Thermococcus kodakarensis is a hyperthermophilic archaeon that can grow at 60 to 100°C. The sequence of tRNATrp from this archaeon was determined by liquid chromatography/mass spectrometry. Fifteen types of modified nucleoside were observed at 21 positions, including 5 modifications at novel positions; in addition, methylwyosine at position 37 was newly observed in an archaeal tRNATrp. The construction of trm11 (Δtrm11) and other gene disruptant strains confirmed the enzymes responsible for modifications in this tRNA. The lack of 2-methylguanosine (m2G) at position 67 in the trm11 trm14 double disruptant strain suggested that this position is methylated by Trm14, which was previously identified as an m2G6 methyltransferase. The Δtrm11 strain grew poorly at 95°C, indicating that archaeal Trm11 is required for T. kodakarensis survival at high temperatures.
Keywords: archaea, gene disruption, mass spectrometry, tRNA methyltransferase, tRNA modification, archaea
ABSTRACT
tRNA m2G10/m22G10 methyltransferase (archaeal Trm11) methylates the 2-amino group in guanosine at position 10 in tRNA and forms N2,N2-dimethylguanosine (m22G10) via N2-methylguanosine (m2G10). We determined the complete sequence of tRNATrp, one of the substrate tRNAs for archaeal Trm11 from Thermococcus kodakarensis, a hyperthermophilic archaeon. Liquid chromatography/mass spectrometry following enzymatic digestion of tRNATrp identified 15 types of modified nucleoside at 21 positions. Several modifications were found at novel positions in tRNA, including 2′-O-methylcytidine at position 6, 2-thiocytidine at position 17, 2′-O-methyluridine at position 20, 5,2′-O-dimethylcytidine at position 32, and 2′-O-methylguanosine at position 42. Furthermore, methylwyosine was found at position 37 in this tRNATrp, although 1-methylguanosine is generally found at this location in tRNATrp from other archaea. We constructed trm11 (Δtrm11) and some gene disruptant strains and compared their tRNATrp with that of the wild-type strain, which confirmed the absence of m22G10 and other corresponding modifications, respectively. The lack of 2-methylguanosine (m2G) at position 67 in the trm11 trm14 double disruptant strain suggested that this methylation is mediated by Trm14, which was previously identified as an m2G6 methyltransferase. The Δtrm11 strain grew poorly at 95°C, indicating that archaeal Trm11 is required for T. kodakarensis survival at high temperatures. The m22G10 modification might have effects on stabilization of tRNA and/or correct folding of tRNA at the high temperatures. Collectively, these results provide new clues to the function of modifications and the substrate specificities of modification enzymes in archaeal tRNA, enabling us to propose a strategy for tRNA stabilization of this archaeon at high temperatures.
IMPORTANCE Thermococcus kodakarensis is a hyperthermophilic archaeon that can grow at 60 to 100°C. The sequence of tRNATrp from this archaeon was determined by liquid chromatography/mass spectrometry. Fifteen types of modified nucleoside were observed at 21 positions, including 5 modifications at novel positions; in addition, methylwyosine at position 37 was newly observed in an archaeal tRNATrp. The construction of trm11 (Δtrm11) and other gene disruptant strains confirmed the enzymes responsible for modifications in this tRNA. The lack of 2-methylguanosine (m2G) at position 67 in the trm11 trm14 double disruptant strain suggested that this position is methylated by Trm14, which was previously identified as an m2G6 methyltransferase. The Δtrm11 strain grew poorly at 95°C, indicating that archaeal Trm11 is required for T. kodakarensis survival at high temperatures.
INTRODUCTION
Adaptor molecule tRNA is required for the conversion of genetic information encoded by nucleic acids to amino acid sequences in proteins. Numerous tRNA modifications are needed for sufficient and correct protein synthesis. To date, more than 100 modified nucleosides have been found in tRNAs from various living organisms (1). In particular, tRNAs from hyperthermophiles contain various modified nucleosides (2–6), which are thought to maintain the functions of tRNA at high temperatures. However, there are only a few examples of a tRNA sequence containing modified nucleosides from hyperthermophilic archaea (i.e., Sulfolobus acidocaldarius initiator tRNAMet [7] and, as published during the preparation of this report, Methanocaldococcus jannaschii tRNAs with several modifications mainly found in anticodon-arms in tRNAs [8]). In general, determining the sequence of tRNA from thermophiles is not so easy, because these tRNAs are structurally very rigid and contain numerous modified nucleosides. In some cases, preparation of standard compounds of modified nucleosides is necessary.
In a recent study, we reported the crystal structure of tRNA m2G10/m22G10 methyltransferase from Thermococcus kodakarensis (9), a hyperthermophilic archaeon that grows at 60 to 100°C (10). Archaeal tRNA m2G10/m22G10 methyltransferase catalyzes the transfer of a methyl group from S-adenosyl-l-methionine to the 2-amino group in guanosine at position 10 (G10) in tRNA and forms N2,N2-dimethylguanosine (m22G) via the intermediate N2-methylguanosine (m2G) (11). Although its eukaryotic counterpart (Trm11) requires another subunit (Trm112) (12, 13) for enzymatic activity (14), the archaeal enzyme does not require a partner subunit (11). Furthermore, the eukaryotic Trm11–Trm112 complex catalyzes a single methyl transfer reaction and forms only m2G10 in tRNA. Therefore, the archaeal enzyme has been called Trm-G10 (11) or Trm-m22G10 (15) to distinguish it from the eukaryotic enzyme. In this study, however, we use the name “archaeal Trm11” instead of Trm-G10 or Trm-m22G10 enzyme, owing to the amino acid sequence similarity between the eukaryotic and archaeal enzymes (9).
Many types of modified nucleoside are specifically formed in individual tRNAs, and they are considered to confer various functional hallmarks on tRNA in a coordinated manner. To gain insight into the molecular and physiological roles of m22G10 and Trm11, it is necessary to reveal the complete sequence of substrate tRNAs for Trm11, including other modified nucleosides. In the present work, we therefore determined the complete sequence of tRNATrp isolated from T. kodakarensis and found several modified nucleosides at novel positions that have not been detected in any tRNA reported so far. Furthermore, established genetic manipulation systems for T. kodakarensis (16–20) enabled us to construct a Tk0981 (trm11) gene disruptant strain (Δtrm11) and additional gene disruptant strains responsible for other modified nucleosides. By analyzing tRNATrp from the disruptant strains, we observed the lack of m22G10 in tRNATrp from the Δtrm11 strain and confirmed that corresponding modified nucleosides were absent in individual gene disruptant strains.
We also studied the growth of the trm11 gene disruptant (Δtrm11) strain at high temperatures. We discuss our findings in terms of the stability of tRNA in hyperthermophilic archaea and the survival of these microbes at high temperatures.
RESULTS
Purification and sequencing of tRNATrp from T. kodakarensis.
To determine all modified nucleosides formed in a substrate tRNA for Trm11 of T. kodakarensis, we used tRNATrp as the target tRNA for the following reasons. First, there is only one tRNATrp gene in the genome; therefore, the gene is causally expressed in T. kodakarensis cells. Second, the sequence of tRNATrp differs considerably from that of other tRNA; therefore, it should be purified relatively easily by the solid-phase DNA probe method (21). Third, given that the nucleosides at positions 6 and 26 in tRNATrp are both C (Fig. 1), it was expected that this tRNA would not be methylated by Trm14 (tRNA m2G6 methyltransferase) (22) or Trm1 (tRNA m2G26/m22G26 methyltransferase) (20, 23–25) at the outset of the study. (As described below, we found that Trm14 can methylate a novel residue, G67, in this study.) Fourth, in our previous study, Trm11 of T. kodakarensis was revealed to methylate G at position 10 to m22G by using in vitro transcribed tRNATrp (9), suggesting that cellular tRNATrp is one of the substrates for Trm11 in vivo. We successfully purified tRNATrp by a solid-phase DNA probe method.
FIG 1.

Cloverleaf structure of tRNATrp from T. kodakarensis. The modified nucleosides are defined in Table 1.
The determined nucleoside sequence of tRNATrp is shown by a cloverleaf structure in Fig. 1 with positions numbered in accordance with the system described in reference 26. The modified nucleosides are defined in Table 1, and their structures are available from the Modomics database (http://modomics.genesilico.pl/) (1). The enzymes predicted to be responsible for the modified nucleosides, together with their genes, are given in Table 2.
TABLE 1.
Abbreviations of modified nucleosides used in this study
| Abbreviation | Modified nucleoside |
|---|---|
| m3C | 3-Methylcytidine |
| m4C | N4-Methylcytidine |
| f5Cm | 5-Formyl-2’-O-methylcytidine |
| D | Dihydrouridine |
| m5U | 5-Methyluridine |
| m1G | 1-Methylguanosine |
| m7G | 7-Methylguanosine |
| Ψm | 2’-O-Methylpseudouridine |
| m1Im | 1,2’-O-Dimethylinosine |
| m22Gm | N2,N2,2′-O-Trimethylguanosine |
| s2U | 2-Thiouridine |
| ac6A | N6-Acetyladenosine |
| Cm | 2′-O-Methylcytidine |
| s4U | 4-Thiouridine |
| m22G | N2,N2-Dimethylguanosine |
| Ψ | Pseudouridine |
| G+ | Archaeosine |
| s2C | 2-Thiocytidine |
| Um | 2′-O-Methyluridine |
| m5Cm | 5,2′-O-Dimethylcytidine |
| mimG | Methylwyosine |
| Gm | 2′-O-Methylguanosine |
| m5C | 5-Methylcytidine |
| m5s2U | 5-Methyl-2-thiouridine |
| m1I | 1-Methylinosine |
| m1A | 1-Methyladenosine |
| m2G | N2-Methylguanosine |
| imG-14 | 4-Demethylwyosine |
| imG2 | Isowyosine |
| imG | Wyosine |
| yW-86 | 7-Aminocarboxypropyldemethylwyosine |
| yW-72 | 7-Aminocarboxypropylwyosine |
| Am | 2ʹ-O-Methyladenosine |
TABLE 2.
Predicted enzymes and genes for tRNATrp nucleoside modificationsa
| Modified nucleoside and position | Enzyme(s) or RNA | Predicted gene ID |
|---|---|---|
| Cm6 | Unknown | Unknown |
| s4U8 | ThiI | Tk0366 |
| m22G10 | Archaeal Trm11 | Tk0981 |
| Ψ13 | TruD | Tk2302 |
| G+15 | ArcTGT, ArcS | Tk0760, Tk2156 |
| s2C17 | Unknown | Unknown |
| Um20 | L7Ae, Nop5, archaeal fibrillarin, C/D-box guide RNA | Tk1311, Tk0184, Tk0183, RNA |
| Ψ22 | Unknown | Unknown |
| m5Cm32 | Unknown methyltransferase, archaeal TrmJ | Unknown, Tk1970 |
| Cm34 | L7Ae, Nop5, archaeal fibrillarin, intron (C/D-box guide RNA) | Tk1311, Tk0184, Tk0183, intron |
| mimG37 | Trm5b, TYW1, TYW3, Trm5a | Tk0497, Tk1671, Tk0175, Tk2223 |
| Cm39 | L7Ae, Nop5, archaeal fibrillarin, intron (C/D-box guide RNA) | Tk1311, Tk0184, Tk0183, intron |
| Gm42 | L7Ae, Nop5, archaeal fibrillarin, C/D-box guide RNA | Tk1311, Tk0184, Tk0183, RNA |
| m5C48 | Archaeal Trm4 | Tk0360 |
| m5C49 | Archaeal Trm4 | Tk0360 |
| m5s2U54 | RumA, TtuA?, TtuB?, α | Tk2134, Tk1556?, Tk1093?, α |
| Ψ55 | Pus10 or archaeal Cbf5 | Tk0903 or Tk1509 |
| Cm56 | Trm56 | Tk0060 |
| m1I57 | Archaeal TrmI, unknown deaminase | Tk1328, unknown |
| m1A58 | Archaeal TrmI | Tk1328 |
| m2G67 | Trm14 | Tk1863 |
?, enzymatic activity of the protein has not been confirmed in archaea.
The sequence shown in Fig. 1 was determined by liquid chromatography-mass spectrometry (LC/MS) analysis of digested tRNATrp from the wild-type strain. The base peak chromatograms of tRNATrp fragments derived from digestion with RNase T1 and RNase A are shown in Fig. S1A and B, respectively, in the supplemental material. The nucleoside composition of each fragment was determined by comparing the measured m/z with the m/z calculated from the primary sequence of tRNATrp with possible modifications (Tables 3 and 4). The sequences of the fragments and modification sites were assigned by collision-induced dissociation (CID) (Fig. S1C). Pseudouridine (Ψ), a mass-silent uridine modification, was identified in a similar way, but with derivatization to 1-cyanoethyl Ψ by acrylonitrile treatment prior to RNase digestion (Fig. S1C). In these analyses, Cm32 was found to be further methylated (RNase A-derived fragment 4). We deduced that the second methylation would be a base methylation: m5Cm has been found specifically in thermophilic archaea (2, 4–6). In humans, the ALKBH1 gene is responsible for f5Cm34 formation in tRNALeuCAA (27): in ALKBH1 knockout cells, the intermediate m5Cm34 is found in tRNALeuCAA instead of the final product (f5Cm34). Here, therefore, we used this modified nucleoside (m5Cm) as a standard marker. We purified tRNALeuCAA from human ALKBH1 knockout cells and tRNATrp from T. kodakarensis and digested them to nucleosides, which were then mixed and analyzed by LC/MS (Fig. 2). The dimethylated C in T. kodakarensis tRNATrp was eluted at the same time as the standard m5Cm by LC (Fig. 2, top), and CID analysis showed that the cytosine base is monomethylated (Fig. 2, bottom). On the basis of these results, we concluded that a portion of Cm32 is modified to m5Cm32 in tRNATrp. All modifications were also confirmed by LC/MS analysis of nucleosides derived from complete digestion of tRNATrp (Fig. S2). All of the fragments detected with modifications are listed in Tables 3 and 4.
TABLE 3.
List of fragments of T. kodakarensis tRNATrp after digestion with RNase T1a
| Fragment no. | Fragment sequence | Mol wt | Monoisotopic m/z |
Charge state | |
|---|---|---|---|---|---|
| Calculated | Observed | ||||
| 1 | CmUCmCAmimGACmCCGmCGp | 4,274.682 | 711.439 | 711.439 | –6 |
| 2 | m5s2UΨCmm1Im1AAUCCCCGp | 3,908.532 | 643.412 | 643.414 | –6 |
| 3 | UmCCAΨCAUCGp | 3,173.420 | 633.676 | 633.676 | –5 |
| 3′ | UCCAΨCAUCGp | 3,159.404 | 630.873 | 630.874 | –5 |
| 4 | CCCCCACCAOH (3′ terminal) | 2,731.438 | 1,363.711 | 1,364.713 | –2 |
| 5 | ΨAG+Cs2CUGp | 2,316.295 | 1,157.134 | 1,157.139 | –2 |
| 5′ | ΨAG+CCUGp | 2,300.318 | 1,149.151 | 1,149.149 | –2 |
| 6 | Am5Cm5CGp | 1,330.224 | 664.104 | 664.103 | –2 |
| 6′ | ACm5CGp | 1,316.209 | 657.097 | 657.099 | –2 |
| 7 | m22GUGp | 1,042.162 | 1,041.154 | 1,041.152 | −1 |
| 8 | s4UGp | 685.060 | 684.053 | 684.051 | −1 |
| 9 | Cm2Gp, CmGp | 682.115 | 681.107 | 681.106 | −1 |
| 8′ | UGp | 669.083 | 668.075 | 668.074 | −1 |
| 10 | CGp | 668.099 | 667.091 | 667.089 | −1 |
Partially modified fragments detected in reasonable quantity are indicated. “OH” and “p” indicate the 3′ terminal hydroxyl group and terminal phosphate, respectively.
TABLE 4.
List of fragments of T. kodakarensis tRNATrp after digestion with RNase Aa
| Fragment no. | Fragment sequence | Mol wt | Monoisotopic m/z |
Charge state | |
|---|---|---|---|---|---|
| Calculated | Observed | ||||
| 1 | pGGGGGCmGs4Up (5′ terminal) | 2,809.321 | 1,403.653 | 1,403.651 | –2 |
| 1′ | pGGGGGCmGUp (5′ terminal) | 2,793.344 | 1,395.664 | 1,395.665 | –2 |
| 2 | GGGGm5s2UΨp | 2,040.244 | 1,019.114 | 1,019.115 | –2 |
| 3 | AmimGACmCp | 1,711.308 | 854.646 | 854.650 | –2 |
| 4 | GGGm5CmUp | 1,692.251 | 845.118 | 845.114 | –2 |
| 4′ | GGGCmUp | 1,678.235 | 838.110 | 838.114 | –2 |
| 5 | Cmm1Im1AAUp | 1,659.266 | 828.625 | 828.627 | –2 |
| 6 | GGAm5Cp | 1,356.215 | 677.100 | 677.097 | –2 |
| 6′ | GGACp | 1,342.199 | 670.092 | 670.095 | –2 |
| 7 | GGUmCp | 1,333.188 | 665.586 | 665.588 | –2 |
| 8 | Gm22GUp | 1,042.162 | 1,041.154 | 1,041.155 | −1 |
| 9 | AG+Cp | 1,038.178 | 1,037.171 | 1,037.170 | −1 |
| 7′ | GGUp | 1,014.131 | 1,013.123 | 1,013.124 | −1 |
| 10 | GmCp | 682.115 | 681.107 | 681.107 | −1 |
| 11 | m2GCp | 682.115 | 681.107 | 681.106 | −1 |
| 12 | GΨp | 669.083 | 668.075 | 668.075 | −1 |
| 13 | GCp | 668.099 | 667.091 | 667.091 | −1 |
| 14 | AUp, AΨp | 653.088 | 652.081 | 652.081 | −1 |
| 15 | ACp | 652.104 | 651.097 | 651.097 | −1 |
| 16 | CmCp | 642.109 | 641.101 | 641.102 | −1 |
Partially modified fragments detected in reasonable quantity are indicated. “OH” and “p” indicate the 3′ terminal hydroxyl group and terminal phosphate, respectively.
FIG 2.
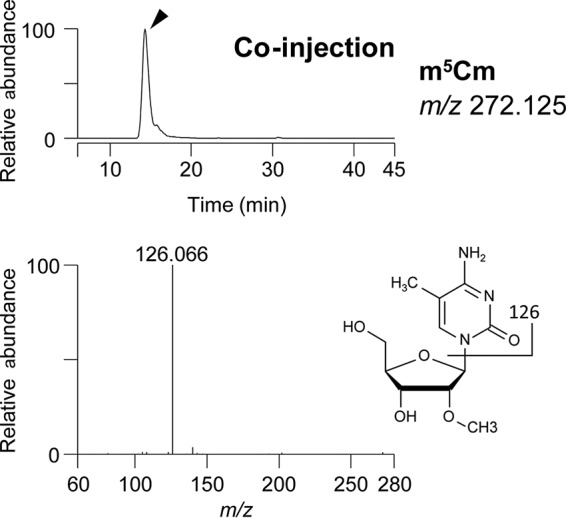
Position 32 is modified to m5Cm in T. kodakarensis tRNATrp. Top, extracted ion chromatography (XIC) showing coelution of the nucleoside modified at position 32 in tRNATrp from T. kodakaraensis and m5Cm in human cytoplasmic tRNALeuCAA from ALKBH1 knockout cells. Bottom, CID spectrum of m5Cm. The cleavage position of the base-related ion is indicated on the chemical structures.
m22G10 formation by Trm11 in vivo and growth phenotype of the trm11 gene disruption.
In the wild-type tRNATrp, m22G was detected in RNase T1-derived fragment 7 and RNase A-derived fragment 8, indicating that m22G is present at position 10. No m2G at position 10, an intermediate of m22G10, was detected in our analysis (data not shown), indicating that m22G10 is efficiently introduced by Trm11 in vivo. To confirm that the trm11 gene is responsible for the m22G10 modification, we constructed a trm11 gene disruptant (Δtrm11) strain (Fig. S3 and S4). The methods for construction of the gene disruptant strain are described in the supplemental material. The m22G nucleoside was not detected in the corresponding fragments of tRNATrp from the Δtrm11 strain (Fig. S5), and the nucleoside at position 10 was confirmed as unmodified G. We therefore concluded that the trm11 gene is responsible for the m22G10 modification in tRNATrp. During the preparation of this paper, it was reported that the m22G content in total tRNA from a T. kodakarensis trm11 gene disruptant strain, which was obtained by transposon random mutagenesis, was decreased relative to that from the wild-type strain (28). Our results provide experimental support for that observation.
We hypothesized that the m22G10 modification might be required for the survival of T. kodakarensis at high temperatures. We therefore measured the growth of the Δtrm11 strain at 85, 90, 93 and 95°C. In addition, we constructed a complemented (Δtrm11 + trm11) strain to confirm that the growth phenotype observed was due to the lack of Trm11. The trm11 gene was reinserted into the chiA (Tk1765; chitinase gene) region in the genomic DNA of the Δtrm11 strain. Deletion of the Tk1765 gene does not cause growth defects unless chitin is used as a carbon source (29). Although its expression level was lower in the complemented strain than in the wild-type strain, Trm11 was expressed in the complemented strain, as determined by Western blotting (Fig. 3A). At 85°C, the wild-type, Δtrm11, and complemented strains showed similar growth curves (Fig. 3B). As the temperature increased, however, the growth of the Δtrm11 strain was clearly slower than that of the wild-type or complemented strain. At 95°C, the Δtrm11 strain showed a considerable growth defect, whereas the complemented strain grew at approximately the same speed as the wild-type strain, indicating that the growth defect of the Δtrm11 strain is due to the lack of archaeal Trm11 protein. The study based on random mutagenesis reported that the trm11 gene product is required for the effective growth of T. kodakarensis at 93°C (28). Although there is a slight difference in the growth speeds between our data and those data at 93°C, this might be due to differences in the culture conditions. Collectively, these observations reveal that Trm11 is required for the survival of T. kodakarensis at high temperatures.
FIG 3.
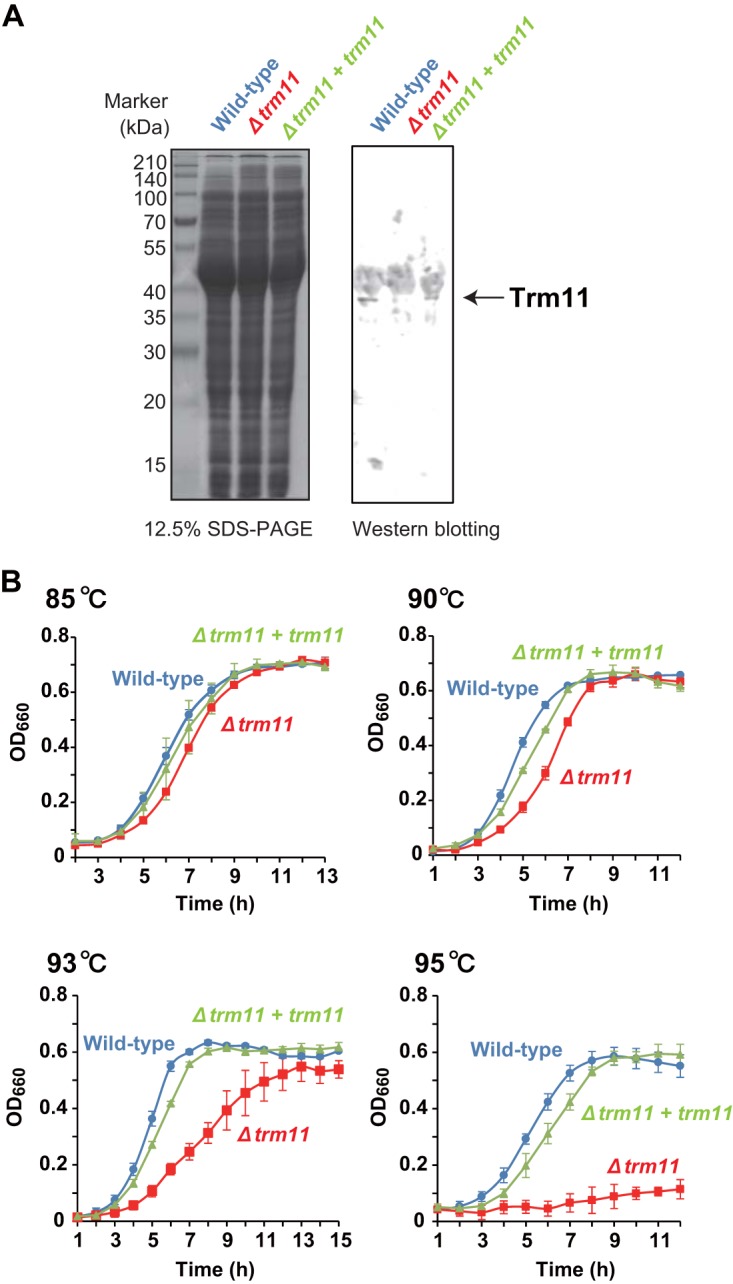
The trm11 gene disruptant strain shows defective growth at high temperature. (A) Western blot confirming the expression of Trm11 protein in the complemented (Δtrm11 + trm11) strain. Left, proteins in the cell extracts from wild-type, Δtrm11, and complemented strains were separated by 12.5% SDS-PAGE. The gel was stained with Coomassie brilliant blue. Right, proteins were transferred to a membrane, and Western blotting was performed. (B) Growth of the wild-type, Δtrm11, and complemented (Δtrm11 + trm11) strains was measured at 85, 90, 93, and 95°C. Error bars indicate the standard deviations of results of three independent culture experiments.
Validation of predicted thiI, rumA, and TYW1 genes.
In general, s4U8 modification in eubacterial and archaeal tRNA is performed by ThiI (30). To determine whether the s4U8 modification in tRNATrp is carried out by ThiI, we analyzed tRNATrp from the ΔthiI strain. Whereas RNase A-derived fragment 1 (pGGGGCmGs4Up) was clearly detected in the wild-type sample (Fig. 4A, left), this fragment was not found in the ΔthiI sample and only RNase A-derived fragment 1′ (pGGGGCmGUp) was detected (Fig. 4A, right). These results confirm that the s4U8 modification in tRNATrp is conferred by ThiI.
FIG 4.

The thiI and rumA genes are responsible for the formation of s4U8 and 5-methylation of U54, respectively, in tRNATrp. (A) XICs of RNase A-digested fragments containing s4U (top) or U (bottom) at position 8 (arrowheads) are shown. The sequences, m/z, and charge states are indicated on the right. n.d., not detected. (B) XICs of an RNase A-digested fragment containing m5s2U (top) or s2U (bottom) at position 54 (arrowheads). The sequence, m/z, values, and charge states are indicated on the right. (C) CID spectrum of the RNase A-derived fragment from the ΔrumA strain. The sequence and assigned signals are shown in the inset (precursor, doubly charged ions of m/z 1,012.1).
S-Adenosyl-l-methionine-dependent tRNA m5U54 methyltransferase activity was previously detected in the cell extract of Pyrococcus furiosus (31), and the responsible rumA-like gene was identified from Pyrococcus abyssi and T. kodakarensis (32). To determine whether the rumA gene (Tk2134) is responsible for the 5-methylation of U54 in T. kodakarensis, we analyzed tRNATrp from the ΔrumA strain. RNase T1-derived fragment 2 (GGGGm5s2UΨp) was detected in the wild-type sample (Fig. 4B, left) but not in the ΔrumA sample, which instead contained a new fragment (GGGGs2UΨp) (Fig. 4B, right, and C). This finding indicated that the rumA gene is responsible for the m5U54 modification and also that s2U54 formation is not dependent on the presence of a 5-methyl group in m5U54.
The mimG (33) nucleoside is one of the final products of the biosynthetic pathway of archaeal wyosine derivatives (Fig. 5A) (34, 35). Traditionally, mimG was thought to exist only in tRNAPhe. Recently, however, it was reported that imG-14 and imG are present at position 37 in several tRNAs from M. jannaschii (8). In that study, the modified nucleoside at position 37 in tRNATrp from M. jannaschii was determined to be m1G37 (8). In our study, however, LC/MS analysis indicated the presence of a modified nucleoside corresponding to mimG (m/z 350.146) at position 37 of tRNATrp. To confirm the presence of mimG in tRNATrp, we analyzed tRNATrp from a ΔTYW1 strain in which the gene encoding TYW1, the enzyme catalyzing the second step of mimG synthesis, was disrupted. We considered that if the modified nucleoside at position 37 is mimG, then m1G37, the first product of the mimG synthesis pathway catalyzed by archaeal Trm5b (Fig. 5A) (36–38), should be detected in tRNATrp from the ΔTYW1 strain. As expected, the modified nucleoside corresponding to mimG (m/z 350.146) was not observed in the nucleosides from the digested tRNATrp from the ΔTYW1 strain (Fig. 5B). Furthermore, RNase A-derived fragment 3 (AmimGACmCp) disappeared and a new RNase A-derived fragment (Am1GACmCp) appeared (Fig. 5C). Taking the results altogether, we concluded that mimG37 is present in tRNATrp from T. kodakarensis.
FIG 5.

Methylwyosine is present at position 37 in tRNATrp. (A) Predicted biosynthetic pathway of wyosine derivatives in T. kodakarensis. This figure is based on data from a report by de Crécy-Lagard et al. (34). The abbreviations of modified nucleotides are listed in Table 1. The predicted enzymes are indicated. (B) Nucleoside analysis of tRNATrp from wild-type and ΔTYW1 strains. mimG is not observed in the ΔTYW1 sample. (C) In the RNase A fragment from the ΔTYW1 strain, m1G is observed at position 37 instead of mimG37. Asterisks show other eluates with almost the same m/z values. n.d., not detected.
The trm14 gene is responsible for the m2G67 modification.
The m2G67 modification was previously found in tRNALys from Loligo bleekeri (39). Furthermore, it has been reported that tRNAArg, tRNAAsn, tRNAGly, tRNAIle, and tRNAVal from M. jannaschii contain m2G67 (8). The modification site (G67) forms a Watson-Crick base pair with C6 in tRNA. Archaeal Trm14 methylates G6 in tRNA and contains a THUMP domain (22, 40), which often recognizes the CCA terminus in tRNA (9, 41, 42). Therefore, we considered that Trm14 may be responsible for the m2G67 modification in tRNATrp. To test this idea, we analyzed tRNATrp from the trm11 trm14 double disruptant (Δtrm11 Δtrm14) strain (Fig. S6): the construction of the Δtrm11 Δtrm14 strain is described in the supplemental text. As shown in Fig. 6, the m2G nucleoside (Fig. 6A) and the RNA fragment (m2GCp) (Fig. 6B) completely disappeared in the sample from the Δtrm11 Δtrm14 strain, demonstrating that the Trm14 is responsible for the m2G67 modification in tRNA.
FIG 6.

The trm14 gene is responsible for m2G67 formation. (A) Nucleoside analysis of tRNATrp from wild-type (WT) and Δtrm11 Δtrm14 (double disruptant) strains. m22G and m2G are absent in the double disruptant strain. (B) XICs tracing an RNase A-digested fragment containing m2G at position 67 (arrowhead). The sequences, m/z, and charge states are indicated on the right. Asterisks show GmCp (Table 4) with the same m/z value as the m2GCp fragment. n.d.; not detected.
DISCUSSION
Our present study revealed the complete sequence of tRNATrp from T. kodakarensis as the first instance of this species. The result that 15 modified nucleosides were found at 21 positions provides insight into their molecular function and their modifying genes or enzymes. Indeed, we successfully confirmed that trm11 is the gene responsible for m22G at position 10 as well as thiI for s4U, rumA for m5U, TYW1 for imG-14, and trm14 for m2G at positions 8, 54, 37, and 67, respectively, by analysis of tRNATrp from gene disruptant strains. Notably, the requirement of trm14 for m2G67 formation has not previously been reported. The functional features and biogenesis of modified nucleosides in the tRNATrp are discussed below in detail.
To our knowledge, Cm6 has not previously been found in tRNAs from archaea, eubacteria, and eukaryotes. However, Am6 formation activity was previously detected in the cell extract of Pyrococcus furiosus (24). Therefore, a novel tRNA 2′-O-methyltransferase, which methylates the 2′-OH of ribose at position 6 in tRNA and does not differentiate between adenine and cytosine, may exist in Thermococcus and Pyrococcus genera. In terms of the other enzymes responsible for the observed 2ʹ-O-methylations, Cm56 is a product of Trm56 (43, 44). The Um20 and Gm42 modifications are likely to be products of L7Ae, Nop5, archaeal fibrillarin (aFib), and the C/D-box guide RNA system (45, 46), with the following predicted C/D-box RNAs: 5′-CCU GAU GAU GAG UAA ACC CGU UGC UGA GAA AAA GAU GAU GAU GGA UGG ACC AGC UGA CC-3′ (coding region, positions 159454 to 159512) for Um20, and 5′-CGG GAU GAU GAG UCU GGA GCC CCC UGA GAG GUG AAG AGG UUU CGC GGG GCU GAC C-3′ (coding region, positions 1371729 to 1371783) for Gm42 (underlining indicates the sequences of the C, D′, C′, and D boxes). Furthermore, Cm34 and Cm39 are also products of L7Ae, Nop5, aFib, and the C/D-box guide RNA system. In this case, an intron in precursor tRNATrp functions as the guide RNA (47–49). Notably, the gene of T. kodakarensis tRNATrp contains a similar intron (50). 2′-O-Methylated nucleosides at multiple positions in tRNA can stabilize the tRNA structure (6). For example, Pyrodictium occultum can grow at 105°C, and various 2′-O-methylated nucleosides such as 2′-O-methylpseudouridine (Ψm), 1,2′-O-dimethylinosine (m1Im), and N2,N2,2′-O-trimethylguanosine (m22Gm) are present in tRNA from this archaeon: however, 2-thiouridine (s2U) and 5-methyl-2-thiouridine (m5s2U) are not found (2, 51). Whereas the melting temperature of the P. occultum tRNAMet transcript is 80°C, that of the native tRNAMet is more than 100°C (52), indicating that the melting temperature of P. occultum tRNA is increased by more than 20°C via a combination of numerous 2′-O-methylated nucleosides. In general, 2′-O-methylation shifts the equilibrium of ribose puckering to the C3′-endo form and enhances the hydrophobic interaction. Thus, 2′-O-methylation is one of the strategies to maintain tRNA structure at high temperatures.
The m5Cm modification has been considered to be specific to thermophilic archaea (2, 4–6). So far, the only exception in mesophiles is the intermediate of f5Cm34 synthesis observed in human tRNALeuCAA. For a long time, however, the position of m5Cm in tRNA from thermophilic archaea has remained unclear, and, to our knowledge, our study is the first to clarify the presence of m5Cm at position 32 in tRNA from these microbes. The 2′-O-methylation of m5Cm32 is probably performed by archaeal TrmJ. The substrate RNA specificity of S. acidocaldarius TrmJ was previously investigated using several mutant tRNA transcripts (53); that study suggests that methylation of ribose of C32 in T. kodakarensis tRNATrp can occur after removal of the intron. It has been reported that C32 in tRNATrp from M. jannaschii is modified to s2C32 (8); the modification pathway of C32 in tRNATrp differs between T. kodakarensis and M. jannaschii. The Cm32 modification is often observed in tRNAs that are used to decode codons in one- and two-codon boxes (54). In terms of archaeal tRNA m5C methyltransferases, for a long time, only Trm4 had been characterized. In 1999, the enzymatic activity of Trm4 was detected in the cell extract of P. furiosus as tRNA m5C49 methyltransferase (31). Subsequently, it was found that Trm4 changes its methylation site in the presence of archease (55). Archaeal Trm4 is now known to be a multiple-site-specific tRNA methyltransferase for m5C48 and m5C49 modifications (55). Therefore, m5C48 and m5C49 modifications in T. kodakarensis tRNATrp can be explained by the enzymatic activity of Trm4. Furthermore, during the preparation of this paper, it was reported that Pyrococcus horikoshii NSUN6 methylates C72 and forms m5C72 in several tRNAs (56). However, a tRNA m5C32 methyltransferase has not been reported in any of the three domains of life. It is possible that the 5-methyl group in m5C32 enhances the stacking effect of C32 with G31 and contributes to stabilizing the anticodon arm at high temperatures.
Three types of sulfur-containing modifications were present in tRNATrp. Although the s4U modification has been observed in unfractionated tRNA nucleosides from several archaea (3), it has not been found in tRNAs from haloarchaea (57–59) or initiator tRNAMet from S. acidocaldarius (7); therefore, the modified position(s) of s4U in tRNA has been confirmed in limited tRNAs (position 8 in Thermoplasma acidophilum elongator tRNAMet [60], Methanosarcina barkeri tRNAPyr [61], and several M. jannaschii tRNAs [8] and positions 8 and 9 in T. acidophilum tRNALeu [62]). In terms of enzymatic properties, ThiI from Methanococcus maripaludis has been recently shown to contain a 3Fe-4S cluster and use inorganic sulfur compounds as sulfur donors (63).
Although the 2-thiocytidine (s2C) nucleoside has been observed in tRNAs from several archaea (2, 3, 5), position 17 represents a novel modification site of s2C. In tRNAs from mesophiles, position 17 is often modified to D17 (64). In Escherichia coli and Saccharomyces cerevisiae, for example, U17 in tRNA is modified to D17 by DusB (65, 66) and Dus1 (67), respectively. Nuclear magnetic resonance (NMR) analyses have suggested that D may destabilize the structure of tRNA by promoting the C2′-endo form of ribose (68). In general, therefore, D is thought to enhance the flexibility of tRNA. In contrast, the 2-thio group in s2C promotes the C3′-endo form of ribose. Thus, the conformation of the D-loop in T. kodakarensis tRNATrp seems to be different from that in tRNA from mesophiles. Possibly, s2C17 is required to maintain the D-loop structure (and interaction of the T and D arms) at high temperatures. The s2C modification is usually found at position 32 in eubacterial tRNAs (e.g., Escherichia coli tRNAArg [69]) and archaeal tRNA (8). In E. coli and Salmonella enterica serovar Typhimurium, the s2C32 modification in tRNA is performed by TtcA (70), which possesses a 4Fe-4S cluster (71). A ttcA-like gene (Tk1821) is included in the T. kodakarensis genome (71); however, the biosynthetic pathway of s2C17 is currently unknown.
It was shown that the biogenesis of m5s2U54 is mediated independently by a methyltransferase RumA and an unidentified 2-thiolation system. This feature is common to the formation of m5s2U54 in eubacterial tRNA (72, 73); however, whereas the methylation step in the m5s2U54 formation of archaeal tRNA is conferred by the S-adenosyl-l-methionine-dependent enzyme (RumA) (32), that of eubacterial tRNA is conferred by a folate- and FAD-dependent enzyme (TrmFO) (74, 75). In T. kodakarensis, two proteins homologous to TtuA (Tk1556 gene product) and TtuB (Tk1093 gene product) may be involved in the 2-thiolation of m5s2U54, as in eubacteria (76). Given that archaea do not possess a homolog of the IscS protein (77), however, the complete 2-thiolation system for the formation of m5s2U54 in T. kodakarensis tRNA remains unknown. The 2-thiolation of U54 has been found only in tRNAs from thermophiles such as Aquifex aeolicus (78) and Thermus thermophilus (72, 79, 80). The m5s2U54 modification forms a reverse Hoogsteen base pair with A58 (or m1A58) in tRNA, like m5U54 and m1Ψ54 (81), and the 2-thio group in m5s2U54 enhances the stacking effect with the G51-C61 base pair (80). Because the 2-thio modification at position 54 increases the melting temperature of tRNA by more than 3°C (72, 76, 79, 80, 82), the m5s2U54 modification probably contributes to stabilization of the tRNA structure even in the case of T. kodakarensis.
In this study, we found mimG37 in tRNATrp. Archaeal Trm5 can recognize the guanine base in an A36G37 sequence (83), and there are no reports of the tRNA specificity of TYW1, TYW2, and TYW3. Thus, our findings do not conflict with the results of previous studies. Because position 36 in T. kodakarensis tRNATrp is an unmodified A nucleoside, mimG37 may contribute to stabilize the base pair between A36 and U in mRNA during the protein synthesis at high temperatures. This idea is consistent with the fact that a mesophilic archaeon, Haloferax volcanii, does not contain wyosine derivatives in tRNA (58, 84).
G+15 (85) is formed by ArcTGT (86) and ArcS (87), and ArcTGT from T. kodakarensis modifies only G15 in tRNA (20). During the preparation of this paper, it was reported that an ArcTGT gene disruptant mutant of T. kodakarensis cannot grow at 93°C (28). The m1I57 modification is produced by deamination of m1A57, which is carried out by archaeal TrmI (88, 89). Although deaminase activity has been detected in the cell extract of H. volcanii (90), the responsible gene(s) has not been identified as yet. The m1A58 modification is a product of archaeal TrmI (88), and a recent random mutation study revealed that the trmI gene disruption strain cannot grow at 93°C (28).
In this study, we confirmed that the gene responsible for m2G67 is trm14. However, it has not been confirmed that T. kodakarensis Trm14 methylates G6 in tRNA like M. jannaschii Trm14. Furthermore, the presence of m2G6 modification in T. kodakarensis tRNAs has not been confirmed. To clarify these issues, further investigation is necessary.
In T. kodakarensis, Trm10 has been reported to methylate both G9 and A9 in tRNA, forming m1G9 and m1A9, respectively (91). Our analysis found, however, that G9 in tRNATrp was unmodified. Therefore, T. kodakarensis Trm10 seems to methylate specific tRNAs. Recently, kinetic analysis of T. kodakarensis Trm10 revealed that the rate-determining step for catalysis involves a conformational change of the substrate tRNA (92).
The Ψ55 modification in tRNATrp is likely to be performed by archaeal Pus10 (93–95) or archaeal Cbf5 (95–98). Because Sulfolobus solfaraticus Pus7 has been reported to possess weak activity for Ψ13 formation (99), the homologous protein (annotated as TruD [100] in the database [54]; Tk2302 gene product) may form Ψ13 in tRNATrp from T. kodakarensis. The enzyme responsible for Ψ22 formation in archaeal tRNA is unknown (101). The contribution of a Ψ13-Ψ22 base pair to tRNA structure has been recently reviewed (102): this base pair may stabilize the D-arm structure at high temperatures.
It is an intriguing finding that the lack of Trm11 impacted the viability of T. kodakarensis at high temperatures. Because m22G does not form a Watson-Crick base pair with C, m22G10 may contribute to the folding of specific tRNAs, such as tRNAPro from P. abyssi (15). In the case of T. thermophilus, an extremely thermophilic eubacterium, tRNA modification enzymes and the modified nucleosides in tRNA form a network (6, 72, 73, 103–106). At high temperatures, three modified nucleosides, m5s2U54 (76), m1A58 (107), and m7G46 (103), are essential for the survival of T. thermophilus. As described above, the 2-thio group in m5s2U54 increases the melting temperature of tRNA. The m1A58 modification is known to be a positive determinant of the sulfur transfer system used for m5s2U54 formation (76). The presence of m7G46 in tRNA increases the activity of several tRNA modification enzymes, such as TrmH (108, 109) for Gm18, TrmD (110) for m1G37, and TrmI (105, 107) for m1A58. It is possible that thermophilic archaea possess a similar network of tRNA modification enzymes and modified nucleosides in tRNA. Indeed, the requirement of the archaeal trmI gene (TrmI produces m1A57 and m1A58) for the m5s2U54 modification in T. kodakarensis has been reported previously (28). We attempted to measure the melting temperature of tRNA mixtures from the Δtrm11 strain, but it was not possible to determine it accurately because it was above 100°C in the presence of 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 100 mM NaCl (data not shown). Therefore, the growth defect of the Δtrm11 strain at 95°C cannot be explained simply by the melting temperature of tRNA. As predicted for tRNA from P. abyssi, the m22G10 modification in tRNA from T. kodakarensis might have an effect on folding of a specific tRNA(s) at high temperatures. To clarify these issues, further studies are required.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The strains of T. kodakarensis used in this study are listed in Table S1 in the supplemental material. The culture methods for T. kodakarensis KUW1 (17), KUWA (20), and gene disruptant strains are described in the supplemental text.
Disruption of trm11 (Tk0981), trm14 (Tk1863), rumA (Tk2135), thiI (Tk0368), and TYW1 (Tk1671) genes.
The plasmids used for gene disruptions are listed in Table S1. The primers used for genetic manipulations are listed in Table S2. The constructions of gene disruptant strain are described in the supplemental text.
Construction of a complemented strain expressing Trm11 in the Δtrm11 strain.
The conditional expression system in T. kodakarensis has been previously described (29). The construction of a complemented strain is described in the supplemental text.
Western blotting.
The recombinant 6×His-tagged Trm11 protein was prepared as described previously (9) and used to immunize rabbits and obtain antibodies (Eurofins Genomics, Inc., Japan). Cell extracts of the wild-type and Δtrm11 strains were prepared from cells grown to an optical density at 660 nm (OD660) of ∼0.6. A 1-ml aliquot of cells was mixed with 10 μl of 2× SDS-PAGE loading buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 2.5% SDS, 0.2% bromophenol blue, and 20% glycerol), boiled for 5 min, and then applied to a 12.5% SDS-PAGE gel. The gel was electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Inc.) in accordance with the manufacturer’s instructions. Trm11 protein was detected by using Alexa Fluor 488–anti-rabbit IgG (Invitrogen) as a secondary antibody and visualized with a Typhoon FLA 7000 laser scanner (GE Healthcare). For the complemented strain (Δtrm11 + trm11), Trm11 expression was analyzed by the same method.
Purification of tRNATrp.
Total RNA was extracted from 2.0 g of cells by using Isogen II (Nippon Gene Co., Ltd.) in accordance with the manufacturer’s protocol. The tRNA fraction was further purified by 10% PAGE (7 M urea). Transfer RNATrp was purified from the tRNA fraction by the solid-phase DNA probe method (21). The sequence of the DNA probe was complementary to G16 to A36 in tRNATrp: 5′-TGG AGC CCG CGA TGA TGG ACC AGG-biotin 3′.
Purification of human tRNALeuCAA.
Human cytoplasmic tRNALeuCAA containing m5Cm was isolated from ALKBH1 knockout cells as described previously (27).
Cyanoethylation of pseudouridines in tRNA.
Five picomoles of isolated tRNA dissolved in 1 μl of Milli-Q water was mixed with 30 μl of 50% (vol/vol) ethanol–1.1 M trimethylammonium acetate (pH 8.6)–1 mM EDTA. After the addition of 4 μl of acrylonitrile (Wako Pure Chemical Industries), the solution was incubated at 70°C for 2 h. Cyanoethylated tRNA was collected by ethanol precipitation. The tRNA sample was treated with RNase T1 digestion and then analyzed by liquid chromatography-mass spectrometry (LC/MS), as described below.
LC/MS.
The isolated tRNAs were digested with RNase T1 (Ambion) or RNase A (Ambion) and subjected to capillary LC–nano-electrospray ionization MS, as previously described (111, 112). For nucleoside analysis, 5 to 10 pmol of tRNA was digested by a 3-step reaction using nuclease P1 (Wako Pure Chemical Industries), phosphodiesterase I (Worthington Biochemical Corporation), and bacterial alkaline phosphatase (Escherichia coli C75) (TaKaRa Bio) (112). The digests were subjected to hydrophilic-interaction LC/MS analysis as described previously (113).
Supplementary Material
ACKNOWLEDGMENTS
We thank Layla Kawarada (University of Tokyo) for providing the ALKBH1 knockout tRNA and Kei Sugiyama and Shinichiro Akichika (University of Tokyo) for technical support.
This work was funded by Grants-in-Aid for Scientific Research (no. 15K06975 and 18K06088 to Akira Hirata, no. 26702035 and 18H02094 to Takeo Suzuki, and no. 16H04763 to Hiroyuki Hori) from the Japan Society for the Promotion of Science (JSPS).
We declare no competing interests with respect to the work performed in the study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00448-19.
REFERENCES
- 1.Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA. 1991. Posttranscriptional modification of tRNA in thermophilic archaea (archaebacteria). J Bacteriol 173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCloskey JA, Graham DE, Zou S, Crain PF, Ibba M, Konisky J, Söll D, Olsen GJ. 2001. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res 29:4299–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R. 2003. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 degrees C) and Stetteria hydrogenophila (Topt, 95 degrees C). J Bacteriol 185:5483–5490. doi: 10.1128/jb.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauerwald A, Sitaramaiah D, McCloskey JA, Söll D, Crain PF. 2005. N6-Acetyladenosine: a new modified nucleoside from Methanopyrus kandleri tRNA. FEBS Lett 579:2807–2810. doi: 10.1016/j.febslet.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Hori H, Kawamura T, Awai T, Ochi A, Yamagami R, Tomikawa C, Hirata A. 2018. Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganisms 6:110. doi: 10.3390/microorganisms6040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuchino Y, Ihara M, Yabusaki Y, Nishimura S. 1982. Initiator tRNAs from archaebacteria show common unique sequence characteristics. Nature 298:684–685. doi: 10.1038/298684a0. [DOI] [PubMed] [Google Scholar]
- 8.Yu N, Jora M, Solivio B, Thakur P, Acevedo-Rocha CG, Randau L, de Crécy-Lagard V, Addepalli B, Limbach PA. 2019. tRNA modification profiles and codon-decoding strategies in Methanocaldococcus jannaschii. J Bacteriol 201:e00690-18. doi: 10.1128/JB.00690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirata A, Nishiyama S, Tamura T, Yamauchi A, Hori H. 2016. Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res 44:6377–6390. doi: 10.1093/nar/gkw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol 60:4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. 2004. N2-Methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J Biol Chem 279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 12.Bourgeois G, Létoquart J, van Tran N, Graille M. 2017. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 7:7. doi: 10.3390/biom7010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourgeois G, Marcoux J, Saliou JM, Cianférani S, Graille M. 2017. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res 45:1971–1982. doi: 10.1093/nar/gkw1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. 2005. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol 25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbonavicius J, Armenguad J, Grosjean H. 2006. Identity elements required for enzymatic formation of N2, N2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J Mol Biol 357:387–399. doi: 10.1016/j.jmb.2005.12.087. [DOI] [PubMed] [Google Scholar]
- 16.Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220. doi: 10.1128/jb.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atomi H, Imanaka T, Fukui T. 2012. Overview of the genetic tools in the Archaea. Front Microbiol 3:337. doi: 10.3389/fmicb.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaoka E, Hidese R, Imanaka T, Fujiwara S. 2013. Importance and determinants of induction of cold-induced DEAD RNA helicase in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 195:3442–3450. doi: 10.1128/JB.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamura T, Hirata A, Ohno S, Nomura Y, Nagano T, Nameki T, Yokogawa T, Hori H. 2016. Multisite-specific archaeosine tRNA-guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo-acidophilic archaeon. Nucleic Acids Res 44:1894–1908. doi: 10.1093/nar/gkv1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokogawa T, Kitamura Y, Nakamura D, Ohno S, Nishikawa K. 2010. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res 38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menezes S, Gaston KW, Krivos KL, Apolinario EE, Reich NO, Sowers KR, Limbach PA, Perona JJ. 2011. Formation of m2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res 39:7641–7655. doi: 10.1093/nar/gkr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Constantinesco F, Benachenhou N, Motorin Y, Grosjean H. 1998. The tRNA(guanine-26, N2-N2) methyltransferase (Trm1) from the hyperthermophilic archaeon Pyrococcus furiosus: cloning, sequencing of the gene and its expression in Escherichia coli. Nucleic Acids Res 26:3753–3761. doi: 10.1093/nar/26.16.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constantinesco F, Motorin Y, Grosjean H. 1999. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J Mol Biol 291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 25.Kawamura T, Anraku R, Hasegawa T, Tomikawa C, Hori H. 2014. Transfer RNA methyltransferases from Thermoplasma acidophilum, a thermoacidophilic archaeon. Int J Mol Sci 16:91–113. doi: 10.3390/ijms16010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. 2017. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res 45:7401–7415. doi: 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orita I, Futatsuishi R, Adachi K, Ohira T, Kaneko A, Minowa K, Suzuki M, Tamura T, Nakamura S, Imanaka T, Suzuki T, Fukui T. 2019. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res 47:1964–1976. doi: 10.1093/nar/gky1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirata A, Kanai T, Santangelo TJ, Tajiri M, Manabe K, Reeve JN, Imanaka T, Murakami KS. 2008. Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive. Mol Microbiol 70:623–633. doi: 10.1111/j.1365-2958.2008.06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Čavužić M, Liu Y. 2017. Biosynthesis of sulfur-containing tRNA modifications: a comparison of bacterial, archaeal, and eukaryotic pathways. Biomolecules 7:27. doi: 10.3390/biom7010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Constantinesco F, Motorin Y, Grosjean H. 1999. Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res 27:1308–1315. doi: 10.1093/nar/27.5.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urbonavicius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H. 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 67:323–335. doi: 10.1111/j.1365-2958.2007.06047.x. [DOI] [PubMed] [Google Scholar]
- 33.McCloskey JA, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, Stetter KO. 1987. Structure determination of a new fluorescent tricyclic nucleoside from archaebacterial tRNA. Nucleic Acids Res 15:683–693. doi: 10.1093/nar/15.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. 2010. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol 27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urbonavičius J, Meškys R, Grosjean H. 2014. Biosynthesis of wyosine derivatives in tRNA(Phe) of Archaea: role of a remarkable bifunctional tRNA(Phe):m1G/imG2 methyltransferase. RNA 20:747–753. doi: 10.1261/rna.043315.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP. 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christian T, Evilia C, Williams S, Hou YM. 2004. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol 339:707–719. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Jia Q, Zeng J, Chen R, Xie W. 2017. Structural insight into the methyltransfer mechanism of the bifunctional Trm5. Sci Adv 3:e1700195. doi: 10.1126/sciadv.1700195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo M, Yokogawa T, Nishikawa K, Watanabe K, Okada N. 1995. Highly specific and efficient cleavage of squid tRNALys catalyzed by magnesium ions. J Biol Chem 270:10097–10104. doi: 10.1074/jbc.270.17.10097. [DOI] [PubMed] [Google Scholar]
- 40.Fislage M, Roovers M, Tuszynska I, Bujnicki JM, Droogmans L, Versées W. 2012. Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res 40:5149–5161. doi: 10.1093/nar/gks163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauhon CT, Erwin WM, Ton GN. 2004. Substrate specificity for 4-thiouridine modification in Escherichia coli. J Biol Chem 279:23022–23029. doi: 10.1074/jbc.M401757200. [DOI] [PubMed] [Google Scholar]
- 42.Neumann P, Lakomek K, Naumann PT, Erwin WM, Lauhon CT, Ficner R. 2014. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res 42:6673–6685. doi: 10.1093/nar/gku249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clouet-d’Orval B, Gaspin C, Mougin A. 2005. Two different mechanisms for tRNA ribose methylation in Archaea: a short survey. Biochimie 87:889–895. doi: 10.1016/j.biochi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Renalier MH, Joseph N, Gaspin C, Thebault P, Mougin A. 2005. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2’-O-methylase or a C/D sRNP. RNA 11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiss-László Z, Henry Y, Kiss T. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J 17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lui L, Lowe T. 2013. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem 54:53–77. doi: 10.1042/bse0540053. [DOI] [PubMed] [Google Scholar]
- 47.Clouet d’Orval B, Bortolin ML, Gaspin C, Bachellerie JP. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res 29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bortolin ML, Bachellerie JP, Clouet-d’Orval B. 2003. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNATrp. Nucleic Acids Res 31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 279:47661–47671. doi: 10.1074/jbc.M408868200. [DOI] [PubMed] [Google Scholar]
- 50.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 52.Ushida C, Muramatsu T, Mizushima H, Ueda T, Watanabe K, Stetter KO, Crain PF, McCloskey JA, Kuchino Y. 1996. Structural feature of the initiator tRNA gene from Pyrodictium occultum and the thermal stability of its gene product, tRNA(imet). Biochimie 79:847–855. doi: 10.1016/S0300-9084(97)84337-4. [DOI] [PubMed] [Google Scholar]
- 53.Somme J, Van Laer B, Roovers M, Steyaert J, Versées W, Droogmans L. 2014. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 20:1257–1271. doi: 10.1261/rna.044503.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosjean H, Westhof E. 2016. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 44:8020–8040. doi: 10.1093/nar/gkw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Auxilien S, El Khadali F, Rasmussen A, Douthwaite S, Grosjean H. 2007. Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J Biol Chem 282:18711–18721. doi: 10.1074/jbc.M607459200. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. 2019. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res 47:2041–2055. doi: 10.1093/nar/gky1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu XR, Nicoghosian K, Cedergren RJ, Wong JT. 1983. Sequences of halobacterial tRNAs and the paucity of U in the first position of their anticodons. Nucleic Acids Res 11:5433–5442. doi: 10.1093/nar/11.16.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem 259:9461–9471. [PubMed] [Google Scholar]
- 59.Mandal D, Köhrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Söll D, RajBhandary UL. 2010. Agmatidine, a modified cytidine in the anticodon of archaeal tRNAIle, base pairs with adenosine but not with guanosine. Proc Natl Acad Sci U S A 107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kilpatrick MW, Walker RT. 1981. The nucleotide sequence of the tRNAMMet from the archaebacterium Thermoplasma acidophilum. Nucleic Acids Res 9:4387–4390. doi: 10.1093/nar/9.17.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srinivasan G, James CM, Krzycki JA. 2002. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 62.Tomikawa C, Ohira T, Inoue Y, Kawamura T, Yamagishi A, Suzuki T, Hori H. 2013. Distinct tRNA modifications in the thermo-acidophilic archaeon, Thermoplasma acidophilum. FEBS Lett 587:3575–3580. doi: 10.1016/j.febslet.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Vinyard DJ, Reesbeck ME, Suzuki T, Manakongtreecheep K, Holland PL, Brudvig GW, Söll D. 2016. A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc Natl Acad Sci U S A 113:12703–12708. doi: 10.1073/pnas.1615732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bishop AC, Xu J, Johnson RC, Schimmel P, de Crécy-Lagard V. 2002. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem 277:25090–25095. doi: 10.1074/jbc.M203208200. [DOI] [PubMed] [Google Scholar]
- 66.Bou-Nader C, Montémont H, Guérineau V, Jean-Jean O, Brégeon D, Hamdane D. 2018. Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario. Nucleic Acids Res 46:1386–1394. doi: 10.1093/nar/gkx1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xing F, Martzen MR, Phizicky EM. 2002. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 8:370–381. doi: 10.1017/s1355838202029825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. 1996. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res 24:1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiesewetter S, Fischer W, Sprinzl M. 1987. Sequences of three minor tRNAsArg from E. coli. Nucleic Acids Res 15:3184. doi: 10.1093/nar/15.7.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jäger G, Leipuviene R, Pollard MG, Qian Q, Björk GR. 2004. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J Bacteriol 186:750–757. doi: 10.1128/jb.186.3.750-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouvier D, Labessan N, Clémancey M, Latour JM, Ravanat JL, Fontecave M, Atta M. 2014. TtcA a new tRNA-thioltransferase with an Fe-S cluster. Nucleic Acids Res 42:7960–7970. doi: 10.1093/nar/gku508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hori H. 2019. Regulatory factors for tRNA modifications in extreme-thermophilic bacterium Thermus thermophilus. Front Genet 10:204. doi: 10.3389/fgene.2019.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamagami R, Tomikawa C, Shigi N, Kazayama A, Asai S, Takuma H, Hirata A, Fourmy D, Asahara H, Watanabe K, Yoshizawa S, Hori H. 2016. Folate-/FAD-dependent tRNA methyltransferase from Thermus thermophilus regulates other modifications in tRNA at low temperatures. Genes Cells 21:740–754. doi: 10.1111/gtc.12376. [DOI] [PubMed] [Google Scholar]
- 74.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. 2005. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucleic Acids Res 33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamagami R, Yamashita K, Nishimasu H, Tomikawa C, Ochi A, Iwashita C, Hirata A, Ishitani R, Nureki O, Hori H. 2012. The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). J Biol Chem 287:42480–42494. doi: 10.1074/jbc.M112.390112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shigi N, Sakaguchi Y, Suzuki T, Watanabe K. 2006. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J Biol Chem 281:14296–14306. doi: 10.1074/jbc.M511675200. [DOI] [PubMed] [Google Scholar]
- 77.Kambampati R, Lauhon CT. 1999. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry 38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 78.Awai T, Kimura S, Tomikawa C, Ochi A, Ihsanawati Bessho Y, Yokoyama S, Ohno S, Nishikawa K, Yokogawa T, Suzuki T, Hori H. 2009. Aquifex aeolicus tRNA (N2, N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J Biol Chem 284:20467–20478. doi: 10.1074/jbc.M109.020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe K, Shinma M, Oshima T, Nishimura S. 1976. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem Biophys Res Commun 72:1137–1144. doi: 10.1016/s0006-291x(76)80250-1. [DOI] [PubMed] [Google Scholar]
- 80.Yokoyama S, Watanabe K, Miyazawa T. 1987. Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv Biophys 23:115–147. doi: 10.1016/0065-227X(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 81.Chatterjee K, Blaby IK, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crécy-Lagard V. 2012. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18:421–433. doi: 10.1261/rna.030841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H. 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res 6:1571–1581. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christian T, Hou YM. 2007. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol 373:623–632. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta R. 1986. Transfer RNAs of Halobacterium volcanii: sequences of five leucine and three serine tRNAs. Syst Appl Microbiol 7:102–105. doi: 10.1016/S0723-2020(86)80131-X. [DOI] [Google Scholar]
- 85.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. 1993. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-beta-d-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J Biol Chem 268:10076–10086. [PubMed] [Google Scholar]
- 86.Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katze JR, Hashizume T, Crain PF, McCloskey JA, Okada N. 1997. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem 272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- 87.Phillips G, Chikwana VM, Maxwell A, El-Yacoubi B, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. 2010. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J Biol Chem 285:12706–12713. doi: 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. 2004. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res 32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamdane D, Guelorget A, Guérineau V, Golinelli-Pimpaneau B. 2014. Dynamics of RNA modification by a multi-site-specific tRNA methyltransferase. Nucleic Acids Res 42:11697–11706. doi: 10.1093/nar/gku820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grosjean H, Constantinesco F, Foiret D, Benachenhou N. 1995. A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res 23:4312–4319. doi: 10.1093/nar/23.21.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kempenaers M, Roovers M, Oudjama Y, Tkaczuk KL, Bujnicki JM, Droogmans L. 2010. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res 38:6533–6543. doi: 10.1093/nar/gkq451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishnamohan A, Dodbele S, Jackman JE. 2019. Insights into catalytic and tRNA recognition mechanism of the dual-specific tRNA methyltransferase from Thermococcus kodakarensis. Genes 10:100. doi: 10.3390/genes10020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34:4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gurha P, Gupta R. 2008. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 14:2521–2527. doi: 10.1261/rna.1276508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crécy-Lagard V, Gupta R. 2011. Pseudouridine formation in archaeal RNAs: the case of Haloferax volcanii. RNA 17:1367–1380. doi: 10.1261/rna.2712811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C. 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Psi55-synthase and RNA-guided RNA:Psi-synthase activities. Nucleic Acids Res 35:5610–5624. doi: 10.1093/nar/gkm606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gurha P, Joardar A, Chaurasia P, Gupta R. 2007. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol 4:101–109. doi: 10.4161/rna.4.2.5177. [DOI] [PubMed] [Google Scholar]
- 98.Kamalampeta R, Kothe U. 2012. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep 2:663. doi: 10.1038/srep00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muller S, Urban A, Hecker A, Leclerc F, Branlant C, Motorin Y. 2009. Deficiency of the tRNATyr:Psi 35-synthase aPus7 in Archaea of the Sulfolobales order might be rescued by the H/ACA sRNA-guided machinery. Nucleic Acids Res 37:1308–1322. doi: 10.1093/nar/gkn1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaya Y, Ofengand J. 2003. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9:711–721. doi: 10.1261/rna.5230603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spenkuch F, Motorin Y, Helm M. 2014. Pseudouridine: still mysterious, but never a fake (uridine)!. RNA Biol 11:1540–1554. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lorenz C, Lünse CE, Mörl M. 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7:35. doi: 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tomikawa C, Yokogawa T, Kanai T, Hori H. 2010. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability through a tRNA modification network. Nucleic Acids Res 38:942–957. doi: 10.1093/nar/gkp1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. 2011. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res 39:2304–2318. doi: 10.1093/nar/gkq1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takuma H, Ushio N, Minoji M, Kazayama A, Shigi N, Hirata A, Tomikawa C, Ochi A, Hori H. 2015. Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI). J Biol Chem 290:5912–5925. doi: 10.1074/jbc.M114.606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kusuba H, Yoshida T, Iwasaki E, Awai T, Kazayama A, Hirata A, Tomikawa C, Yamagami R, Hori H. 2015. In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium. J Biochem 158:431–521. [DOI] [PubMed] [Google Scholar]
- 107.Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. 2003. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res 31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Persson BC, Jäger G, Gustafsson C. 1997. The spoU gene of Escherichia coli, the fourth gene of the spot operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res 25:3969–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hori H, Suzuki T, Sugawara K, Inoue Y, Shibata T, Kuramitsu S, Yokoyama S, Oshima T, Watanabe K. 2002. Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells 7:259–272. doi: 10.1046/j.1365-2443.2002.00520.x. [DOI] [PubMed] [Google Scholar]
- 110.Byström AS, Björk GR. 1982. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol Gen Genet 188:440–446. doi: 10.1007/bf00330046. [DOI] [PubMed] [Google Scholar]
- 111.Suzuki T, Suzuki T. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Asano K, Suzuki T, Saito A, Wei FY, Ikeuchi Y, Numata T, Tanaka R, Yamane Y, Yamamoto T, Goto T, Kishita Y, Murayama K, Ohtake A, Okazaki Y, Tomizawa K, Sakaguchi Y, Suzuki T. 2018. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res 46:1565–1583. doi: 10.1093/nar/gky068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sakaguchi Y, Miyauchi K, Kang BI, Suzuki T. 2015. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol 560:19–28. doi: 10.1016/bs.mie.2015.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


