Nat. Commun. 9, 1165 10.1038/s41467-018-03544-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stabell A. C., Hawkins J., Li M., Gao X., David M., Press W. H., and Sawyer S. L. (2016) Non-human primate Schlafen11 inhibits production of both host and viral proteins. PLoS Pathog. 12, e1006066 10.1371/journal.ppat.1006066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan P. P., and Lowe T. M. (2009) GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 10.1093/nar/gkn787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdel-Mohsen M., Raposo R. A., Deng X., Li M., Liegler T., Sinclair E., Salama M. S., Ghanem Hel-D., Hoh R., Wong J. K., David M., Nixon D. F., Deeks S. G., and Pillai S. K. (2013) Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 10, 106 10.1186/1742-4690-10-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murai J., Tang S.-W., Leo E., Baechler S. A., Redon C. E., Zhang H., Al Abo M., Rajapakse V. N., Nakamura E., Jenkins L. M. M., Aladjem M. I., and Pommier Y. (2018) SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol. Cell 69, 371–384.e6 10.1016/j.molcel.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mu Y., Lou J., Srivastava M., Zhao B., Feng X. H., Liu T., Chen J., and Huang J. (2016) SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 17, 94–109 10.15252/embr.201540964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiernan R. E., Vanhulle C., Schiltz L., Adam E., Xiao H., Maudoux F., Calomme C., Burny A., Nakatani Y., Jeang K. T., Benkirane M., and Van Lint C. (1999) HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 18, 6106–6118 10.1093/emboj/18.21.6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bollen M., Peti W., Ragusa M. J., and Beullens M. (2010) The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 35, 450–458 10.1016/j.tibs.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Küntziger T., Landsverk H. B., Collas P., and Syljuåsen R. G. (2011) Protein phosphatase 1 regulators in DNA damage signaling. Cell Cycle 10, 1356–1362 10.4161/cc.10.9.15442 [DOI] [PubMed] [Google Scholar]
- 18. Tang X., Hui Z. G., Cui X. L., Garg R., Kastan M. B., and Xu B. (2008) A novel ATM-dependent pathway regulates protein phosphatase 1 in response to DNA damage. Mol. Cell. Biol. 28, 2559–2566 10.1128/MCB.01711-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supporting information files) or are available from the corresponding author upon reasonable request.