Abstract
Protein kinase signaling networks stringently regulate cellular processes, such as proliferation, motility, and cell survival. These networks are also central to the evolution and progression of cancer. Accordingly, genetically encoded fluorescent biosensors capable of directly illuminating the spatiotemporal dynamics of kinase signaling in live cells are being increasingly used to investigate kinase signaling in cancer cells and tumor tissue sections. These biosensors enable visualization of biological processes and events directly in situ, preserving the native biological context and providing detailed insight into their localization and dynamics in cells. Herein, we first review common design strategies for kinase activity biosensors, including signaling targets, biosensor components, and fluorescent proteins involved. Subsequently, we discuss applications of biosensors to study the biology and management of cancer. These versatile molecular tools have been deployed to study oncogenic kinase signaling in living cells and image kinase activities in tumors or to decipher the mechanisms of anticancer drugs. We anticipate that the diversity and precision of genetically encoded biosensors will expand their use to further unravel the dysregulation of kinase signaling in cancer and the modes of actions of cancer-targeting drugs.
Keywords: biosensor, fluorescence resonance energy transfer (FRET), phosphorylation, cancer, cell signaling, fluorescent protein, in vivo imaging, kinase signaling, posttranslational modification
Introduction
Kinases are of central importance in cellular signaling networks. In humans, 535 protein kinases have been identified, which can be further subclassified into seven major eukaryotic protein kinase families, as well as atypical and other kinases, based on primary sequence (1). In terms of residues that are targeted by phosphorylation, kinases mostly fall into two major groups: tyrosine kinases and serine/threonine kinases. Tyrosine kinases further comprise two classes, receptor tyrosine kinases (RTKs)2 and nonreceptor tyrosine kinases (NRTKs), which play important roles in cell–cell signaling and transmembrane signal transduction (2). Intracellular signal transduction invariably requires Ser/Thr kinase cascades and their downstream targets. The most recent report shows that nearly 300,000 phosphosites in humans have been experimentally identified and curated in PhosphoSitePlus (www.phosphosite.org),3 (61) which represents over 65% of all posttranslational modification sites (3). This reflects the importance of Tyr/Ser/Thr kinases in controlling nearly every aspect of cellular life, such as the cell cycle, proliferation, differentiation, motility, and cell death or survival.
Under normal physiological conditions, kinase signaling must be precisely coordinated and integrated, and properly regulated differentiation signals are critical for preventing neoplasia, a type of abnormal and excessive tissue growth. In contrast, dysregulation of kinase signaling is frequently associated with oncogenesis. In tumor cells, it is common that key kinases are no longer adequately controlled, and excessive phosphorylation sustains signal transduction pathways in an activated state. Most tyrosine kinases remain stringently and negatively regulated in the absence of specific signals. When altered through gain-of-function mutation or chromosomal translocation, or overexpression and genomic amplification, they can become potent oncogenes by delivering continuous or enhanced signaling. For example, RTKs such as EGFR regulate cell growth and proliferation by signaling through intracellular MAP kinase cascades, and aberrant RTK activation is a frequent driver of malignancies, including breast and lung cancer (4). Meanwhile, direct mutational activation of MAP kinase cascades, such as the Ras-Raf-MEK-ERK MAP kinase signaling cascade, is arguably one of the most important oncogenic drivers of human cancers, such as in skin melanoma, thyroid, and colorectal cancer (5). NRTKs are also involved in integrating signal transduction through interactions with multiple receptors. Deregulation and overexpression of NRTKs such as Src have also been implicated in several human cancers, including melanoma, colon and breast carcinoma, and prostate cancer, with elevated Src activity promoting tumor growth as well as migratory and invasive potential (6, 7). In addition to canonical MAP kinase pathways, dysregulation of special signaling modules such as the Hippo pathway is frequently observed in human cancers (8, 9). The Hippo pathway is a key regulator of organ size and tissue homeostasis by limiting cell growth. In the activated Hippo pathway, MST1/2 activates LATS1/2, which directly phosphorylates and inactivates the effectors YAP/TAZ. Mutations or gene fusions involving these kinases and regulators in the Hippo pathway can lead to persistent YAP/TAZ activation, which induces gene transcription and cancer initiation and development (10). Understanding the molecular mechanisms regulating kinase signaling is therefore essential to understanding their role in cancer. To this end, new and more powerful tools for interrogating kinases in fundamental research and clinical diagnosis, as well as drug discovery, will be extremely useful.
Over the past 2 decades, the study of protein kinase signaling has been greatly enhanced by the design and application of various genetically encoded fluorescent biosensors capable of effectively monitoring kinase activities in living cells (11). The rapid and spatially precise optical readout of fluorescent biosensors makes it possible to visualize biological analytes and events directly in situ, preserving the native biological context and providing more detailed spatiotemporal information than can be obtained in vitro. For instance, the rapid dynamics of RTK activity can be easily monitored in live cells using biosensors but are difficult to capture using biochemical approaches, such as Western blotting. Furthermore, biosensors can be used to observe variations in kinase activity at the single-cell or even subcellular level. Whereas biosensors have already been widely used to study key cancer-associated kinases in normal cells and tissues, they are now increasingly being applied in cancer models. In this review, we first provide a brief summary of common biosensor design strategies. We then describe various studies that have used fluorescent biosensors to probe different types of kinase activities, especially in cancers.
Common biosensor design strategies
Genetically encoded fluorescent biosensors for kinase activity typically include two essential components: a sensing unit to detect changes in the signaling event and a reporting unit to translate the state of the sensing unit into a quantifiable readout. The sensing unit generally comes from a whole or central part of a substrate protein that is sensitive to the kinase of interest and functions as a “molecular switch” to control the behavior of the reporting unit. The reporting unit typically consists of one or more fluorescent protein (FP) variants, although recombinant luciferases can also be used depending on the requirements of the assay, yielding biosensors that variously change their photophysical properties or localization in response to phosphorylation. More details of biosensor design, including signaling targets, sensing components, and FPs, were recently reported by Greenwald et al. (12) and can also be found in our Fluorescent Biosensor Database (biosensordb.ucsd.edu).3 Although the development and optimization of such reporters commonly require intensive efforts, more researchers can adapt these tools using an established pipeline for kinase activity reporter development (11). Furthermore, based on feedback from applications, more and more sensitive and effective tools will continue to be generated using various approaches, including screening-based methods (13), to further expand our understanding of physiological and pathological processes.
FRET-based biosensors
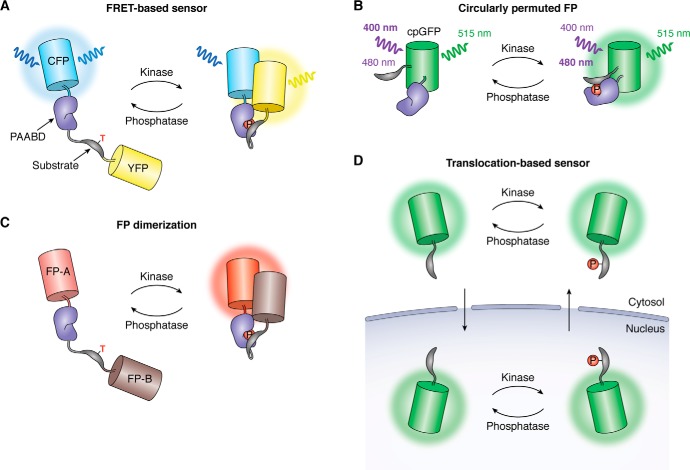
The most commonly used biosensors for measuring kinase activity dynamics in single cells are based on FRET. FRET-based biosensors are engineered by sandwiching the sensing unit between two FPs that act as a FRET pair and utilize the phosphorylation-induced conformational change in the sensing unit to modulate nonradiative energy transfer between the FPs (Fig. 1A). At the turn of the millennium, Zhang et al. (14) developed a first-generation protein kinase A activity reporter (AKAR1), in which a consensus phosphorylation sequence for protein kinase A (PKA; LRRASLP) was tethered by a flexible linker to the phospho-amino acid–binding domain (PAABD) 14-3-3τ, which is capable of specifically recognizing and binding the phosphorylated peptide. Importantly, phosphorylation of the substrate is determined by the balance between kinase and phosphatase activity, with PKA-mediated phosphorylation expected to increase the intramolecular binding between 14-3-3τ and the substrate, whereas dephosphorylation by cellular phosphatases should reverse this effect. Because kinase activity typically changes against a backdrop of relatively constant phosphatase activity, the resulting conformationally induced FRET changes between a flanking CFP and YFP pair should allow this design to faithfully report net increases and decreases in kinase activity. However, the high affinity of the 14-3-3τ domain was found to protect AKAR1 from dephosphorylation, yielding a largely irreversible FRET response, and all subsequent variants (e.g. AKAR2 (15), AKAR3 (16), and AKAR4 (17)) have instead utilized the lower-affinity forkhead-associated 1 (FHA1) domain, in conjunction with a modified PKA substrate (LRRATLVD), to more accurately capture PKA activity dynamics. Since then, a diverse family of FRET-based sensors have been developed based on the same general design for monitoring the activity of a multitude of different kinases, including but not limited to the insulin receptor (18), Abl (19), protein kinase C (PKC) (20), Src (21), Akt (22), and p38 (23). However, not all efforts to develop kinase biosensors based on this chimeric molecular switch will be successful, in which case an intrinsic conformational change in a substrate protein may alternatively be used as the sensing unit. For instance, Zhou et al. (24) took advantage of the native phosphorylation-induced conformational change in full-length 4EBP1 when designing the sensing unit for their mTOR complex 1 activity reporter (TORCAR). Furthermore, this design has also been adapted to yield a specific phosphatase activity biosensor for calcineurin, in which case acutely regulated phosphatase activity drives the changes in FRET (25).
Figure 1.
Genetically encoded biosensors for visualizing kinase activity in living cells. A, FRET-based biosensors utilize a kinase-inducible molecular switch to modulate the relative distance and orientation of a pair of FPs in response to changes in biosensor phosphorylation, thereby coupling endogenous kinase activity to biosensor FRET efficiency. As shown here, the molecular switch is often constructed by fusing a substrate peptide for a specific kinase to a PAABD, such that recognition of the phosphorylated substrate by the PAABD drives a conformational change in the biosensor. B, single-fluorophore kinase biosensors have recently been generated in which the substrate peptide and PAABD are inserted within the sequence of a cpFP, whereby the recognition of the phosphorylated substrate by the PAABD distorts the barrel of the cpFP, thus altering its photophysical behavior. In the example shown here, phosphorylation of the sensor leads to a shift in the maximum excitation wavelength of cpGFP from 400 to 480 nm, producing an excitation-ratiometric response. C, an alternative strategy for constructing single-fluorophore biosensors retains the same overall configuration of FRET-based sensors but replaces the FRET FP pair with a pair of dimerization-dependent FPs. This design also relies on the molecular switch to modulate the proximity of the dimerization-dependent FPs, as bringing FP-A and B into close proximity results in enhanced fluorescence emission by FP-A and has been used to develop red-fluorescent kinase sensors. D, instead of relying on a conformational change, translocation-based kinase biosensors use phosphorylation to control the behavior of nuclear localization and export signals that are incorporated within the substrate sequence. At rest, the dephosphorylated biosensor accumulates in the nucleus, whereas elevations in kinase activity drive the translocation of the biosensor into the cytosol, whereby the ratio of nuclear-to-cytoplasmic fluorescence serves as a dynamic readout of kinase activity.
FRET increases the occurrence of donor-sensitized emission by the acceptor while quenching direct donor fluorescence. Responses from FRET-based biosensors can therefore be measured using the acceptor/donor emission ratio, with the fixed 1:1 ratio of the donor and acceptor FPs enabling these sensors to provide a quantitative readout. However, FRET also results in a shortening of the fluorescence lifetime (τ) of the donor FP, which can be measured via fluorescence lifetime imaging microscopy (FLIM). Thus, FLIM represents an alternative approach to monitoring FRET-based biosensors. Notably, because FLIM is less sensitive to the loss of donor emission intensity caused by scattering in tissues, FLIM-FRET can be effectively applied in vivo. For example, Weitsman et al. (26) generated a FLIM-FRET–compatible version of the EGFR activity biosensor phosphorylation indicator of CrkII chimeric unit (Picchu) (27) by replacing YFP and CFP with mRFP1 and EGFP, respectively, and used the resulting Picchu-FLIM sensor to investigate EGFR activity in tumor xenografts. Another benefit of FLIM-FRET is that it does not require fluorescence emission by the acceptor, which can potentially alleviate some challenges of ratiometric imaging, such as spectral bleed-through. To this end, Chen et al. (28) recently modified AKAR3 to generate FLIM-AKAR, which they imaged using 2-photon FLIM to monitor PKA activity in brain tissue. This sensor contains monomeric EGFP as a bright donor and cpsREACh, which displays a high absorption coefficient but low quantum yield, as a “dark” acceptor, which provides a better signal/noise ratio and less bleed-through from the acceptor into the donor channel. Similar strategies have recently been applied to AKAR3EV and AKAR4 to yield the FLIM-FRET–based sensors AKARet (29) and AKAR5 (30), respectively.
Single-fluorophore biosensors
Along with FRET-based biosensors, single-fluorophore biosensors mainly based on circularly permuted fluorescent proteins (cpFPs) have emerged as another popular biosensor design strategy. Circular permutation involves rearranging the linear sequence of a typical FP to relocate the N and C termini to be within the rigid β-barrel so that the conformational change of the embedded molecular switch can more easily alter the conformation and fluorescence of the cpFP. Single-fluorophore biosensors based on cpFPs have advantages of a smaller spectral footprint. However, although numerous single-fluorophore sensors have been developed to monitor different cellular analytes, such as calcium (31, 32), cAMP (33), and metabolic products (34), the application of this sensor design to visualize kinase activity has been largely unexplored. To this end, Mehta et al. (35) recently engineered a series of single-fluorophore biosensors in which cpGFP is sandwiched between a kinase substrate peptide and PAABD, giving rise to a novel suite of excitation ratiometric kinase activity reporters (ExRai-KARs), wherein the maximum excitation peak of cpGFP undergoes a phosphorylation-dependent shift from ∼400 to ∼488 nm, with ∼515-nm (green) emission, in response to elevated kinase activity (Fig. 1B). The resulting sensors showed a substantial improvement compared with best-in-class FRET-based reporters, such as an almost 2-fold higher dynamic range and signal/noise ratio in the case of ExRai-AKAR. With its high sensitivity and robustness to submaximal kinase stimulation, this design may thus represent a promising new strategy for developing high-performance kinase activity sensors.
Continuing with this same substrate-cpFP-PAABD configuration, Mehta et al. (35) were also able to develop single-color KARs based on cp-T-sapphire, which exhibits single excitation and emission maxima at ∼400 and ∼513 nm, respectively, and cpBFP, which displays single excitation and emission peaks at ∼385 and ∼450 nm, respectively. By utilizing an alternative design based on FP dimerization (36), they were further able to develop red fluorescent single-color KARs (Fig. 1C). Given that complex cell behaviors are regulated by highly integrated kinase signaling networks, these single-color kinase sensors are highly desirable tools for simultaneously examining multiple kinase activities, as well as the cross-regulation between kinases, in single cells. Indeed, through the use of their single-color KARs, Mehta et al. (35) were able to successfully perform 4–6-parameter multiplexed imaging to simultaneously monitor changes in kinases activities, as well as cAMP and Ca2+ elevations, in single cells.
Translocation-based biosensors
In contrast to the above biosensors, translocation-based biosensors are entirely different in design, which depends on relocalizing the sensor to a predetermined subcellular site rather than directly altering FP photophysics. The basic design principle for kinase translocation reporters stems from the endogenous sequences found in kinase substrates that contain a nuclear localization signal (NLS) and/or nuclear export signal (NES) neighboring the phosphorylation sites. Phosphorylation and dephosphorylation of these sites alter the recognition of these NLS and NES motifs, which results in translocation in and out of the nucleus (Fig. 1D). For instance, Spencer et al. (37) initially developed a biosensor of CDK2 activity by fusing Venus (YFP) to the C-terminal domain of human DNA helicase B (HDHB), which exits the nucleus in response to CDK2-dependent phosphorylation. Gross et al. (38) similarly generated an Akt translocation sensor by fusing full-length FoxO1 to the green FP Clover, and Maryu et al. (39) divided the central region of FoxO3a, containing the Akt phosphorylation site and NLS and NES sequences, to generate their own Akt kinase sensor. In contrast to these endogenous sequences, Regot et al. (40) have demonstrated the generalizability of this approach by engineering a family of kinase translocation reporters based on a minimal translocation domain that contains a kinase-specific substrate peptide fused in tandem to a bipartite NLS and an NES, such that phosphorylation inhibits the NLS and activates the NES. This design, which relies on cellular machinery such as nuclear import to function, has been applied to PKA, c-Jun N-terminal kinase, p38, and ERK activity.
Bioluminescence-based biosensors
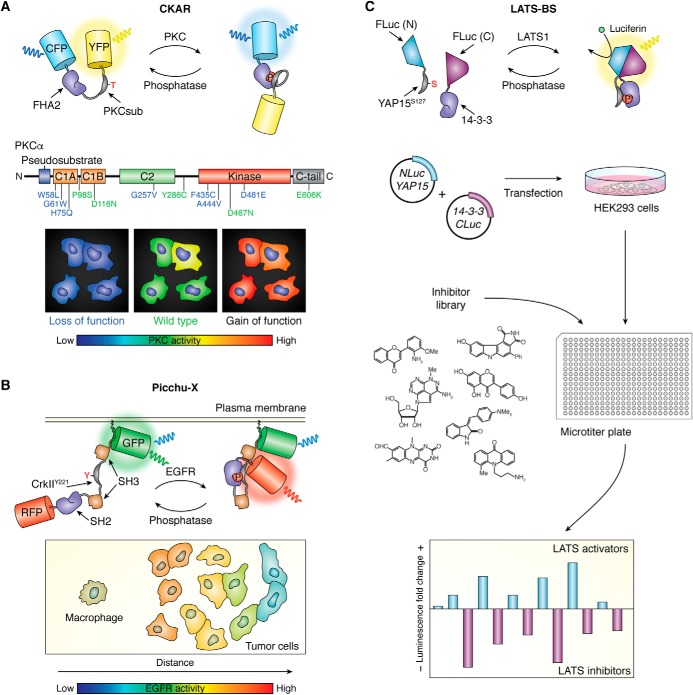
An alternative biosensing strategy is to develop bioluminescence-based biosensors that incorporate luminescent proteins as the reporting unit. Luciferases are a well-known group of luminescent proteins that emit light as a byproduct of the catalytic oxidation of a luciferin substrate. Because these enzymes do not rely on external illumination to produce a signal, bioluminescence-based readouts are not affected by the autofluorescence, photobleaching, or phototoxicity that can occur during fluorescence imaging and thus have been considered a potential way to enhance biosensor sensitivity and versatility. A typical strategy for designing luminescent sensors is to utilize the reversible fragment complementation of a split luciferase as a means of dynamically controlling light output. For example, Herbst et al. (41) developed a luminescent PKA activity reporter (lumAKAR) based on a biomolecular design in which the kinase-inducible molecular switch from FRET-based AKAR is divided into two fragments (substrate peptide and PAABD) that are fused to complementary N- and C-terminal portions of Renilla luciferase (RLuc). Elevations in PKA activity cause this bimolecular switch to form a complex, which reconstitutes catalytically active RLuc. This generalizable design was also applied to monitor the dynamics of other kinases, such as PKC (41). RLuc fragment complementation was also used by Stefan et al. (42) to investigate dynamic protein kinase complexes, wherein the regulatory and catalytic subunits of PKA were fused to complementary N- and C-terminal RLuc fragments. Azad et al. also recently developed a bioluminescence-based LATS biosensor (LATS-BS) consisting of complementary N- and C-terminal luciferase fragments fused to a 15-amino acid YAP peptide (YAP15), which contains the Ser-127 LATS phosphorylation site, and a 14-3-3τ domain (43) (Fig. 2C).
Figure 2.
Using genetically encoded kinase biosensors to study cancer. A, studying cancer-associated kinases at the single-cell level. PKC has long been considered to be an oncogene due to the ability of phorbol esters, which potently activate PKC, to promote tumorigenesis. However, when Antal et al. (44) probed the role of PKC in cancer by examining the effect of various cancer-associated mutations on PKC activity using the FRET-based PKC activity reporter CKAR, most mutations were found to either have no effect on PKC activity or be loss-of-function mutations. For example, among 12 PKCα mutations tested, seven exhibited reduced activity compared with WT PKCα, whereas the remaining five showed no difference; no gain-of-function mutants were observed. Their results strongly suggest that PKC is in fact a tumor suppressor and highlight the importance of using biosensors to dissect cancer-associated signaling in the native context of living cells. B, illuminating heterogenous signaling in tumor tissues. Tumors are highly complex and intricately organized tissues. Interactions among cells within different parts of the tumor can form diverse microenvironments associated with unique signaling dynamics, and genetically encoded biosensors can be powerful tools for elucidating the mechanisms of this heterogeneity. For example, by using the FRET-based EGFR biosensor Picchu-X to visualize EGFR signaling in tumor xenografts, Weitsman et al. (27) were able to observe substantial heterogeneity in EGFR activation within the tumor. Because they were imaging biosensor responses in intact tissue, they were further able to correlate the responses to the proximity of different tumor structures and cell types, revealing that proximity to infiltrating macrophages was strongly associated with elevated EGFR activity in tumor cells. C, biosensors enable high-content screening for pathway regulators. Because they can directly and sensitively report on endogenous kinase activities, genetically encoded biosensors are increasingly being used in high-content studies to dissect signaling pathways and identify drug targets. Azad et al. (43) recently applied this approach using their newly developed bioluminescent LATS1 sensor to identify upstream regulators of the Hippo pathway. By culturing biosensor-transfected HEK293 cells in microtiter plates and treating them with a large panel of kinase inhibitors, they were able to identify both activators and inhibitors of LATS1 kinase, thus providing new details of Hippo pathway regulation.
Applications of biosensors in cancer
From their introduction nearly 20 years ago, genetically encoded kinase biosensors have provided numerous insights into the regulation of kinase signaling in physiological processes. In light of this success, the use of biosensors to study kinase signaling in cancer has attracted increasing interest in recent years. From cultured cancer cell lines to tumor tissue, biosensors provide direct in situ measurements of the spatiotemporal dynamics of kinase activity and play an increasingly crucial role in diverse studies, ranging from the molecular dissection of kinase signaling pathways in cancer to monitoring the effects of cancer drugs on specific kinase pathways and targets for improved cancer therapy and drug discovery.
Dissecting oncogenic kinase signaling in living cells
Cell function and behavior are dynamically regulated by kinase signaling pathways, which often play a key role in oncogenesis. The use of genetically encoded biosensors to rapidly and sensitively probe changes in kinase activities at the single-cell level can thus provide novel and valuable insights into the mechanisms of aberrant kinase signaling in cancer biology.
PKC has long been regarded as an oncogenic kinase based on the ability of various PKC isozymes to act as high-affinity intracellular receptors for tumor-promoting phorbol esters. However, a 2015 study in which Antal et al. (44) used the genetically encoded PKC activity sensor CKAR to systematically characterize 46 cancer-associated mutations known to alter PKC signaling has helped to largely overturn this long-standing dogma. By co-expressing CKAR in COS7 cells along with different PKC variants bearing mutations in the regulatory or kinase domains and monitoring the FRET ratio change in response to pharmacological stimulation of PKC, the authors found ∼61% of PKC mutations to be loss-of-function, with most either reducing or abolishing PKC activity, whereas none were activating (Fig. 2A). Their analysis of cancer-associated mutations from diverse cancers and throughout the PKC family revealed that PKC isozymes are generally inactivated in cancer, supporting a tumor-suppressive function for this kinase. Further supporting the role of PKC as a tumor suppressor, correcting a loss-of-function PKCβ mutation via CRISPR-mediated genome editing in a patient-derived colon cancer cell line suppressed anchorage-independent growth and reduced tumor growth in a xenograft model. Based on these results, it is not difficult to see why over 30 years of clinical trials using PKC inhibitors to treat cancer would not only have failed, but in some instances also worsened patient outcomes. Thus, we can only hope that biosensor-based studies will play an even greater role in guiding clinical studies in the future.
Generally, biosensor development and optimization are focused on ensuring the specificity of a biosensor for a single kinase. Nevertheless, Midde et al. (45) recently presented an intriguing counterexample to this strategy, in which a substrate protein shared by multiple kinases was applied to develop a biosensor to indicate metastatic potential. Specifically, the authors based their biosensor design on the Tyr-phosphorylated protein CCDC88A (GIV/Girdin), which they had revealed through phosphoproteomics analyses to be a key metastasis-specific phosphoevent across a variety of solid tumors. Furthermore, because GIV integrates prometastatic signals from multiple oncogenic receptors, these multimodular biosensors were named integrators of metastatic potential (IMPs). Using these IMP sensors, the authors were successfully able to capture the heterogeneity of metastatic potential within primary lung and breast tumor cells at steady state, sensitively detect the few cells with the highest metastatic potential, and even track the metastatic potential of tumor cells during metastatic progression and the development of drug resistance. These findings indicate that IMPs are effective tools for measuring the diversity and plasticity of metastatic potential of tumor cells in a sensitive and unbiased way.
Imaging kinase activities in tumor tissues
Tumors are complex tissues that engage in multifaceted interactions with the surrounding host environment, which ultimately shape the course of disease progression. Applying biosensors at the tissue level is therefore an extremely exciting and powerful approach to studying the role of kinase-mediated signaling in the occurrence and development of tumors.
Kinase activity can be influenced by changes in both intracellular signaling networks and the external environment. Particularly within the complex tumor microenvironment, the regulation of kinase signaling can be quite intricate, and kinase activities are often heterogeneous. Intratumoral heterogeneity is considered a major mechanism underlying treatment failure using molecularly targeted therapies. For example, EGFR is overexpressed or aberrantly activated in most common solid tumors, including non-small-cell lung cancer and cancers of the breast, prostate, and colon, yet despite the development of numerous EGFR-targeted drugs with specific mechanisms of action in recent years, clinical testing has revealed relatively poor response rates. Importantly, the failure of cells to respond to drugs could be linked to differences in the external microenvironment and not to EGFR expression. Therefore, Weitsman et al. (26) utilized their FLIM-FRET-based EGFR biosensor Picchu-FLIM, which was delivered into a murine model of basal-like breast cancer in vivo via liposomes, to examine the intratumoral heterogeneity of EGFR activity. This work revealed substantial heterogeneity in both EGFR activity and EGFR inhibitor efficacy in a murine breast cancer model. Furthermore, the proximity of macrophages to tumor cells was positively associated with tumor cell EGFR activity and was found to at least partially account for the observed intratumoral heterogeneity (Fig. 2B). This result was consistent with the demonstration that tumor cells and macrophages interacted through a paracrine loop involving the macrophage growth factor CSF-1 released from tumor cells and the reciprocal release of EGF by macrophages (46). This association was also found to persist in the presence of an EGFR inhibitor. The same effect of macrophage infiltration on EGFR activation was also seen in colorectal cancer xenografts, but not in non-small-cell lung cancer xenografts expressing a constitutively activated EGFR mutant that could not be affected by macrophages (26).
Vascular permeability, or the extent to which a blood vessel wall allows the flow of small molecules or whole cells in and out of the vessel, is markedly increased in tumor tissues, and this “hyperpermeability” is associated with angiogenesis and metastasis. In microvascular endothelial cells, the cAMP/PKA pathway mediates endothelial barrier function by suppressing vascular permeability. Yamauchi et al. (47) utilized the PKA activity reporter AKAR3EV, which is an improved AKAR3 containing a flexible long Eevee linker (48), to understand the contribution of PKA to vascular hyperpermeability in tumor tissue. Through a combination of in vivo two-photon excitation microscopy and AKAR3EV-expressing transgenic mice, the level of PKA activity was revealed to be significantly lower in intratumoral endothelial cells compared with subcutaneous endothelial cells. Meanwhile, a modified Miles assay, in which higher extravasation of Evans blue dye was observed in tumor tissue, confirmed the hyperpermeability of the tumor vessels. PKA activation with a cAMP analogue further alleviated tumor vascular hyperpermeability, suggesting that low PKA activity in endothelial cells may be responsible for vascular hyperpermeability in tumor tissues.
Screening for kinase inhibitors and evaluating drug effects
Because they offer a direct, real-time readout of kinase activity, kinase biosensors can potentially be used to monitor drug-targeting efficacy as well as to understand the spatiotemporal changes of kinase activity in response to inhibitors. For example, imatinib was developed to treat chronic myelogenous leukemia (CML), which is mediated by the Bcr-Abl gene fusion product, yet acquired resistance to imatinib has prompted the search for novel tyrosine kinase inhibitors directed against Bcr-Abl mutants. As such, there is an emerging need for in vivo tools that can rapidly and sensitively monitor Bcr-Abl activity and evaluate drug resistance as well as guide clinical therapies. With this in mind, Tunceroglu et al. (49) utilized Picchu (26) to profile inhibitor efficacy and thus monitor drug resistance in CML cells. However, because Picchu can also respond to both Abl and EGFR activity, Mizutani et al. (50) instead developed a more specific and sensitive FRET-based biosensor based on CrkL, which is the most characteristic Bcr-Abl substrate. In the resulting phosphorylation indicator of CrkL en substrate (Pickles), CrkL is fused to an SH2 domain and sandwiched between Venus and ECFP, so that intramolecular binding of the SH2 domain to Tyr-207–phosphorylated CrkL increases FRET. Using this sensor, they assessed the effect of imatinib on Bcr-Abl activity in CML and evaluated the influence of second-generation drugs on Bcr-Abl mutants, finding that the T315I mutation conferred resistance to both nilotinib and dasatinib, whereas the G250E mutation showed dose-dependent resistance to nilotinib. By applying this sensor in patient cells, the authors were also able to monitor disease status during imatinib therapy.
Src up-regulation and activation governed by the tumor microenvironment may affect drug targeting in pancreatic cancer. To probe the spatiotemporal regulation of Src activity in pancreatic cancer and determine the activity changes in response to the anti-invasive Src inhibitor dasatinib, Nobis et al. (21) constructed a FLIM-FRET–based version of a previously described Src biosensor to visualize Src activity in pancreatic cancer (51). In both 3D organotypic cultures and live pancreatic tumors, a spatially distinct gradient of Src activity was observed within invading tumor cells, which was greatly enhanced at the invasive border relative to the tumor cortex. The authors observed that upon treatment with dasatinib, cells at the invasive borders switched from a predominantly active to an inactive cell state, which correlated with impaired metastatic capacity in vivo. They also found that the extent to which Src activity responded to dasatinib treatment was correlated by the distance of tumor cells from the host vasculature, further illustrating how biosensors can be used to uncover drug penetrance in vivo and thereby map areas of poor drug-targeting efficiency within specific tumor microenvironments.
Kinase biosensors can also be applied to directly screen for more effective inhibitors. For example, Azad et al. (43) developed a bioluminescence-based LATS biosensor (LATS-BS, described above) to monitor the activity of the Hippo core component LATS kinase, which they used to perform a screen for upstream kinases capable of modulating LATS kinase activity. By applying a library of 80 small-molecule kinase inhibitors to treat HEK293A cells expressing this sensor, the authors identified six kinase inhibitors that activated the biosensor, in addition to six inhibitors that reduced the biosensor signal (Fig. 2C). Of these “activating” inhibitors, the vascular endothelial growth factor receptor (VEGFR) inhibitor SU4312 showed the most dramatic effect on LATS. Further verification using multiple VEGFR inhibitors suggested that VEGFR signaling was indeed a regulator of the Hippo pathway. Specifically, activated VEGFR was found to inhibit LATS and activate the downstream Hippo effectors YAP and TAZ, which participates in regulating blood vessel formation in both physiological and pathological settings.
Conclusions and perspectives
Genetically encoded kinase activity biosensors have emerged as powerful tools to illuminate kinase signaling networks in both physiological and pathological settings. The commonly used biosensor designs described above are all capable of rapidly and sensitively visualizing dynamic changes in kinase activity, thereby providing important new insights into the dysregulation of kinase signaling in cancer as well as into the molecular mechanisms of cancer-targeting drugs.
The use of biosensors to perform tissue-level imaging of kinase signaling in whole tumors represents an extremely exciting new area of cancer research. However, in vivo fluorescence imaging generally suffers from problems such as phototoxicity and tissue penetrability of visible light. The development of biosensors incorporating various recently engineered IR fluorescent proteins (IFPs) (52), which are capable of excitation and emission at wavelengths from 650 to 900 nm (e.g. the near-IR window (53)), has thus opened a window onto in vivo activity imaging. For example, Tchekanda et al (54). have developed an IFP-based reporter for detecting the spatiotemporal dynamics of protein–protein interactions, wherein a protein fragment complementation assay based on IFP1.4 was used to monitor β2-adrenergic receptor signaling in U2OS cells. Continued engineering efforts have also produced improved IFPs with much higher brightness and quantum yield and have even been used to carry out in vivo tumor imaging in intact mouse brains (55, 56).
Even with these advances, whole-tumor imaging of kinase signaling, such as efforts to carefully delineate relationships between kinase activity and the tumor microenvironment, represents a tremendous technical challenge, which is made even more daunting by the real-time, dynamic nature of biosensor imaging. Alternatively, biosensors that are capable of “memorizing” differences in kinase activities across a heterogeneous and complex tumor could enable retrospective examination of chronic signaling differences over the course of tumor development and evolution. In neuroscience, the concept of such an activity “integrator” has recently been realized for the study of neuronal activity and neuromodulator signaling in the brain (57–59). Many of these snapshot reporters share a similar design in which co-application of an external (e.g. light) and internal (e.g. Ca2+) stimulus are required to trigger an AND-gate that drives the tobacco etch virus (TEV) protease-mediated cleavage and release of a transcription factor to induce the expression of an FP reporter gene. Thus, any cell that experiences the internal stimulus (e.g. Ca2+ elevation) during the recording period, which is defined by the external stimulus (e.g. illumination), will be permanently marked with fluorescence. In a more recent example, Wintgens et al. (60) applied a split TEV protease system to monitor the recruitment of adapter proteins to ligand-activated RTKs, including the epidermal growth factor receptor (ERBB), the insulin receptor (INSR), and the hepatocyte growth factor receptor (HGFR) families. Binding of the activated RTK and adaptor reconstitutes the split TEV, causing cleavage of a TEV substrate sequence between the RTK and the Gal-VP16 transcription factor, which then migrates into the nucleus to initiate luciferase expression. Using this integrator, the entire history of RTK activation during ligand treatment is recorded and converted into a luminescent signal. In addition, these RTK-split-TEV recruitment assays were validated in dose-dependent inhibition assays using ERBB family–selective antagonists. Although lacking the AND-gate design of neuronal activity integrators, this work hints at the potential for developing kinase activity integrators to assist with whole-tumor recording of kinase activities in cancer research.
Together, IFP-based biosensors and “snapshot” integrators are effective new tools that promise to deepen our understanding of kinase signaling in cancer by enabling the study of complex specimens. We expect that far more comprehensive and detailed future studies, including investigations of kinase activation, heterogeneity, and responses to drug treatment, will be made possible using these tools.
This work was supported by National Institutes of Health Grants R01 MH111516, R35 CA197622, R01 DK073368, and R01 GM111665 (to J. Z.) and Air Force Office of Scientific Research Grant FA9500-18-1-0051 (to J. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RTK
- receptor tyrosine kinase
- NRTK
- nonreceptor tyrosine kinase
- MAP
- mitogen-activated protein
- FP
- fluorescent protein
- PKA
- protein kinase A
- PKC
- protein kinase C
- PAABD
- phospho-amino acid–binding domain
- FLIM
- fluorescence lifetime imaging microscopy
- cpFP
- circularly permuted fluorescent protein
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- ERK
- extracellular signal–regulated kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase
- RLuc
- Renilla luciferase
- LATS-BS
- LATS biosensor
- IMP
- integrator of metastatic potential
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- CML
- chronic myelogenous leukemia
- SH2
- Src homology 2
- VEGFR
- vascular endothelial growth factor receptor
- IFP
- IR fluorescent protein
- TEV
- tobacco etch virus.
References
- 1. Wilson L. J., Linley A., Hammond D. E., Hood F. E., Coulson J. M., MacEwan D. J., Ross S. J., Slupsky J. R., Smith P. D., Eyers P. A., and Prior I. A. (2018) New perspectives, opportunities, and challenges in exploring the human protein kinome. Cancer Res. 78, 15–29 10.1158/0008-5472.CAN-17-2291 [DOI] [PubMed] [Google Scholar]
- 2. Hunter T. (2009) Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146 10.1016/j.ceb.2009.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hornbeck P. V., Kornhauser J. M., Latham V., Murray B., Nandhikonda V., Nord A., Skrzypek E., Wheeler T., Zhang B., and Gnad F. (2019) 15 years of PhosphoSitePlus® integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 47, D433–D441 10.1093/nar/gky1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Du Z., and Lovly C. M. (2018) Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 17, 58 10.1186/s12943-018-0782-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roskoski R., Jr. (2019) Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 142, 151–168 10.1016/j.phrs.2019.01.039 [DOI] [PubMed] [Google Scholar]
- 6. Frame M. C. (2002) Src in cancer: deregulation and consequences for cell behaviour. Biochim. Biophys. Acta 1602, 114–130 10.1016/s0304-419x(02)00040-9 [DOI] [PubMed] [Google Scholar]
- 7. Cai H., Smith D. A., Memarzadeh S., Lowell C. A., Cooper J. A., and Witte O. N. (2011) Differential transformation capacity of Src family kinases during the initiation of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 6579–6584 10.1073/pnas.1103904108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plouffe S. W., Hong A. W., and Guan K. L. (2015) Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 21, 212–222 10.1016/j.molmed.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeung B., Yu J., and Yang X. (2016) Roles of the Hippo pathway in lung development and tumorigenesis. Int. J. Cancer 138, 533–539 10.1002/ijc.29457 [DOI] [PubMed] [Google Scholar]
- 10. Yu F. X., Zhao B., and Guan K. L. (2015) Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oldach L., and Zhang J. (2014) Genetically encoded fluorescent biosensors for live-cell visualization of protein phosphorylation. Chem. Biol. 21, 186–197 10.1016/j.chembiol.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenwald E. C., Mehta S., and Zhang J. (2018) Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794 10.1021/acs.chemrev.8b00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belal A. S., Sell B. R., Hoi H., Davidson M. W., and Campbell R. E. (2014) Optimization of a genetically encoded biosensor for cyclin B1-cyclin dependent kinase 1. Mol. Biosyst. 10, 191–195 10.1039/C3MB70402E [DOI] [PubMed] [Google Scholar]
- 14. Zhang J., Ma Y., Taylor S. S., and Tsien R. Y. (2001) Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc. Natl. Acad. Sci. U.S.A. 98, 14997–15002 10.1073/pnas.211566798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Hupfeld C. J., Taylor S. S., Olefsky J. M., and Tsien R. Y. (2005) Insulin disrupts β-adrenergic signalling to protein kinase A in adipocytes. Nature 437, 569–573 10.1038/nature04140 [DOI] [PubMed] [Google Scholar]
- 16. Allen M. D., and Zhang J. (2006) Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem. Biophys. Res. Commun. 348, 716–721 10.1016/j.bbrc.2006.07.136 [DOI] [PubMed] [Google Scholar]
- 17. Depry C., Allen M. D., and Zhang J. (2011) Visualization of PKA activity in plasma membrane microdomains. Mol. Biosyst. 7, 52–58 10.1039/C0MB00079E [DOI] [PubMed] [Google Scholar]
- 18. Sato M., Ozawa T., Inukai K., Asano T., and Umezawa Y. (2002) Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat. Biotechnol. 20, 287–294 10.1038/nbt0302-287 [DOI] [PubMed] [Google Scholar]
- 19. Ting A. Y., Kain K. H., Klemke R. L., and Tsien R. Y. (2001) Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. U.S.A. 98, 15003–15008 10.1073/pnas.211564598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Violin J. D., Zhang J., Tsien R. Y., and Newton A. C. (2003) A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161, 899–909 10.1083/jcb.200302125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Botvinick E. L., Zhao Y., Berns M. W., Usami S., Tsien R. Y., and Chien S. (2005) Visualizing the mechanical activation of Src. Nature 434, 1040–1045 10.1038/nature03469 [DOI] [PubMed] [Google Scholar]
- 22. Gao X., and Zhang J. (2008) Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol. Cell 19, 4366–4373 10.1091/mbc.e08-05-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomida T., Takekawa M., and Saito H. (2015) Oscillation of p38 activity controls efficient pro-inflammatory gene expression. Nat. Commun. 6, 8350 10.1038/ncomms9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou X., Clister T. L., Lowry P. R., Seldin M. M., Wong G. W., and Zhang J. (2015) Dynamic visualization of mTORC1 activity in living cells. Cell Rep. 10, 1767–1777 10.1016/j.celrep.2015.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newman R. H., and Zhang J. (2008) Visualization of phosphatase activity in living cells with a FRET-based calcineurin activity sensor. Mol. Biosyst. 4, 496–501 10.1039/b720034j [DOI] [PubMed] [Google Scholar]
- 26. Weitsman G., Mitchell N. J., Evans R., Cheung A., Kalber T. L., Bofinger R., Fruhwirth G. O., Keppler M., Wright Z. V. F., Barber P. R., Gordon P., de Koning T., Wulaningsih W., Sander K., Vojnovic B., et al. (2017) Detecting intratumoral heterogeneity of EGFR activity by liposome-based in vivo transfection of a fluorescent biosensor. Oncogene 36, 3618–3628 10.1038/onc.2016.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurokawa K., Mochizuki N., Ohba Y., Mizuno H., Miyawaki A., and Matsuda M. (2001) A pair of fluorescent resonance energy transfer-based probes for tyrosine phosphorylation of the CrkII adaptor protein in vivo. J. Biol. Chem. 276, 31305–31310 10.1074/jbc.M104341200 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Saulnier J. L., Yellen G., and Sabatini B. L. (2014) A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front. Pharmacol. 5, 56 10.3389/fphar.2014.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang S., and Yasuda R. (2017) Imaging ERK and PKA activation in single dendritic spines during structural plasticity. Neuron 93, 1315–1324.e3 10.1016/j.neuron.2017.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tillo S. E., Xiong W. H., Takahashi M., Miao S., Andrade A. L., Fortin D. A., Yang G., Qin M., Smoody B. F., Stork P. J. S., and Zhong H. (2017) Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 19, 617–629 10.1016/j.celrep.2017.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baird G. S., Zacharias D. A., and Tsien R. Y. (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11241–11246 10.1073/pnas.96.20.11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagai T., Sawano A., Park E. S., and Miyawaki A. (2001) Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc. Natl. Acad. Sci. U.S.A. 98, 3197–3202 10.1073/pnas.051636098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kitaguchi T., Oya M., Wada Y., Tsuboi T., and Miyawaki A. (2013) Extracellular calcium influx activates adenylate cyclase 1 and potentiates insulin secretion in MIN6 cells. Biochem. J. 450, 365–373 10.1042/BJ20121022 [DOI] [PubMed] [Google Scholar]
- 34. Zhao Y., Hu Q., Cheng F., Su N., Wang A., Zou Y., Hu H., Chen X., Zhou H. M., Huang X., Yang K., Zhu Q., Wang X., Yi J., Zhu L., et al. (2015) SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 21, 777–789 10.1016/j.cmet.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehta S., Zhang Y., Roth R. H., Zhang J. F., Mo A., Tenner B., Huganir R. L., and Zhang J. (2018) Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat. Cell Biol. 20, 1215–1225 10.1038/s41556-018-0200-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alford S. C., Abdelfattah A. S., Ding Y., and Campbell R. E. (2012) A fluorogenic red fluorescent protein heterodimer. Chem. Biol. 19, 353–360 10.1016/j.chembiol.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spencer S. L., Cappell S. D., Tsai F. C., Overton K. W., Wang C. L., and Meyer T. (2013) The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 10.1016/j.cell.2013.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gross S. M., and Rotwein P. (2015) Akt signaling dynamics in individual cells. J. Cell Sci. 128, 2509–2519 10.1242/jcs.168773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maryu G., Matsuda M., and Aoki K. (2016) Multiplexed fluorescence imaging of ERK and Akt activities and cell-cycle progression. Cell Struct. Funct. 41, 81–92 10.1247/csf.16007 [DOI] [PubMed] [Google Scholar]
- 40. Regot S., Hughey J. J., Bajar B. T., Carrasco S., and Covert M. W. (2014) High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 10.1016/j.cell.2014.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Herbst K. J., Allen M. D., and Zhang J. (2011) Luminescent kinase activity biosensors based on a versatile bimolecular switch. J. Am. Chem. Soc. 133, 5676–5679 10.1021/ja1117396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefan E., Aquin S., Berger N., Landry C. R., Nyfeler B., Bouvier M., and Michnick S. W. (2007) Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 16916–16921 10.1073/pnas.0704257104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Azad T., Janse van Rensburg H. J., Lightbody E. D., Neveu B., Champagne A., Ghaffari A., Kay V. R., Hao Y., Shen H., Yeung B., Croy B. A., Guan K. L., Pouliot F., Zhang J., Nicol C. J. B., and Yang X. (2018) A LATS biosensor screen identifies VEGFR as a regulator of the Hippo pathway in angiogenesis. Nat. Commun. 9, 1061 10.1038/s41467-018-03278-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Antal C. E., Hudson A. M., Kang E., Zanca C., Wirth C., Stephenson N. L., Trotter E. W., Gallegos L. L., Miller C. J., Furnari F. B., Hunter T., Brognard J., and Newton A. C. (2015) Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell 160, 489–502 10.1016/j.cell.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Midde K., Sun N., Rohena C., Joosen L., Dhillon H., and Ghosh P. (2018) Single-cell imaging of metastatic potential of cancer cells. iScience 10, 53–65 10.1016/j.isci.2018.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wyckoff J., Wang W., Lin E. Y., Wang Y., Pixley F., Stanley E. R., Graf T., Pollard J. W., Segall J., and Condeelis J. (2004) A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 64, 7022–7029 10.1158/0008-5472.CAN-04-1449 [DOI] [PubMed] [Google Scholar]
- 47. Yamauchi F., Kamioka Y., Yano T., and Matsuda M. (2016) In vivo FRET imaging of tumor endothelial cells highlights a role of low PKA activity in vascular hyperpermeability. Cancer Res. 76, 5266–5276 10.1158/0008-5472.CAN-15-3534 [DOI] [PubMed] [Google Scholar]
- 48. Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y., and Matsuda M. (2011) Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656 10.1091/mbc.e11-01-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tunceroglu A., Matsuda M., and Birge R. B. (2010) Real-time fluorescent resonance energy transfer analysis to monitor drug resistance in chronic myelogenous leukemia. Mol. Cancer Ther. 9, 3065–3073 10.1158/1535-7163.MCT-10-0623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mizutani T., Kondo T., Darmanin S., Tsuda M., Tanaka S., Tobiume M., Asaka M., and Ohba Y. (2010) A novel FRET-based biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clin. Cancer Res. 16, 3964–3975 10.1158/1078-0432.CCR-10-0548 [DOI] [PubMed] [Google Scholar]
- 51. Nobis M., McGhee E. J., Morton J. P., Schwarz J. P., Karim S. A., Quinn J., Edward M., Campbell A. D., McGarry L. C., Evans T. R., Brunton V. G., Frame M. C., Carragher N. O., Wang Y., Sansom O. J., et al. (2013) Intravital FLIM-FRET imaging reveals dasatinib-induced spatial control of Src in pancreatic cancer. Cancer Res. 73, 4674–4686 10.1158/0008-5472.CAN-12-4545 [DOI] [PubMed] [Google Scholar]
- 52. Chernov K. G., Redchuk T. A., Omelina E. S., and Verkhusha V. V. (2017) Near-infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 117, 6423–6446 10.1021/acs.chemrev.6b00700 [DOI] [PubMed] [Google Scholar]
- 53. Weissleder R. (2001) A clearer vision for in vivo imaging. Nat. Biotechnol. 19, 316–317 10.1038/86684 [DOI] [PubMed] [Google Scholar]
- 54. Tchekanda E., Sivanesan D., and Michnick S. W. (2014) An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat. Methods 11, 641–644 10.1038/nmeth.2934 [DOI] [PubMed] [Google Scholar]
- 55. Yu D., Gustafson W. C., Han C., Lafaye C., Noirclerc-Savoye M., Ge W. P., Thayer D. A., Huang H., Kornberg T. B., Royant A., Jan L. Y., Jan Y. N., Weiss W. A., and Shu X. (2014) An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 5, 3626 10.1038/ncomms4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu D., Baird M. A., Allen J. R., Howe E. S., Klassen M. P., Reade A., Makhijani K., Song Y., Liu S., Murthy Z., Zhang S. Q., Weiner O. D., Kornberg T. B., Jan Y. N., Davidson M. W., and Shu X. (2015) A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 12, 763–765 10.1038/nmeth.3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang W., Wildes C. P., Pattarabanjird T., Sanchez M. I., Glober G. F., Matthews G. A., Tye K. M., and Ting A. Y. (2017) A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol. 35, 864–871 10.1038/nbt.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee D., Hyun J. H., Jung K., Hannan P., and Kwon H. B. (2017) A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 35, 858–863 10.1038/nbt.3902 [DOI] [PubMed] [Google Scholar]
- 59. Lee D., Creed M., Jung K., Stefanelli T., Wendler D. J., Oh W. C., Mignocchi N. L., Lüscher C., and Kwon H. B. (2017) Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat. Methods 14, 495–503 10.1038/nmeth.4234 [DOI] [PubMed] [Google Scholar]
- 60. Wintgens J. P., Wichert S. P., Popovic L., Rossner M. J., and Wehr M. C. (2019) Monitoring activities of receptor tyrosine kinases using a universal adapter in genetically encoded split TEV assays. Cell Mol. Life Sci. 76, 1185–1199 10.1007/s00018-018-03003-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]