Abstract
IMP dehydrogenase (IMPDH) is an essential enzyme that catalyzes the rate-limiting step in the de novo guanine nucleotide biosynthetic pathway. Because of its involvement in the control of cell division and proliferation, IMPDH represents a therapeutic for managing several diseases, including microbial infections and cancer. IMPDH must be tightly regulated, but the molecular mechanisms responsible for its physiological regulation remain unknown. To this end, we recently reported an important role of adenine and guanine mononucleotides that bind to the regulatory Bateman domain to allosterically modulate the catalytic activity of eukaryotic IMPDHs. Here, we have used enzyme kinetics, X-ray crystallography, and small-angle X-ray scattering (SAXS) methodologies to demonstrate that adenine/guanine dinucleoside polyphosphates bind to the Bateman domain of IMPDH from the fungus Ashbya gossypii with submicromolar affinities. We found that these dinucleoside polyphosphates modulate the catalytic activity of IMPDHs in vitro by efficiently competing with the adenine/guanine mononucleotides for the allosteric sites. These results suggest that dinucleoside polyphosphates play important physiological roles in the allosteric regulation of IMPDHs by adding an additional mechanism for fine-tuning the activities of these enzymes. We propose that these findings may have important implications for the design of therapeutic strategies to inhibit IMPDHs.
Keywords: allosteric regulation, X-ray crystallography, small-angle X-ray scattering (SAXS), conformational change, structural biology, Bateman domain, conformational switch, dinucleoside polyphosphates, IMP dehydrogenase, molecular sensor
Introduction
Dinucleoside polyphosphates are ubiquitous molecules in which two nucleosides are linked by a chain of two to seven phosphate moieties. Dinucleoside polyphosphates naturally found in biological systems are those formed by two adenosines (ApnA), two guanines (GpnG), or the combination of adenine and guanine in the same molecule (ApnG), most commonly linked by three to five phosphates (pn = 3–5).
These compounds were originally discovered in the late sixties (1, 2), but the first indication of their putative physiological role came a decade later when a correlation between the concentration of Ap4A and the proliferative status of mammalian cells was demonstrated (3). At present, dinucleoside polyphosphates have been described to participate in an increasing variety of cellular processes, including DNA replication and repair (4), cell division (5), neurotransmission (6), apoptosis (7), analgesia (8), vasoconstriction (9), and platelet aggregation (10) among others. Accordingly, dinucleoside polyphosphates have been described to interact with several target proteins, including adenylate kinase (11), purinergic receptors (12), heat shock proteins (13), 5′-nucleotidase II (14), and poly(A) polymerase (4) among others. Interestingly, diadenosine polyphosphates have also been recently reported to bind to the Bateman domains of some bacterial pyrophosphatases (15) as well as to bacterial IMP dehydrogenases (16, 17), which raises the interesting question of whether Bateman domains are physiological targets of dinucleoside polyphosphates in vivo.
Bateman domains are regulatory protein modules composed of pairs of cystathionine β-synthase motifs, which are present in organisms belonging to all kingdoms of life. They either exist as isolated proteins or associated to a variety of enzymes where they allosterically modulate their functions in response to the binding of different molecules. Thereby, Bateman domains sense the cellular energy status, metal ion concentration, or ionic strength and regulate protein function accordingly (18). The physiological relevance of Bateman domains is stressed by the fact that missense mutations within them have been associated to a variety of human hereditary diseases that include the Wolff–Parkinson–White syndrome, congenital myotonia, homocystinuria, and certain retinopathies such as Leber congenital amaurosis and retinitis pigmentosa (19).
Among the proteins that contain Bateman domains, IMP dehydrogenase (IMPDH)4 is the enzyme that catalyzes the rate-limiting step in the de novo guanine nucleotide biosynthetic pathway and, thereby, is involved in the control of cell division and proliferation. IMPDH is composed of an archetypal triose-phosphate isomerase (TIM) barrel catalytic domain that contains a Bateman regulatory module inserted within a loop. The basic units of IMPDH in solution are homotetramers that can dimerize in different ways to form octamers, linear oligomers, and mesoscale polymers, denoted as cytoophidia (20). Despite the fact that IMPDH is a widely studied therapeutic target, it has been only since the last few years that we are beginning to understand its molecular mechanisms of allosteric regulation.
The Bateman domain of eukaryotic IMPDHs has three nucleotide-binding sites that operate coordinately to allosterically modulate the catalytic activity. Canonical sites 1 and 2 are present in all IMPDH Bateman domains and can be occupied by either adenine (ATP, ADP, and AMP) or guanine (GTP and GDP) nucleotides. In contrast, the noncanonical site 3 is exclusive of eukaryotic IMPDHs and can only be occupied by guanine (GTP and GDP) nucleotides (Fig. 1). Adenine nucleotide binding to sites 1 and 2 induces catalytically active, extended octamers, whereas the coordinated binding of GTP/GDP to sites 2 and 3 induces the compaction of these octamers into an inhibited conformation (Fig. 1).
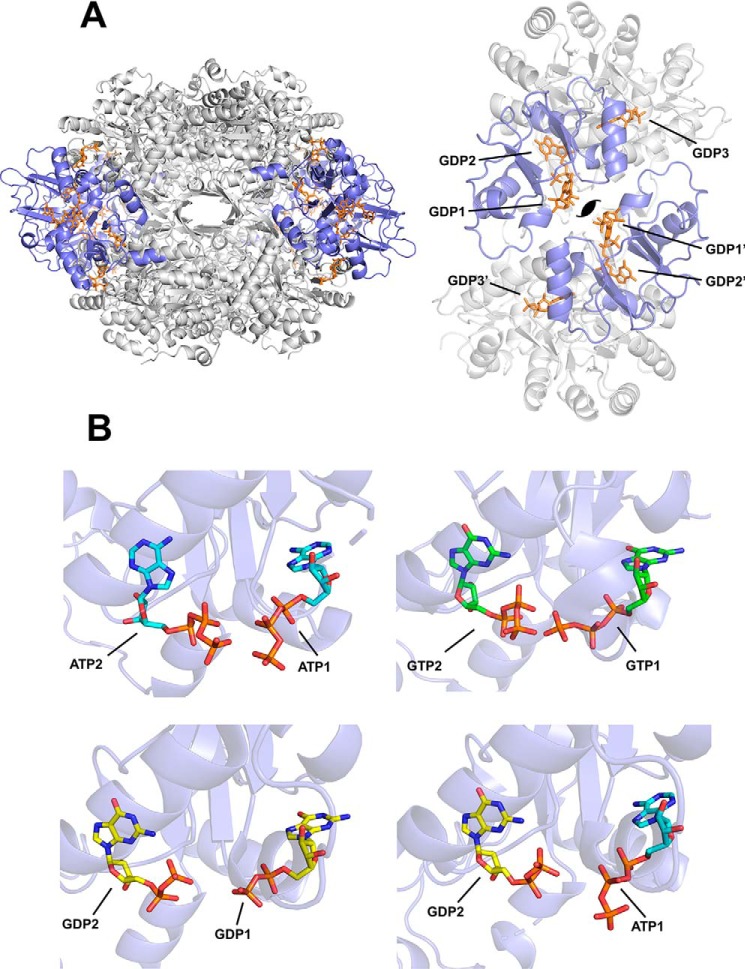
Figure 1.
Nucleotide binding to the Bateman domain of IMDPH. A, cartoon representation of the structure of AgIMPDH octamers formed in the presence of GDP (orange sticks). The catalytic domain is shown in light gray, and the Bateman regulatory domain is colored in blue. The right panel corresponds to a close-up view of two interacting Bateman domains within the octamer, showing the disposition of the three bound GDP molecules. The approximate position of the symmetry axis is indicated. B, close-up views of the adenine and guanine nucleotides (shown in sticks) bound to the two canonical sites of AgIMPDH (represented in semitransparent blue cartoons). Upper left panel, AgIMPDH–ATP (PDB code 5MCP). Upper right panel, HsIMPDH–GTP (PDB code 6I0O). Lower left panel, AgIMPDH–GDP (PDB code 4Z87). Lower right panel, AgIMPDH–ATP/GDP (PDB code 5TC3). In all cases, it can be observed that the β- and γ-phosphates of the nucleotides bound to the canonical sites face each other. The canonical binding sites (1 and 2) are indicated after the corresponding nucleotides; i.e. ATP1 means ATP bound to the canonical site 1.
Strong cooperativity between allosteric sites 2 and 3 must exist because the incubation of IMPDH with a mixture of ATP and GDP resulted in a complex with ATP bound to site 1 and GDP bound to sites 2 and 3. GDP binding to site 3 is obvious because this site is exclusive for GTP/GDP, but GDP binding to site 2 must be favored by the occupancy of site 3 given that GDP has 20-fold less apparent affinity than ATP (21). These data imply that sites 2 and 3 are linked.
Moreover, given that site 3 is exclusive for guanine nucleotides and the binding of these to site 2 is tightly linked to the occupancy of site 3, ATP cannot efficiently displace GTP/GDP from sites 2 and 3, and thereby the nucleotide-controlled conformational switch of IMPDH has been reported to be unidirectional. That is, GTP/GDP are able to inhibit the active octamers induced by ATP, but in contrast, ATP cannot activate the inhibited octamers induced by GTP/GDP (20, 21). Altogether, the occupancy of the second canonical and the third noncanonical nucleotide-binding sites of the Bateman domain determines the conformation of IMPDH octamers and, therefore, their catalytic activity.
In this work, we demonstrate that adenine/guanine dinucleoside polyphosphates bind with submicromolar affinities to the Bateman domains and modulate the catalytic activity of IMPDHs in vitro by efficiently competing with the adenine/guanine mononucleotides for the canonical sites. We further report the crystallographic structure of the inhibited ternary complex of IMPDH bound to Ap5G and GDP that indicates that dinucleoside polyphosphates occupy both canonical sites simultaneously. Altogether, our results strongly suggest that dinucleoside polyphosphates might play important physiological roles by adding an extra level for fine-tuning the allosteric regulation of IMPDHs. Furthermore, the reported data might have important implications for the design of novel therapeutic strategies to inhibit IMPDHs.
Results and discussion
Adenine/guanine dinucleoside polyphosphates efficiently bind to the Bateman domain of IMPDH
An analysis of the published structures of the human (HsIMPDH) and Ashbya gossypii IMPDH (AgIMPDH) enzymes bound to GTP (20), ATP, ATP/GDP (21), and GDP (22) showed that the β- and γ-phosphates of the mononucleotides bound at canonical sites 1 and 2 are facing each other (Fig. 1B). Thus, we speculated that adenine/guanine dinucleoside polyphosphates could bind simultaneously to canonical sites 1 and 2 and, thereby, could efficiently compete with adenine and guanine mononucleotides. Following this hypothesis, we tested the effects of several commercially available dinucleoside polyphosphates on the catalytic activity of AgIMPDH.
First, we assayed Ap4A, Ap5A, and Ap6A and compared them side-by-side with ATP. All these nucleotides bind to AgIMPDH and slightly activate its catalytic activity. Ap6A and Ap5A bind to AgIMPDH with high affinity with an approximate K1/2 value of 0.3 μm, which is about 2 orders of magnitude higher than that of ATP (Fig. 2A).
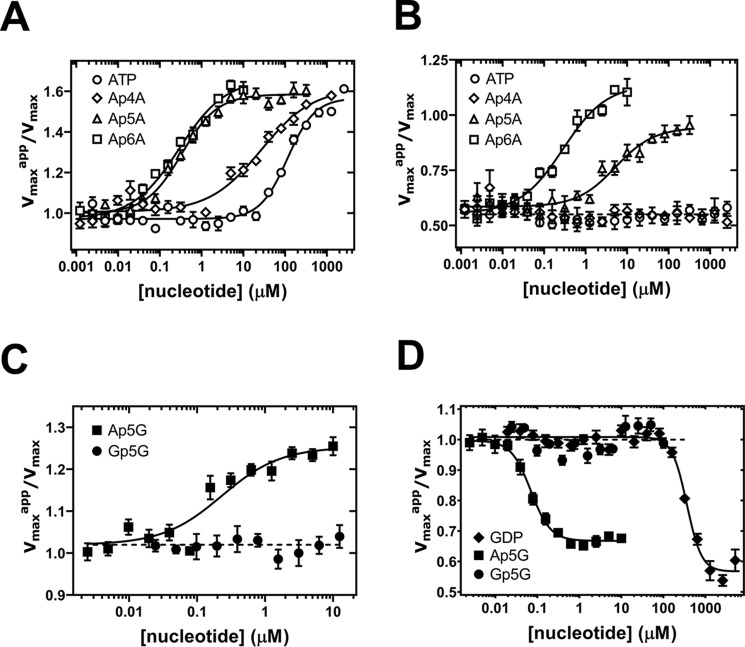
Figure 2.
Effects of dinucleoside polyphosphates on the catalytic activity of AgIMPDH in vitro. Graphs show the normalized Vmax values (Vmaxapp in the presence of nucleotide divided by the Vmax in the absence of nucleotide) versus nucleotide concentration. A and C show the effects of the indicated dinucleoside polyphosphates alone on the AgIMPDH enzyme, whereas B and D show their effects on the enzyme in the presence of GDP (B, inhibited enzyme at 3 mm GDP (21); D, subsaturating GDP concentration of 0.3 mm, which cannot inhibit the enzyme significantly (21)). Experimentally determined Vmax values were fitted to a dose-response function (four parameters, variable slope) as implemented in GraphPad Prism software (continuous lines). Error bars represent S.E.
Remarkably, both Ap5A and Ap6A are able to revert the strong allosteric inhibition of AgIMPDH mediated by millimolar amounts of GDP (3 mm) in contrast to Ap4A and ATP (Fig. 2B), which cannot activate the GDP-inhibited enzyme even when Ap4A and ATP bind to AgIMPDH 2 orders of magnitude more efficiently than GDP (21). This observation is indeed interesting because it implies that submicromolar concentrations of Ap5A and Ap6A could activate the enzyme even at high concentrations of GTP and GDP. We speculate that this might constitute a mechanism that can be used by the cell to maintain higher steady-state levels of guanine nucleotides that might be required under certain circumstances, such as high cell proliferation rates (23).
The high binding affinity of Ap5A and Ap6A to AgIMPDH suggests that these are occupying both canonical sites simultaneously, in contrast to Ap4A that seems to bind as a surrogate of ATP, that is, one molecule of Ap4A per canonical site. These results agree with those previously reported stating that Ap4A binds the Escherichia coli and Pseudomonas aeruginosa IMPDH enzymes (EcIMPDH and PaIMPDH) with only 5-fold higher affinity than ATP, and thereby this interaction is physiologically irrelevant (16). In contrast, our results suggest that Ap5A and Ap6A might bind with much higher affinity than ATP (and Ap4A) to these bacterial enzymes. We corroborated this hypothesis by monitoring the ATP-induced activation of PaIMPDH and observed a potent activation mediated by Ap6A and partially Ap5A but not Ap4A and ATP that needed much higher concentrations to activate the enzyme (Fig. S1). Altogether, these data demonstrate that adenine dinucleotides are in vitro common ligands of the Bateman domains of bacterial and eukaryotic IMPDHs.
We next assayed the effects of Ap5G and Gp5G on the enzymatic activity of AgIMPDH in vitro and observed a slight activator effect of Ap5G and no effect at all for Gp5G (Fig. 2C). However, submicromolar amounts of Ap5G were able to significantly inhibit the enzyme in combination with a subsaturating concentration of GDP (300 μm), which lies within the expected range of nucleotide intracellular concentrations (24); Gp5G, in contrast, showed no effect at all (Fig. 2D). Thereby, our data show that submicromolar amounts of Ap5G, but not Gp5G, are able to hypersensitize AgIMPDH to GTP/GDP-mediated allosteric inhibition.
These observations suggest that the guanine moieties of Gp5G cannot bind simultaneously to the two canonical sites of the Bateman domain of AgIMPDH, opposite to Ap5G that occupies both canonical sites simultaneously with submicromolar affinity. Nonetheless, Ap5G alone has no effect on the catalytic activity because the noncanonical binding site 3 needs to be occupied by GTP/GDP to inhibit the enzyme; that is, all three allosteric sites must be occupied to inhibit efficiently the enzyme (22).
Structure of the ternary complex AgIMPDH–Ap5G–GDP
The biochemical data reported in the previous section suggest that Ap5A, Ap6A, and Ap5G bind simultaneously to the two canonical nucleotide-binding sites of the Bateman domain. To corroborate this hypothesis, we aimed to determine the crystal structure of IMPDH bound to a dinucleoside polyphosphate and succeeded at solving the ternary complex AgIMPDH–Ap5G–GDP at a maximum resolution of 2.1 Å despite the strong anisotropy of the experimental data (details are given under “Experimental procedures” and in Table 1). The crystal belongs to space group I 4 2 2 and contains only one IMPDH monomer in the asymmetric unit, but the octameric species, expected from the small-angle X-ray scattering (SAXS) experiments, can be reconstructed from the crystallographic contacts (Fig. S2A). The overall protein structure is essentially identical to that of AgIMPDH–GDP (PDB code 4Z87; all-atom r.m.s.d., 1.08 Å) and AgIMPDH–ATP/GDP (PDB code 5TC3; all-atom r.m.s.d., 0.78 Å); both of them compacted and inhibited octamers (21) as shown in Fig. S2B. The electron density for Ap5G and GDP is clear and allows modeling these nucleotides unequivocally.
Table 1.
X-ray crystallography data collection and refinement statistics
Statistics for the highest-resolution shell are shown in parentheses. Diffraction intensities were indexed and integrated by using the autoPROC toolbox (28), which makes use of the STARANISO software to deal with data anisotropy (29). CC, Pearson correlation coefficient.
| AgIMPDH–Ap5G–GDP | |
|---|---|
| Crystal parameters | |
| PDB code | 6RPU |
| Resolution range (Å) | 85.10–2.11 (2.26–2.11) |
| Ellipsoidal resolution limits (Å) | 2.10, 2.10, 3.13a |
| Resolution I/Iσ > 2.0 (overall) | 2.43 |
| Resolution I/Iσ > 2.0 (along h) | 2.26 |
| Resolution I/Iσ > 2.0 (along k) | 2.26 |
| Resolution I/Iσ > 2.0 (along l) | 3.30 |
| Resolution CC1/2 > 0.3 (overall) | 2.16 |
| Resolution CC1/2 > 0.3 (along h) | 2.08 |
| Resolution CC1/2 > 0.3 (along k) | 2.08 |
| Resolution CC1/2 > 0.3 (along l) | 3.05 |
| Space group | I 4 2 2 |
| Unit cell a, b, c/α, β, γ(Å/°) | 148.54, 148.54, 103.83/90, 90, 90 |
| Unique reflections | 22,408 (1,106) |
| Multiplicity | 23.3 (23.4) |
| Completeness spherical (%) | 66.4 (17.8) |
| Completeness ellipsoidal (%) | 95.8 (68.4) |
| Mean I/σ(I) | 21.5 (1.7) |
| Rmerge | 0.09 (2.41) |
| Rmeas | 0.10 (2.46) |
| Rpim | 0.02 (0.51) |
| CC1/2 | 0.999 (0.704) |
| Refinement statistics | |
| Rwork | 0.27 (0.40) |
| Rfree | 0.29 (0.31) |
| Number of non-hydrogen atoms | 3,325 |
| Macromolecules | 3,171 |
| Ligands | 102 |
| Solvent | 52 |
| Protein residues | 432 |
| r.m.s.bonds | 0.013 |
| r.m.s.angles | 1.78 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 3.34 |
| Clashscore | 29.79 |
| Average B-factor | 61.69 |
| Macromolecules | 61.97 |
| Ligands | 65.05 |
| Solvent | 38.22 |
a Experimental data showed strong anisotropy with ellipsoidal resolution limits defined at 2.10 (a), 2.10 (b), and 3.13 (c) Å by STARANISO using as threshold I/σI ≥ 1.2.
Remarkably, Ap5G binds simultaneously to the two canonical sites, with the adenine and guanine moieties bound to sites 1 and 2, respectively (Figs. 3A and S3) and the polyphosphate chain buried in a highly charged positive groove (Fig. S3B). The binding modes for the adenosine and guanosine moieties of Ap5G are identical to those observed for ATP and GDP in the ATP/GDP complex structure (Fig. 3B), as it happens with the GDP molecule that occupies the noncanonical site 3 (21).
Figure 3.
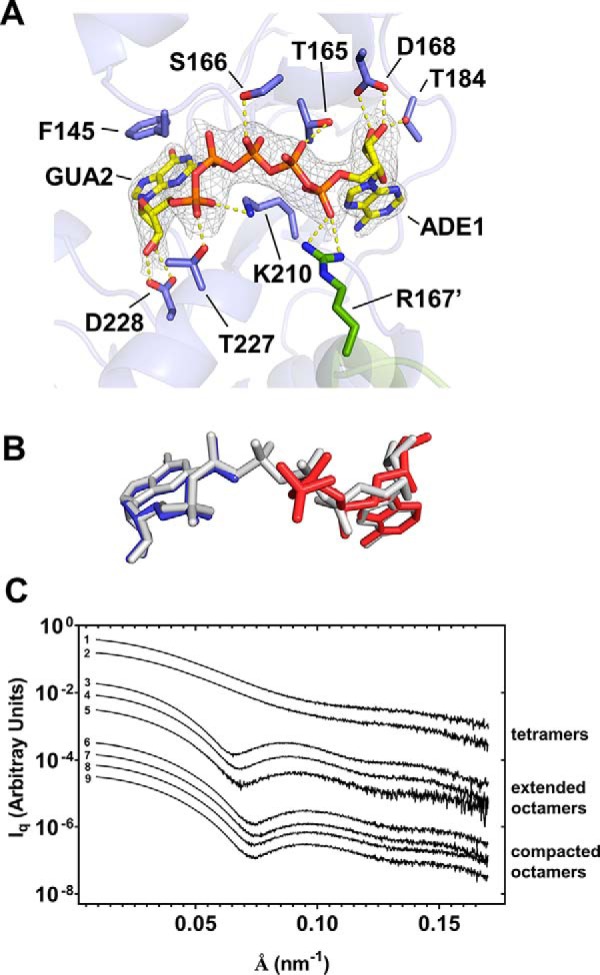
Structure of AgIMPDH bound to dinucleoside polyphosphates. A, crystal structure of the ternary complex AgIMPDH–Ap5G–GDP. Shown is a close-up view of Ap5G (shown in sticks) simultaneously bound to the two canonical sites of the Bateman domain. The protein is represented in semitransparent blue cartoons with the side chain of key interacting residues shown in sticks. The adjacent monomer and the side chain of residue Arg-167′ are shown in green cartoon and sticks, respectively. The gray mesh around Ap5G represents the 2mFo − DFc electron density map contoured at the 1.3σ level (a stereoview of the final Fo − Fc omit map is shown in Fig. S2A). Key protein–nucleotide atomic interactions are represented as yellow dashed lines. B, ATP and GDP mononucleotide structures bound to AgIMPDH (PDB code 5TC3 (21)) compared with the bound structure of Ap5G (PDB code 6RPU, this work). The structural alignment was generated by superimposing the backbone atoms of the Bateman domains (residues 130–210) of both structures. C, SAXS profiles of AgIMPDH in the presence of different mononucleotides and dinucleoside polyphosphates. To facilitate visualization, the plots have been conveniently displaced along the y axis and grouped according to three different conformations: tetramers, extended, and compacted octamers. The profiles shown correspond to: (i) tetramers: 1, control; 2, 0.3 mm GDP; (ii) extended octamers: 3, 3 mm ATP; 4, 0.1 mm Ap6A; 5, 0.1 mm Ap6A + 3 mm GDP; (iii) compacted octamers: 6, 3 mm GDP; 7, 0.1 mm Ap5G + 0.3 mm GDP; 8, 0.1 mm Ap4A + 3 mm GDP; 9, 3 mm ATP + 3 mm GDP.
The structure demonstrates that purine dinucleoside polyphosphates can bind simultaneously the two canonical binding sites in the Bateman domain, which explains why their affinities are 2 orders of magnitude higher than the corresponding adenine and guanine mononucleotides. The large increase in affinity presumably comes from the simultaneous reduction of the entropic penalty of binding, due to the molecularity change, and the electrostatic repulsion.
This structure, together with the biochemical data reported above, further corroborates our previous observation that the canonical site 2 and the noncanonical site 3 are tightly linked. This is also evidenced by their spatial proximity and the common residues that both sites share (20, 21). Thereby, the occupancy of site 3 facilitates the binding of guanine mononucleotides to site 2, but the binding of adenine mononucleotides is difficulted (21, 22). Given that Ap5A/Ap6A occupy both canonical sites 1 and 2 with affinities that are 3 orders of magnitude higher than GTP/GDP, they can readily displace GTP/GDP from site 2 and, subsequently, from site 3, reverting the GTP/GDP-mediated inhibition of IMPDH. Moreover, the fact that Ap5G hypersensitizes the IMPDH enzyme to the GDP-mediated allosteric inhibition indicates that the occupancy of site 2 by the guanosine moiety of Ap5G favors the binding of GDP to site 3.
Dinucleoside polyphosphates binding to IMPDH is highly specific in the sense that it requires an appropriate polyphosphate chain length to allow the two purine nucleoside moieties to be accommodated simultaneously into the canonical binding sites. In this respect, the binding of the guanine ring at the canonical site 1 occurs with a shift of about 3 Å with respect to adenine (Fig. S4A), thus imposing a different register to the polyphosphate chain. This observation explains the lack of effect of Gp5G, in contrast to Ap5G, on the catalytic activity of AgIMPDH. We speculate that Gp6G might efficiently bind to AgIMPDH, but this remains to be experimentally demonstrated (Gp6G is not commercially available so far). Additionally, slight variations in the primary sequence among organisms and/or isoforms might completely change the binding specificity as will be discussed below.
Dinucleoside polyphosphates modulate the conformational switch of AgIMPDH
We studied next the effects of the dinucleoside polyphosphates on the quaternary structure and conformation of AgIMPDH in solution by using SAXS. AgIMPDH has been extensively characterized by using this methodology, and it has been reported that AgIMPDH exists as a tetramer in solution in the absence of nucleotides (profile 1), and the binding of adenine (ATP, ADP and AMP) and guanine (GTP and GDP) nucleotides in the Bateman domain induces extended-active (profile 3, ATP) and compacted-inhibited octamers (profile 6, GDP), respectively (Ref. 21 and Fig. 3C).
We then corroborated that Ap6A (profile 4) induces the association of AgIMPDH tetramers to form octamers highly similar to those induced by ATP (profile 3). Moreover, Ap6A (profile 5), but not Ap4A (profile 8) or ATP (profile 9), is able to expand the compacted octamers formed in the presence of millimolar amounts of GDP (Fig. 3C), reverting the GDP-induced inhibition of the catalytic activity. Additionally, as expected, Ap5G (profile 7) in the presence of a subsaturating concentration of GDP (0.3 mm GDP (profile 2); unable to induce a significant proportion of octamers) induces compacted, inhibited octamers identical to those formed in the presence of millimolar amounts of GDP (profile 6; Fig. 3C). A more detailed SAXS-data analysis, including the estimated structural parameters for each profile, can be found in Table S1.
Thereby, as for the mononucleotides, the conformations of AgIMPDH induced by the binding of dinucleoside polyphosphates correlate with their catalytic activities. Ap6A induces the formation of extended octamers that are catalytically active, whereas Ap5G in combination with subsaturating concentrations of GTP/GDP induces the formation of compacted octamers that show a significant reduction of the catalytic activity. Moreover, Ap6A is able to revert the GDP-mediated inhibition of the IMPDH catalytic activity by expanding the GDP-induced compacted octamers.
Dinucleoside polyphosphates modulate the activity of the human IMPDH isoforms
Several lines of experimental evidence indicating that the mechanism for allosteric regulation, initially described for AgIMPDH (22), also operates in other eukaryotic IMPDHs, including the two human isoforms, have been reported (20, 23, 25). We therefore tested next whether dinucleoside polyphosphates might also modulate the catalytic activity of human IMPDHs in vitro.
We first assayed the effects of diadenosine polyphosphates on HsIMPDH1 and observed no significant effect on its catalytic activity (Fig. 4A). However, Ap5A was able to efficiently revert the GDP-induced inhibition of HsIMPDH1, in contrast to Ap6A that showed no effect (Fig. 4A). These results are in contrast to those found for AgIMPDH where Ap6A efficiently activates the GDP-inhibited enzyme (Fig. 2B). These data are indeed unexpected given the high similarity between the structures of both organisms (20, 22). Nonetheless, the comparison of the human and fungal structures of IMPDH bound to GDP showed a displacement of about 2 Å in the guanine ring bound to the canonical site 1, which might thus impose a different optimal polyphosphate chain length (Fig. S4B). These data indicate the existence of sequence determinants that dictate the binding specificity of different dinucleoside polyphosphates. These sequence determinants might serve to specifically adapt the regulation of IMPDH to the metabolic requirements of each organism as anticipated above.
Figure 4.
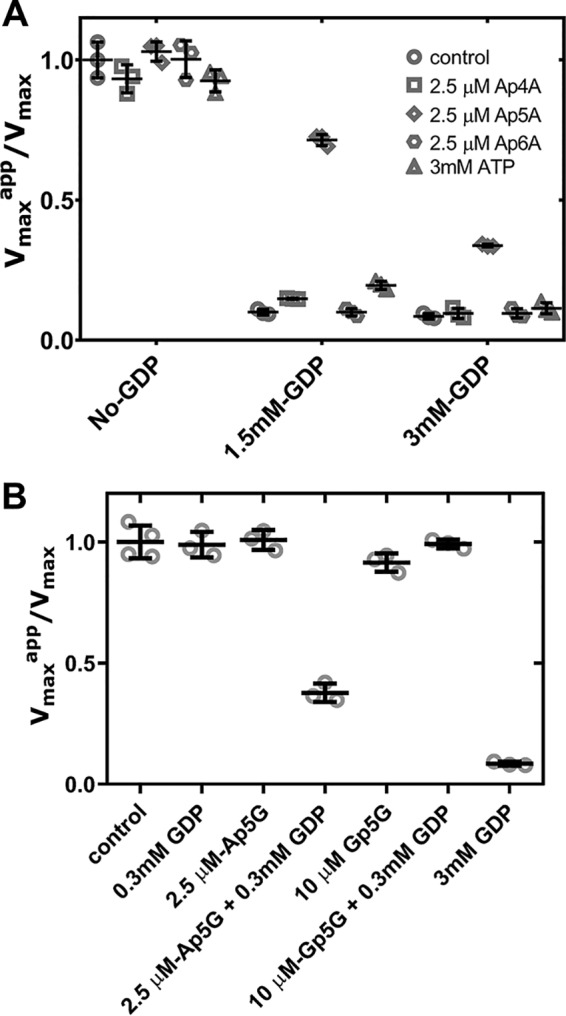
Effects of dinucleoside polyphosphates on the catalytic activity of HsIMPDH1 in vitro. Scatter plots show the normalized Vmax values (Vmaxapp in the presence of nucleotide divided by the Vmax in the absence of nucleotide) of several adenine dinucleoside polyphosphates compared with ATP (A) as well as Ap5G and Gp5G alone and in combination with GDP at a subsaturating concentration of 0.3 mm (B). Vmax values are derived from the Michaelis–Menten analysis of the experimental data. The empty symbols (light gray) are the values obtained from independent experiments, and the error bars (black) represent S.E.
We next assayed the effects of Ap5G and Gp5G on the enzymatic activity of HsIMPDH1 in vitro and observed no significant effect for both nucleotides alone (Fig. 4B). However, as observed for AgIMPDH, Ap5G was able to inhibit the enzyme in combination with a subsaturating concentration of GDP; Gp5G, in contrast, showed no effect at all (Fig. 4B). Thereby, these data further corroborate the results obtained with the fungal protein, showing that Ap5G hypersensitizes the IMPDH enzyme to the GTP/GDP-mediated allosteric inhibition.
Altogether, the data reported here show that submicromolar amounts of dinucleoside polyphosphates modulate the conformational switch and the catalytic activity of IMPDHs in vitro. These concentrations are in the same range as the reported intracellular concentrations (26), which indicates that dinucleoside polyphosphates might have an important role in the physiological regulation of IMPDH. These results significantly contribute to the understanding of the regulation of IMPDH and yield relevant information for potential new pharmacological approaches to target IMPDH that definitively deserve further investigation.
Experimental procedures
Proteins and nucleotides
Expression and purification of AgIMPDH and HsIMPDH proteins were performed as described previously (20, 22). PaIMPDH was overexpressed in E. coli (BL21) cells fused to an N-terminal His8 tag and purified following standard immobilized metal affinity chromatography (HisTrap FF Crude column) and size-exclusion chromatography (HiPrep 16/60 Sephacryl S-300 HR column). All proteins were stored frozen at −80 °C in buffer (20 mm Tris-HCl, 500 mm KCl, 1 mm DTT, pH 8.0). Mononucleotides and dinucleoside polyphosphates were purchased from Sigma-Aldrich and Jena Bioscience.
Enzyme kinetics experiments
IMPDH activity was assayed at 28 (AgIMPDH) or 32 °C (HsIMPDH and PaIMPDH) using 384-well microtiter plates by monitoring the reduction of NAD+ to NADH and the subsequent increase in absorbance at 340 nm. IMPDH enzymes (15–55 μg/ml) in reaction buffer (100 mm Tris-HCl, 100 mm KCl, 2 mm DTT, 1 mm free MgCl2, pH 8.0) were assayed using 0.5 mm NAD+ and 0.039–5 mm IMP as substrates in the presence of different mononucleotide and dinucleoside polyphosphate concentrations. Experimental data were fitted by nonlinear regression to Michaelis–Menten and dose-response (four parameters, variable slope) equations to estimate the Km or K1/2 and Vmax values using the software GraphPad Prism.
Crystallization and structure determination
Crystals of the ternary complex AgIMPDH–Ap5G–GDP were grown at 22 °C using the vapor-diffusion method by mixing a protein solution at 10 mg/ml in 10 mm Tris-HCl, 50 mm KCl, 1 mm DTT, 0.5 mm Ap5G, 3 mm GDP, 5 mm total MgCl2, pH 8.0, with an equal volume of mother liquor consisting of 20% PEG-6000, 0.1 m sodium acetate, pH 5.0, 0.2 m sodium chloride. Protein crystals were cryoprotected by immersion in NVH oil before being flash cooled in liquid nitrogen. Diffraction data were collected at 100 K using monochromatic X-rays of ≈1-Å wavelength at the XALOC beamline (27) in the ALBA Synchrotron (Spain).
Diffraction intensities were indexed and integrated with the autoPROC toolbox (28), which makes use of STARANISO software (29) to deal with data anisotropy. Indeed, the experimental data showed strong anisotropy with ellipsoidal resolution limits defined by STARANISO at 2.10 (a), 2.10 (b), and 3.13 (c) Å using as threshold I/σI ≥ 1.2. The anisotropically truncated data were phased by molecular replacement using the program Phaser (30) and the structure of the complex AgIMPDH–ATP–GDP (PDB code 5TC3) as template. The initial structure obtained by Phaser was iteratively refined by alternating manual modeling with Coot (31) and automated refinement with the PHENIX crystallographic software suite (32) using rigid-body, gradient-driven positional, restrained individual isotropic B-factor and TLS (33). The R and Rfree factors of the final model are 0.27 and 0.29, respectively, which seem relatively high given the maximum resolution of the data. However, it must be taken into account that the data are markedly anisotropic and the R factors in anisotropic refinement tend to be high, mostly due to the inclusion of numerous poorly measured reflections in anisotropic data sets.
Small angle X-ray scattering
SAXS measurements were performed at the B21 beamline in the Diamond Light Source. Buffer (20 mm Tris-HCl, pH 8.0, 150 mm KCl, 3 mm DTT, 2 mm free MgCl2) and proteins at a concentration of 2.5 mg/ml were measured by flowing the solutions through a quartz capillary at 10 °C to minimize radiation damage. The beam energy was set to 12.4 keV, and the distance from the sample to the detector (Eiger 4M) was fixed at 4.04 m, which allows measurement of a scattering vector (q) range from 0.0032 to 0.38 Å−1. Data reduction and analysis were performed by standard methodologies using the PRIMUS program (34) within the ATSAS software package (35).
Author contributions
D. F.-J., R. P., and R. M. B. formal analysis; D. F.-J. and R. M. B. investigation; D. F.-J. and R. M. B. methodology; R. P., J. L. R., and R. M. B. writing-original draft; J. L. R. and R. M. B. conceptualization; J. L. R. and R. M. B. validation; J. L. R. and R. M. B. project administration; J. L. R. and R. M. B. writing-review and editing; R. M. B. funding acquisition; R. M. B. visualization.
Supplementary Material
Acknowledgments
We thank María Dolores Sánchez, Silvia Domínguez, and Marta Santos for excellent technical help. Protein crystallography experiments were performed at the XALOC beamline at ALBA Synchrotron with the collaboration of the ALBA staff. We also acknowledge Diamond Light Source for time on Beamline B21, where the SAXS experiments were performed, under Proposal Infrastructure for NMR, EM and X-rays for Translational Research (iNEXT) number 8311.
This work was supported by Spanish Ministerio de Ciencia, Innovación y Universidades Grants BFU2016-79237-P (to R. M. B.) and BIO2017-88435-R (to J. L. R.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
The atomic coordinates and structure factors (code 6RPU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- IMPDH
- IMP dehydrogenase
- SAXS
- small-angle X-ray scattering
- PDB
- Protein Data Bank
- Hs
- Homo sapiens
- Ag
- A. gossypii
- Ec
- E. coli
- Pa
- P. aeruginosa
- r.m.s.d.
- root mean square deviation.
References
- 1. Finamore F. J., and Warner A. H. (1963) The occurrence of P1,P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J. Biol. Chem. 238, 344–348 [PubMed] [Google Scholar]
- 2. Zamecnik P. C., Stephenson M. L., Janeway C. M., and Randerath K. (1966) Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 91–97 10.1016/0006-291X(66)90415-3 [DOI] [PubMed] [Google Scholar]
- 3. Rapaport E., and Zamecnik P. C. (1976) Presence of diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) in mammalian cells in levels varying widely with proliferative activity of the tissue: a possible positive “pleiotypic activator”. Proc. Natl. Acad. Sci. U.S.A. 73, 3984–3988 10.1073/pnas.73.11.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sillero M. A., De Diego A., Osorio H., and Sillero A. (2002) Dinucleoside polyphosphates stimulate the primer independent synthesis of poly(A) catalyzed by yeast poly(A) polymerase. Eur. J. Biochem. 269, 5323–5329 10.1046/j.1432-1033.2002.03253.x [DOI] [PubMed] [Google Scholar]
- 5. Nishimura A., Moriya S., Ukai H., Nagai K., Wachi M., and Yamada Y. (1997) Diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 2, 401–413 10.1046/j.1365-2443.1997.1300328.x [DOI] [PubMed] [Google Scholar]
- 6. Gómez-Villafuertes R., Pintor J., Gualix J., and Miras-Portugal M. T. (2004) GABA modulates presynaptic signalling mediated by dinucleotides on rat synaptic terminals. J. Pharmacol. Exp. Ther. 308, 1148–1157 10.1124/jpet.103.061564 [DOI] [PubMed] [Google Scholar]
- 7. Vartanian A. A., Suzuki H., and Poletaev A. I. (2003) The involvement of diadenosine 5′,5‴-P1,P4-tetraphosphate in cell cycle arrest and regulation of apoptosis. Biochem. Pharmacol. 65, 227–235 10.1016/S0006-2952(02)01481-8 [DOI] [PubMed] [Google Scholar]
- 8. Giraldez L., Díaz-Hernandez M., Gomez-Villafuertes R., Pintor J., Castro E., and Miras-Portugal M. T. (2001) Adenosine triphosphate and diadenosine pentaphosphate induce [Ca2+]i increase in rat basal ganglia aminergic terminals. J. Neurosci. Res. 64, 174–182 10.1002/jnr.1063 [DOI] [PubMed] [Google Scholar]
- 9. Conant A. R., Theologou T., Dihmis W. C., and Simpson A. W. (2008) Diadenosine polyphosphates are selective vasoconstrictors in human coronary artery bypass grafts. Vascul. Pharmacol. 48, 157–164 10.1016/j.vph.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 10. Lüthje J., Baringer J., and Ogilvie A. (1985) Effects of diadenosine triphosphate (Ap3A) and diadenosine tetraphosphate (Ap4A) on platelet aggregation in unfractionated human blood. Blut 51, 405–413 10.1007/BF00320727 [DOI] [PubMed] [Google Scholar]
- 11. Lienhard G. E., and Secemski I. I. (1973) P1,P5-Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J. Biol. Chem. 248, 1121–1123 [PubMed] [Google Scholar]
- 12. Jankowski V., van der Giet M., Mischak H., Morgan M., Zidek W., and Jankowski J. (2009) Dinucleoside polyphosphates: strong endogenous agonists of the purinergic system. Br. J. Pharmacol. 157, 1142–1153 10.1111/j.1476-5381.2009.00337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnstone D. B., and Farr S. B. (1991) AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10, 3897–3904 10.1002/j.1460-2075.1991.tb04959.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marques A. F., Teixeira N. A., Gambaretto C., Sillero A., and Sillero M. A. (1998) IMP-GMP 5′-nucleotidase from rat brain: activation by polyphosphates. J. Neurochem. 71, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 15. Anashkin V. A., Salminen A., Tuominen H. K., Orlov V. N., Lahti R., and Baykov A. A. (2015) Cystathionine β-synthase (CBS) domain-containing pyrophosphatase as a target for diadenosine polyphosphates in bacteria. J. Biol. Chem. 290, 27594–27603 10.1074/jbc.M115.680272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Despotović D., Brandis A., Savidor A., Levin Y., Fumagalli L., and Tawfik D. S. (2017) Diadenosine tetraphosphate (Ap4A)—an E. coli alarmone or a damage metabolite? FEBS J. 284, 2194–2215 10.1111/febs.14113 [DOI] [PubMed] [Google Scholar]
- 17. Guo W., Azhar M. A., Xu Y., Wright M., Kamal A., and Miller A. D. (2011) Isolation and identification of diadenosine 5′,5‴-P1,P4-tetraphosphate binding proteins using magnetic bio-panning. Bioorg. Med. Chem. Lett. 21, 7175–7179 10.1016/j.bmcl.2011.09.070 [DOI] [PubMed] [Google Scholar]
- 18. Ereño-Orbea J., Oyenarte I., and Martínez-Cruz L. A. (2013) CBS domains: ligand binding sites and conformational variability. Arch. Biochem. Biophys. 540, 70–81 10.1016/j.abb.2013.10.008 [DOI] [PubMed] [Google Scholar]
- 19. Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., and Hardie D. G. (2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 113, 274–284 10.1172/JCI19874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández-Justel D., Nuñez R., Martín-Benito J., Jimeno D., González-López A., Soriano E. M., Revuelta J. L., and Buey R. M. (2019) A nucleotide-dependent conformational switch controls the polymerization of human IMP dehydrogenases to modulate their catalytic activity. J. Mol. Biol. 431, 956–969 10.1016/j.jmb.2019.01.020 [DOI] [PubMed] [Google Scholar]
- 21. Buey R. M., Fernández-Justel D., Marcos-Alcalde Í., Winter G., Gómez-Puertas P., de Pereda J. M., and Luis Revuelta J. (2017) A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 7, 2648 10.1038/s41598-017-02805-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buey R. M., Ledesma-Amaro R., Velázquez-Campoy A., Balsera M., Chagoyen M., de Pereda J. M., and Revuelta J. L. (2015) Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 6, 8923 10.1038/ncomms9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duong-Ly K. C., Kuo Y. M., Johnson M. C., Cote J. M., Kollman J. M., Soboloff J., Rall G. F., Andrews A. J., and Peterson J. R. (2018) T cell activation triggers reversible inosine-5′-monophosphate dehydrogenase assembly. J. Cell Sci. 131, jcs223289 10.1242/jcs.223289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]
- 25. Anthony S. A., Burrell A. L., Johnson M. C., Duong-Ly K. C., Kuo Y. M., Simonet J. C., Michener P., Andrews A., Kollman J. M., and Peterson J. R. (2017) Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell 28, 2600–2608 10.1091/mbc.E17-04-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garrison P. N., and Barnes L. D. (1992) Determination of dinucleoside polyphosphates, in Ap4A and Other Dinucleoside Polyphosphates (McLennan A. G., ed) pp. 29–61, CRC Press Inc., Boca Raton, FL [Google Scholar]
- 27. Juanhuix J., Gil-Ortiz F., Cuní G., Colldelram C., Nicolás J., Lidón J., Boter E., Ruget C., Ferrer S., and Benach J. (2014) Developments in optics and performance at BL13-XALOC, the macromolecular crystallography beamline at the ALBA synchrotron. J. Synchrotron Radiat. 21, 679–689 10.1107/S160057751400825X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tickle I. J., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., and Bricogne G. (2018) STARANISO, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winn M. D., Isupov M. N., and Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 10.1107/S0907444900014736 [DOI] [PubMed] [Google Scholar]
- 34. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and D.I., S. (2003) PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 10.1107/S0021889803012779 [DOI] [Google Scholar]
- 35. Franke D., Petoukhov M. V., Konarev P. V., Panjkovich A., Tuukkanen A., Mertens H. D. T., Kikhney A. G., Hajizadeh N. R., Franklin J. M., Jeffries C. M., and Svergun D. I. (2017) ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 10.1107/S1600576717007786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.