Abstract
Human tankyrase-1 (TNKS) is a member of the poly(ADP-ribose) polymerase (PARP) superfamily of proteins that posttranslationally modify themselves and target proteins with ADP-ribose (termed PARylation). The TNKS ankyrin repeat domain mediates interactions with a growing number of structurally and functionally diverse binding partners, linking TNKS activity to multiple critical cell processes, including Wnt signaling, Golgi trafficking, and telomere maintenance. However, some binding partners can engage TNKS without being modified, suggesting that separate parameters influence TNKS interaction and PARylation. Here, we present an analysis of the sequence and structural features governing TNKS interactions with two model binding partners: the PARylated partner telomeric repeat-binding factor 1 (TRF1) and the non-PARylated partner GDP-mannose 4,6-dehydratase (GMD). Using a combination of TNKS-binding assays, PARP activity assays, and analytical ultracentrifugation sedimentation analysis, we found that both the specific sequence of a given TNKS-binding peptide motif and the quaternary structure of individual binding partners play important roles in TNKS interactions. We demonstrate that GMD forms stable 1:1 complexes with the TNKS ankyrin repeat domain; yet, consistent with results from previous studies, we were unable to detect GMD modification. We also report in vitro evidence that TNKS primarily directs PAR modification to glutamate/aspartate residues. Our results suggest that TNKS-binding partners possess unique sequence and structural features that control binding and PARylation. Ultimately, our findings highlight the binding partner:ankyrin repeat domain interface as a viable target for inhibition of TNKS activity.
Keywords: ADP-ribosylation; protein-protein interaction; post-translational modification (PTM); enzyme mechanism; enzyme structure; GDP-mannose dehydratase, GMD; poly(ADP-ribose) polymerase; tankyrase; telomere repeat factor 1, TRF1
Introduction
Human tankyrase-1 (TNKS)3 is a member of the poly(ADP-ribose) polymerase (PARP) superfamily of proteins. PARPs posttranslationally modify themselves and target proteins with mono- or poly(ADP-ribose) using NAD+ as a substrate (termed MARylation or PARylation, respectively) (1, 2). PARP family members possess conserved catalytic (CAT) domains, yet their unique regulatory domains endow individual PARPs with specific cellular functions (1). The regulatory domains of the founding family member, PARP-1, facilitate interaction with damaged DNA, allowing PARP-1 to perform several functions in DNA damage repair pathways. In contrast, TNKS regulatory domains facilitate interaction with a growing number of structurally and functionally diverse binding partners, linking TNKS to several critical cellular processes, including Wnt signaling, telomere maintenance, Golgi trafficking, and apoptosis (3–7). Although the regulatory mechanism of PARP-1 has been well-studied (8, 9), a gap persists in our understanding of how the TNKS regulatory domains control TNKS activity.
TNKS regulatory domains consist of a histidine-, proline-, and serine-rich (HPS) domain, a sterile α motif (SAM) domain, and an ankyrin repeat domain (Fig. 1A). No function has yet been identified for the HPS, and this domain is not present in the closely related tankyrase-2 (TNKS2), indicating that the HPS does not serve a core function in the overlapping roles of TNKS and TNKS2. TNKS can multimerize through self-assembly of its SAM domain, which can form head-to-tail helical polymers with roughly seven SAM monomers per turn (10–13). Although the mechanisms influencing TNKS self-assembly and disassembly are not yet understood, disruption of the SAM polymer interface reduces TNKS Wnt signaling function (12, 13). The ankyrin repeat domain consists of 25 ankyrin repeats and mediates interactions with TNKS-binding partners. Unlike other ankyrin repeat proteins, the TNKS ankyrin repeat domain is segmented into five ankyrin repeat clusters (ARCs 1–5). Each ARC, with the exception of ARC3, possesses a binding pocket that engages TNKS-binding motifs (TBMs) within target proteins, identified in early studies as the consensus sequence “RXXPDG” (14). More recent structural analysis has defined the arginine at position 1 (R1) and the glycine at position 6 (G6) to be the critical binding determinants of a TBM (15). The five ARCs are expected to adopt similar folds, yet low conservation of peptide-interacting residues within the binding pocket endow each ARC with different peptide-binding capacities. Notably, multiple substitutions within the ARC3-binding pocket relative to the other ARCs ablate peptide interaction (15, 16).
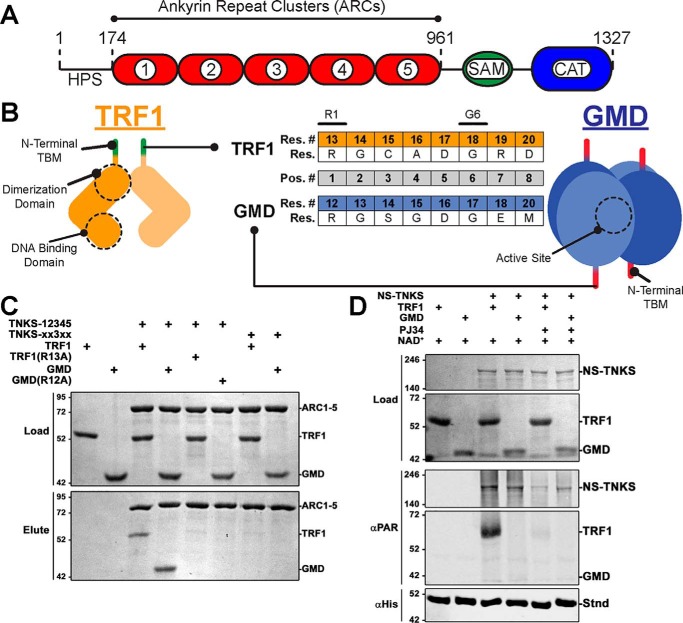
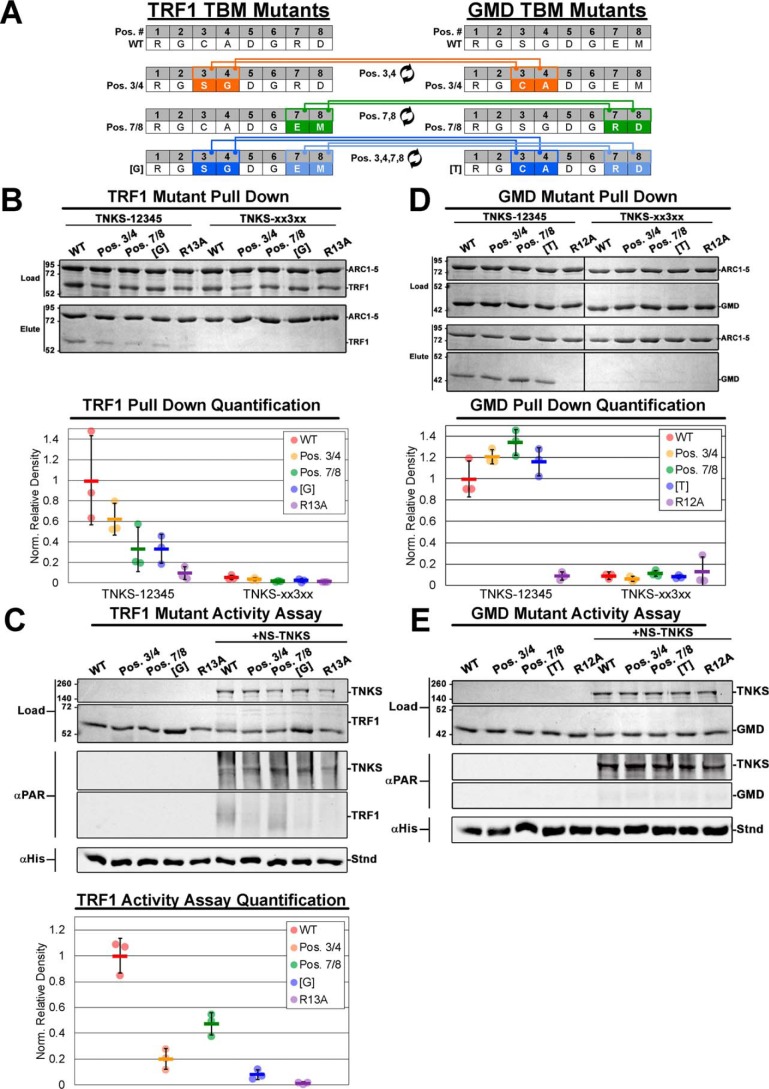
Figure 1.
TRF1 and GMD as example TNKS-binding partners. A, schematic of TNKS domain architecture and regulatory domains. B, cartoon depictions of TRF1 and GMD quaternary structures illustrating TBMs, functional domains, and active sites. The TBM sequences (Res.) and residue numbering (Res. #) of TRF1 and GMD are compared (center). Strictly required residues are noted (R1 and G6), and the standard TBM position numbering (Pos. #) is shown (gray). C, pulldown assay utilizing histidine-tagged ARC1–5 constructs and untagged full-length TRF1 or GMD. TNKS constructs consisted of WT TNKS-12345 and quadruple ARC mutant construct TNKS-xx3xx. Binding partner constructs consisted of WT or TBM mutants R13A and R12A for TRF1 and GMD, respectively. Input proteins (Load) and elution fractions (Elute) were analyzed by SDS-PAGE. D, PARP activity assay utilizing NS-TNKS and full-length TRF1 and GMD. The designated reactions mixtures were analyzed by SDS-PAGE (Load) and by Western blot analysis of PARylation (αPAR). A His-tagged protein (Stnd) was included in each reaction to assess transfer efficiency of the blot; the protein was detected with an anti-His antibody (αHis). NS-TNKS was loaded at loaded at 0.5 μm, TRF1 and GMD were loaded at 1 μm, and PARP inhibitor PJ34 was used at 100 μm.
One of the most perplexing aspects of TNKS regulation is that, although binding the ankyrin repeat domain is required for PARylation, some binding partners are able to interact with TNKS without being modified (5, 17). TNKS thus appears capable of promiscuous binding while simultaneously deciding which binding partners are PARylated. The underlying mechanisms driving TNKS regulation have, however, remained elusive due to the complexity of TNKS:binding partner interactions. This knowledge gap is due largely to the adaptability of TNKS, which tolerates significant sequence and structural variation among its binding partners.
Binding partner variation is evident even at the core TBM:ARC interface. The consensus TBM is only loosely conserved across binding partners, and as many as 8 amino acids can contribute to stabilizing canonical motifs (15). Structural studies have also identified noncanonical TBMs spanning as many as 13 amino acid residues (18, 19), suggesting that the ARCs can engage even longer sequences with large insertions between the critical R1 and G6 contact points. Thus, although some sequence rules have been established, the ARCs appear to accommodate significant variation in TBM sequence and length. Furthermore, binding partners engage TNKS using multiple TBMs, either by possessing multiple consecutive TBMs or by forming homomultimeric structures that thus present multiple copies of the same TBM in a configuration determined by their quaternary structures. We have previously shown that the five ARCs of TNKS dynamically sample multiple conformations to facilitate binding, suggesting that the ARCs can accommodate variation in binding partner structure as well as variations in the TBM sequence (16). The plasticity of the ankyrin repeat domain is undoubtedly indispensable for a master scaffolding protein with multiple binding partners, such as TNKS. However, TNKS-binding partners demonstrate such remarkable diversity that a pattern has yet to emerge that can fully explain the relative importance of structural and sequence contributions to binding affinity and to PARylation. Furthermore, our understanding of TNKS regulation is also limited by the lack of PARylation site mapping. As a result, it is not known whether potential sequence preferences exist for the CAT domain to engage and modify substrates that are bound to the ankyrin repeats of TNKS, which could represent another layer of regulation.
The quantitative study of binding partner interactions has primarily been limited to the ARC:peptide interface, with only limited studies performed with full-length binding partners. To determine sequence and structural features that are important for TNKS-binding partner interaction and PARylation, we analyzed TNKS interaction with two example full-length binding partners: the PARylated binding partner telomeric repeat-binding factor 1 (TRF1) and the non-PARylated binding partner GDP-mannose 4,6-dehydratase (GMD). TRF1 regulates telomere length (6, 20), and TNKS-mediated PARylation of TRF1 causes it to dissociate from telomeric DNA and to be degraded through the ubiquitin–proteasome pathway (21). It is therefore likely that TNKS and TRF1 interact for only a defined period of time. In contrast, GMD, which functions in de novo fucose synthesis (17, 22), is bound to TNKS throughout interphase, only dissociating during mitosis (17). GMD binding has been shown to decrease TNKS catalysis of poly(ADP-ribose) (17). The formation of a stable TNKS:GMD complex may therefore represent a strategy for sequestering TNKS into an inactive complex during interphase, with release at a later time enabling its multiple mitotic functions. Thus, although TRF1 and GMD both engage the TNKS ankyrin repeat domain, some aspect of their interactions with TNKS yields different outcomes. A better understanding of how TRF1 and GMD interact with TNKS will not only aid in elucidating the factors that influence binding but may also provide insight into how TNKS activity is regulated.
In this study, we examine the sequence and structural parameters that affect TNKS interaction with binding partners GMD and TRF1. Using TNKS-binding analysis, PARylation activity assays, and mutants of GMD and TRF1, we found that both the specific sequence of the TBM and the quaternary structure of the binding partner play important roles in the interaction with TNKS. Using analytical ultracentrifugation, we demonstrated that GMD forms stable 1:1 complexes with the TNKS ankyrin repeats. Together with a compact globular fold, we propose that these features underly GMD resistance to PARylation. We also report in vitro evidence that TNKS primarily directs PAR modification to glutamate/aspartate residues and not serine residues.
Results
TRF1 and GMD as example TNKS-binding partners
TRF1 and GMD are both soluble constitutive homomultimers with TBMs at their extreme N termini (Fig. 1B). Within the 8-amino acid–binding footprint, the TBMs of TRF1 and GMD differ at four positions: 3, 4, 7, and 8 (Fig. 1B, middle). We first demonstrated interactions between TNKS and the example binding partners using pulldown assays. A histidine-tagged version of the ankyrin repeat region (TNKS-12345, residues 174–961), the minimal domain that is necessary and sufficient for binding partner interactions, was mixed with untagged full-length constructs of TRF1 and GMD. As expected, both binding partners exhibited a robust interaction with TNKS-12345 (Fig. 1C). Substituting Ala for Arg at position 1 of the TBM (R13A in TRF1; R12A in GMD) effectively disrupted the pulldown interactions, consistent with studies illustrating the importance of R1 (15, 18). Similarly, we have demonstrated that binding can be disrupted by introducing mutations into the ARCs that prevent binding to G6 (16). Introducing these mutations into all four functional ARCs 1, 2, 4, and 5 (construct TNKS-xx3xx where “x” designates a mutated ARC) (16) ablated interaction with TRF1 and GMD. Thus, TRF1 and GMD interaction with TNKS requires functional TBMs and functional ARCs.
Upon binding TNKS, TRF1 is strongly PARylated (6, 23), yet GMD has been shown to bind TNKS without being modified (17). To examine these previous observations in our reconstituted system, we produced a recombinant version of human TNKS containing residues 174–1327 with an N-terminal NusA-SMT tag to aid with the production and solubility of the enzyme (see “Experimental procedures”; referred to as NS-TNKS). This construct lacks the unstructured N-terminal HPS domain, for which no function has been identified, to improve production and purification properties. TNKS and TNKS2 share overlapping binding partners and functions despite the absence of an HPS in TNKS2 (24–26), suggesting that the HPS is not essential for TNKS function. An ADP-ribosylation assay using NS-TNKS demonstrated that TRF1 is indeed PARylated (Fig. 1D), and the PAR signal significantly decreases in the presence of PARP inhibitor PJ34. However, GMD was not PARylated under the same conditions. Thus, both binding partners engage the same ankyrin repeat domain but yield different outcomes in terms of modification with ADP-ribose.
TRF1 and GMD interaction with TNKS-12345
TBMs exhibit a fairly broad range of affinities for TNKS-12345, with typical values in the low micromolar range (e.g. ∼0.1 to ∼10 μm) and with the differences in affinities likely resulting from sequence variations within the TBMs (15, 16). We thus assessed the relative binding affinities of the TBMs of TRF1 and GMD in a fluorescence polarization binding assay utilizing fluorescently labeled peptides (5FAM-TRF1, residues 9–24; 5FAM-GMD, residues 8–23). 5FAM-TRF1 bound to TNKS-12345 with an apparent KD of 0.87 ± 0.07 μm (Fig. 2A), whereas 5FAM-GMD bound with an apparent KD of 1.42 ± 0.01 μm (Fig. 2B), both within the anticipated range of binding affinities. As a control, neither peptide bound to an ARC1–5 construct in which all TBM-binding sites had been inactivated (Fig. 2, A and B, TNKS-xx3xx).
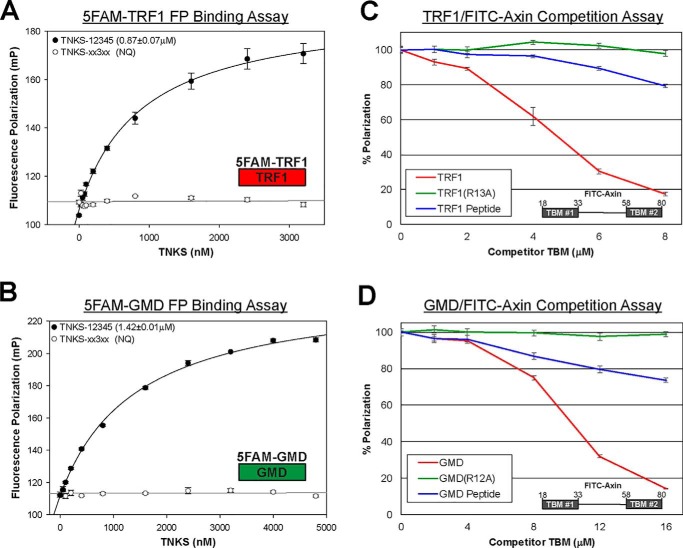
Figure 2.
TNKS binding to TRF1 and GMD. A and B, FP binding analysis of 5FAM-TRF1 (A) and 5FAM-GMD (B) peptide interaction with TNKS-12345 (ankyrin repeat region) and TNKS-xx3xx (ankyrin repeat region with no functional ARCs). Binding was considered NQ if the binding curve did not allow for robust KD determination. Reactions that exhibited NQ binding were analyzed in at least two separate experiments, and all other KD values were determined from a minimum of three different experiments. Data represent the mean, error bars represent S.E. C and D, competition fluorescence polarization binding analysis of full-length TRF1 (C) and full-length GMD (D). A fluorescently labeled Axin peptide and TNKS-12345 were maintained at saturating concentrations (27 nm and 1.5μm, respectively) as increasing amounts of competing binding partners were added. TRF1 and GMD constructs consisted of full-length WT and TBM mutants R13A and R12A for TRF1 and GMD, respectively. For comparison, unlabeled peptides representing their TBMs were evaluated (GMD peptide and TRF1 peptide). Competitor concentrations represent the total TBM concentration. Thus, TRF1 and GMD are reported at their monomer concentrations to directly compare the relative efficiency of full-length proteins versus peptides. Data represent the mean, error bars represent S.D. mP, milli-polarization units.
We next used a competition binding assay to assess TNKS-12345 interaction with full-length TRF1 and GMD (Fig. 2, C and D). The TRF1 homodimer structure presents two TBMs, and the GMD tetramer structure presents four TBMs (Fig. 1B) (20, 27). Multimeric full-length TRF1 and GMD may therefore potentially interact with multiple ARCs simultaneously and exhibit higher affinity than the single-peptide interactions alone. In the competition experiment, TNKS-12345 was mixed with full-length TRF1 or GMD and a fluorescently labeled peptide representing the bipartite TNKS-binding domain of Axin1 (FITC-Axin, residues 18–80). FITC-Axin interacts with multiple ARCs and is capable of binding multiple ARC pairs simultaneously, including ARCs 1/2, 4/5, and 2/5 (16). TNKS-12345 binding to FITC-Axin was maintained at near-saturation levels to yield a high starting level of fluorescence polarization (FP) signal, which was set to 100% (Fig. 2, C and D). TRF1 and GMD were then added at increasing concentrations, and FP measurements were taken at equilibrium. The decrease in signal was interpreted as TRF1 or GMD effectively outcompeting FITC-Axin peptide for binding to TNKS-12345. For comparison with the full-length proteins, unlabeled TBM peptides of TRF1 and GMD were also assessed in the competition experiment (Fig. 2, C and D, peptides alone are in blue). In Fig. 2, C and D, competitor concentrations are plotted as monomers of TRF1 and GMD, thus representing the total number of TBMs and reflecting the competitiveness per TBM. When plotted as concentrations of dimer TRF1 and tetramer GMD (Fig. S1A), both proteins effectively competed for TNKS-12345 binding over a similar concentration range. As expected, mutations to the TBMs of both TRF1 (TRF1(R13A)) and GMD (GMD(R12A)) abrogated their ability to compete with FITC-Axin for binding to TNKS-12345 (Fig. 2, C and D). Both TRF1 and GMD exhibited more efficient competition than their corresponding TBM peptides alone, suggesting that their quaternary structures present TBMs in orientations that allow them to simultaneously engage ARCs in ways that a single peptide cannot. Based on these results, we surmised that the competitive efficiency of TRF1 and GMD is a composite of the individual TBM affinities, and the total number and relative accessibility of TBMs are determined by quaternary structure.
The effects of quaternary structure alteration
We further tested the contribution of TRF1 and GMD quaternary structure to TNKS interaction using site-directed mutagenesis to perturb their multimeric states. Mutations A74D/A75P and W77P within the TRF1 dimerization domain disrupt DNA binding, presumably through disruption of the dimer interface (28) (Fig. S2A); however, the effect of these mutations on the multimeric state of TRF1 has not been directly assessed. Using size-exclusion chromatography and sedimentation analysis, we demonstrated that TRF1 constructs A74D/A75P and W77P are monomers in solution (Fig. S2, B–G). In contrast to TRF1, there are no published mutations that disrupt the dimer-of-dimers quaternary structure of GMD. The GMD dimer consists of two antiparallel monomers with a domain swap region that aids in forming the active sites where cofactors bind (Fig. S3A). Two GMD dimers form a tetramer, stabilized by two patches of hydrophobic residues. We introduced two sets of mutations designed to disrupt the tetramerization interface: A125D/G129A and A181W/Y185D (Fig. S3A, center). Size-exclusion chromatography demonstrated that both constructs elute at later volumes than WT GMD, indicating that they are tetramerization-deficient (Fig. S3B).
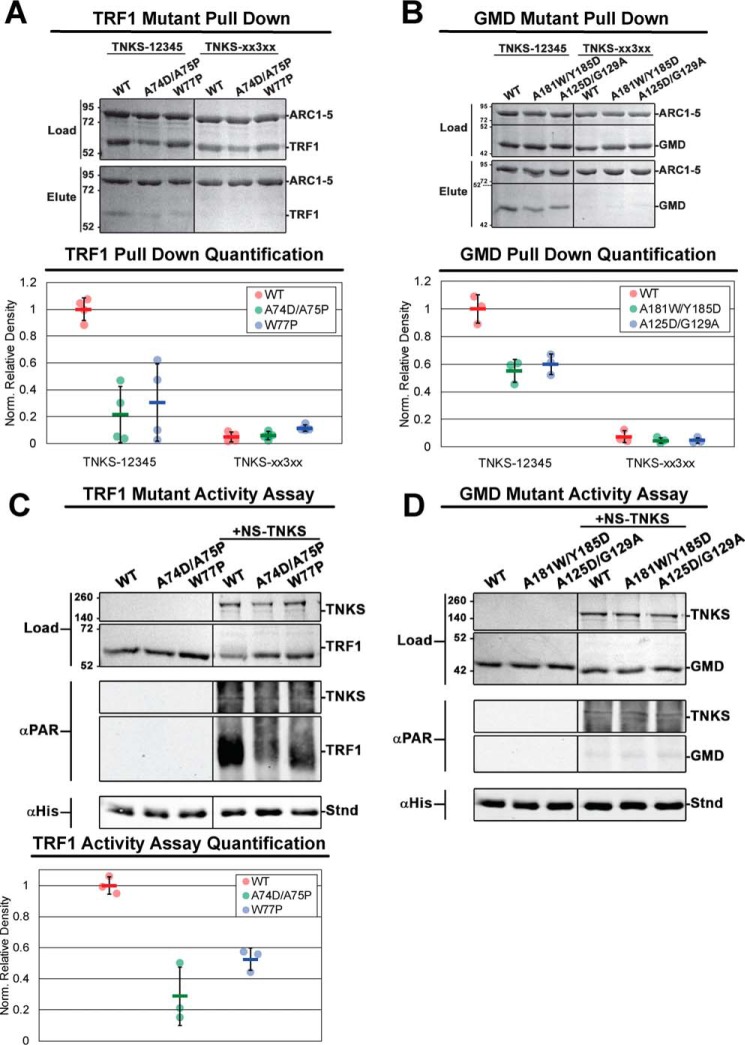
Dimerization-deficient TRF1 constructs showed a significant decrease in binding relative to WT TRF1 in a pulldown assay with TNKS-12345 (Fig. 3A), consistent with dimerization contributing to the overall affinity of the interaction. Tetramerization-deficient GMD constructs also showed an approximate 50% decrease in binding (Fig. 3B). The dimer-deficient TRF1 mutants and the tetramer-deficient GMD mutants were slightly worse competitors than their WT counterparts in competition binding experiments (Fig. S1, B and C). PARP activity assays demonstrated that dimer-deficient TRF1 constructs are PARylated significantly less than dimeric WT TRF1 (Fig. 3C), consistent with a weakened interaction. Similar to WT GMD, tetramer-deficient GMD constructs were not PARylated (Fig. 3D), despite maintaining a notable level of interaction with TNKS (Fig. 3B). This result indicated that disruption of the GMD quaternary structure does not reveal or expose potential sites of modification. Based on the deficiencies in binding (for GMD and TRF1) and PAR modification (for TRF1) resulting from mutations targeting quaternary structure, we conclude that avidity plays an important role in TNKS interaction with GMD and TRF1, allowing these proteins to engage multiple ARCs.
Figure 3.
The effects of quaternary structure alteration on TNKS binding to and PARylation of TRF1 and GMD. A and B, results and quantification of pulldown assays analyzing the binding of TNKS-12345 to multimerization-deficient TRF1 (A) and GMD (B) constructs. Histidine-tagged TNKS-12345 or TNKS-xx3xx (binding-deficient control) were used to pull down untagged TRF1 and GMD constructs with multimerization-disrupting mutations. For quantification, binding partner density was normalized to the density of TNKS-12345 in each respective reaction. C and D, TNKS PARP activity assays analyzing the PARylation of multimerization-deficient TRF1 (C) and GMD (D) constructs. The quantification of PARylation of TRF1 constructs is shown in the bottom plot of C. The designated reaction mixtures were analyzed by SDS-PAGE (Load) and by Western blot analysis of PARylation (αPAR). A His-tagged protein (Stnd) was added to each reaction after quenching to assess transfer efficiency of the blot; the protein was detected with an anti-His antibody (αHis). NS-TNKS, TRF1, and GMD were each loaded at 0.5μm for the TNKS activity assays. Data represent the mean, error bars represent S.D. The black divider lines in the center of each gel and Western blot designate where the image has been sliced for presentation. For C and D, see Fig. S3, C and D, for complete lanes for Western blots.
ARC bias and binding distribution of TRF1 and GMD
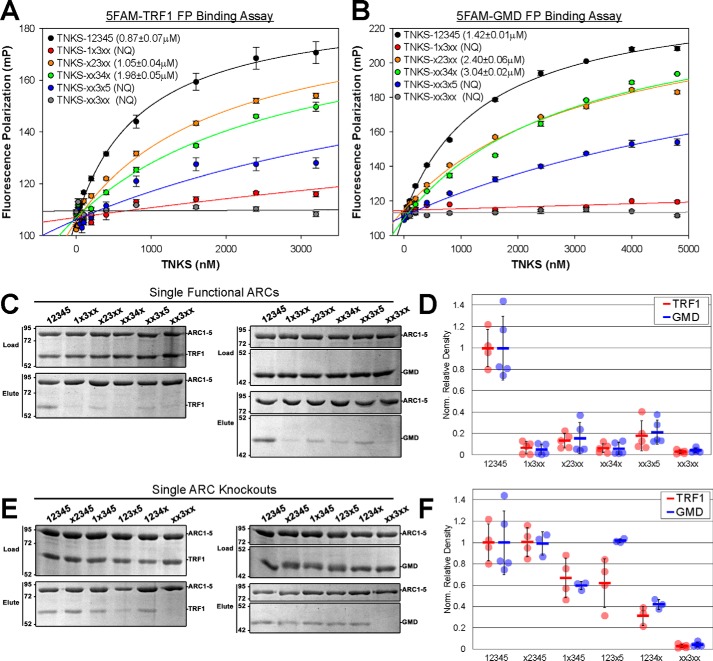
Given that TRF1 and GMD both appear to engage multiple ARCs yet yield different outcomes in PARylation assays, we sought to determine whether they exhibited different binding distributions across the ARCs or whether their binding was biased toward specific and potentially different ARCs. To determine whether the sequence variations within the TBMs of GMD and TRF1 impose an ARC bias, we used the FP binding assay to measure the affinity of 5FAM-TRF1 and 5FAM-GMD for a panel of TNKS-12345 constructs bearing only a single functional ARC (1, 2, 4, or 5) (Fig. 4, A and B). Both TBMs demonstrated similar binding preferences, with the highest affinities toward ARC2 and ARC4 and lower relative affinities for ARC5 and ARC1 (Fig. 4, A and B). Axin1 and insulin-regulated aminopeptidase peptides demonstrated similar ARC binding preferences (16), suggesting that the ARCs of TNKS have a defined hierarchy of binding capacities for TBMs.
Figure 4.
TNKS binding footprint of TRF1 and GMD. A and B, FP binding analysis of TRF1 (A) and GMD (B) peptide interaction with TNKS-12345 and TNKS-12345 constructs bearing a single functional ARC. Binding was deemed NQ if the binding curve did not allow for robust KD determination. TNKS variants that exhibited NQ binding were analyzed in at least two separate experiments, and all other KD values were determined from a minimum of three different experiments. Data represent the mean, error bars represent S.E. C and D, results and quantification of pulldown experiments analyzing the binding of untagged TRF1 and GMD to TNKS-12345 constructs with single functional ARCs. Input proteins (Load) and elution fractions (Elute) were analyzed by SDS-PAGE. E and F, results and quantification of pulldown experiment analyzing the binding of untagged TRF1 and GMD to TNKS constructs where each individual ARC has been rendered nonfunctional. Input proteins (Load) and elution fractions (Elute) were analyzed by SDS-PAGE. Binding data for WT construct TNKS-12345 and TNKS-xx3xx are used as a reference in D and F and are the collective result of multiple experiments. Binding partner density was normalized to the density of TNKS in each reaction. Data represent the mean, error bars represent S.D. mP, milli-polarization units.
To determine whether the ARC bias is preserved in the context of full-length proteins, we used pulldown analysis to evaluate the ability of GMD and TRF1 to interact with TNKS-12345 constructs with ARC mutations (Fig. 4, C–F). Neither GMD nor TRF1 demonstrated robust interaction with TNKS-12345 constructs with single functional ARCs compared with fully functional TNKS-12345, suggesting that neither binding partner prefers any single ARC (Fig. 4, C and D). Pulldown analysis using a panel of TNKS-12345 constructs in which each ARC had individually been disrupted showed a similar profile for GMD and TRF1 (Fig. 4E and F), with a slight difference observed when ARC4 was mutated, where TRF1 binding decreased but GMD binding remained unchanged (Fig. 4F, see TNKS-123x5). Overall, the pulldown analysis suggested a similar dependence on any given ARC. Interestingly, a previous study found that ARC5 is required for TNKS interaction with TRF1 (23), and we observed that loss of ARC5 significantly decreased binding of both TRF1 and GMD (Fig. 4, E and F, see TNKS-1234x). Although ARC5 alone does not exhibit robust interaction with the TBM of TRF1 or GMD, it clearly has a role in the observed interaction with full-length proteins, highlighting that the full-length proteins engage multiple ARCs. Moreover, the analysis indicates that no single ARC bears the majority of the binding affinity, and correspondingly, the knockout of any individual ARC does not abrogate the interaction. Based on the FP and pulldown binding analyses, the interaction modes of TRF1 and GMD appear to utilize similar ARC footprints despite possessing different TBM sequences and quaternary structures.
The effect of exchanging the TBMs of GMD and TRF1
The analysis in the previous section suggested that the TBMs of TRF1 and GMD engage a similar distribution of ARCs. However, these observations do not necessarily exclude the possibility that the different TBMs direct interactions with TNKS in distinct ways that might differentially influence the capacity to be PARylated. We therefore exchanged the TBMs of GMD and TRF1 to assess whether this swapping of sequences might alter aspects of the TNKS interaction and/or PARylation potential of either protein. Over the 8-amino-acid TBM-binding footprint, GMD and TRF1 differ at positions 3, 4, 7, and 8 (Fig. 5A). Mutagenesis was performed such that positions 3 and 4 were exchanged to yield constructs TRF1(Pos. 3/4) and GMD(Pos. 3/4), positions 7 and 8 were exchanged to yield TRF1(Pos. 7/8) and GMD(Pos. 7/8), and all differing positions were exchanged to yield a complete swapping of the two TBM sequences, constructs TRF1[G] and GMD[T] (Fig. 5A).
Figure 5.
The effect of TBM alteration on TNKS binding to and PARylation of TRF1 and GMD. A, schematic of TBM mutations wherein specific residues between the TRF1 and GMD TBMs were exchanged. B, results and quantification of pulldown experiments analyzing the binding of TRF1 constructs with TBM swapping mutants. Histidine-tagged TNKS constructs were either WT TNKS-12345 or TNKS-xx3xx. Input proteins (Load) and elution fractions (Elute) were analyzed by SDS-PAGE. Binding partner density was normalized to the density of TNKS in each reaction. Data represent the mean, error bars represent S.D. C, TNKS PARP activity assays and quantification analyzing the effect of TBM mutation on PARylation of TRF1. The designated reaction mixtures were analyzed by SDS-PAGE (Load) and by Western blot analysis of PARylation (αPAR). A His-tagged protein (Stnd) was added to each reaction after quenching to assess transfer efficiency of the blot; the protein was detected with an anti-His antibody (αHis). Data represent the mean, error bars represent S.D. D, results and quantification of pulldown experiments performed as in B except using GMD constructs with TBM mutations. E, TNKS PARP activity assay performed as in C except using GMD constructs with TBM mutations. For activity assays, NS-TNKS and GMD were loaded at 0.5μm, and TRF1 was loaded at 1μm to better resolve the subtle differences in PARylation density. The black divider lines in the center of the gel in D designate where the image has been sliced for presentation. For C and E, see Fig. S4, A and B, for the complete Western blot images.
Pulldown analysis of the TRF1 TBM mutant constructs demonstrated that all sequence alterations reduced binding (Fig. 5B). Binding of both TRF1(Pos. 7/8) and TRF1[G] was near background levels, suggesting significant inhibition of binding. Exchanging the TBM of TRF1 with the lower-affinity TBM of GMD therefore decreases interaction with TNKS, and consistent with the TBM footprint spanning over 8 residues (15), residues at positions 3/4 and 7/8 both make contributions to overall binding. In agreement with the pulldown interactions, PARylation assays utilizing NS-TNKS showed decreased PAR signal for each of the TBM exchange mutants relative to WT TRF1 (Fig. 5C). The simplest interpretation of these results is that the TBM swap has largely influenced the affinity of TRF1 interaction with TNKS, rather than imposing a new TBM binding distribution that negatively influences PARylation capacity.
The TBM exchange mutants of GMD had no major impact on TNKS-12345 binding relative to that of WT GMD, although there was a general tendency toward increased interaction that correlates with the introduction of the higher-affinity TBM of TRF1 (Fig. 5D). Furthermore, whereas the TRF1 TBM exchange mutant had only a small effect in the competition binding assay (Fig. S1D), the GMD TBM exchange mutants showed an enhanced ability to compete with FITC-Axin binding (Fig. S1E). However, the apparent increase in GMD binding did not lead to a change in the PARylation of GMD in that WT GMD and each of the TBM exchange mutants showed no indications of being PARylated (Fig. 5E). We thus concluded that the TBM alone does not dictate the capacity for a TNKS-binding partner to be modified and that the modification capacity is more likely determined through inherent structural properties of the binding partner.
GMD forms stable 1:1 complexes with TNKS-12345
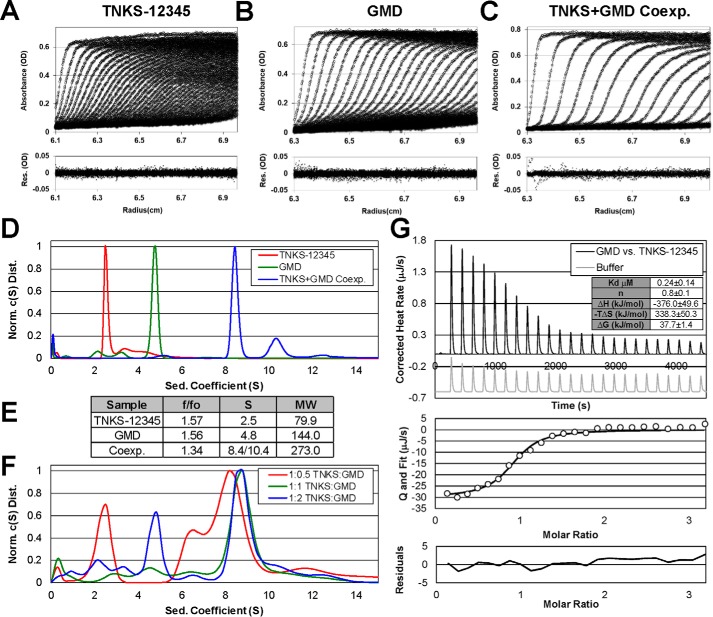
To examine whether there were underlying structural aspects that might explain the lack of GMD PARylation despite its interaction with TNKS, we assessed the complex formed between GMD and TNKS-12345. Indeed, we were able to isolate a stable complex through coexpression of His-tagged TNKS-12345 and untagged GMD followed by copurification using Ni2+-affinity and size-exclusion chromatography. Size-exclusion chromatography yielded a single peak containing both proteins at an apparent ratio of 1:1 based on Coomassie staining (Fig. S5, A and B). We used sedimentation velocity analytical ultracentrifugation (SV-AUC) to further evaluate the complex relative to the individually purified components (Fig. 6, A–E). The c(S) distributions demonstrate that TNKS-12345 and GMD sediment primarily as single species at 2.5S and 4.8S, respectively (Fig. 6, A, B, and D). The approximated molecular masses depict TNKS-12345 as a monomer (79.0 versus 88.2 kDa theoretical), as we have shown previously (16), and GMD as a tetramer (144 versus 169.6 kDa theoretical) (Fig. 6E), as expected based on the crystal structure of GMD (Protein Data Bank (PDB) code 5IN5). The coexpressed complex of TNKS-12345 and GMD sediments as two species, a primary species at 8.4S and a secondary, albeit minor, species at 10.4S (Fig. 6D). These data suggest that TNKS-12345 and GMD sediment primarily as one stable species, yet higher-order complexes also form to some degree. Unfortunately, the higher-order species precluded analysis with structural techniques such as small-angle X-ray scattering, and we have not been able to obtain crystals of the complex for X-ray structure analysis.
Figure 6.
Analysis of TNKS-12345 and GMD complex formation. A–C, SV-AUC analysis of TNKS-12345 (A), GMD (B), or the coexpressed complex of TNKS-12345 and GMD (C). The top panels show absorbance data (circles) and associated c(S) model fit (lines). The bottom panels show residuals for the fit. TNKS was loaded at 9.1 μm (0.8 mg/ml) in A, GMD was loaded at 2.4 μm (tetramer concentration, 9.6 μm monomer, 0.4 mg/ml) in B, and TNKS-12345:GMD coexpressed complex was loaded at 0.5 mg/ml (resulting in 2 μm for a 1:1 TNKS-12345:GMD tetramer ratio) in C. D and E, normalized c(S) distributions (D) and solution parameters (E) derived from sedimentation velocity experiments (A–C). F, normalized c(S) distribution derived from sedimentation velocity experiments where TNKS-12345 and GMD were mixed at different ratios (see Fig. S5, C–E and I, for data and model fitting to the data). G, ITC analysis of the interaction between TNKS-12345 and full-length GMD (reported as tetramer concentration). Binding and thermodynamic parameters represent mean ± S.D. of three independent experiments.
The approximated molecular mass of the primary TNKS-12345:GMD peak corresponds to a complex of one TNKS-12345 molecule and a tetramer of GMD (273.0 versus 257.8 kDa theoretical), thus suggesting a 1:1 complex. Indeed, mixing individually purified TNKS-12345 and GMD at a 1:1 ratio (monomer:tetramer) yielded a c(S) distribution very similar to the coexpressed complex (Figs. 6F and S5, D and I). Shifting the mixing ratio to 1:0.5, thereby decreasing the amount of GMD relative to TNKS-12345, resulted in a new peak in the c(S) distribution that corresponds to unbound TNKS-12345 (Fig. S5, C and I). Similarly, shifting the ratio to 1:2, thereby increasing the amount of GMD relative to TNKS-12345, resulted in a new peak that corresponds to unbound GMD (Fig. S5, E and I). A 1:1 mixture of TNKS-12345 with the TBM mutant GMD(R12A) that disrupts the interaction yielded a c(S) distribution that corresponded to unbound GMD and TNKS-12345 (Fig. S5, F, H, and I), demonstrating that complex formation requires a functional TBM. Similarly, a 1:1 mixture of TNKS-12345 and a GMD mutant with the TRF1 TBM (GMD[T]) obtained the same distribution observed for WT GMD, demonstrating that alteration of the TBM does not have a major impact on complex formation (Fig. S5, G–I), suggesting that the TBM sequence does not influence TNKS:GMD stoichiometry. Isothermal titration calorimetry (ITC) analysis of the TNKS-12345:GMD binding interaction also indicated an approximate 1:1 stoichiometry (n = 0.8 ± 0.1) (Fig. 6G). Ultimately, these data suggest that the TNKS ankyrin repeat domain forms a stable 1:1 complex with a tetramer of GMD.
SV-AUC analysis of TRF1 demonstrated that the unbound protein sediments at 3.7S as a single species with the approximated molecular mass of a dimer (79 versus 100.5kDa theoretical) (Fig. S2, C, F, and G). SV-AUC analysis of mixtures of TNKS-12345 and TRF1 at different ratios yielded multipeak c(S) distributions that extended as far out as 30S, indicating formation of large aggregates that precluded further analysis (Fig. S6, A–D and H). Reducing the concentration of both TNKS-12345 and TRF1 and the TNKS:TRF1 ratio reduced the aggregation tendency, yielding a two-peak distribution: a 2.4S peak that corresponded to a TNKS-12345 monomer and a 4.2S species (Fig. S6, E–H). The identity of this latter species could not be established because the c(S) distribution was unaffected by changing the TNKS-12345:TRF1 ratio, possibly due to aggregation and/or solubility issues.
To determine the effect of concentration on aggregate formation, we conducted a solubility assay wherein TNKS-12345 was mixed with an increasing concentration of TRF1. TNKS-12345:TRF1 mixtures were incubated for 30 min at room temperature and then spun. The supernatant and pellet were evaluated by SDS-PAGE. The solubility assay demonstrated that the TNKS-12345 distribution was shifted to the insoluble pellet as the TRF1 concentration increased (Fig. S7, A–C). This redistribution was not observed with the mutant TRF1(R13A), suggesting that the shift in TNKS-12345 solubility was due to direct interaction with TRF1. The formation of aggregates therefore appears to be due to TRF1 drawing TNKS-12345 into the pelleted fraction. However, whether the TNKS-12345:TRF1 interaction is inherently less soluble or TNKS-12345 preferentially binds a lower-solubility form of TRF1 could not be resolved in this analysis.
Thus, the comparative sedimentation analysis indicated that TRF1 and GMD behave quite differently when complexed with TNKS-12345, with GMD primarily forming a stable and reasonably well-defined 1:1 complex and TRF1 forming large aggregates under most conditions. Because TNKS catalytic activity is coupled to self-assembly through the SAM domain, these differences could potentially represent fundamental properties that relate to modification capacity. For example, GMD may favor complexation with a single ankyrin repeat scaffold and thus avoid modification by the TNKS polymer, whereas TRF1 may complex in a manner that spans multiple tankyrase-1 molecules and is thus easily accessible for modification.
TNKS primarily targets glutamate/aspartate residues for PARylation in vitro
In trying to understand the physical properties of GMD that might underlie the resistance to PARylation, we considered the recent observation that PARP-1 and PARP-2 can target specific residues for modification. Although glutamate and aspartate are commonly targeted for modification, PARPs 1 and 2 can also target serine residues for modification under specific conditions regulated by the protein histone PARylation factor 1 (HPF1) (29). Although the underlying mechanism of this switch is not understood, PARP-specific residue targeting could fundamentally alter our understanding of PARP regulation. For instance, if TNKS only modifies specific residues, reduced solvent exposure of the targeted residues could allow GMD to interact with TNKS without being PARylated.
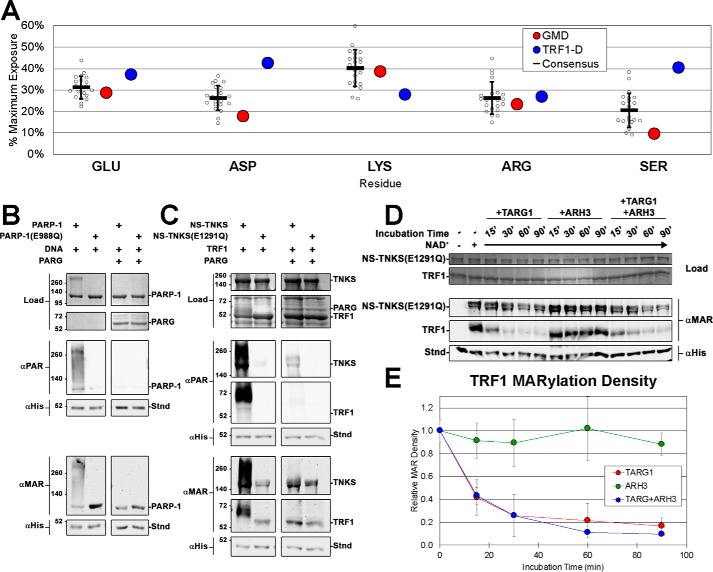
We thus explored the relative solvent accessibility of several PAR-accepting residues in GMD and TRF1 crystal structures. Solvent accessibility was calculated for common PAR-accepting residues using the program Surface Racer (30). These values were normalized to the maximum possible solvent accessibility for each free amino acid residue in solution (31), yielding percent maximum exposures. GMD and the TRF1 dimerization domain (TRF1-D), the largest fragment of TRF1 crystallized to date, were compared with each other as well as with a group of 20 other homomultimeric proteins (Table S1). The 20 consensus structures, GMD, and TRF1-D all demonstrated similar solvent accessibility for PAR-accepting residues glutamate, lysine, and arginine (Fig. 7A and Table S1). Exposure of GMD aspartate and serine residues was somewhat lower than the consensus, and there was a noticeable paucity of serine residues on the surface of GMD (Fig. S8, A–D). TRF1-D, in contrast, demonstrated higher exposure for both aspartate and serine residues, with serine residues being almost 2-fold more exposed than the consensus. Thus, if TNKS is a serine-targeting PARP, the reduced GMD solvent accessibility could prevent GMD from being PARylated. However, it is noteworthy that the pattern of increased serine and aspartate exposure noted for TRF1 was not observed in crystal structures of other modified TNKS-binding partners (Fig. S8E).
Figure 7.
TNKS as a glutamate-targeting PARP in vitro. A, solvent accessibility of common PARylated residues in TRF1-D (DNA-binding domain) and GMD crystal structures compared with a consensus residue exposure derived from a collection of homomultimeric crystal structures (see Table S1). B and C, activity assays examining the effects of PARG treatment of PAR/MAR-modified PARP-1 (B) and PAR/MAR-modified NS-TNKS with TRF1 (C). PARP-1 constructs consisted of WT PARP-1 and NS-TNKS as well as mutants PARP-1(E988Q) and NS-TNKS(E1291Q) that are confirmed to primarily synthesize MAR. The designated reaction mixtures were analyzed by SDS-PAGE (Load) and by Western blot analysis of PARylation (αPAR) and MARylation (αMAR). A His-tagged protein (Stnd) was added to each reaction after quenching to assess transfer efficiency of the blot; the protein was detected with an anti-His antibody (αHis). D, NS-TNKS(E1291Q) and TRF1 were mixed, incubated for 30 min (with or without NAD as indicated), and then quenched by the addition of PARP inhibitor rucaparib. MAR-modified NS-TNKS(E1291Q) was then treated with the hydrolases TARG1 and ARH3 (alone and in combination) over a time course to measure their ability to remove MAR. Reactions were quenched by the addition of Laemmli sample buffer. The designated reaction mixtures were analyzed by SDS-PAGE (Load) and by Western blot analysis of MARylation (αMAR). A His-tagged protein (Stnd) was added to each reaction after quenching to assess transfer efficiency of the blot; the protein was detected with an anti-His antibody (αHis). See Fig. S9, D and E, for the complete SDS-PAGE and Western blot images. E, quantification of the TRF1 band intensity from D. Quantification of the band intensity of NS-TNKS(E1291Q) is shown in Fig. S9A. Data represent the mean, error bars represent S.D.
Targeted PARylation of specific residues may explain how GMD can resist modification; however, the amino acid preferences of TNKS have not been explored. To address possible amino acid preferences, we analyzed the in vitro target sites of TRF1 and TNKS itself. PAR is a transient posttranslational modification that is rapidly removed from target proteins by mono- and poly(ADP-ribosyl) hydrolases, with certain hydrolases removing ADP-ribose from specific amino acid types (32). Residue-specific hydrolases in combination with anti-PAR antibodies and MAR-binding reagents are proving to be invaluable tools in the exploration of PARP residue targeting (33). Poly(ADP-ribose) glycohydrolase (PARG) efficiently degrades PAR, yet it is extremely inefficient at removing the last MAR moiety. A PARP activity assay demonstrated that PARG treatment can remove PAR from PARP-1, NS-TNKS, and TRF1, yet a residual MAR signal remains (Fig. 7, B and C). In addition, the mutant construct PARP-1(E988Q) that only catalyzes MARylation (34, 35) and its TNKS-equivalent NS-TNKS(E1291Q) also retained a MAR signal following PARG treatment (Fig. 7, B and C). In contrast to PARG, the hydrolases named terminal ADP-ribose protein glycohydrolase 1 (TARG1) and ADP-ribosyl hydrolase 3 (ARH3) cleave specific amino acid–linked ADP-ribose moieties, allowing them to remove the final MAR. TARG1 can cleave MAR as well as whole PAR chains from glutamate and aspartate residues (36), and ARH3 degrades PAR chains similarly to PARG but can also remove MAR from serine residues (33).
To examine whether TNKS is a serine-targeting PARP, we conducted a hydrolase time series with TARG1, ARH3, and both hydrolases in combination. The NS-TNKS(E1291Q) construct was utilized in these experiments to better observe the residue-specific de-MARylation activities of the hydrolases and to improve quantification of assay results. NS-TNKS(E1291Q) and TRF1 mixtures were incubated for 30 min, the reaction was quenched with the PARP inhibitor rucaparib, and hydrolases were then added. Samples were taken at specific times, and these reactions were quenched by the addition of Laemmli sample buffer. Over the time series, TARG1 was able to significantly reduce TRF1 and NS-TNKS(E1291Q) MAR density, consistent with removal of MAR from glutamate residues (Figs. 7, D and E, and S9A). However, TARG1 could not entirely remove the MAR signal, suggesting that TNKS targets other residues as secondary acceptors of PAR. ARH3 did not significantly reduce either TRF1 or TNKS MAR signal, and combining ARH3 and TARG1 did not remove more MAR than TARG1 alone. These data suggest that glutamate/aspartate, and not serine, is the primary residue targeted by TNKS for PARylation in vitro. Notably, the TNKS MAR signal decreased to a lesser extent than that of TRF1. TNKS automodification may therefore target residues other than glutamate/aspartate and serine, or TNKS sites may simply be less accessible to the hydrolases used in this experiment. We also confirmed that HPF1 does not alter the substrate specificity of TNKS, in contrast to its action on PARP-1 (Fig. S9, B and C).
Discussion
The adaptability of the TNKS ankyrin repeat domain allows it to accommodate a considerable amount of sequence and structural variation in binding partners. Structural plasticity is undoubtedly critical for TNKS function as a scaffolding protein. However, TNKS-binding partners demonstrate such remarkable heterogeneity that the parameters that influence binding and PARylation are not well-understood. In this study, we utilize the full-length versions of binding partners TRF1 and GMD to expand our understanding of the parameters that contribute to interaction with and modification by TNKS.
Using mutagenesis in combination with binding and activity assays, we demonstrate that both the quaternary structure and TBM sequence of TRF1 contribute to TNKS interaction as well as the capacity for TRF1 to be modified by TNKS. Disrupting TRF1 dimerization decreased interaction with TNKS significantly and resulted in a subsequent decrease in PARylation. In consideration of the TRF1 TBM sequence, the critical importance of TBM residues at position 1 (Arg) and position 6 (Gly) is well-established. However, our analysis of TBM exchange mutants of TRF1 that altered positions 3/4, 7/8, and 3/4/7/8 indicates that changes at less stringently conserved positions within the TBM can also affect TNKS interaction with TRF1. Structural analysis of a TRF1 peptide bound to ARC2 revealed that the side chains of the residues at positions 3, 4, and 7 do not engage the ARC pocket (37). In contrast, the side chain of the Asp-20 residue at position 8 interacts with an Arg residue within TNKS. The TBM exchange mutant affecting positions 7/8 substitutes polar Asp-20 for a nonpolar Met residue, most likely disrupting this Asp–Arg contact and significantly reducing TRF1 binding and PARylation. The decrease in binding and PARylation of the TRF1(Pos. 3/4) construct suggests that these residues have an indirect contribution to TNKS interaction. Furthermore, the analysis of TNKS interaction with a library of TBM peptides established sequence preferences for each of the eight positions in the context of binding partner 3BP2 (15). According to those preferences, the TRF1 TBM exchange mutations used in this study should have increased binding; however, in the context of the TBM of TRF1, each of the mutants reduced interaction with TRF1. The observed variation in TBM sequences may thus represent a mechanism for precisely modulating the specific affinity of each TNKS-binding partner, rather than modulating a single “ideal” binding sequence.
Substituting the TBM of GMD with the higher-affinity sequence of TRF1 did not have a pronounced impact on binding, and it did not result in PARylation of GMD. As a tetramer with four TBMs, avidity may facilitate higher-affinity interactions between GMD and TNKS-12345, perhaps overriding subtle changes in the affinity of the TBM sequence. Consistently, tetramer-deficient GMD constructs demonstrated significantly reduced binding to TNKS-12345 relative to WT GMD, suggesting that the GMD:TNKS binding mode involves several TBMs on a single GMD tetramer. We further observed that GMD forms a stable 1:1 complex with TNKS-12345, suggesting that the multiple TBMs of a GMD tetramer primarily interact with a single ankyrin repeat region of TNKS. In contrast, we were unable to obtain insights into the stoichiometry and structure of the TRF1 complex with TNKS, largely due to solubility issues that arose after mixing the otherwise reasonably well-behaved samples. We anticipate that the formation of insoluble aggregates of TRF1 and TNKS could be indicative of the type of complex these proteins might form in cells when multiple copies of TRF1 coating telomeres and multimeric forms of TNKS interact.
Using residue-specific hydrolases, we showed that TNKS primarily targets glutamate/aspartate residues as acceptors of ADP-ribose on itself and TRF1 in vitro. We also demonstrated that the newly identified HPF1, which steers PARP-1/PARP-2 toward modifying serine residues, does not have the same impact on TNKS modification. The mechanisms underlying PARP residue targeting, let alone its potential implications for TNKS activity, are not well-understood. TNKS may not undergo this level of regulation, or perhaps there are other cellular factors that control TNKS in this manner that have yet to be identified.
Nevertheless, despite possessing surface-exposed glutamate/aspartate and forming a stable complex with the ankyrin repeat domain, GMD was not detectably modified. Although the absence of modification is consistent with previously published findings (17), the constraints that suppress GMD PARylation remain enigmatic. In PARP-1, PARylation is autoinhibited by a helical domain within the catalytic region. When PARP-1 DNA-binding domains engage damaged DNA, a series of interdomain contacts destabilize the autoinhibited conformation, thus enabling PARylation. Although the autoinhibitory helical domain is absent in the CAT of TNKS, direct and specific interactions between binding partners and the CAT could serve as an additional layer of regulating whether a given TNKS-binding partner is modified. A recent study has suggested that TNKS activity is regulated by the formation of a CAT:CAT dimer (38). In the proposed model, SAM-mediated self-assembly causes the CAT domains of neighboring TNKS molecules to assemble along a specific interface. The formation of the dimer interface is proposed to induce a more open conformation that is more accessible to NAD+. In support of this model, disruption of the SAM:SAM interface, or proposed CAT:CAT interface, greatly reduced catalytic activity (38). The propensity for GMD to form 1:1 complexes with the TNKS ankyrin repeat domain suggests that GMD preferentially engages monomeric, and perhaps inactive, TNKS. Using this mechanism, GMD could avoid being PARylated and play a role in restraining TNKS activity in cells. GMD binding could also potentially shield the ankyrin repeat region from other binding partners, perhaps buffering TNKS activity in certain stages of the cell cycle.
Although we could produce a form of catalytically active TNKS suitable for biochemical analysis (NS-TNKS), the protein was not of suitable quality to answer detailed structural questions regarding the role of GMD on TNKS self-assembly or whether GMD has some other structural influence that regulates catalytic output (e.g. locking the CAT in an inactive or sequestered conformation). However, it is notable that the tetramer-deficient mutants still bound to TNKS at some level but did not undergo PARylation, arguing against the GMD tetramer having a specific structural impact on TNKS that modulates activity. Alternatively, GMD may possess structural features that are resistant to modification. PARP-1 automodification sites have been mapped utilizing MS (39). Aside from a few sites near the CAT domain, PARP-1 primarily modifies glutamate, aspartate, and lysine residues that are on flexible linkers or adjacent domains. Flexibility or even specific surface and sequence features may therefore be a critical determinant for sites of PARylation. Crystal structures of GMD demonstrate that individual monomers adopt globular, condensed cylindrical structures that assemble into tightly packed tetramers, which could represent the physical basis for the lack of GMD PARylation. It is also important to note that although GMD is not PARylated, TNKS may impact GMD function by binding GMD and then PARylating subsequent GMD-binding partners or even simply blocking substrate-binding sites.
Pharmacological inhibition of PARP-1 catalytic activity is a potent treatment for BRCA-deficient cancers (40). Similarly, inhibition of TNKS catalytic activity has applications in fibrosis, colorectal cancer, and gastric cancer (4, 41, 42, 44). However, the current PARP inhibition strategy involves using small molecules to compete with NAD+ for the catalytic site (45). Although effective for PARP-1, this method of inhibition has been shown to simultaneously inhibit multiple TNKS functions (46, 47). CAT-targeted inhibition strategies may therefore affect all TNKS functions, and as some of these functions include apoptosis and Golgi trafficking, the current PARP inhibition strategy may have undesirable off-target effects. Alternatively, we have demonstrated that the interaction between TRF1 and the ankyrin repeat domain is extremely sensitive to TBM swapping and changes in multimeric state. Alteration of the TBM sequence strongly decreased TNKS:TRF1 interaction and PARylation without disrupting TRF1 architecture. Targeted disruption of specific ARC:TBM interactions may therefore represent a novel TNKS inhibition strategy that does not target the CAT, and this strategy has recently been shown to be an effective means for inhibiting TNKS Wnt signaling function (48). Importantly, due to the high structural similarity of CAT domains within the PARP family, existing PARP inhibitors can inhibit multiple PARP family members. Indeed, the least selective Food and Drug Administration–approved PARP inhibitor, rucaparib, is a potent TNKS inhibitor (49). The ability to target a domain that is unique to TNKS may facilitate TNKS inhibition without affecting other family members, allowing for a better understanding of the cellular functions of TNKS as well as the physiological effects of TNKS inhibition therapy.
Experimental procedures
Gene cloning
Human TNKS constructs were expressed from a synthetic TNKS gene (GenScript) in pET47 (ARC1–5, residues 174–961) or pET50 (NS-TNKS, residues 174–1327) expression vectors. Human GMD and TRF1 were expressed in pET28 and were kind gifts from Dr. Susan Smith (New York University) and Dr. David Chen (University of Texas Southwestern), respectively. TNKS, TRF1, and GMD mutants were created using QuikChange mutagenesis. Human HPF1 was produced as a fusion to an N-terminal SUMO tag from a pET28 expression vector (BioBasic). Human PARG (residues 488–976) was produced from a pET28b expression vector, a kind gift from Dr. Ivan Ahel (University of Oxford, Cambridge, UK).
Protein and peptide preparation
ARC1–5 constructs were expressed as His6 fusion proteins as described previously as was fluorescently labeled Axin peptide (16). NS-TNKS constructs were expressed as NusA-SUMO (SUMO)-tagged fusion proteins in Escherichia coli strain BL21(DE3). Cells were grown to 0.8 OD in LB medium with 10 mm benzamide and induced with 200 μm β-d-thiogalactopyranoside at 16 °C for 20 h. Cells were pelleted and resuspended in 25 mm HEPES, pH 8.0, 500 mm NaCl, 0.5 mm tris(2-carboxyethyl)phosphine (TCEP), and 5% glycerol and stored at −20 °C. Cell pellets were thawed in a room temperature water bath and adjusted to 0.1% Nonidet P-40 (NP-40), and the following protease inhibitors: 1 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin A, 0.5 μg/ml antipain, and 0.5 μg/ml aprotinin. Samples were lysed using a cell homogenizer (Avestin) and then spun for 2 h at 40,000 × g at 4 °C. Supernatants were loaded onto a 5-ml HP chelating column (GE Healthcare) charged with Ni(II) pre-equilibrated with lysis buffer without NP-40. The column was washed with a low-salt buffer (500 mm NaCl, 25 mm HEPES, pH 8.0, 0.5 mm TCEP, 20 mm imidazole, 5% glycerol, and protease inhibitors), high-salt buffer (1 m NaCl, 25 mm HEPES, pH 8.0, 0.5 mm TCEP, 20 mm imidazole, 5% glycerol, and protease inhibitors), and low-salt buffer again and eluted with elution buffer (500 mm NaCl, 25 mm HEPES, pH 8, 0.5 mm TCEP, 400 mm imidazole, 5% glycerol, and protease inhibitors). Samples were then diluted 1:1 in 50 mm Tris, pH 7.5, 5% glycerol, 1 mm EDTA, and 0.1 mm TCEP and loaded onto a 5-ml HP heparin column (GE Healthcare) equilibrated with 250 mm NaCl, 25 mm Tris, pH 7.5, 5% glycerol, 0.1 mm TCEP, and 1 mm EDTA. Proteins were eluted with an increasing gradient of NaCl, pooled, concentrated, and stored at −80 °C. Protein concentrations were measured using absorbance at 280 nm and confirmed visually on SDS-PAGE versus a BSA standard. GMD and TRF1 constructs were expressed in E. coli strain Rosetta II (DE3), grown, and pelleted as with the NS-TNKS constructs although without the addition of benzamide. Cells expressing GMD and TRF1 were lysed and run over a nickel column the same as with NS-TNKS. TRF1 purification also included heparin chromatography. TRF1 and GMD were concentrated and then passed over a Sephacryl 200 gel filtration column (GE Healthcare) in 20 mm HEPES, pH 8.0, 150 mm NaCl, 0.1 mm TCEP, 0.1 mm EDTA, and 5% glycerol. Fractions containing TRF1 or GMD were pooled, mixed with SUMO-protease ULP1, and dialyzed for a minimum of 2 × 2 h against 2 liters of 25 mm HEPES, pH 8.0, 150 mm NaCl, 30 mm imidazole, 0.1 mm TCEP, and 5% glycerol at 4 °C. Samples were then loaded onto a 5-ml chelating column pre-equilibrated with dialysis buffer, and untagged proteins were collected in the flow-through. Proteins were then concentrated as with NS-TNKS. TNKS-12345:GMD complexes were obtained by simultaneously expressing both proteins in Rosetta II cells. Stable complexes were purified by chelating column and size-exclusion chromatography and concentrated as described above. PARG was expressed in BL21(DE3) cells and purified over Ni(II) and gel-filtration columns as described above. SUMO-HPF1 was expressed in E. coli Rosetta II cells and purified over a Ni(II) column as described for TNKS above but in the absence of glycerol. The SUMO tag was then digested using ULP1 protease, and the cut sample was passed again over a Ni(II) column. The untagged HPF1 that flowed through the column was collected and diluted to 50 mm NaCl and then loaded consecutively onto heparin and gel-filtration columns as described above. PARP-1 constructs were expressed and purified as described previously (8). Fluorescently labeled and unlabeled TRF1 (N-APSPRGCADGRDADPT-C) and GMD (N-CPSARGSGDGEMGKPR-C) were ordered from GenScript. Lyophilized peptides were resuspended in 20 mm HEPES, pH 8.0, 100 mm NaCl, 0.1 mm TCEP, and 1 mm EDTA and then stored at −80 °C. Purified TARG1 and ARH3 proteins were a kind gift from Dr. Ivan Ahel and purified as described previously (36, 50).
Pulldown binding analysis
ARC1–5 constructs (1 μm) were mixed with TRF1 (1 μm) or GMD (2 μm) constructs in a buffer consisting of 25 mm HEPES, pH 8.0, 150 mm NaCl, 0.1 mm TCEP, 5% glycerol, and 0.2% NP-40 and incubated for 30 min at room temperature. Imidazole was added to 50 mm final concentration, and samples were loaded onto 10 μl Ni(II)-Sepharose beads in Wizard mini spin columns (Promega) pre-equilibrated with 25 mm HEPES, pH 8.0, 500 mm NaCl, 0.1 mm TCEP, 5% glycerol, 0.2% NP-40, and 50 mm imidazole. Samples were incubated for 10 min at room temperature and then spun for 1 min at 10,000 × g. Samples were then washed four times by incubating in equilibration buffer for 5 min at room temperature followed 1 min spins at 10,000 × g. Samples were treated with 25 mm HEPES, pH 8.0, 500 mm NaCl, 0.1 mm TCEP, 5% glycerol, 0.2% NP-40, and 400 mm imidazole; incubated for 10 min; and eluted by spinning down. The flow-through was then resolved by SDS-PAGE. TNKS and binding partner densities were quantified using ImageJ software (51). Binding partner density was normalized to the density of the TNKS construct in the same pulldown reaction. All pulldown data represent results from a minimum of three experiments.
Analytical gel filtration
Proteins were diluted to 3 mg/ml in 20 mm HEPES, pH 8.0, 0.1 mm TCEP, 0.1 mm EDTA, and 5% glycerol. Samples were then spun for 10 min at top speed on a tabletop centrifuge at 4 °C, and 100 μl of each sample was loaded onto a Superdex 200 10/300 column. Molecular weight estimates were determined from a calibration curve derived from the apparent elution volumes of gel-filtration standards (Bio-Rad).
PARP activity assay Western blotting
PARP-1 (0.25 μm) constructs and NS-TNKS (0.5 μm) constructs were mixed with NAD+ (250 μm and 5 mm, respectively) with equimolar concentrations of DNA (18-bp oligonucleotide) or 0.5 or 1 μm of TRF1/GMD constructs (see figure legends), respectively, in a buffer consisting of 20 mm HEPES, pH 8.0, 150 mm NaCl, 0.1 mm TCEP, 0.1 mm EDTA, 5% glycerol, and 5 mm MgCl2. Mixtures were incubated for 30 min and quenched with addition of 6× concentrated Laemmli sample buffer. A His6-tagged ARC1–3 (residues 174–649) construct was included after quenching with Laemmli sample buffer at 0.2 μm as a loading and transfer control. Reactions were incubated for 10 min at 100 °C and resolved by 7.5% SDS-PAGE. The gel was transferred onto nitrocellulose membranes (Pierce) and blocked for 1 h in Tris-buffered saline with Tween (TBST; 20 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Tween 20) supplemented with 5% evaporated cow's milk. Blots were incubated overnight with 1:2000 mouse anti-His6 (Invitrogen) and 1:3000 rabbit anti-PAR (Trevigen) antibodies at 4 °C overnight. Blots were washed with TBST and Tris-buffered saline (20 mm Tris, pH 7.5, and 150 mm NaCl) and then incubated with 1:15,000 IRDye®800CW donkey anti-rabbit (LI-COR Biosciences) and 1:20000 IRDye680 goat anti-mouse (LI-COR Biosciences) for 1 h at room temperature. Blots were washed again and imaged on a LI-COR Odyssey gel imager (ODY-2477). Hydrolase cleavage assays were performed by mixing the PARP-1 (mixture described above) or NS-TNKS(E1291Q) (2 μm NS-TNKS(E1291Q), 2 μm TRF1, and 20 mm NAD+) mixtures and incubating them for 30 min at room temperature. The reactions were quenched with 3.5 μm rucaparib and incubated for 10 min. Hydrolase (PARG, TARG1, or ARH3) was added at 1 μm. Reactions containing PARG were quenched after 30 min with Laemmli sample buffer, and time series reactions containing TARG1 or ARH3 were quenched at the times indicated. The samples were resolved, transferred, and developed as before except a MAR-binding reagent fused to a rabbit Fc tag (Millipore, catalog number MABE1076) was used at a dilution of 1:2000 instead of the rabbit anti-PAR. PAR and MAR densities were quantified using ImageJ software. All data represent a minimum of three separate experiments.
Fluorescence polarization binding and competition assays
Binding and competition reactions were performed and fit as described previously (16). Binding was classified as nonquantifiable (NQ) if the binding curve did not allow for a robust KD determination. TNKS variants that exhibited NQ binding were analyzed in at least two separate experiments, and all other KD values were determined from a minimum of three separate experiments. Competition experiments contained 27 nm Axin1 peptide and 1.5 μm TNKS-12345. Competitor concentrations represent multimers or monomers, as designated in figure legends. All competition data represent a minimum of three experiments.
Isothermal titration calorimetry
All titrations were performed at 22 °C using a Nano-ITC (TA Instruments). Protein samples were dialyzed twice against 500 ml of a buffer containing 50 mm Tris, pH 8.5, 150 mm NaCl, 2 mm TCEP, and 5% glycerol at 4 °C. TNKS-12345 was loaded into the 350-μl cell at 5 μm and titrated with 45 μm GMD (tetramer concentration). The titration began with a 0.4-μl injection followed by 24 subsequent injections of 2 μl at 3-min intervals. The data were analyzed, and thermodynamic parameters were determined using NanoAnalyze software (TA Instruments) with a one-site independent model. GMD ITC data represent three separate experiments.
Analytical ultracentrifugation
AUC samples were dialyzed for 2 × 2 h in 500 ml of buffer containing 50 mm Tris, pH 8.5, 150 mm NaCl, 2 mm TCEP, and 5% glycerol at 4 °C. All samples except TNKS-12345 and TRF1 mixtures were spun at 10 °C in a microcentrifuge at maximum speed for 10 min prior to loading. Samples were analyzed in an XL-I analytical centrifuge (Beckman) in a 50Ti rotor pre-equilibrated at 10 °C. Sedimentation velocity analysis was performed in two-sector cells with quartz windows. Absorbance scans at 280 nm were taken at ∼3-min intervals for ∼10 h while spinning at 50,000 rpm. Continuous sedimentation coefficient and c(S) plots and frictional ratios (f/f0) were generated using SEDFIT by fitting the Lamm equation to the absorbance boundaries (52). Theoretical molecular weights, extinction coefficients, partial specific volumes, buffer density (r, 1.0074 g/cm3), and buffer viscosity (h, 0.01 poise) were calculated using SEDNTERP (43).
Solubility assay
TNKS-12345 and TRF1 constructs were mixed with TNKS-12345 at 0.4 μm and TRF1 added at increasing concentrations and incubated at room temperature for 30 min in 20 mm HEPES, pH 8.0, 0.1 mm TCEP, 0.1 mm EDTA, and 5% glycerol. Samples were then spun for 10 min on a tabletop centrifuge at full speed. The supernatant was removed to a separate tube, and the pellet was resuspended in an equal volume of buffer. Reactions were resolved by 12.5% SDS-PAGE and stained using Imperial protein stain (Pierce). Protein densities were quantified using ImageJ software. Data represent three separate experiments.
Solvent accessibility
Solvent accessibility was calculated using Surface Racer (30) using the Richards' van der Waals radii sets and a probe radius of 1.4 Å. Surface accessibility data from the output files are averaged by amino acid type. Average accessibility was averaged for all consensus structures and compared with the maximum calculated solvent exposure in Tien et al. (31).
HPF1 assay
PARP-1 (1 μm) was incubated with DNA (1 μm) and HPF1 (1 μm) where indicated for 10 min at room temperature in 20 mm Tris, pH 7.5, 50 mm NaCl, 5 mm MgCl2, and 0.1 mm TCEP. NAD+ was added to a final concentration of 0.5 mm for 5 min. Reactions were stopped by the addition of PARP inhibitor veliparib (0.5 mm). Hydroxylamine (1 m) was added when indicated for 3 h at room temperature, and the treatment was quenched by the addition of Laemmli sample buffer. The reactions were resolved by 12% SDS-PAGE and visualized using Imperial stain. NS-TNKS reactions were performed in a similar manner but using 0.35 μm NS-TNKS and TRF1 and with an incubation time of 30 min for the initial mixtures followed by a 30-min incubation with 2.5 mm NAD+. The reactions were resolved by SDS-PAGE, transferred onto a nitrocellulose membrane, and detected using the anti-pan-ADP-ribose binding reagent from Millipore (MABE1016) at a 1:1500 dilution.
Author contributions
T.E. and J.M.P. conceptualization; T.E. formal analysis; T.E. and M.-F.L. investigation; T.E. visualization; T.E., M.-F.L., and J.M.P. methodology; T.E. writing-original draft; M.-F.L. resources; M.-F.L. and J.M.P. writing-review and editing; J.M.P. supervision; J.M.P. funding acquisition; J.M.P. project administration.
Supplementary Material
Acknowledgments
This manuscript includes work carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interactions facility, which is supported in part by NCI, National Institutes of Health Grant P30 CA56036 and NIGMS, National Institutes of Health Grant OD017987. We thank Dr. Susan Smith and Dr. David Chen for the kind gifts of the GMD and TRF1 plasmids, respectively. We thank Dr. Ivan Ahel for the kind gift of the human PARG expression plasmid and purified TARG1 and ARH3 proteins. We also thank Dr. Karen Knudsen for access to the LI-COR Biosciences gel imager.
This work was supported in part by Canadian Institutes of Health Research Grant PJT374609 (to J. M. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9 and Table S1.
- TNKS
- tankyrase-1
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- TRF1
- telomeric repeat-binding factor 1
- GMD
- GDP-mannose 4,6-dehydratase
- MAR
- mono(ADP-ribose)
- CAT
- catalytic
- HPS
- histidine-, proline-, and serine-rich
- SAM
- sterile α motif
- TNKS2
- tankyrase-2
- ARC
- ankyrin repeat cluster
- TBM
- TNKS-binding motif
- SUMO
- small ubiquitin-like modifier
- NS
- NusA-SMT
- 5FAM
- 5-carboxyfluorescein
- FP
- fluorescence polarization
- Pos.
- position
- SV-AUC
- sedimentation velocity analytical ultracentrifugation
- ITC
- isothermal titration calorimetry
- HPF1
- histone PARylation factor 1
- TRF1-D
- TRF1 dimerization domain
- PARG
- poly(ADP-ribose) glycohydrolase
- TARG1
- terminal ADP-ribose protein glycohydrolase 1
- ARH3
- ADP-ribosyl hydrolase 3
- TCEP
- tris(2-carboxyethyl)phosphine
- NP-40
- Nonidet P-40
- NQ
- nonquantifiable
- R1
- arginine at position 1 of TBM
- G6
- glycine at position 6 of TBM.
References
- 1. Amé J. C., Spenlehauer C., and de Murcia G. (2004) The PARP superfamily. Bioessays 26, 882–893 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- 2. Smith S. (2001) The world according to PARP. Trends Biochem. Sci. 26, 174–179 10.1016/S0968-0004(00)01780-1 [DOI] [PubMed] [Google Scholar]
- 3. Chi N. W., and Lodish H. F. (2000) Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275, 38437–38444 10.1074/jbc.M007635200 [DOI] [PubMed] [Google Scholar]
- 4. Huang S. M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., et al. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- 5. Bae J., Donigian J. R., and Hsueh A. J. (2003) Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 278, 5195–5204 10.1074/jbc.M201988200 [DOI] [PubMed] [Google Scholar]
- 6. Smith S., Giriat I., Schmitt A., and de Lange T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 10.1126/science.282.5393.1484 [DOI] [PubMed] [Google Scholar]
- 7. Smith S., and de Lange T. (2000) Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302 10.1016/S0960-9822(00)00752-1 [DOI] [PubMed] [Google Scholar]
- 8. Langelier M. F., Planck J. L., Roy S., and Pascal J. M. (2012) Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 10.1126/science.1216338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langelier M. F., and Pascal J. M. (2013) PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 23, 134–143 10.1016/j.sbi.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Rycker M., Venkatesan R. N., Wei C., and Price C. M. (2003) Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 372, 87–96 10.1042/bj20021450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Rycker M., and Price C. M. (2004) Tankyrase polymerization is controlled by its sterile α motif and poly(ADP-ribose) polymerase domains. Mol. Cell. Biol. 24, 9802–9812 10.1128/MCB.24.22.9802-9812.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mariotti L., Templeton C. M., Ranes M., Paracuellos P., Cronin N., Beuron F., Morris E., and Guettler S. (2016) Tankyrase requires SAM domain-dependent polymerization to support Wnt-β-catenin signaling. Mol. Cell 63, 498–513 10.1016/j.molcel.2016.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Riccio A. A., McCauley M., Langelier M. F., and Pascal J. M. (2016) Tankyrase sterile α motif domain polymerization is required for its role in Wnt signaling. Structure 24, 1573–1581 10.1016/j.str.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sbodio J. I., and Chi N. W. (2002) Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1: NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277, 31887–31892 10.1074/jbc.M203916200 [DOI] [PubMed] [Google Scholar]
- 15. Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., and Sicheri F. (2011) Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 147, 1340–1354 10.1016/j.cell.2011.10.046 [DOI] [PubMed] [Google Scholar]
- 16. Eisemann T., McCauley M., Langelier M. F., Gupta K., Roy S., Van Duyne G. D., and Pascal J. M. (2016) Tankyrase-1 ankyrin repeats form an adaptable binding platform for targets of ADP-ribose modification. Structure 24, 1679–1692 10.1016/j.str.2016.07.014 [DOI] [PubMed] [Google Scholar]
- 17. Bisht K. K., Dudognon C., Chang W. G., Sokol E. S., Ramirez A., and Smith S. (2012) GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity. Mol. Cell. Biol. 32, 3044–3053 10.1128/MCB.00258-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morrone S., Cheng Z., Moon R. T., Cong F., and Xu W. (2012) Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc. Natl. Acad. Sci. U.S.A. 109, 1500–1505 10.1073/pnas.1116618109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DaRosa P. A., Klevit R. E., and Xu W. (2018) Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs. Protein Sci. 27, 1057–1067 10.1002/pro.3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bianchi A., Smith S., Chong L., Elias P., and de Lange T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794 10.1093/emboj/16.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang W., Dynek J. N., and Smith S. (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17, 1328–1333 10.1101/gad.1077103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Becker D. J., and Lowe J. B. (2003) Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R 10.1093/glycob/cwg054 [DOI] [PubMed] [Google Scholar]
- 23. Seimiya H., Muramatsu Y., Smith S., and Tsuruo T. (2004) Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol. Cell. Biol. 24, 1944–1955 10.1128/MCB.24.5.1944-1955.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiang Y. J., Hsiao S. J., Yver D., Cushman S. W., Tessarollo L., Smith S., and Hodes R. J. (2008) Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One 3, e2639 10.1371/journal.pone.0002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sbodio J. I., Lodish H. F., and Chi N. W. (2002) Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361, 451–459 10.1042/0264-6021:3610451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cook B. D., Dynek J. N., Chang W., Shostak G., and Smith S. (2002) Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22, 332–342 10.1128/MCB.22.1.332-342.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allen J. G., Mujacic M., Frohn M. J., Pickrell A. J., Kodama P., Bagal D., San Miguel T., Sickmier E. A., Osgood S., Swietlow A., Li V., Jordan J. B., Kim K. W., Rousseau A. C., Kim Y. J., Caille S., et al. (2016) Facile modulation of antibody fucosylation with small molecule fucostatin inhibitors and cocrystal structure with GDP-mannose 4,6-dehydratase. ACS Chem. Biol. 11, 2734–2743 10.1021/acschembio.6b00460 [DOI] [PubMed] [Google Scholar]
- 28. Fairall L., Chapman L., Moss H., de Lange T., and Rhodes D. (2001) Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8, 351–361 10.1016/S1097-2765(01)00321-5 [DOI] [PubMed] [Google Scholar]
- 29. Bonfiglio J. J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., and Matic I. (2017) Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e6 10.1016/j.molcel.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsodikov O. V., Record M. T. Jr., and Sergeev Y. V. (2002) Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 23, 600–609 10.1002/jcc.10061 [DOI] [PubMed] [Google Scholar]
- 31. Tien M. Z., Meyer A. G., Sydykova D. K., Spielman S. J., and Wilke C. O. (2013) Maximum allowed solvent accessibilities of residues in proteins. PLoS One 8, e80635 10.1371/journal.pone.0080635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crawford K., Bonfiglio J. J., Mikoč A., Matic I., and Ahel I. (2018) Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 53, 64–82 10.1080/10409238.2017.1394265 [DOI] [PubMed] [Google Scholar]
- 33. Fontana P., Bonfiglio J. J., Palazzo L., Bartlett E., Matic I., and Ahel I. (2017) Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife 6, e28533 10.7554/eLife.28533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marsischky G. T., Wilson B. A., and Collier R. J. (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation: evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 270, 3247–3254 10.1074/jbc.270.7.3247 [DOI] [PubMed] [Google Scholar]
- 35. Rolli V., O'Farrell M., Ménissier-de Murcia J., and de Murcia G. (1997) Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 36, 12147–12154 10.1021/bi971055p [DOI] [PubMed] [Google Scholar]
- 36. Sharifi R., Morra R., Appel C. D., Tallis M., Chioza B., Jankevicius G., Simpson M. A., Matic I., Ozkan E., Golia B., Schellenberg M. J., Weston R., Williams J. G., Rossi M. N., Galehdari H., et al. (2013) Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 10.1038/emboj.2013.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li B., Qiao R., Wang Z., Zhou W., Li X., Xu W., and Rao Z. (2016) Crystal structure of a tankyrase 1-telomere repeat factor 1 complex. Acta Crystallogr. F Struct. Biol. Commun. 72, 320–327 10.1107/S2053230X16004131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fan C., Yarravarapu N., Chen H., Kulak O., Dasari P., Herbert J., Yamaguchi K., Lum L., and Zhang X. (2018) Regulation of tankyrase activity by a catalytic domain dimer interface. Biochem. Biophys. Res. Commun. 503, 1780–1785 10.1016/j.bbrc.2018.07.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chapman J. D., Gagné J. P., Poirier G. G., and Goodlett D. R. (2013) Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12, 1868–1880 10.1021/pr301219h [DOI] [PubMed] [Google Scholar]
- 40. Lupo B., and Trusolino L. (2014) Inhibition of poly(ADP-ribosyl) ation in cancer: Old and new paradigms revisited. Biochim. Biophys. Acta 1846, 201–215 10.1016/j.bbcan.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 41. Distler A., Deloch L., Huang J., Dees C., Lin N. Y., Palumbo-Zerr K., Beyer C., Weidemann A., Distler O., Schett G., and Distler J. H. (2013) Inactivation of tankyrases reduces experimental fibrosis by inhibiting canonical Wnt signalling. Ann. Rheum. Dis. 72, 1575–1580 10.1136/annrheumdis-2012-202275 [DOI] [PubMed] [Google Scholar]
- 42. Wang C., Zhu H., Sun Z., Xiang Z., Ge Y., Ni C., Luo Z., Qian W., and Han X. (2014) Inhibition of Wnt/β-catenin signaling promotes epithelial differentiation of mesenchymal stem cells and repairs bleomycin-induced lung injury. Am. J. Physiol. Cell Physiol. 307, C234–C244 10.1152/ajpcell.00366.2013 [DOI] [PubMed] [Google Scholar]
- 43. Laue T. M., Shah B., Ridgeway T. M., Pelletier S. L., Harding S. E., Rowe A. J., and, Horton J. C. (1992) Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., and Horton J. C., eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 44. Seimiya H., Muramatsu Y., Ohishi T., and Tsuruo T. (2005) Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell 7, 25–37 10.1016/j.ccr.2004.11.021 [DOI] [PubMed] [Google Scholar]
- 45. Steffen J. D., Brody J. R., Armen R. S., and Pascal J. M. (2013) Structural implications for selective targeting of PARPs. Front. Oncol. 3, 301 10.3389/fonc.2013.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lupo B., Vialard J., Sassi F., Angibaud P., Puliafito A., Pupo E., Lanzetti L., Comoglio P. M., Bertotti A., and Trusolino L. (2016) Tankyrase inhibition impairs directional migration and invasion of lung cancer cells by affecting microtubule dynamics and polarity signals. BMC Biol. 14, 5 10.1186/s12915-016-0226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kulak O., Chen H., Holohan B., Wu X., He H., Borek D., Otwinowski Z., Yamaguchi K., Garofalo L. A., Ma Z., Wright W., Chen C., Shay J. W., Zhang X., and Lum L. (2015) Disruption of Wnt/β-catenin signaling and telomeric shortening are inextricable consequences of tankyrase inhibition in human cells. Mol. Cell. Biol. 35, 2425–2435 10.1128/MCB.00392-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu W., Lau Y. H., Fischer G., Tan Y. S., Chattopadhyay A., de la Roche M., Hyvönen M., Verma C., Spring D. R., and Itzhaki L. S. (2017) Macrocyclized extended peptides: inhibiting the substrate-recognition domain of tankyrase. J. Am. Chem. Soc. 139, 2245–2256 10.1021/jacs.6b10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thorsell A. G., Ekblad T., Karlberg T., Löw M., Pinto A. F., Trésaugues L., Moche M., Cohen M. S., and Schüler H. (2017) Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J. Med. Chem. 60, 1262–1271 10.1021/acs.jmedchem.6b00990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kernstock S., Koch-Nolte F., Mueller-Dieckmann J., Weiss M. S., and Mueller-Dieckmann C. (2006) Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of human ARH3, the first eukaryotic protein-ADP-ribosylhydrolase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 224–227 10.1107/S1744309106003435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abràmoff M. D., Magalhães P. J., and Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 52. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.