Streptococcus pneumoniae strains produce hydrogen peroxide (H2O2) to kill bacteria in the upper airways, including pathogenic Staphylococcus aureus strains. The targets of S. pneumoniae-produced H2O2 have not been discovered, in part because of a lack of knowledge about the underlying molecular mechanism. We demonstrated that an increased density of S. pneumoniae kills S. aureus by means of H2O2 produced by two enzymes, SpxB and LctO. We discovered that SpxB/LctO-produced H2O2 is converted into a hydroxyl radical (·OH) that rapidly intoxicates and kills S. aureus. We successfully inhibited the toxicity of ·OH with three different scavengers and detected ·OH in the supernatant. The target(s) of the hydroxyl radicals represents a new alternative for the development of antimicrobials against S. aureus infections.
KEYWORDS: Staphylococcus aureus, Streptococcus pneumoniae, eradication, hydrogen peroxide, hydroxyl radicals
ABSTRACT
Streptococcus pneumoniae rapidly kills Staphylococcus aureus by producing membrane-permeable hydrogen peroxide (H2O2). The mechanism by which S. pneumoniae-produced H2O2 mediates S. aureus killing was investigated. An in vitro model that mimicked S. pneumoniae-S. aureus contact during colonization of the nasopharynx demonstrated that S. aureus killing required outcompeting densities of S. pneumoniae. Compared to the wild-type strain, isogenic S. pneumoniae ΔlctO and S. pneumoniae ΔspxB, both deficient in production of H2O2, required increased density to kill S. aureus. While residual H2O2 activity produced by single mutants was sufficient to eradicate S. aureus, an S. pneumoniae ΔspxB ΔlctO double mutant was unable to kill S. aureus. A collection of 20 diverse methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) strains showed linear sensitivity (R2 = 0.95) for S. pneumoniae killing, but the same strains had different susceptibilities when challenged with pure H2O2 (5 mM). There was no association between the S. aureus clonal complex and sensitivity to either S. pneumoniae or H2O2. To kill S. aureus, S. pneumoniae produced ∼180 μM H2O2 within 4 h of incubation, while the killing-defective S. pneumoniae ΔspxB and S. pneumoniae ΔspxB ΔlctO mutants produced undetectable levels. Remarkably, a sublethal dose (1 mM) of pure H2O2 incubated with S. pneumoniae ΔspxB eradicated diverse S. aureus strains, suggesting that S. pneumoniae bacteria may facilitate conversion of H2O2 to a hydroxyl radical (·OH). Accordingly, S. aureus killing was completely blocked by incubation with scavengers of ·OH radicals, dimethyl sulfoxide (Me2SO), thiourea, or sodium salicylate. The ·OH was detected in S. pneumoniae cells by spin trapping and electron paramagnetic resonance. Therefore, S. pneumoniae produces H2O2, which is rapidly converted to a more potent oxidant, hydroxyl radicals, to rapidly intoxicate S. aureus strains.
IMPORTANCE Streptococcus pneumoniae strains produce hydrogen peroxide (H2O2) to kill bacteria in the upper airways, including pathogenic Staphylococcus aureus strains. The targets of S. pneumoniae-produced H2O2 have not been discovered, in part because of a lack of knowledge about the underlying molecular mechanism. We demonstrated that an increased density of S. pneumoniae kills S. aureus by means of H2O2 produced by two enzymes, SpxB and LctO. We discovered that SpxB/LctO-produced H2O2 is converted into a hydroxyl radical (·OH) that rapidly intoxicates and kills S. aureus. We successfully inhibited the toxicity of ·OH with three different scavengers and detected ·OH in the supernatant. The target(s) of the hydroxyl radicals represents a new alternative for the development of antimicrobials against S. aureus infections.
INTRODUCTION
Streptococcus pneumoniae and Staphylococcus aureus colonize the upper airways of humans, forming persistent biofilms (1–9). Once in the nasopharynx, S. pneumoniae forms a biofilm that increases resistance to desiccation and antibiotic resistance and also provides a source of planktonic bacteria that migrate to the ears, lower respiratory tract, circulation, heart, and meninges, causing pneumococcal disease, the burden of which is extremely high in the human population (5, 6, 10–13). S. aureus strains colonize the skin of >30% of the human population but also reside in the nasopharynx, causing severe pathologies, including bacteremia and pneumonia (1, 3, 7, 11, 14, 15).
Over the last few years, our laboratories and others have conducted carriage studies of important human pathogens in the nasopharynxes of children of different ethnicities. These studies demonstrated a negative association for the concurrent carriage of S. pneumoniae and S. aureus (3, 7, 16). Soon after pneumococcal conjugate vaccines (PCV) became available, a potential mechanistic competition between S. pneumoniae and S. aureus for the colonization of the upper airways was observed. Some of the first studies showed that nasopharyngeal carriage of S. aureus increased in children who had received PCV. The increased S. aureus colonization was attributed to the decreased carriage of pneumococcal serotypes targeted by PCV (1, 7, 8). It is therefore clear that S. pneumoniae in vivo interferes with colonization by S. aureus.
Although evidence that S. pneumoniae was capable of killing S. aureus was published over 100 years ago (17, 18), studies of the molecular mechanism(s) behind these epidemiological observations were reinitiated when the pneumococcal vaccine was licensed in early 2000 in developed countries. Pericone et al. (19), and then other investigators, demonstrated that pneumococcal strains isolated from disease or carriage interfered with the growth of S. aureus in broth cultures. The proposed mechanism involved the production of hydrogen peroxide (H2O2) that was released by S. pneumoniae into the supernatant (20). This H2O2-mediated killing of S. aureus occurred within 6 h post-inoculation of S. pneumoniae, but it was inhibited in cocultures with catalase added; by incubating these cocultures in an anaerobic chamber; or by a mutation within the spxB gene, encoding the enzyme streptococcal pyruvate oxidase, which endogenously produces H2O2 during conversion of acetylphosphate from pyruvate (19–23). Notably, SpxB accounts for ∼85% of the membrane-permeable H2O2 that is released by the bacteria into the supernatant (24, 25). A second contributor to the pool of H2O2 released by bacteria is the enzyme lactate dehydrogenase (LctO), which converts lactate to pyruvate (24, 26). While the mechanism by which S. pneumoniae kills S. aureus strains has been related to production of H2O2, only spxB mutants have been assessed (20, 27).
SpxB-produced H2O2 has also been involved in inducing cytotoxicity to lung cells, apoptosis, and the toxic events observed when S. pneumoniae invades the central nervous system and heart, albeit the specific mechanism(s) mediating this damage is still to be clarified (12, 13). Moreover, S. pneumoniae mutants in the spxB gene produced less capsule, due to the lack of acetylated capsule precursors, and were attenuated for virulence in mouse models of pneumococcal disease (25, 28). The attenuated virulence phenotype can be explained in part by a recent publication showing that endogenously produced H2O2 was required to release the toxin pneumolysin (29).
In contrast to the in vitro evidence presented above, studies conducted using an animal model of colonization demonstrated that S. aureus colonized the nasal cavity of neonatal rats even when it was inoculated concurrently with S. pneumoniae strain TIGR4 or with an H2O2-deficient TIGR4 ΔspxB mutant (30). When the TIGR4 wild type (wt) or an isogenic TIGR4 ΔspxB mutant was inoculated along with S. aureus in animals, S. aureus colonization densities were similar whether S. pneumoniae produced hydrogen peroxide or not (30, 31). Therefore, the role of S. pneumoniae-produced H2O2 in interfering with S. aureus growth has been debated (32). Killing of S. aureus by incubation with pure H2O2, however, has already been documented (33, 34). A dose of ∼10 mM H2O2 was required to kill S. aureus bacteria (19), whereas preloading S. aureus with iron reduced the bactericidal dose to ∼1 mM (33, 34). The presence of intracellular iron was required to generate, by the Fenton reaction, the hydroxyl radical (·OH) (34), which is a stronger oxidant than H2O2 itself (35). Other bacterial species are also susceptible to H2O2 at a concentration similar to that killing S. aureus, with ∼2.5 mM H2O2 showing the maximal killing rate for Escherichia coli (35–37).
We recently demonstrated that the S. pneumoniae-induced killing of S. aureus biofilms, including those formed by methicillin-resistant S. aureus (MRSA) strains, was enhanced by physical contact (23). Complete eradication of ∼109 S. aureus bacteria within S. pneumoniae-S. aureus biofilms occurred within 4 h of incubation. Furthermore, washed S. pneumoniae bacteria were more lethal to S. aureus strains than their H2O2-containing supernatants, suggesting pneumococcal cells may be required to convert H2O2 into a more potent intoxicant (23). Moreover, our studies and those of others (19, 38) demonstrated that, when S. aureus has been completely killed, S. pneumoniae produced significantly less H2O2 (e.g., TIGR4, <200 μM) than the demonstrated minimal bactericidal concentration (MBC) of pure H2O2 (10 mM) for S. aureus strains (19, 38).
In this study, we used in vitro models mimicking S. pneumoniae-S. aureus cocolonization of the upper airways and demonstrated that an outcompeting density of S. pneumoniae was necessary to kill S. aureus. We also demonstrated that the interaction between S. pneumoniae and S. aureus stimulates the conversion of hydrogen peroxide into the strongest oxidative radical, hydroxyl (·OH), which reacts at nearly diffusion rates with most substrates, inducing DNA degradation and leading to the intoxication and death of S. aureus bacteria. The target(s) of the ·OH radicals represents an exciting new alternative for the development of therapeutics against S. aureus infections.
RESULTS
Contact-mediated killing of S. aureus by S. pneumoniae requires a threshold pneumococcal density.
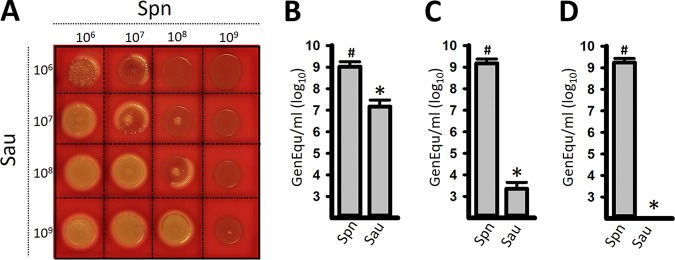
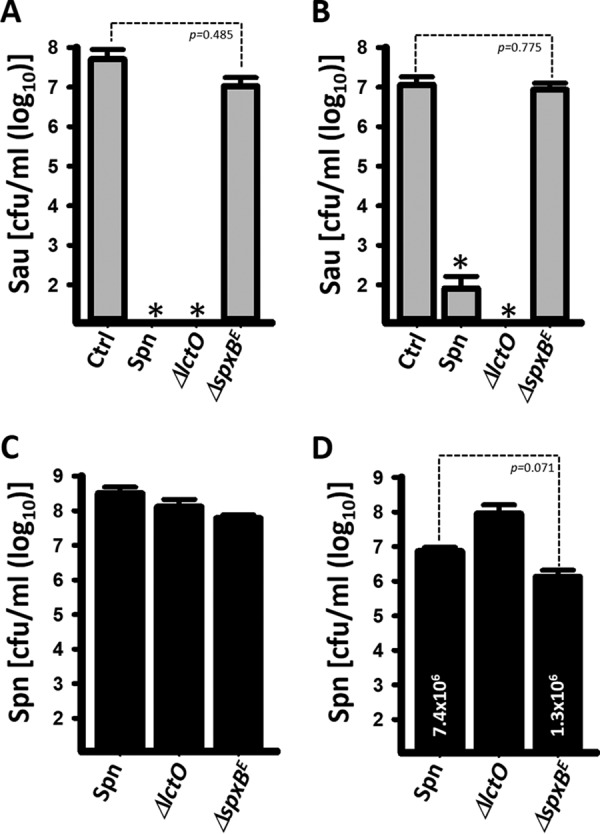
We previously demonstrated that killing of S. aureus strains by S. pneumoniae in liquid cultures required physical contact (23). We therefore reasoned that killing was likely to occur on solid media and designed a contact-mediated killing assay on blood agar plates. In this assay, we inoculated increasing densities of early-log-phase cultures of S. pneumoniae with different densities of S. aureus (i.e., 106 CFU/ml of S. aureus versus 106, 107, 108, or 109 CFU/ml of S. pneumoniae), and the plates were incubated overnight. In most mixtures where the density of S. pneumoniae outcompeted that of S. aureus by at least 2 log units (i.e., S. pneumoniae, 108 CFU/ml, and S. aureus, 106 CFU/ml), S. pneumoniae completely eradicated S. aureus (Fig. 1A). S. pneumoniae inoculated at 109 CFU/ml eradicated all S. aureus inocula. Similar results were obtained when two other S. aureus strains, NRS170 and NRS408, were assessed (not shown). To confirm the killing of S. aureus and the observed loss of chromosomal DNA observed by confocal microscopy (explained below), we isolated DNA from each mixture presented in Fig. 1A or single cultures (control). The DNA was used as a template in quantitative PCRs (qPCRs) targeting either S. pneumoniae or S. aureus. As expected, the number of genome equivalents (GenEqu) of S. aureus per milliliter did not change when the density outcompeted that of S. pneumoniae (not shown). When DNA was isolated from experiments where the S. pneumoniae density was greater than that of S. aureus, a density-dependent decrease of S. aureus GenEqu per milliliter was observed to a point where DNA from S. aureus was no longer detected (Fig. 1B to D; see Fig. S1 in the supplemental material). The number of GenEqu per milliliter of S. pneumoniae DNA (median, 1.4 × 109 GenEqu/ml) was not affected by incubation with any density of S. aureus. Density-dependent killing of S. aureus was also confirmed by culture (see Fig. S2 in the supplemental material).
FIG 1.
Contact-, and density-dependent killing of S. aureus (Sau) by S. pneumoniae (Spn). (A) S. pneumoniae strain TIGR4 and S. aureus strain Newman were inoculated concurrently at the indicated densities (CFU per milliliter). Once inoculated, the plates were incubated for 24 h at 37°C. (B to D) To quantify genome equivalents per milliliter of S. pneumoniae or S. aureus, bacteria growing on spots inoculated with 108 CFU/ml of S. pneumoniae and 108 (B), 107 (C), or 106 (D) CFU/ml of S. aureus were collected; DNA was extracted; and the DNA was used as a template in species-specific qPCRs. The error bars represent the standard errors of the means calculated using data from at least three independent experiments. *, P < 0.05 compared to S. aureus control incubated alone; #, P > 0.67 compared to S. pneumoniae control inoculated alone.
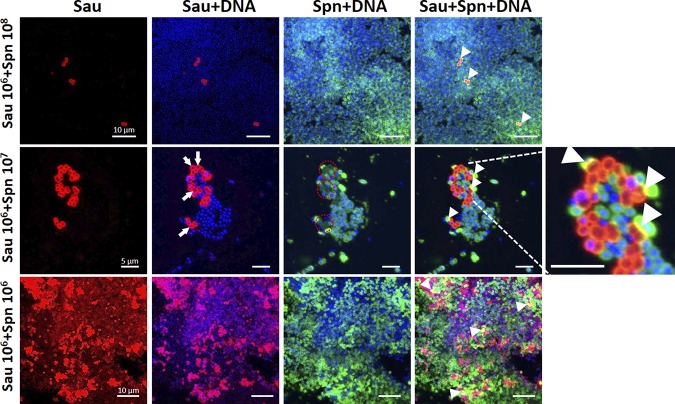
Confocal micrographs using antibodies against their capsules showed that, when inoculated at similar densities (i.e., ∼106 CFU/ml), S. pneumoniae and S. aureus were observed intact and with areas of strong colocalization (Fig. 2, bottom row, arrowheads). DAPI (4′,6-diamidino-2-phenylindole) staining showed DNA from both species. In micrographs where S. pneumoniae outcompeted S. aureus (e.g., S. pneumoniae, ∼107 CFU/ml versus S. aureus, ∼106 CFU/ml), only a few S. aureus cells were observed in comparison to the abundant pneumococci (Fig. 2, middle and top rows). Moreover, three-dimensional (3D) reconstruction of z stacks revealed that, in the majority of S. aureus cells in mixtures with outcompeting S. pneumoniae, the DAPI signal was absent, suggesting that the DNA had been degraded (Fig. 2, arrows). Absence of DNA in S. aureus particularly coincided with the bacteria colocalizing with S. pneumoniae (Fig. 2, enlarged image). Together, these results identified dose- and contact-dependent killing of S. aureus by S. pneumoniae that included DNA degradation.
FIG 2.
S. pneumoniae (Spn) contact-dependent killing of S. aureus (Sau) induces loss of DNA signal. Bacteria growing on blood agar plates from the experiments presented in Fig. 1, inoculated with the specific density of each species shown on the left, were imprinted onto glass slides. The preparations were fixed with paraformaldehyde, and S. aureus bacteria were stained with an anti-S. aureus antibody followed by an anti-rabbit Alexa Fluor 555-labeled antibody. S. pneumoniae was stained with an Alexa Fluor 488-labeled anti-S. pneumoniae antibody, while the DNA was stained with DAPI. The preparations were analyzed with a confocal microscope. Shown are 3D reconstructions of z stacks obtained from xy optical sections. The specific channel of each panel is shown at the top. The arrows point to S. aureus bacteria stained red with a loss of DNA signal, while the red dashed circles indicate areas where DNA signal is missing, corresponding to the arrows. The arrowheads show physical colocalization (yellow) of S. aureus and S. pneumoniae. The dimensions of the scale bars shown in the left column apply to all the images in the same row. The image on the far right was digitally enlarged to show details of the area indicated by the dashed lines.
Differential sensitivities of S. aureus strains to killing by S. pneumoniae.
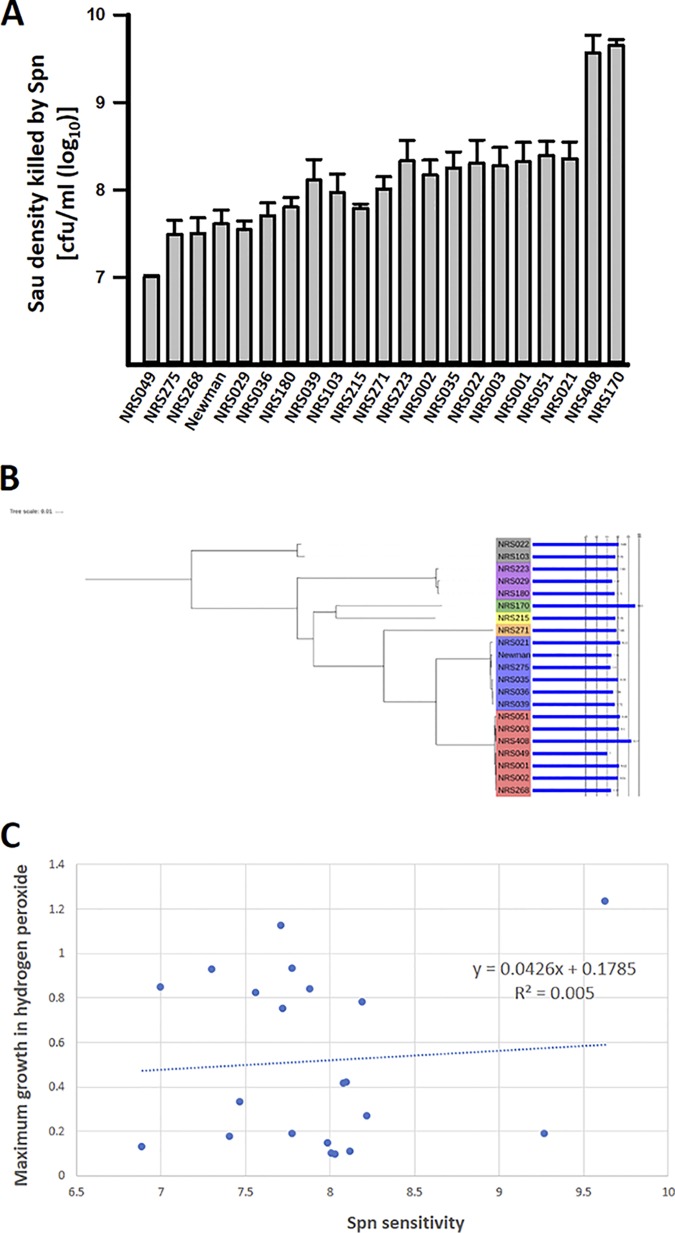
Different S. aureus strains have distinct sensitivities to H2O2 killing; thus, it was possible that the results described above were not representative. We therefore tested a collection of 20 MRSA (including vancomycin-intermediate S. aureus [VISA]) and methicillin-susceptible S. aureus (MSSA) strains from seven clonal complexes (see Table S1 in the supplemental material) for their sensitivities to killing when incubated along with S. pneumoniae. To quantify the maximum density of S. aureus killed by S. pneumoniae, we utilized a microplate model with 4 h of coculture incubation at 37°C. All the S. aureus strains were killed by S. pneumoniae, but we noted statistically significant differences across strains (P = 0.002) (Fig. 3A). The most sensitive strain, NRS170, had a 426-fold difference (P = 0.008) in sensitivity to S. pneumoniae compared to the most resistant strain, NRS049 (Fig. 3A; see Table S1). Increased sensitivity of NRS170 to S. pneumoniae killing was also observed using the plate-killing model (not shown). The rest of the strains, including S. aureus strain Newman, showed linear distributions in their sensitivities to S. pneumoniae (R2 = 0.95) that spanned an ∼30-fold range. The variability among this group (excluding NRS170 and NRS408) was also statistically significant (P = 0.05). Surprisingly, there was no association between the clonal complex and sensitivity to S. pneumoniae (Fig. 3B).
FIG 3.
Variability in S. aureus (Sau) strain sensitivity to S. pneumoniae (Spn). (A) Decreasing densities of the indicated S. aureus strains spanning ∼1 × 1010 and ∼1 × 106 CFU/ml were cocultured with 1.5 × 107 CFU/ml of S. pneumoniae in THY and incubated for 4 h at 37°C. Cultures were serially diluted and plated on TSA supplemented with optochin. The maximum S. aureus inoculum completely killed by 1.5 × 107 CFU/ml of S. pneumoniae was then determined from the maximum concentration killed. The standard errors of the mean of three independent experiments are shown. (B) Maximum-likelihood phylogeny of strains tested with log10 bactericidal efficiency of S. pneumoniae plotted as a bar chart. The clonal complex designation of each strain is shown by the color range. (C) Maximum growth (OD600 measured after 12 h of incubation at 37°C) in hydrogen peroxide (2.5 mM)-supplemented TSB plotted against S. pneumoniae sensitivity of the corresponding strain. The values represent averages from at least three replicates for each strain.
Hydrogen peroxide has been implicated as the main factor produced by S. pneumoniae to kill S. aureus strains (14, 20). We hypothesized that the level of sensitivity of an S. aureus strain to H2O2 correlated with sensitivity to S. pneumoniae killing. Differential sensitivity to H2O2 was observed in our experiments, with the growth of some strains (i.e., NRS3 and NRS21) completely inhibited by H2O2 whereas a subset of strains were not susceptible at all to challenge with even 5 mM H2O2 (not shown). However, contrary to our hypothesis, the level of S. pneumoniae killing did not correlate with retardation of growth by H2O2 (Fig. 3C). These results showed that there was a complex genetic relationship between the ability to grow in the presence of hydrogen peroxide and the degree of sensitivity to S. pneumoniae killing. The finding that the genetic background (clonal complex) was not strongly associated with the level of killing suggests that recently acquired mutations may play a major role in determining the level of susceptibility of each individual S. aureus strain.
Resistant S. aureus strains can protect sensitive strains from killing by S. pneumoniae.
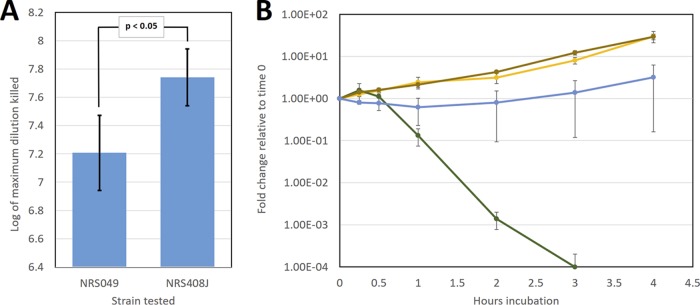
To study the effect of S. pneumoniae sensitivity on S. aureus strain selection, we utilized one of the most sensitive strains (NRS408) and one of the most resistant strains (NRS049). Strain NRS049 was resistant to tetracycline, and we isolated an NRS408-derived rifampin-resistant mutant (NRS408J) to track the growth of the strain. There was a significant difference between strains NRS049 and NRS408J (P < 0.05) in S. pneumoniae sensitivity (Fig. 4A). There was no significant difference in NRS049 S. pneumoniae sensitivity measurements or NRS408 relative to NRS408J measurements between results presented in Fig. 3A and 4A. We then competed the resistant NRS049 strain and the sensitive NRS408J strain (5 × 106 CFU/ml of each strain, the minimum concentration at which NRS049 was predicted to survive) in the presence of TIGR4 (1.5 × 107 CFU/ml). While this dose of S. pneumoniae killed NRS408J but not the NRS049 strain, coincubation of the two S. aureus strains led to survival of both under S. pneumoniae challenge (Fig. 4B). Given that killing of S. aureus is density dependent, to determine whether this competition outcome was affected by the S. aureus strain density, we performed several endpoint S. aureus growth assays over a range of NRS049 and NRS408J densities (see Table S2 in the supplemental material). The sensitive NRS408J survived under all conditions under which the resistant NRS049 strain survived, regardless of whether the total S. aureus dose was 5 × 106 CFU/ml or more (see Table S2).
FIG 4.
Competition between S. pneumoniae-sensitive and S. pneumoniae-resistant S. aureus strains in the presence of S. pneumoniae. (A) Sensitivities of S. aureus strains NRS049 and NRS408J to S. pneumoniae (TIGR4) killing. Sensitivity was measured as described in the legend to Fig. 3A. The results represent three biological replicates and two independent experiments. The results are presented with two standard errors above and below the mean. (B) Competition experiments between resistant (NRS049) and sensitive (NRS408J) S. aureus strains (5e6 CFU/ml) in the presence of S. pneumoniae (TIGR4; 1.5e7 CFU/ml). Strains NRS049, NRS408J, and TIGR4 at the previously stated doses were cocultured in THY for 4 h at 37°C without agitation. Coculture samples were collected at 0 min, 15 min, 30 min, 1 h, 2 h, 3 h, and 4 h. Coculture sample dilutions were then spotted on TSA supplemented with 16 μg/ml tetracycline or 4 μg/ml rifampin and grown overnight at 37°C to determine the concentrations of NRS049 and the NRS408 rifampin-resistant mutant (NRS408J) at corresponding time points. The fold change of the rifampin-resistant mutant NRS408J relative to time zero is shown with TIGR4 alone (green), TIGR4 and NRS049 (blue), NRS049 (brown), and the mutant itself alone (yellow). The results represent three biological replicates and are presented with one standard error above and one below the mean.
Mutations in spxB and lctO are required to inhibit S. pneumoniae killing of S. aureus.
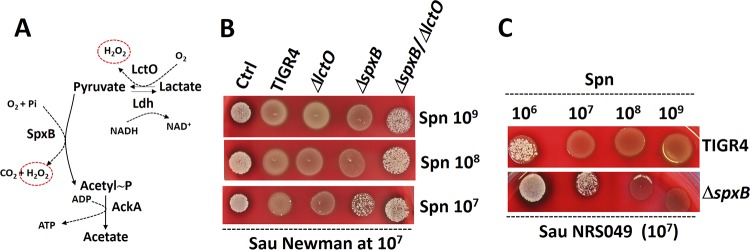
The experiments shown in Fig. 3 suggested that killing of S. aureus strains was not the sole consequence of exposure to H2O2. In S. pneumoniae, most H2O2 (∼85%) is produced during the oxidation of pyruvate to acetyl-phosphate (acetyl∼P) by SpxB (Fig. 5A). Although to a lesser extent, hydrogen peroxide is also produced during the oxidation of lactate to pyruvate by the enzyme LctO (Fig. 5A) (26). To gain insight into this contact-dependent, molecular-mechanism-mediated killing of S. aureus, we generated single ΔspxB and ΔlctO mutants and a ΔspxB ΔlctO double mutant in TIGR4. We then assessed killing of S. aureus by these mutants using our density-controlled experimental models. When incubated along with ∼107 CFU/ml S. aureus in the contact-dependent plate model, the TIGR4 ΔlctO mutant killed S. aureus to the same extent as the TIGR4 wild-type strain at all tested S. pneumoniae densities (Fig. 5B). At a density of ∼107 CFU/ml, TIGR4 ΔspxB did not kill S. aureus. Surprisingly, TIGR4 ΔspxB killed S. aureus when we increased the challenge density to >108 CFU/ml (Fig. 5B). S. pneumoniae strain Pn20, isolated from the nasopharynx of a child (20), and its ΔspxB mutant derivative were also tested with essentially similar results (i.e., an increased density killed S. aureus [data not shown]). Furthermore, a similar 100-fold-increased density of TIGR4 ΔspxB killing S. aureus, in comparison to the TIGR4 wild type, was observed when another S. aureus strain, NRS049, was challenged (Fig. 5C). These results suggested that H2O2 generated by LctO was sufficient to induce killing of S. aureus. Confirming this, the TIGR4 ΔspxB ΔlctO mutant was unable to kill S. aureus even at a high density of ∼109 CFU/ml (Fig. 5B).
FIG 5.
S. pneumoniae (Spn) contact-mediated killing of S. aureus (Sau) requires enzymes SpxB and LctO. (A) Oxidation of pyruvate to acetyl∼P by the enzyme pyruvate oxidase (SpxB). The reaction uses molecular O2 and inorganic phosphate (Pi), producing CO2 and H2O2 (circled). Acetyl∼P is then converted to acetyl coenzyme A by acetate kinase (AckA) in a reaction that produces ATP. The enzyme lactate oxidase (LctO) catalyzes the formation of pyruvate from lactate, producing H2O2. (B) S. pneumoniae strains TIGR4, TIGR4 ΔlctO, TIGR4 ΔspxB, and TIGR4 ΔspxB ΔlctO were inoculated on blood agar plates at the densities indicated on the right (CFU per milliliter) concurrently with S. aureus strain Newman, which was inoculated at a density of ∼107 CFU/ml. (C) S. pneumoniae strains were inoculated at the densities shown at the top concurrently with S. aureus strain NRS049, inoculated at ∼107 CFU/ml. The agar plates were incubated overnight at 37°C.
We then used the microplate model to quantitatively assess S. aureus killing by these mutant strains. Experiments revealed that neither planktonic S. aureus nor biofilm S. aureus cells were killed by TIGR4 ΔspxB in comparison with the wild-type strain (Fig. 6A and B). The same phenotype was observed when two other independent TIGR4-derived ΔspxB mutants were tested (see Fig. S3 in the supplemental material). Similar to what we observed using the plate model, the TIGR4 ΔlctO mutant killed S. aureus strains at rates similar to that of wild-type TIGR4 (Fig. 6A and B). As expected, as a mutation in spxB was enough to block S. aureus killing in this model, the ΔspxB ΔlctO double mutant was unable to kill S. aureus strain Newman (not shown). Under the culture conditions utilized (i.e., incubation in Todd-Hewitt broth containing 0.5% [wt/vol] yeast extract [THY], with environmental oxygen and 5% CO2), S. pneumoniae strains TIGR4 and TIGR4 ΔlctO produced, after 4 h of incubation, ∼180 μM and ∼140 μM H2O2, respectively (Table 1). Cultures of three different spxB mutants, however, yielded undetectable levels of hydrogen peroxide (Table 1). Overall, our experiments demonstrated that both H2O2-producing enzymes, SpxB and LctO, contribute to the contact-dependent killing of S. aureus strains.
FIG 6.
A mutation in spxB, but not in lctO, renders S. pneumoniae (Spn) unable to kill S. aureus (Sau) in a microplate model. S. aureus strain Newman (∼1 × 106 CFU/ml) was inoculated alone (Ctrl) or along with the indicated S. pneumoniae strains (∼1 × 106 CFU/ml) in microplates containing THY and incubated for 4 h at 37°C. Bacteria were harvested and then diluted and plated onto SMA (A and B) or BAP with gentamicin (C and D) to obtain counts of S. aureus planktonic cells (A), S. aureus biofilms (B), S. pneumoniae planktonic cells (C), or S. pneumoniae biofilms (D). The error bars represent the standard errors of the means calculated using data from at least three independent experiments. (A and B) *, P < 0.05 compared to S. aureus control incubated alone. (D) For comparison, the median (CFU per milliliter) is shown inside two of the bars.
TABLE 1.
Production of H2O2 by TIGR4 and isogenic derivative mutants
| Strain | Production of H2O2 at (h): |
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 4 | |
| TIGR4 | 5.7 μM | 35.6 μM | 50.3 μM | 179.6 μM |
| TIGR4 ΔlctO | <50 nMa | 9.9 μM | 45.1 μM | 136.3 μM |
| TIGR4 ΔspxBU | <50 nM | <50 nM | <50 nM | <50 nM |
| TIGR4 ΔspxBE | <50 nM | <50 nM | <50 nM | <50 nM |
| TIGR4 ΔspxBE ΔlctO | <50 nM | <50 nM | <50 nM | <50 nM |
Limit of detection.
A hydroxyl radical (·OH) is generated during the interaction between S. pneumoniae and S. aureus to rapidly kill S. aureus.
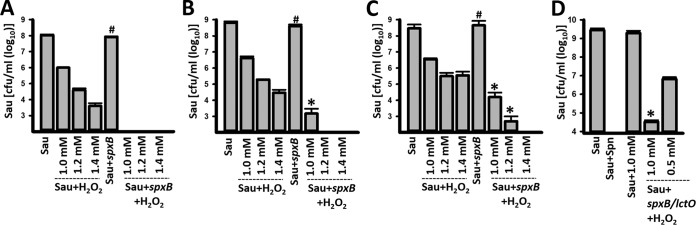
Given that our experiments demonstrated that even ∼5 mM pure H2O2 did not affect the viability of some S. aureus strains (Fig. 3) but that cultures of the same strains were killed by S. pneumoniae producing ∼36-fold less H2O2 (i.e., ∼140 μM), the possibility was raised that SpxB/LctO-produced H2O2 was converted into the ·OH radical. To test this hypothesis, we first conducted a dose-response study to identify three sublethal doses of H2O2 for S. aureus, 1.0, 1.2, and 1.4 mM (Fig. 7). For example, 1 mM H2O2 allowed the survival of >1 × 106 CFU/ml S. aureus when challenged against three different S. aureus strains (Fig. 7). We reasoned that if H2O2 is converted to a hydroxyl radical, then incubating S. aureus, TIGR4 ΔspxB (which does not produce significant amounts of H2O2), and a sublethal dose of H2O2 would allow killing. As shown in Fig. 7, the density of any of the three S. aureus strains incubated with TIGR4 ΔspxB was similar to the density in control wells containing S. aureus alone (Fig. 7A to C). Incubation of S. aureus; TIGR4 ΔspxB; and 1.0, 1.2, or 1.4 mM H2O2 was sufficient to completely eradicate cultures of S. aureus strain Newman, NRS408, and NRS049, respectively. Experiments with S. aureus strain Newman incubated with TIGR4 ΔspxB ΔlctO and 1 mM H2O2 showed essentially the same result (Fig. 7D). These experiments strengthened our hypothesis that H2O2 was converted into a hydroxyl radical (·OH).
FIG 7.
S. pneumoniae (Spn) produces a stronger oxidant from H2O2 to kill S. aureus (Sau) strains. S. aureus strain Newman (A and D), NRS408 (B), or NRS049 (C) was incubated alone at a density of ∼1 × 106 CFU/ml, with the indicated molarity of H2O2, TIGR4 ΔspxB (∼1 × 106 CFU/ml), TIGR4 ΔspxB (∼1 × 106 CFU/ml) and H2O2 or with TIGR4 ΔspxB lctO (∼1 × 106 CFU/ml). Bacteria were incubated for 4 h at 37°C and then harvested, diluted, and plated on SMA to obtain counts of S. aureus. The error bars represent the standard errors of the means calculated using data from at least three independent experiments. *, P < 0.05 compared to the corresponding concentration of H2O2; #, P > 0.30 compared to S. aureus control incubated alone.
Thiourea (15 mM), sodium salicylate, and dimethyl sulfoxide (Me2SO [300 mM]), are specific ·OH scavengers (34); thiourea and Me2SO reduced H2O2 killing of S. aureus by 98% and 38%, respectively (34). As shown in Fig. 8A, incubating S. aureus strain Newman, S. pneumoniae, and 10 mM thiourea was enough to significantly inhibit killing of S. aureus, whereas 20 mM and 40 mM completely inhibited H2O2-mediated killing. The density of S. pneumoniae was not affected by incubation with any amount of thiourea (Fig. 8B). Similar protection from challenge with S. pneumoniae was conferred on S. aureus by incubating the two species, along with a scavenger of hydroxyl radicals, Me2SO (Fig. 8C and D) or sodium salicylate (see Fig. S4 in the supplemental material).
FIG 8.
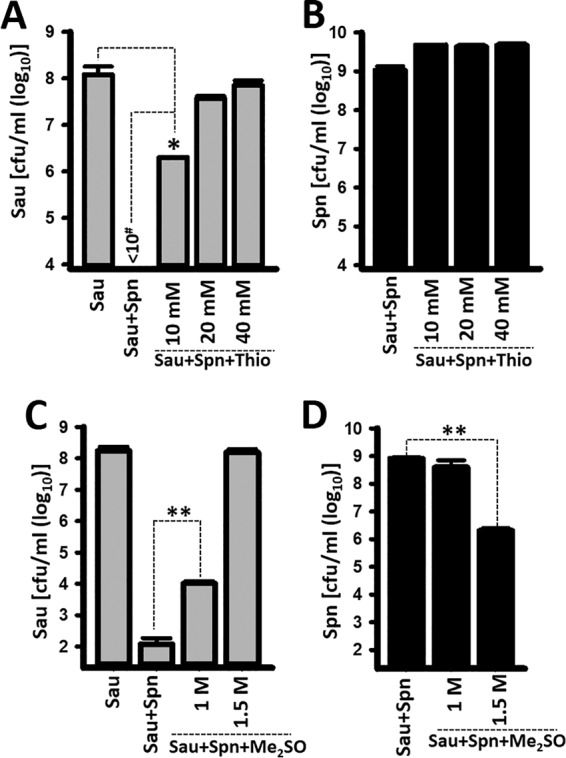
Scavengers of ·OH block S. pneumoniae (Spn)-induced killing of S. aureus (Sau). S. aureus strain Newman was inoculated (∼1 × 106 CFU/ml) alone, with S. pneumoniae TIGR4 (∼1 × 106 CFU/ml), with S. pneumoniae and thiourea (Thio), or with S. pneumoniae and Me2SO. After 4 h of incubation at 37°C, bacteria were harvested, diluted, and plated to obtain counts of S. aureus (A and C) or S. pneumoniae (B and D). The error bars represent the standard errors of the mean calculated using data from at least three independent experiments. *, P < 0.05 compared with S. aureus control; **, P < 0.0005 compared to the density of S. aureus incubated with S. pneumoniae; #, limit of detection.
To identify the in vivo formation of hydroxyl radicals, we utilized the α-(4-pyridyl-1-oxide)-N-tert-butyl nitrone (4-POBN)–ethanol spin-trapping system. Figure 9 shows a marked increase in the hydroxyethyl radical spin adduct in bacterial cells of the TIGR4 wt in comparison to signals from a reaction mixture containing TIGR4 wt cells and a reaction mixture with bacterial cells harvested from cultures of the TIGR4 ΔspxB ΔlctO double mutant.
FIG 9.
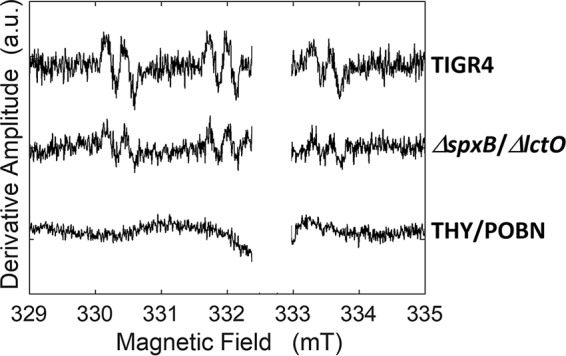
Detection of ·OH radicals in pneumococci by 4-POBN and DETAPAC-ethanol spin trapping. Shown are the EPR spectra of 4-POBN spin trapping in wild-type and double-mutant samples and, for comparison, a medium-only sample. The unassigned signal near the free-electron g value of 2.0023 (333.0 mT) was deleted. The EPR conditions were as follows: microwave frequency, 9.346 GHz; microwave power, 10 mW; modulation amplitude, 0.2 mT; modulation frequency, 100 kHz; temperature, 295 K. The spectra of wild-type and double-mutant samples represent an average of 96 scans minus the bacterial-medium spectrum. The bacterial-medium spectrum represents an average of 224 scans, with polynomial baseline correction. The spectra were baseline corrected using a polynomial function.
DISCUSSION
We have demonstrated in this study that the interaction between S. pneumoniae and S. aureus stimulates the conversion of H2O2 into a stronger oxidant, the hydroxyl radical, ·OH, to rapidly kill S. aureus bacteria. The toxic effects of several pneumococcal strains against S. aureus have been documented (14, 19, 20, 23, 27, 38). Hydrogen peroxide had long been believed to be the killing factor, but contrasting data suggesting that H2O2 was not required for killing have been published in the last few years (23, 30–32). Compelling evidence within this study has now identified highly reactive ·OH radicals generated from H2O2, because of interaction between the species, as the effector of such a mechanism. Given that H2O2 is permeable but a sublethal dose of H2O2 killed S. aureus strains when incubated along with H2O2-deficient S. pneumoniae strains, the conversion to ·OH radicals may be facilitated by pneumococcal cells. Certainly, other possibilities exist, including increased free Fe2+ in S. aureus due to the interaction with S. pneumoniae, which could be the result of stimulated iron uptake. The ·OH generated reacts at nearly diffusion-limited rates near the site of its generation (22, 39).
Although S. aureus can also produce H2O2, its potent catalase fully scavenges the H2O2 before it can cross the cytoplasmic membrane, and therefore, levels of hydrogen peroxide in the supernatant are undetectable (19). Intracellular levels of H2O2 in S. aureus have not been investigated, but in E. coli, these levels are maintained at 0.1 to 0.2 μM during aerobic growth (40). Therefore, the experiments in this study, along with data from other laboratories (14, 19, 20, 31, 41), support a model where S. pneumoniae secretes large amounts of H2O2 near S. aureus bacteria—but beyond a concentration that catalase would successfully scavenge—and then, due to a still unknown mechanism, it is rapidly converted into ·OH radicals that intoxicate S. aureus cells. We hypothesize that the target(s) of ·OH radicals on S. aureus cells is absent in pneumococci, and therefore, S. pneumoniae is less susceptible to these radicals (see below for more details). S. aureus intoxication with ·OH radicals may be accompanied by degradation of DNA, as confocal micrographs and quantitative PCRs demonstrated absence of DNA in S. aureus bacteria incubated with H2O2-producing S. pneumoniae. Hydroxyl radicals attack at the sugar or the base of the DNA, leading to sugar fragmentation, base loss, and a strand break with a terminal fragmented sugar residue (42). The resistance of S. pneumoniae to DNA damage has been speculated to occur by sequestration of Fe2+, required to produce ·OH through the Fenton reaction, away from its DNA (22).
Animal studies demonstrated that S. pneumoniae (TIGR4) and S. aureus (Newman) cohabited in the nasal cavities of rats when the strains were inoculated at the same density (30, 31). One would expect that in this animal cocolonization model, only S. pneumoniae would be able to colonize, but this outcome did not occur. Instead, coinoculation of wild-type S. pneumoniae did not affect colonization by S. aureus, and neither a mutation in the pneumococcal spxB gene nor a mutation in the S. aureus katG gene (encoding catalase) had a positive or negative effect, respectively, on S. aureus density. If S. pneumoniae kills S. aureus, why did they cocolonize? Experiments in this study offer an explanation. When TIGR4 and S. aureus strain Newman were inoculated at similar densities, S. aureus in fact survived the challenge with S. pneumoniae (Fig. 1). However, with an outcompeting density of S. pneumoniae, staphylococci succumbed to the challenge. The killing was negated when S. aureus was incubated with the hydrogen peroxide production-defective S. pneumoniae ΔspxB ΔlctO mutant. We speculate that, in the surface-bound environment of the nasal cavity of rats or in the plate model, the close proximity of S. pneumoniae to S. aureus allows inactivation of H2O2 by the S. aureus-produced catalase but that increased production of H2O2 (i.e., by outcompeting pneumococci) overcomes the catalase-mediated inactivation, and thus, H2O2 is converted into ·OH radicals. We hypothesize that the conflicting results obtained in population studies where a negative association, or no association, has been demonstrated between concurrent carriage (i.e., colonization) of S. pneumoniae and S. aureus would have been resolved if the density of the strains had been taken into consideration.
The finding that the level of sensitivity is variable across S. aureus strains (Fig. 3A) and the surprising result that S. pneumoniae and H2O2 sensitivity in S. aureus are not correlated (Fig. 3B) suggest that other, undiscovered factors modulate S. aureus killing. We found that S. aureus strains can apparently cross-protect in mixtures, suggesting that a diffusible molecule is possibly involved. This result needs to be followed up in future work. Since the phenotype is variable across S. aureus strains without being closely linked to a particular clade, it may be possible to identify the genetic loci responsible by using a hypothesis-free genome-wide association study (GWAS) approach.
Studies conducted with E. coli demonstrated that H2O2-mediated killing occurred only in actively metabolizing cells (35, 42). Exogenously added H2O2 also kills S. aureus strains with a calculated 90% lethal dose (LD90) and MBC of ∼10 mM (19, 34). A 10-fold decrease in the sublethal concentration was determined and utilized in our studies (presented in Fig. 7) against strain Newman and two MRSA strains. The membrane-permeable H2O2 enters bacterial cells, reacting with the intracellular Fe2+ by Fenton reactions to produce ·OH radicals (34). The concentration of H2O2 quantified in the supernatant of S. pneumoniae to reach the LD90, however, was ∼7-fold lower (∼140 μM) than a sublethal dose of exogenous H2O2 and ∼70-fold lower than the LD90 of exogenously added H2O2 (∼10 mM). This decreased amount of H2O2 in the S. pneumoniae supernatant, the fact that an H2O2 production-defective strain, when incubated with a sublethal dose of H2O2 (∼1 mM), killed S. aureus strains, our experiments showing that S. aureus killing was blocked by hydroxyl scavengers, and spin-trapping experiments supported the hypothesis that production of ·OH radicals was stimulated by the interaction between pneumococcal cells and S. aureus.
Given that H2O2 targets metabolically active (i.e., respiring) E. coli cells (35) and that S. aureus undergoes cellular respiration whereas S. pneumoniae does not encode proteins of the respiratory chain (43), we speculate that hydroxyl radicals either target a component(s) of the respiratory chain or require a reducing equivalent from the respiratory chain to generate toxic hydroxyl radicals. In fact, the pneumococcus could be intoxicated by incubating it with increasing amounts of H2O2, but it required at least 10 mM H2O2 to completely eradicate S. pneumoniae bacteria. A challenge with such a large amount of H2O2 was partially inhibited by incubation with the ·OH scavenger thiourea (see Fig. S4 in the supplemental material).
An investigation by Selva et al. suggested that H2O2-mediated interference was triggered by lysogenic S. aureus phages, although in their study they obtained a 3- to 4-log-unit reduction in S. aureus density, but not eradication, under similar culture conditions (41). Whereas some S. aureus strains utilized in the current study may be lysogenic, we tested a nonlysogenic S. aureus strain (RN4220) and obtained density-dependent, SpxB/LctO-dependent killing (data not shown). The detailed mechanism(s) and its target(s) are under active investigation in our laboratories. The target(s) of the ·OH radicals represents an exciting new alternative for the development of therapeutics against S. aureus infections.
MATERIALS AND METHODS
Bacterial strains and culture media.
The S. pneumoniae and S. aureus wild-type strains and mutant derivatives utilized in this study are listed in Table 2. S. pneumoniae strains were cultured on blood agar plates (BAP) or BAP with 25 μg/ml gentamicin, whereas S. aureus strains were grown on salt mannitol agar (SMA) plates or tryptic soy agar (TSA) plates with or without 5 μg/ml optochin or on Luria-Bertani agar (LBA) (1% tryptone [Becton-Dickinson], 0.5% yeast extract, 1% NaCl, and 1.5% agar [Becton-Dickinson]). THY was utilized in all the experiments.
TABLE 2.
Pneumococcal and staphylococcal strains used in this study
| Strain | Descriptiona | Reference(s) or source |
|---|---|---|
| TIGR4 | Invasive clinical isolate; phenotype CSP2; capsular serotype 4 | 43 |
| TIGR4 ΔspxBH | TIGR4 with an insertion within the spxB gene; spxB::kan-rpsL+ | 20, 27 |
| TIGR4 ΔspxBU | TIGR4 with a deletion of the spxB gene by transformation of erm(B); Eryr cassette | 13 |
| SPJV29 (TIGR4 ΔspxBE) | TIGR4 with a deletion of the spxB gene by transformation of erm(B); Eryr cassette | This study |
| TIGR4 ΔlctO | TIGR4 with a deletion of the lctO gene by transformation of a spectinomycin cassette | This study |
| TIGR4 ΔspxB ΔlctO | TIGR4 ΔspxBU with a deletion of the lctO gene by transformation of a spectinomycin cassette | This study |
| Pn20 | Serotype 35B; nasopharyngeal human isolate | 20 |
| Pn20ΔspxB | Pn20 ΔspxB::kan-rpsL+ by transformation with PCR product of TIGR4 ΔspxBH | 20 |
| S. aureus Newman | NCTC 8178, ATCC 13420 | 56 |
| NRS408J | S. aureus NRS408 spontaneous rifampin-resistant mutant | This study |
| RN4220 | S. aureus Hla− nonlysogenic strain | 57 |
Eryr, erythromycin resistance; H, spxB mutant strain obtained from M. Lipsitch at Harvard; U, spxB mutant strain prepared by C. J. Orihuela's group at UAB; E, spxB mutant prepared at Emory University (SPJV29).
Preparation of inoculum for experiments.
The inoculum was prepared essentially as previously described (44, 45). Briefly, an overnight BAP (for S. pneumoniae) or LBA (for S. aureus) culture was used to prepare a cell suspension in THY broth to an optical density at 600 nm (OD600) of ∼0.08. This suspension was incubated at 37°C in a 5% CO2 atmosphere until the culture reached an OD600 of ∼0.2 (early log phase). Then, glycerol was added to give a final 10% (vol/vol) concentration, and the suspension was stored at −80°C until it was used. An aliquot of these stocks was further diluted and plated to obtain bacterial counts (CFU per milliliter).
Mixed-culture killing assay on plates.
Blood agar plates were coinoculated with both S. pneumoniae and S. aureus strains at different densities ranging from ∼106 through ∼109 CFU/ml. The inoculated plates were then incubated at 37°C in a 5% CO2 atmosphere overnight. A creamy-yellow color on the BAP culture and the presence of a beta-hemolytic halo around each culture indicated growth of S. aureus. To quantify the densities of strains, cultures were harvested and an aliquot was diluted and plated to obtain bacterial counts, whereas DNA was extracted from another aliquot using a QIAamp DNA minikit (Qiagen) according to the manufacturer’s instructions. DNA preparations were eluted with 100 μl of elution buffer, quantified using a Nanodrop spectrophotometer, and stored at –80°C until they were used.
Quantitative PCRs.
Strain-specific qPCRs were performed to measure the densities of strains. Primers, probes, and the concentrations utilized are listed in Table 3. The total S. pneumoniae density was quantified using the panpneumococcus lytA assay (46), and detection of the nuc gene was used to quantify S. aureus density (47). Reactions were run along with serially diluted DNA standards corresponding to 4.29 × 105, 4.29 × 104, 4.29 × 103, 4.29 × 102, 4.29 × 101, and 2.14 × 101 genome equivalents of S. pneumoniae or 3.29 × 105, 3.29 × 104, 3.29 × 103, 3.29 × 102, 3.29 × 101, and 3.14 × 101 genome equivalents of S. aureus. Reactions were carried out using a Bio-Rad CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA) and the following cycling parameters: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The final numbers of genome equivalents per milliliter were calculated using the CFX software (Bio-Rad, Hercules, CA).
TABLE 3.
Primers and probes used in this study
| Target | Primer or probe sequence (5′–3′)a | Reference |
|---|---|---|
| S. pneumoniae lytA | F, ACGCAATCTAGCAGATGAAGCA | 46 |
| R, TCGTGCGTTTTAATTCCAGCT | ||
| Probe, FAM-TGCCGAAAACGCTTGATACAGGGAG | ||
| S. aureus nuc | F, GTTGCTTAGTGTTAACTTTAGTTGTA | 47 |
| R, AATGTCGCAGGTTCTTTATGTAATTT | ||
| Probe, HEX-AAGTCTAAGTAGCTCAGCAAATGCA | ||
| UP_spxB_DN | F, TATCAATCACGCTCCTGC | 13 |
| R, CTCGTTATGGACAATGCT | ||
| XbaI-erm(B)-XhoI | F, CAGTCTAGAAAAAATTTGTAATTAAGAAGGAGT | This study |
| R, CAGCTCGAGCCAAATTTACAAAAGCGACTCA | ||
| lctO_UP-FW | TGGAAAGTAGGCATCAGC | This study |
| lctO_UP (Spl_KnSpc)-RV | TCCTCCTCACTATTTTGATAAACTGTCCTCCTCG | This study |
| lctO_DN (Spl_KnSpc)-FW | TGGAAACACTTCGTGAAGACTTAAAATTGTATTG | This study |
| lctO_DN-RV | CGTAATTCCACTTGATCC | This study |
| SpcK7-FW | CAAAATAGTGAGGAGGA | This study |
| SpcK7-RV | TTCACGAAGTGTTTCCA | This study |
F, forward; R, reverse; FAM, 6-carboxyfluorescein; HEX, 6-carboxy-2,4,4,5,7,7-hexachlorofluorescein. Sequences overlapping the spectinomycin resistance cassette are in boldface.
Determination of S. aureus strain sensitivity to S. pneumoniae.
Inocula of S. aureus strains prepared as stated above, with known densities, were serially diluted in THY to generate inocula. Decreasing densities of S. aureus strains were inoculated along with 1.5 × 107 CFU/ml of S. pneumoniae and incubated for 4 h in 96-well microplates. The cultures were diluted and plated on TSA supplemented with optochin (5 μg/ml). The highest density of each S. aureus strain killed by 1.5 × 107 CFU/ml of S. pneumoniae within 4 h was then determined.
Competition between resistant and sensitive S. aureus strains under S. pneumoniae selection.
For the competition experiments, we prepared an NRS408-derived rifampin-resistant mutant (NRS408J) by plating an overnight tryptic soy broth (TSB) culture concentrated 10-fold on a TSA plate supplemented with 4 μg/ml rifampin. S. pneumoniae strain TIGR4 (1.5 × 107 CFU/ml) was then cocultured in THY with three different mixtures of S. aureus strains (5 × 106 CFU/ml of each S. aureus strain). A mixture contained NRS049, NRS408J, or both strains NRS049 and NRS408J. Experiments with negative controls without S. pneumoniae were performed for each of the S. aureus strain mixtures. All the cultures (3 ml each) were incubated without shaking at 37°C in a 5% CO2 atmosphere. Cocultures were sampled at 0-, 15-, and 30-min and 1-, 2-, 3-, and 4-h time points. Sample dilutions were then spotted on TSA plates supplemented with 16 μg/ml tetracycline or 4 μg/ml rifampin to detect NRS049 and the NRS408 rifampin-resistant mutant (NRS408J), respectively. The following day, spot colonies were enumerated to calculate the number of residual CFU per milliliter at each time point.
Growth curves and hydrogen peroxide sensitivity.
Strains were grown at 37°C in TSB or TSB supplemented with 0.1, 2.5, or 5.0 mM H2O2. The initial ODs were normalized to between 0.1 and 0.2 (roughly a 1/100 dilution of the overnight culture) readings from a microplate reader (Eon; BioTek, Inc.) before the growth curves were performed. Growth curves (OD600) were determined in a plate reader collecting data every 10 minutes for up to 12 h.
Confocal-microscopy studies.
Bacteria grown in experiments performed using the plate model (Fig. 1A) were imprinted onto rounded glass slides and immediately fixed with 2% paraformaldehyde (PFA) for 15 min at room temperature. The fixed bacteria were then blocked with 1% bovine serum albumin (BSA) for 30 min at 37°C and incubated first with a rabbit polyclonal anti-S. aureus antibody (4 μg/ml; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature, followed by phosphate-buffered saline (PBS) washes and 1 h of incubation with a secondary Alexa Fluor 555-labeled goat anti-rabbit antibody (20 μg/ml; Molecular Probes). The preparation was then washed with sterile PBS and incubated for 30 min with anti-S. pneumoniae antibodies raised in rabbit (Statens Serum Institute) that had been previously labeled with Alexa Fluor 488 (50 μg/ml; Molecular Probes). The stained preparations were finally washed twice with PBS, mounted with ProLong Diamond antifade mountant with DAPI (Molecular Probes), and analyzed with an Olympus FV1000 confocal microscope. The confocal images were analyzed with Image J version 1.49k (National Institutes of Health).
Preparation of TIGR4-derived ΔspxB, ΔlctO, and ΔspxB ΔlctO mutants.
Isogenic spxB mutant derivatives of S. pneumoniae strain TIGR4 (43) were prepared as described in our recent publication (13). A deletion within the lctO gene in the wild-type TIGR4, or TIGR4 ΔspxB, was generated using a cassette containing lctO upstream (lctO_UP-FW and lctO_UP-RV) and downstream (lctO_DN-FW and lctO_DN-RV) sequences and the spectinomycin resistance gene (SpcK7-FW and SpcK7-RV). This cassette was prepared by splicing overlap extension PCR with primers (in parentheses above and listed in Table 3) (48, 49) and transformed into pneumococci using standard procedures (50). BAP with spectinomycin (100 μg/ml) or erythromycin (0.5 μg/ml) was used to select lctO or spxB mutants, respectively. All deletions were confirmed by PCR and sequencing.
Microplate model to investigate killing of S. aureus by S. pneumoniae strains.
An S. pneumoniae strain was inoculated along with S. aureus strain Newman at a density of ∼1 × 106 CFU/ml in a 6-well microplate containing THY and incubated for 4 h at 37°C in a 5% CO2 atmosphere. Control wells were inoculated with only S. pneumoniae or only S. aureus. In another set of experiments, S. aureus was inoculated, along with increasing amounts of hydrogen peroxide (Sigma), with the TIGR4 ΔspxBE (SPJV29) mutant alone or with TIGR4 ΔspxBE and hydrogen peroxide. Technical duplicates were included throughout these experiments. At the end of the incubation, planktonic cells were removed, diluted, and plated onto BAP with gentamicin to obtain the number of CFU of S. pneumoniae per milliliter or onto LBA plates with optochin to obtain the number of CFU of S. aureus per milliliter. The biofilms were washed once with PBS, resuspended in 1 ml of sterile PBS, and sonicated for 15 s using a Bransonic ultrasonic water bath (Branson, Danbury, CT), followed by extensive pipetting to remove the remaining attached biofilm bacteria. The biofilms were then diluted and plated as described above. Experiments were repeated three times.
Quantifying production of hydrogen peroxide.
S. pneumoniae strains were inoculated at a density of ∼1 × 106 CFU/ml in a 6-well plate containing THY and incubated at 37°C in a 5% CO2 atmosphere. The supernatant containing planktonic bacteria was collected 0, 1, 2, or 4 h postinoculation and then centrifuged at 8,000 rpm for 10 min to separate the planktonic cells; the supernatant was further filtered through a 0.45-μm syringe filter. The concentration of H2O2 present in each cell-free supernatant was assessed using an Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen) following the manufacturer’s instructions.
Detection of hydroxyl radicals by spin trapping.
A spin-trapping system was utilized to detect the formation of hydroxyl radicals essentially as described previously (38, 51). Briefly, pneumococci were grown in 100 ml of THY broth to an OD600 of 0.4 to 0.5. The pellets were harvested and washed twice with Hanks balanced salt solution lacking calcium and magnesium (HBSS) and finally resuspended in 500 μl of HBSS. The reaction mixtures included 200 μl of cell suspension, 100 μM diethylenetriaminepentaacetic acid (DETAPAC), 10 mM 4-POBN, 170 mM ethanol, and HBSS for a final volume of 1 ml. The reaction mixture was incubated for 10 min at room temperature and immediately transferred to a quartz capillary tube (2-mm outer diameter), and X-band electron paramagnetic resonance (EPR) spectra were acquired under the following conditions: microwave frequency, 9.346 GHz; microwave power, 10 mW; modulation amplitude, 0.2 mT; modulation frequency, 100 kHz; temperature, 295 K; the spectra represent an average of 92 scans minus a medium baseline (224 scans).
S. aureus strain sequence and phylogeny.
In brief, Nextera random shotgun libraries were prepared, and 300-bp paired-end reads were sequenced to >20 to 100× coverage. Sequence types were ascribed based on BLASTN against the BIGSdb database (52, 53). The phylogeny was created using Parsnp (54) based on mapping to the Newman reference. The tree was midpoint rooted and visualized using iTOL (55).
Statistical analysis.
Differences between S. aureus strain sensitivities to S. pneumoniae were calculated by a Kruskal-Wallis test for overall variability in the collection. The test was performed for all strains with or without the two most sensitive strains (NRS408 and NRS170). All other statistical analysis was performed using a two-tailed Student t test and the software SigmaPlot version 14.0.
Accession number(s).
The data were deposited in NCBI BioProject under accession no. PRJNA289526 as part of a larger study that will be described in more detail elsewhere.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the National Institutes of Health (NIH) (R21AI112768-01A1 and 1R21AI144571-01 to J.E.V.). The confocal studies were in part supported by funds from the Integrated Cellular Imaging (ICI) pediatric core and the Emory+Children’s Pediatric Research Center to J.E.V. Funds from the Raymond F. Schinazi International Exchange Program (SIEP), a faculty exchange program between the Read laboratory at Emory University and the Hadassah Medical Organization to promote collaborative research and education, supported O.G. EPR spectroscopy was supported by NIH grants R01DK054514 and NIH RR17767 (to K.W.). U.A.A.-Z. thanks CONACyT for mobility scholarship 291250.
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
We thank Faidad Khan (Emory University) for his assistance in some experiments and Marc Lipsitch (Harvard T. H. Chan School of Public Health) for providing us with TIGR4 and Pn20 hydrogen peroxide-deficient mutants.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00474-19.
REFERENCES
- 1.Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, Rahav G, Rubinstein E. 2004. Association between carriage of Streptococcus pneumoniae, and Staphylococcus aureus in children. JAMA 292:716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- 2.Bakaletz LO. 2007. Bacterial biofilms in otitis media: evidence and relevance. Pediatr Infect Dis J 26:S17–S19. doi: 10.1097/INF.0b013e318154b273. [DOI] [PubMed] [Google Scholar]
- 3.Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, Satzke C. 2013. Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine 31:2333–2342. doi: 10.1016/j.vaccine.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Shak JR, Cremers AJ, Gritzfeld JF, de Jonge MI, Hermans PW, Vidal JE, Klugman KP, Gordon SB. 2014. Impact of experimental human pneumococcal carriage on nasopharyngeal bacterial densities in healthy adults. PLoS One 9:e98829. doi: 10.1371/journal.pone.0098829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shak JR, Vidal JE, Klugman KP. 2013. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21:129–135. doi: 10.1016/j.tim.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP. 2013. Quorum-sensing systems LuxS/Autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81:1341–1353. doi: 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, Griffin MR, Verastegui H, Hartinger SM, Gil AI, Lanata CF, Klugman KP. 2013. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 32:72–77. doi: 10.1097/INF.0b013e318270d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rumke HC, Verbrugh HA, Hermans PW. 2004. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 9.Chao Y, Marks LR, Pettigrew MM, Hakansson AP. 2014. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect Microbiol 4:194. doi: 10.3389/fcimb.2014.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gritzfeld JF, Cremers AJ, Ferwerda G, Ferreira DM, Kadioglu A, Hermans PW, Gordon SB. 2014. Density and duration of experimental human pneumococcal carriage. Clin Microbiol Infect 20:O1145–1151. doi: 10.1111/1469-0691.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/jb.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenoy AT, Brissac T, Gilley RP, Kumar N, Wang Y, Gonzalez-Juarbe N, Hinkle WS, Daugherty SC, Shetty AC, Ott S, Tallon LJ, Deshane J, Tettelin H, Orihuela CJ. 2017. Streptococcus pneumoniae in the heart subvert the host response through biofilm-mediated resident macrophage killing. PLoS Pathog 13:e1006582. doi: 10.1371/journal.ppat.1006582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brissac T, Shenoy AT, Patterson LA, Orihuela CJ. 2018. Cell invasion and pyruvate oxidase derived H2O2 are critical for Streptococcus pneumoniae mediated cardiomyocyte killing. Infect Immun 86:e00569-17. doi: 10.1128/IAI.00569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regev-Yochay G, Malley R, Rubinstein E, Raz M, Dagan R, Lipsitch M. 2008. In vitro bactericidal activity of Streptococcus pneumoniae and bactericidal susceptibility of Staphylococcus aureus strains isolated from cocolonized versus noncocolonized children. J Clin Microbiol 46:747–749. doi: 10.1128/JCM.01781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya M, Wozniak DJ, Stoodley P, Hall-Stoodley L. 2015. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev Anti-Infect Ther 13:1499–1516. doi: 10.1586/14787210.2015.1100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chochua S, D'Acremont V, Hanke C, Alfa D, Shak J, Kilowoko M, Kyungu E, Kaiser L, Genton B, Klugman KP, Vidal JE. 2016. Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLoS One 11:e0167725. doi: 10.1371/journal.pone.0167725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan HJ, Avery OT. 1924. Growth-inhibitory substances in pneumococcus cultures. J Exp Med 39:335–346. doi: 10.1084/jem.39.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery OT, Morgan HJ. 1924. The occurrence of peroxide in cultures of Pneumococcus. J Exp Med 39:275–287. doi: 10.1084/jem.39.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pericone CD, Bae D, Shchepetov M, McCool T, Weiser JN. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J Bacteriol 184:4392–4399. doi: 10.1128/jb.184.16.4392-4399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. 2013. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol 21:187–195. doi: 10.1016/j.tim.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Khan F, Wu X, Matzkin GL, Khan MA, Sakai F, Vidal JE. 2016. Streptococcus pneumoniae eradicates preformed Staphylococcus aureus biofilms through a mechanism requiring physical contact. Front Cell Infect Microbiol 6:104. doi: 10.3389/fcimb.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisher JP, Tsui HT, Ramos-Montanez S, Hentchel KL, Martin JE, Trinidad JC, Winkler ME, Giedroc DP. 2017. Biological and chemical adaptation to endogenous hydrogen peroxide production in Streptococcus pneumoniae D39. mSphere 2:e00291-16. doi: 10.1128/mSphere.00291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos-Montanez S, Tsui HC, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham LT, Winkler ME. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol 67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- 26.Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, Yoshida S. 2008. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol 190:3572–3579. doi: 10.1128/JB.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park B, Nizet V, Liu GY. 2008. Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J Bacteriol 190:2275–2278. doi: 10.1128/JB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Echlin H, Frank MW, Iverson A, Chang TC, Johnson MD, Rock CO, Rosch JW. 2016. Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLoS Pathog 12:e1005951. doi: 10.1371/journal.ppat.1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant JC, Dabbs RC, Oswalt KL, Brown LR, Rosch JW, Seo KS, Donaldson JR, McDaniel LS, Thornton JA. 2016. Pyruvate oxidase of Streptococcus pneumoniae contributes to pneumolysin release. BMC Microbiol 16:271. doi: 10.1186/s12866-016-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolis E. 2009. Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J Bacteriol 191:571–575. doi: 10.1128/JB.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolis E, Yates A, Levin BR. 2010. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol 10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiss-Mandel A, Regev-Yochay G. 2016. Staphylococcus aureus and Streptococcus pneumoniae interaction and response to pneumococcal vaccination: myth or reality? Hum Vaccin Immunother 12:351–357. doi: 10.1080/21645515.2015.1081321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoepelman IM, Bezemer WA, Vandenbroucke-Grauls CM, Marx JJ, Verhoef J. 1990. Bacterial iron enhances oxygen radical-mediated killing of Staphylococcus aureus by phagocytes. Infect Immun 58:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repine JE, Fox RB, Berger EM. 1981. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J Biol Chem 256:7094–7096. [PubMed] [Google Scholar]
- 35.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 36.Quillin SJ, Hockenberry AJ, Jewett MC, Seifert HS. 2018. Neisseria gonorrhoeae exposed to sublethal levels of hydrogen peroxide mounts a complex transcriptional response. mSystems 3:e00156-18. doi: 10.1128/mSystems.00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong SM, Alugupalli KR, Ram S, Akerley BJ. 2007. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol 64:1375–1390. doi: 10.1111/j.1365-2958.2007.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185:6815–6825. doi: 10.1128/jb.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Flecha B, Demple B. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem 270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 41.Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, Novick RP, Penadés JR. 2009. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci U S A 106:1234–1238. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 43.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 44.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Jacobs NT, Bozio C, Palm P, Lattar SM, Hanke CR, Watson DM, Sakai F, Levin BR, Klugman KP, Vidal JE. 2017. Competitive dominance within biofilm consortia regulates the relative distribution of pneumococcal nasopharyngeal density. Appl Environ Microbiol 83:e00953-17. doi: 10.1128/AEM.00953-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho MDGS, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilic A, Muldrew KL, Tang YW, Basustaoglu AC. 2010. Triplex real-time polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci and determination of methicillin resistance directly from positive blood culture bottles. Diagn Microbiol Infect Dis 66:349–355. doi: 10.1016/j.diagmicrobio.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 50.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park S, You X, Imlay JA. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci U S A 102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595–510. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petit RA III, Read TD. 2018. Staphylococcus aureus viewed from the perspective of 40,000+ genomes. Peer J 6:e5261. doi: 10.7717/peerj.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524–510. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boake WC. 1956. Antistaphylocoagulase in experimental staphylococcal infections. J Immunol 76:89–96. [PubMed] [Google Scholar]
- 57.Nair D, Memmi G, Hernandez D, Bard J, Beaume M, Gill S, Francois P, Cheung AL. 2011. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol 193:2332–2335. doi: 10.1128/JB.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.