Spore-forming bacteria are problematic for the food industry, as spores can survive decontamination procedures and subsequently revive in food products, with the risk of food spoilage and foodborne disease. The Ynd and GerA germination receptors (GRs) cooperate in triggering efficient germination of Bacillus licheniformis spores when nutrients are present in the surrounding environment. This study shows that the single B subunit of GerA is essential for the cooperative function between Ynd and GerA, while the three B subunits of the Ynd GR are dispensable. The ability of GRs lacking individual subunits to stimulate germination together with other GRs could explain why ger operons lacking GR subunit genes are maintained in genomes of spore-forming species.
KEYWORDS: Bacillus licheniformis, spore germination, germination receptor, endospore, germinant, Ynd, GerK, GerA
ABSTRACT
Germination of Bacillus spores is triggered by the binding of specific nutrients to germinant receptors (GRs) located in the spore’s inner membrane. The GRs typically consist of A, B, and C subunits, encoded by tricistronic ger operons. The Bacillus licheniformis genome contains the gerA family operons gerA, ynd, and gerK. In contrast to the ABC(D) organization that characterizes gerA operons of many Bacillus species, B. licheniformis genomes contain a pentacistronic ynd operon comprising the yndD, yndE3, yndE2, yndF1, and yndE1 genes encoding A, B, B, C, and B GR subunits, respectively (subscripts indicate paralogs). Here we show that B. licheniformis spores can germinate in the absence of the Ynd and GerK GRs, although cooperation between all three GRs is required for optimal germination with amino acids. Spores carrying an incomplete set of Ynd B subunits demonstrated reduced germination efficiencies, while depletion of all three Ynd B subunits restored germination of the spore population to levels only slightly lower than those of wild-type spores at high germinant concentrations. This suggests that the presence of an incomplete set of Ynd B subunits exhibits a dominant negative effect on germination and that the A and C subunits of the Ynd GR are sufficient for the cooperative functionality between Ynd and GerA. In contrast to the B subunits of Ynd, the B subunit of GerA was essential for amino acid-induced germination. This study provides novel insights into the role of individual GR subunits in the cooperative interaction between GRs in triggering spore germination.
IMPORTANCE Spore-forming bacteria are problematic for the food industry, as spores can survive decontamination procedures and subsequently revive in food products, with the risk of food spoilage and foodborne disease. The Ynd and GerA germination receptors (GRs) cooperate in triggering efficient germination of Bacillus licheniformis spores when nutrients are present in the surrounding environment. This study shows that the single B subunit of GerA is essential for the cooperative function between Ynd and GerA, while the three B subunits of the Ynd GR are dispensable. The ability of GRs lacking individual subunits to stimulate germination together with other GRs could explain why ger operons lacking GR subunit genes are maintained in genomes of spore-forming species.
INTRODUCTION
When starved for nutrients, many Bacillus species differentiate into the endospore (spore) form, which is a metabolically dormant, highly stress-resistant, and nonreproductive differentiation state (1, 2). The spores can stay dormant for long periods and survive environmental stressors that will kill vegetative bacteria (3). When survival conditions improve, the bacteria will initiate their metabolism and rapidly return to vegetative growth through the process of spore germination (3).
In Bacillus species, spore germination can be induced by exposure to biochemical and physical stimuli (3). However, in nature, spore germination is most likely triggered by exposure to nutrient compounds (germinants) such as amino acids, nucleosides, and sugars (3). In Bacillus spores, germinants are recognized by the GerA family of germinant receptors (GRs), located in the spore’s inner membrane (3). Activation of GRs by their cognate nutrient germinant(s) induces subsequent germination events, including the release of spore’s large depot of Ca2+-dipicolinic acid (CaDPA), core rehydration, hydrolysis of the spore’s peptidoglycan cortex, and, finally, outgrowth of the vegetative cell (3).
There is a large diversity among GRs with regard to their recognition pattern and responsiveness to germinants, and most Bacillus species carry genes encoding several different GerA family GRs (2, 4, 5). Some GRs function alone, while others require cooperation with another GR to trigger spore germination in response to a single germinant compound or a combination of germinant compounds (5). However, it is still largely unknown how the different GRs interact and how the recognition of germinants ultimately leads to spore germination (3). In Bacillus subtilis, GRs localize in germinosomes, and the colocalization appears to be essential for efficient spore germination (6, 7). Notably, the germinosomes have always been visualized in defective cotE and gerE mutants to reduce autofluorescence of the spore’s coat layers, but it is not known if this could have influenced the organization of GRs in the spore inner membrane (6, 7).
Molecular, genetic, and biochemical data indicate that GRs are composed of A, B, and C subunits (8). GR genes are usually organized in tricistronic operons, encoding the A, B, and C subunits. However, gerA family genes are also found as single genes, in dicistronic operons, or in operons containing more than one gene encoding homologous GR subunits (2, 4). Some Bacillus species also encode an additional D subunit, but the function of the D subunit is currently unknown (2, 4). In Bacillus species, all A, B, and C subunits appear to be required for a functional GR, but the specific role of each subunit is poorly understood (9). The A and B subunits are predicted to contain 5 and 10 transmembrane domains, respectively, and are therefore almost certainly integral membrane proteins. Recently, the crystal structure of the N-terminal domain of the A subunit of the Bacillus megaterium GerK3 GR was resolved (10). Structural analyses revealed that this protein shares structural similarity to substrate binding proteins that serve as receptors for membrane-associated small-molecule transporters and signal transducers. As B. megaterium GerK3 is not functional in germination, experimental verification was done in B. subtilis gerAA subunits and later in B. cereus gerIA. These analyses suggested that the N-terminal domain of GerA is involved directly in the recognition and binding of germinant molecules (10). The B subunit shows homology to proteins of the amino acid/polyamine/organocation (APC) superfamily of single-component membrane transporters (11). However, there is no evidence as to whether the B subunit functions in molecular transport. Evidence based on cross-homologue chimeric constructs and site-directed mutagenesis suggests that the B subunit contains the germinant recognition site, but the identity of the germinant binding site and the mechanism of binding are currently unknown (12–14). In B. megaterium, B subunits encoded by different ger operons can be used interchangeably in the GerU GR complex, which provides an extended range of recognized germinants (15). In cases where GR operons consist of more than three genes, the B subunit gene is often present in multiple copies. For example, in B. megaterium QM B1551, there is an atypical cluster of GR-associated genes, comprised of two B-subunit genes separated by a putative D-subunit gene (16). GR operons possessing multiple B-subunit genes have also been found in the genomes of Bacillus cereus E33, Bacillus halodurans, Bacillus cytotoxicus, Bacillus licheniformis, Clostridium botulinum, Clostridium sporogenes, and Clostridium acetobutylicum (4). However, the functional significance of multiple B-subunit genes in the same GR operon remains to be elucidated. The C subunit is a membrane-anchored lipoprotein whose structure has been resolved to a 2.3-Å resolution, but unfortunately, there is still no knowledge on its function (17).
B. licheniformis is widespread in nature and a common food spoilage bacterium (18–24). It is closely related to B. subtilis and has occasionally been associated with disease in humans and abortions in cattle (25–31). B. licheniformis carries three gerA family operons (gerA, gerK, and ynd), and some strains also carry a monocistronic yndF2 gene (TRNA_RS32565) (4, 32, 33). In contrast to the ABC(D) organization that characterizes gerA family operons in B. subtilis, the B. licheniformis type strain ATCC 14580/DSM13 possesses a pentacistronic ynd operon with the gene organization yndD (TRNA_RS32310), yndE3 (TRNA_RS32305), yndE2 (TRNA_RS32300), yndF1 (TRNA_RS32295), and yndE1 (TRNA_RS32290), encoding the GR A, B, B, C, and B subunits, respectively (subscripts indicate paralogs). B. licheniformis spores germinate in response to a range of different amino acids as well as glucose, but alanine, cysteine, and valine are the most potent germinants (34). Mutational analyses have shown that GerA, Ynd, and GerK GRs are all functional in nutrient-induced germination of B. licheniformis spores (34). GerA appears to function as the primary GR in amino acid-induced germination, but it seems to require intact Ynd and GerK GRs to stimulate efficient germination in response to the above-mentioned germinants (34). A B. licheniformis ΔgerAA-C mutant (lacking the entire gerA operon; the hyphen indicates “from A to C”) showed no detectable germination with 100 mM alanine and valine and only very weak germination with 100 mM cysteine, which suggests that Ynd and GerK do not function alone in triggering efficient germination with the tested amino acids (34). A ΔgerAA ΔgerKA-C mutant, which still expresses the intact Ynd GR and the B and C subunits of the GerA GR, showed detectable germination with 100 mM alanine and cysteine (34). The same study showed that deletion of yndD in the ΔgerAA ΔgerKA-C mutant background resulted in spores that showed no germination. No functional interdependence between the Ynd and GerK GRs has been found, but similar to GerK in B. subtilis and B. megaterium, GerK is required for d-glucose-induced germination of B. licheniformis spores. However, d-glucose functions as only a weak germinant for B. licheniformis spores (34).
The purpose of this study is to further examine the cooperative function between GerA and Ynd. In particular, by using mutational analyses, this study addresses the functional role of the three paralogous B-subunit genes in the ynd operon in the interplay between the Ynd and GerA GRs. The functional importance of the orphan yndF2 gene, encoding a C subunit of GerA family GRs, was also investigated.
RESULTS
GerA functions in spore germination in the absence of other germination receptors, and the GerA B subunit is essential for its functionality.
It has previously been shown that Ynd requires GerA to function in B. licheniformis spore germination. However, it is not known whether GerA can function individually or is dependent on Ynd or GerK to be functional. To examine this further, a B. licheniformis mutant strain lacking both the entire ynd and gerK operons (ΔyndDE3E2F1E1 ΔgerKACB) was constructed. Spores of this mutant (strain NVH-1412), which expresses only a functional GerA GR, still germinated with l-alanine, l-cysteine, and l-valine although with a much lower efficiency than for spores of the wild-type background strain (Table 1). To test the importance of the B subunit of GerA in the functionality of the GR, a mutant strain carrying a gerAB in-frame deletion was constructed (NVH-1389). Spores of this mutant showed no detectable germination response after 2 h of germinant exposure (Table 1), which indicates that the B subunit of the GerA GR is essential for B. licheniformis spore germination with l-alanine, l-cysteine, and l-valine.
TABLE 1.
Germination properties of B. licheniformis wild-type and mutant strainsa
| Strain | Genotype | Protein(s) present | Mean no. of spores by microscopic count ± SDb |
Mean Gmax ± SDc |
||||
|---|---|---|---|---|---|---|---|---|
| l-Ala | l-Cys | l-Val | l-Ala | l-Cys | l-Val | |||
| WTd | Ynd, GerA, GerK | 96 ± 1 | 97 ± 1 | 78 ± 13 | −1.5 ± 0.3 | −1.9 ± 0.4 | −0.7 ± 0.2 | |
| NVH-1387 | ΔyndDE3E2F1E1 | GerA, GerK | 47 ± 7 | 36 ± 10 | 52 ± 5 | −0.4 ± 0.2 | −0.1 ± 0.0 | −0.4 ± .0.1 |
| NVH-1412 | ΔyndDE3E2F1E1 ΔgerKACB | GerA | 46 ± 2 | 39 ± 0 | 47 ± 2 | −0.3 ± 0.1 | −0.2 ± 0.0 | −0.3 ± .0.0 |
| NVH-1389 | ΔgerAB | Ynd, GerK, GerAA, GerAC | 2 ± 2 | 4 ± 4 | 2 ± 1 | −0.1 ± 0.0 | −0.1 ± 0.0 | −0.1 ± .0.0 |
| NVH-1369 | ΔyndE3 | YndDE2F1E1, GerA, GerK | 58 ± 6 | 48 ± 8 | 57 ± 1 | −0.4 ± 0.0 | −0.4 ± 0.0 | −0.4 ± .0.0 |
| NVH-1378 | ΔyndE3E2 | YnDF1E1, GerA, GerK | 50 ± 4 | 41 ± 3 | 53 ± 6 | −0.5 ± 0.1 | −0.4 ± 0.2 | −0.5 ± .0.2 |
| NVH-1405 | ΔyndE2E3E1 | YndDF1, GerA, GerK | 86 ± 5 | 76 ± 7 | 85 ± 5 | −0.9 ± 0.3 | −0.6 ± 0.1 | −0.8 ± .0.2 |
| NVH-1371 | ΔyndF2 | Ynd, GerA, GerK | 95 ± 4 | 98 ± 1 | 70 ± 12 | −1.5 ± 0.4 | −1.8 ± 0.2 | −0.5 ± .0.2 |
| NVH-1404 | NVH-1387 ΔgerKA-C | GerA, GerKB | 47 ± 5 | 42 ± 8 | 55 ± 3 | −0.4 ± 0.1 | −0.3 ± 0.1 | −0.4 ± .0.1 |
| Complementations | ||||||||
| cis | ||||||||
| NVH-1421 | NVH-1369::yndE3 | 95 ± 2 | 97 ± 1 | 75 ± 24 | −1.3 ± 0.2 | −1.3 ± 0.3 | −0.7 ± .0.1 | |
| NVH-1416 | NVH-1405::yndE3E2 | 85 ± 6 | 81 ± 4 | 80 ± 5 | −0.7 ± 0.0 | −0.6 ± 0.0 | −0.6 ± 0.1 | |
| NVH-1417 | NVH-1405::yndE3E2E1 | 95 ± 4 | 94 ± 4 | 89 ± 6 | −1.4 ± 0.3 | −1.0 ± 0.0 | −0.9 ± .0.0 | |
| NVH-1413 | NVH-1404::yndDE2F1 | 47 ± 5 | 48 ± 5 | 37 ± 3 | −0.5 ± 0.0 | −0.4 ± 0.0 | −0.3 ± .0.0 | |
| NVH-1427 | NVH-1412::yndDF1 | 69 ± 6 | 60 ± 3 | 71 ± 4 | −0.4 ± 0.0 | −0.3 ± 0.0 | −0.4 ± 0.0 | |
| trans | ||||||||
| NVH-1474 | NVH-1412/pHT315 | 28 ± 5 | 14 ± 6 | 27 ± 1 | −0.2 ± 0.0 | −0.1 ± 0.0 | −0.2 ± .0.0 | |
| NVH-1438 | NVH-1412/pHT315-yndDF1 | 49 ± 9 | 36 ± 11 | 55 ± 10 | −0.4 ± 0.1 | −0.2 ± 0.1 | −0.4 ± .0.1 | |
| NVH-1473 | NVH-1412/pHT315-yndD | 24 ± 3 | 14 ± 4 | 27 ± 4 | −0.2 ± 0.0 | −0.1 ± 0.0 | −0.2 ± .0.0 | |
All data are presented as means from three biological replications.
The percentages of germinated (phase-dark) spores were determined after 120 min of exposure to 100 mM germinant compounds.
Gmax is the maximum rate of germination (ΔOD600 units per minute).
The wild-type (WT) strain shows 1% ± 1% germination with just buffer (no germinants added), as determined by microscopic counts.
The ynd operon is highly conserved among B. licheniformis strains.
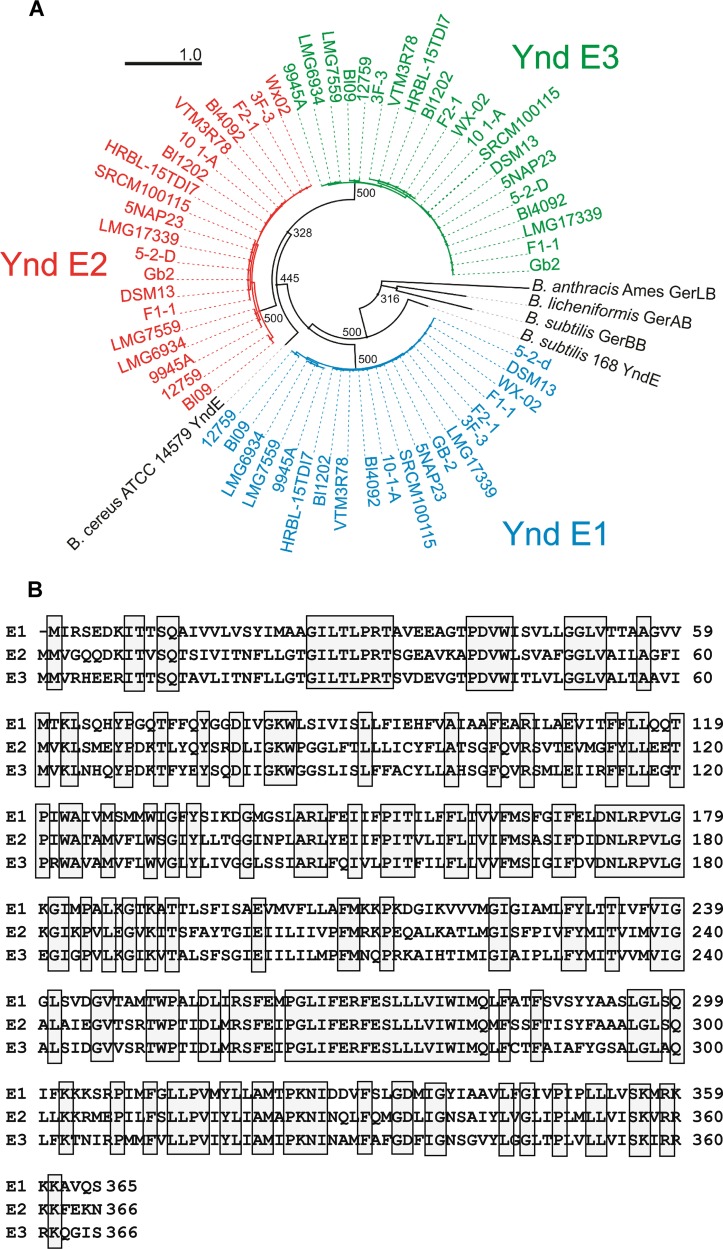
B. licheniformis and Bacillus paralicheniformis genomes were explored for the presence of the ynd operon using the complete yndD nucleotide sequence from B. licheniformis ATCC 14580/DSM13 as the seed sequence. Pentacistronic ynd operons were found in all 25 B. licheniformis strains investigated. However, five genomes carried ynd operons with premature stop codons in the yndE genes (4/5) or in the yndF1 gene (1/5) (results not shown). These genomes were excluded from the phylogenetic analysis. All remaining 17 B. licheniformis and 3 B. paralicheniformis (9945a, BL09, and 12759) strains possessed atypical pentacistronic ynd operons with a yndD, yndE3, yndE2, yndF1, and yndE1 gene organization similar to the one found in the B. licheniformis type strain ATCC 14580/DSM13. A maximum likelihood phylogram based on alignment of the deduced amino acid sequences of yndE genes from the 17 B. licheniformis and 3 B. paralicheniformis strains revealed a monophyletic clade with three distinct branches corresponding to YndE1, YndE2, and YndE3 (Fig. 1A).
FIG 1.
(A) Maximum likelihood phylogram for YndE1, YndE2, and YndE3 of 20 strains of B. licheniformis as well as of B. cereus and B. subtilis. Bootstrap support values above 50% are indicated at the nodes; however, no bootstrap support values for nodes within the clades for the three B. licheniformis Ynd genes are given. GerLB, GerAB, and GerBB from B. anthracis, B. licheniformis, and B. subtilis, respectively, were used as the outgroups. (B) Amino acid sequence alignment of the Ynd B subunits encoded by the yndE3, yndE2, and yndE1 genes of B. licheniformis DSM13/ATCC 14580.
The amino acid sequences of the three B. licheniformis YndE subunits are resolved as monophyletic groups, each with 100% bootstrap support. YndE2 and YndE3 appear to be more closely related to each other than to YndE1, as determined by phylogenetic analyses of the yndE genes of the B. licheniformis strains (Fig. 1A). Accordingly, alignments of B. licheniformis DSM13/ATCC 14580 amino acid sequences showed that YndE3 and YndE2 shared more identity (64%) than YndE1 and YndE2 or YndE3 (52% and 54%, respectively) (Fig. 1B; see also Table S2 in the supplemental material).
All ynd genes are expressed during sporulation.
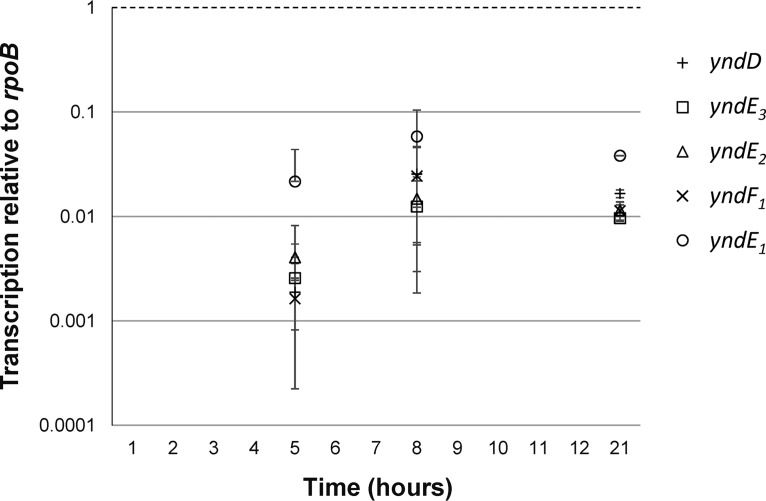
Expression analysis of B. licheniformis grown in sporulation medium revealed that the yndE3 and yndE2 genes were expressed at approximately the same level as yndD and yndF1 during sporulation of the type strain derivative MW3 (Fig. 2). The impression during the analysis was that yndE1 was transcribed at a level slightly higher than those of the other genes of the operon; however, it was statistically significant only by pairwise comparisons using two-tailed paired Student t tests (P < 0.05) at 21 h. The transcription level of the individual ynd genes increased approximately five times from 5 h (late exponential growth) to 8 h (early stationary growth) after inoculation and was between 0.01 and 0.1 times the expression of rpoB at 21 h, when there were about 50% spores in the culture, as observed by phase-contrast microscopy (Fig. 2).
FIG 2.
Transcription levels of yndD, yndE3, yndE2, yndF1, and yndE1 relative to rpoB determined by RT-qPCR during 21 h of growth of B. licheniformis MW3. At 21 h, there were about 50% spores in the cultures, as observed by phase-contrast microscopy. Whiskers represent standard deviations of the means based on data from three independent experiments.
The Ynd AC subunits are functional in germination in the absence of the Ynd B subunits.
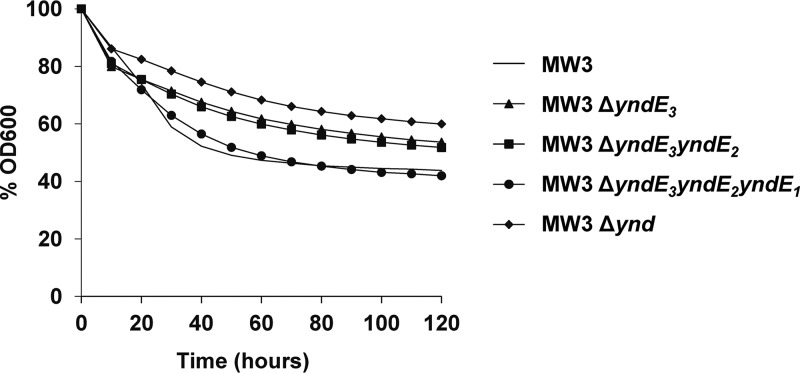
To examine the contribution of the Ynd B subunits to spore germination, spores of a mutant carrying in-frame deletions of all three yndE genes (NVH-1405) were tested for germination with l-alanine, l-cysteine, and l-valine. Despite the absence of all B subunits, ΔyndE3E2E1 mutant spores germinated in a manner similar to that of wild-type spores with 100 mM germinants, and their germination rate was only slightly lower than that of wild-type spores (Fig. 3 and Table 1). Wild-type and ΔyndE3E2E1 mutant spores demonstrated similar germination levels when lower concentrations of l-alanine and l-cysteine were used, but the mutant spores showed slightly improved germination compared to wild-type spores with l-valine at all concentrations tested (Table 2).
FIG 3.
Spore germination of yndE1, yndE1E2, and yndE3E2E1 deletion mutants and of the mutant lacking the entire ynd operon. All mutants were derived from the MW3 strain. Germination was measured by the decrease in the OD600 over a period of 120 min after the addition of the germinant.
TABLE 2.
Germination of wild-type and ΔyndE2E3E1 mutant spores at different concentrations of germinants
| Amino acid concn (mM) | Mean germination rate (%) ± SDa |
|||||
|---|---|---|---|---|---|---|
| WT |
ΔyndE2E3E1 mutant |
|||||
| l-Ala | l-Cys | l-Val | l-Ala | l-Cys | l-Val | |
| 100 | 96 ± 1 | 97 ± 1 | 79 ± 10 | 88 ± 2 | 84 ± 6 | 88 ± 4 |
| 10 | 73 ± 16 | 80 ± 16 | 35 ± 12 | 70 ± 7 | 69 ± 2 | 65 ± 3 |
| 5 | 56 ± 20 | 51 ± 22 | 19 ± 9 | 60 ± 7 | 40 ± 1 | 49 ± 1 |
| 1 | 44 ± 21 | 3 ± 1 | 3 ± 2 | 54 ± 4 | 4 ± 2 | 21 ± 4 |
| 0.5 | 30 ± 14 | 2 ± 1 | 1 ± 0 | 43 ± 7 | 2 ± 1 | 6 ± 2 |
All data are presented as means from three biological replications. The percentages of germinated (phase-dark) spores were determined by phase-contrast microscopy after 120 min of exposure to decreasing concentrations of germinants.
It was previously shown that compounds other than l-alanine, l-cysteine, and l-valine functions as moderate (l-serine, l-isoleucine, and l-aspartic acid) and weak (d-glucose, l-methionine, and l-lysine) germinants for B. licheniformis MW3 spores (34). When ΔyndE3E2E1 mutant spores were tested for germination with 100 mM these compounds, they demonstrated a slightly improved germination response toward l-methionine compared to wild-type spores (Table 3); however, the improvement was not statistically significant. The similar germination efficiencies of spores of the wild type and of the ΔyndE3E2E1 mutant after 2 h of exposure to germinants (Table 2) suggest that the Ynd B subunits do not play an important role in germination with these compounds. When ΔyndE3E2E1 mutant spores were cis complemented with yndE3E2E1 (NVH-1417), the germination responses with l-alanine, l-cysteine, and l-valine were similar to those of wild-type spores (Table 1).
TABLE 3.
Germination of wild-type and ΔyndE2E3E1 mutant spores in response to other amino acids
| Germinant (100 mM) | Mean germination rate (%) ± SDa |
|
|---|---|---|
| WT | ΔyndE3E2E1 mutant | |
| l-Methionine | 16 ± 7 | 48 ± 22 |
| d-Glucose | 2 ± 1 | 9 ± 7 |
| l-Lysine | 1 ± 0 | 3 ± 2 |
| l-Serine | 48 ± 28 | 33 ± 22 |
| l-Isoleucine | 53 ± 10 | 76 ± 22 |
All data are presented as means from three biological replications. The percentages of germinated (phase-dark) spores were determined by phase-contrast microscopy after 120 min of exposure to 100 mM germinant compounds.
The more efficient germination observed for ΔyndE3E2E1 spores than for ΔyndDE3E2F1E1 spores suggests that the A and C subunits of Ynd are sufficient for an effective germination response. Deletion of the whole gerK operon (ΔgerKACB) in the ΔyndDE3E2F1E1 background (NVH-1412) did not alter the germination properties compared to the background strain (Table 1), which indicates that GerK does not contribute significantly to the observed germination. To test whether both the A and C subunits of Ynd are required for the cooperation between GerA and Ynd, attempts were made to construct a yndE3E2F1E1-null mutant, but despite several attempts, we were not able to construct this mutant. Several attempts to make cis complementations with yndD and yndDF1 in the ΔyndDE3E2F1E1 mutant also failed for an unknown reason. However, trans complementation of the ΔyndDE3E2F1E1 ΔgerKACB mutant (NVH-1412) with yndD and yndDF1 gave some new insight. NVH-1412 spores carrying the empty pHT315 vector showed a reduced maximum germination rate (Gmax), and the number of germinated spores as measured by microscopic counts was reduced about 50% compared to spores of the background NVH-1412 strain, indicating that the plasmid had a negative effect on germination (Table 1). Accordingly, in order to avoid bias due to the presence or absence of the plasmid, germination properties of the complemented spores were compared to those of spores of NVH-1412 carrying the empty pHT315 vector. trans complementation of NVH-1412 with yndD (NVH-1473, encoding the A subunit) resulted in spores that showed the same germinability as NVH-1412 spores carrying the empty vector, while complementation with yndDF1 (NVH-1438, encoding the A and C subunits) resulted in spores showing germination approximately twice as effective as that of NVH-1412 spores with the empty pHT315 vector (Table 1). This indicates that at least the C subunit of Ynd is needed to trigger efficient germination in cooperation with GerA and that the A subunit alone is not sufficient for the cooperative functionality between Ynd and GerA. cis complementation of NVH-1412 with yndDF1 also demonstrated an increased germination response compared to that of NVH-1412 (Table 1).
The individual Ynd B subunits contribute differently to spore germination.
The separate phylogenetic clustering of YndE1, YndE2, and YndE3 (Fig. 1A) suggests that they may perform different functions. To explore the function of individual B subunits, mutants lacking one or several Ynd B subunits were tested for germination with 100 mM l-alanine, l-cysteine, and l-valine. Spores of the ΔyndE3 mutant showed germination that was reduced to levels comparable to those of ΔyndDE3E2F1E1 mutant spores lacking the whole ynd operon (Table 1 and Fig. 3). Wild-type levels of spore germination were restored when the ΔyndE3 mutant was cis complemented with the yndE3 gene (NVH-1421) (Table 1). No further reduction in total germination compared to the ΔyndE3 mutant spores was observed when both yndE2 and yndE3 were deleted simultaneously (NVH-1378) (Table 1). Next, the mutant where the entire ynd operon was deleted was cis complemented with yndDE2F1. Spores of the resulting NVH-1413 strain, which express YndDE2F1, showed severely reduced germinability (Table 1). Together, these results suggest that the YndE3 subunit plays an important role in germination with the tested germinants, while YndE2 plays a minor role, if any. For an unknown reason, we were not able to construct a ΔyndE1 mutant. Therefore, to investigate the role of YndE1 in germination, a ΔyndE3E2E1 mutant (NVH-1405) was cis complemented with ΔyndE3E2. Spores of the resulting strain (NVH-1416) (Table 1), which lack only the YndE1 subunit of the Ynd GR, demonstrated slightly reduced total germination after 2 h of germinant exposure and a 2-fold-reduced Gmax compared to wild-type spores. These results suggest that YndE1 also contributes to germination with l-alanine, l-cysteine, and l-valine but is less important than YndE3. Notably, spores of strain NVH-1405, which lacks all three YndE subunits, germinated markedly more efficiently than did spores lacking either or both of the YndE2 and YndE3 subunits.
Analysis of YndF2.
The orphan yndF2 gene was present in 12 of the 20 B. licheniformis genomes analyzed in this study. The deduced sequence of YndF2 from ATCC 14580/DSM13 is 184 amino acids long and shows 60% identity and 79% similarity to the C-terminal region of YndF1 (Table S2). However, in the majority of the 12 investigated genomes where YndF2 is present, it is 401 amino acids long, which strongly suggests that YndF2 from ATCC 14580/DSM13 is truncated at the N-terminal end. We were unable to identify a promoter upstream of yndF2 in strain ATCC 14580/DSM13; however, analysis of its expression by reverse transcriptase quantitative PCR (RT-qPCR) revealed that yndF2 is transcribed at approximately the same level as the yndE genes (data not shown). Therefore, to evaluate its potential role in spore germination, we constructed a mutant strain carrying an in-frame deletion of yndF2 (NVH-1371). Spores of the resulting mutant demonstrated germination efficiencies similar to those of wild-type spores (Table 1), suggesting that YndF2 is not important for germination with the tested amino acids.
DISCUSSION
The main objective of the present study was to increase the understanding of the functional interaction occurring between cooperating GRs during nutrient-triggered spore germination, using B. licheniformis GerA and Ynd as models. This work also attempts to characterize the role of the paralogous B subunits encoded by the yndE3, yndE2, and yndE1 genes in B. licheniformis spore germination.
B. licheniformis is so far the only species known to contain a functional Ynd GR. The ynd operon is highly conserved among the B. licheniformis and B. paralicheniformis strains investigated in this study. In contrast, the selection pressure for maintaining this GR seems to be much lower among B. subtilis strains (35). Phylogenetic examination of yndE genes from 17 B. licheniformis and 3 B. paralicheniformis strains showed that YndE1, YndE2, and YndE3 form three separate phylogenetic clusters, suggesting that individual B subunits play different functional roles. Interestingly, the B. cereus yndE gene forms a clade with B. licheniformis yndE2 and yndE3. Two scenarios can explain the observed phylogeny. A putative horizontally transferred copy of the B. cereus yndE gene might have been acquired and subsequently duplicated into yndE2 and yndE3 in B. licheniformis. Alternatively, duplication of an ancestral B. licheniformis yndE gene resulted in yndE1 and an ancestral paralogue that subsequently duplicated into yndE2 and yndE3. In this scenario, B. cereus might have received the ancestral paralogue from B. licheniformis. B. subtilis YndE also has an uncertain placement due to low support values but is either sister to B. licheniformis YndE3 or YndE2 and B. cereus YndE or is sister to B. licheniformis YndE1.
Transcriptional analyses revealed that the yndE3, yndE2, and yndE1 genes are expressed during sporulation. The absence of yndE3 or both yndE3 and yndE2 resulted in weaker germination responses with l-alanine, l-cysteine, and l-valine than for wild-type spores, indicating that the Ynd GR could not function optimally in the absence of YndE3 in triggering germination in cooperation with GerA. Deletion of yndE1 also seemed to have a negative effect on germination although not as pronounced as what was seen for spores of mutants lacking YndE3. Remarkably, simultaneous deletion of the yndE1, yndE2, and yndE3 genes restored the germination rate and total germination of the spore population to levels only slightly lower than those of wild-type spores. While spores carrying an incomplete set of Ynd B subunits showed reduced germination efficiencies, spores lacking all three Ynd B subunits germinated almost as efficiently as wild-type spores (Tables 1 and 2). This indicates that the presence of an incomplete set of Ynd B subunits exhibits a dominant negative effect on germination properties. Since all B. licheniformis and B. paralicheniformis genomes investigated in this study encode the GerD protein, which facilitates germinosome formation in B. subtilis (at least in defective cotE and gerE mutants), it is likely that GerA, Ynd, and GerK are colocalized in germinosomes. However, there is very little knowledge on the heteromeric organization and potential clustering of GRs in the spore’s inner membrane, and therefore, the interpretation of the physiological basis for our results remains speculative. Assuming that GerA and Ynd form a heteromeric complex in the spore’s inner membrane, the absence of one or more of the encoded Ynd B subunits may obstruct the organization of the GR complex, resulting in a dominant negative effect on the cooperative function between Ynd and GerA (Tables 1 and 2). However, when all Ynd B subunits are absent, the remaining GerAB subunit alone may support the cooperative function between Ynd and GerA. The B subunit of the GerA GR was in contrast to the B subunits of the Ynd GR indispensable for B. licheniformis spore germination with l-alanine, l-cysteine, and l-valine, which indicates that the absence of GerAB cannot be functionally complemented with B subunits in the Ynd or GerK GR. Similarly, some amino acid substitutions in B. subtilis gerAB have been shown to result in a loss of spore germination with l-alanine and an apparent loss of the GerA GR as judged by the loss of the GerAC protein from crude spore extracts (36).
B. licheniformis GerA is necessary and sufficient for germination with all tested germinant compounds, and the presence of an intact Ynd GR is important for wild-type levels of germination with l-alanine, l-cysteine, and l-valine, especially at low germinant concentrations (34, 37). Previously, we have shown that a ΔgerAA ΔgerKA-C mutant, which still expressed the intact Ynd GR and the B and C subunits of the GerA GR, showed detectable germination with 100 mM alanine and cysteine (34). The same study revealed that deletion of yndD in the ΔgerAA ΔgerKA-C mutant background resulted in spores that showed no germination at all. Together, these results indicate that the Ynd GR can complement the absence of the GerA A subunit to some extent and that the functions of these GRs are tightly interconnected. The interdependence between the GerA and the Ynd GRs is important for a robust germination response and could be a major determinant for maintaining the ynd operon in B. licheniformis genomes. The B subunit of GRs has been suggested to contain the germinant binding site, as amino acid substitutions in B. megaterium GerVB changed the germinant recognition specificity (12, 38). However, the three B subunits in the B. licheniformis Ynd GR do not appear to play a major role in germination with the tested nutrients, as ΔyndE3E2E1 mutant spores, which lack all Ynd B subunits, did not differ from wild-type spores in the repertoire of nutrients recognized as germinants (Tables 2 and 3). It is likely, however, that the structure of the Ynd GR in B. licheniformis reflects an ecological specialization, and it could have a more pronounced function in complex natural soil environments where the spores can be exposed a wide range of different nutrient compounds.
Mutant spores containing the GerA GR and the A and C subunits of the Ynd GR (ΔyndE3E2E1) demonstrated near-wild-type efficiencies of spore germination with l-alanine, l-cysteine, and l-valine, suggesting that the A and C subunits encoded by the ynd operon are sufficient for the functional cooperation between Ynd and GerA. Further study of spores lacking the entire ynd operon (ΔyndDE3E2F1E1) but complemented with the yndD gene showed that they germinated with the same efficiency as ΔyndDE3E2F1E1 spores, which lack the whole Ynd GR, suggesting that at least the C subunit is essential for functional cooperation between Ynd and GerA. Dicistronic operons encoding A and C, A and B, and B and C subunits and monocistronic operons encoding single A, B, and C subunits are found in many Bacillales and Clostridiales genomes (2). Whether a GR encoded by such “atypical” gerA family operons can be capable of functioning alone or whether they function only in cooperation with other GRs remains to be clarified. However, the ability of the A and C subunits of the B. licheniformis Ynd GR to mediate a functional interaction with GerA suggests that GRs containing only the A and C subunits might also be functional in other species, at least in cooperation with other GRs. A situation where a GR does not require all subunits to function in triggering spore germination has been described in C. perfringens, which contains a bicistronic gerK operon encoding an A subunit and a C subunit, which can function in the absence of a B subunit (39, 40). Interestingly, the C subunit was found to be essential for C. perfringens spore germination, whereas the A subunit was dispensable (41). Additional studies are needed to address whether this is also the case for the A and C subunits of the B. licheniformis Ynd GR.
The YndF2 subunit was found to be dispensable for B. licheniformis spore germination with all tested germinant compounds. However, yndF2 appears to be truncated in strain ATCC 14580/DSM13, while it is present as a full-length (1,200-bp) gene in most other B. licheniformis genomes. It is therefore possible that it plays a functional role in other B. licheniformis strains, but this has yet to be determined.
The methods to assess germination used in this work determine the average germination kinetics and total level of germination for the entire spore population. These methods are useful for studying the germination behaviors of a range of different GR mutants with different germinant compounds, but since spore populations generally are very heterogeneous, they do not provide information about the behavior of individual spores or differentiate between different germination phases. However, single-spore analyses such as live imaging, laser tweezers Raman spectroscopy, and/or flow cytometry analysis are more laborious but will provide more detailed information about the behavior of individual spores and differentiates better between the time to start germination and the time of the germination process itself (42–44).
MATERIALS AND METHODS
Strains.
The B. licheniformis strains used in this study are listed in Table 4. Strain MW3 carries deletions in the hsdR loci encoding two type I restriction-modification systems and is therefore a more readily transformable derivative of the B. licheniformis type strain DSM13 (45).
TABLE 4.
Strains used in this study
| Strain | Description or genotype | Reference(s) |
|---|---|---|
| ATCC 14580/DSM13 | Type strain | 32, 33 |
| MW3 | DSM13 ΔhsdR1 ΔhsdR2 | 45 |
| NVH-1387 | ΔyndDE3E2F1E1 | This study |
| NVH-1412 | Δynd ΔgerKACB | This study |
| NVH-1389 | ΔgerAB | This study |
| NVH-1369 | ΔyndE3 | This study |
| NVH-1378 | ΔyndE3E2 | This study |
| NVH-1405 | ΔyndE2E3E1 | This study |
| NVH-1371 | ΔyndF2 | This study |
| NVH-1404 | NVH-1387 ΔgerKA-C | This study |
| NVH-1421 | NVH-1369::yndE3 | This study |
| NVH-1416 | NVH-1405::yndE3E2 | This study |
| NVH-1417 | NVH-1405::yndE3E2E1 | This study |
| NVH-1413 | NVH-1404::yndDE2F1 | This study |
| NVH-1427 | NVH-1412::yndDF1 | This study |
| NVH-1474 | NVH-1412/pHT315 | This study |
| NVH-1438 | NVH-1412/pHT315-yndDF1 | This study |
| NVH-1473 | NVH-1412/pHT315-yndD | This study |
Bioinformatic analyses.
The ynd gene sequences of B. licheniformis type strain DSM13/ATCC 14580, B. subtilis strain 168, B. cereus strain ATCC 14579, and the panel of 20 B. licheniformis and B. paralicheniformis strains were acquired from the NCBI database, including both complete and draft assembled genomes (www.ncbi.nlm.nih.gov/). The ynd genes were identified using nBLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), using the ynd genes from strain DSM13/ATCC 14580 as query sequences. To investigate the evolutionary relationships between yndE genes, we performed a phylogenetic analysis. The deduced amino acid sequences of the three yndE genes from each of the 20 strains of B. licheniformis and B. paralicheniformis were aligned using ClustalX, followed by manual inspection in BioEdit (46). We also included the deduced amino acid sequences of the yndE genes of B. subtilis and B. cereus. As outgroups, we used GerLB from B. anthracis, GerAB from B. licheniformis, and GerBB from B. subtilis. Based on the Akaike information criterion calculated in SMS (47), we estimated maximum likelihood phylogenies from the data under the LG+G+F substitution model using PhyML (48) with 500 bootstrap replicates.
Analyses of relative gene expression using RT-qPCR.
The relative expression levels of yndE1, yndE2, yndE3, yndF2, and yndD relative to rpoB were determined using reverse transcriptase quantitative PCR (RT-qPCR). Sporulation for RNA extraction, cDNA synthesis, and RT-qPCR analysis was performed as described previously (34, 49). RT-qPCR was performed in triplicates on three biological replicates, and the results were analyzed as described previously (34, 49). All primers used for RT-qPCR analyses are listed in Table S1 in the supplemental material.
Construction of deletion mutants.
In-frame deletion mutants were constructed by replacing the target gene(s) with the nucleotide sequence 5′-ATGTAR-3′ (where R is A or G) using a markerless gene replacement method (50), as described previously (37). This method results in an in-frame deletion of the target gene and ensures that the up- and downstream flanking sequences, including the promoter region, are intact. Briefly, the deletion mutants were constructed by amplifying an ∼500-bp segment upstream of the target gene, using primers A and B, and an ∼500-bp segment downstream of the target gene, using primers C and D. Primers B and C contain a sequence overlap enabling fusion of the AB and CD PCR products by sequence- and ligation-independent cloning (SLIC)-PCR. The resulting AD fragment contains the upstream and downstream sequences of the target gene. The AD fragment was then cloned into a thermosensitive shuttle vector, pMAD (51), containing an I-SceI restriction site (52). The resulting plasmid, pMAD-I-SceI, carrying the gene deletion construct was then transformed into electrocompetent B. licheniformis cells (53). Here the whole plasmid construct was expected to integrate into the chromosome by a single crossover. The plasmid pBKJ233, encoding the I-SceI enzyme, was then introduced by electroporation. The I-SceI restriction enzyme makes a double-stranded cut at its recognition site, which allows for recombinational repair, resulting in a second crossover where the target gene is deleted. Deletion of the target gene was confirmed by PCR and sequencing (GATC Biotech). All primers used for construction of the mutant strains are listed in Table S2. All PCR products used for making the deletion mutants were produced using Phusion high-fidelity DNA polymerase (Finnzymes, Finland) according to the manufacturer’s instructions. All PCRs were performed using an Eppendorf Mastercycler ep-Gradient S instrument.
Complementation tests.
The complementing constructs used for cis complementation, comprising the ynd promoter region (570 bp) followed by the complementing gene(s), were cloned into pMAD-NotI. The NotI site was introduced into the EcoRI site of pMAD by SLIC-PCR, comprising an upstream part (404 bp) and a downstream part (252 bp) of B. licheniformis amyL joined by a NotI site (primers are listed in Table S1). The purpose of including the amyL sequence was to achieve homologous recombination of the plasmid into the amyL gene of B. licheniformis. The complementing sequences were constructed by PCR (ordinary or SLIC) using AccuPrime high-fidelity Taq polymerase (Invitrogen) and the primers listed in Table S1. The pMAD vector carrying the complementing sequence was transformed into B. licheniformis mutant strains by electroporation (37, 53). The whole plasmid construct was integrated into the chromosome by a single crossover caused by a temperature shift, which influences the temperature-sensitive replicon of pMAD. All complementations were verified by PCR and sequencing.
For trans complementation, selected genes were carried by the low-copy-number shuttle vector pHT315 (54). The respective genes and their associated regulatory sequences were amplified by PCR using primers listed in Table S1 and AccuPrime Taq DNA polymerase (Thermo Fisher Scientific) according to the manufacturer’s instructions. The amplicons were cloned into pHT315, and the resulting constructs were used to transform electrocompetent B. licheniformis deletion mutants as described previously (53). The presence of the correct plasmid construct was verified by PCR and sequencing.
Germination assays.
Spores were prepared, washed, and stored for at least 7 days prior to use, as described previously (49). All germination assays were performed on pure (>98%), heat-activated (20 min at 65°C) spore suspensions as described previously (49). This purification protocol gives a homogeneous suspension of phase-bright spores without traces of vegetative cells. L-Amino acids (Sigma-Aldrich) were added, and the decrease in the optical density at 600 nm (OD600) of the spore population was monitored. The germination rate was assessed as the rate of phase darkening of the whole spore population, which most likely represents an average rate of individual spore germination. The maximum germination rate (Gmax) was calculated from the curves obtained from measuring the decrease in the OD600 using DMFit (DM is dynamic modeling) (55). Phase-contrast microscopy was also used to monitor the level of germinated spores after 120 min of exposure to germinants. The number of phase-dark (germinated) spores was determined for at least 400 spores in each experiment by counting spores in 10 random fields of view, and the average percentages of germinated spores were calculated from three independent spore batches. Spore suspensions with Milli-Q water were used as negative controls in all germination assays.
Supplementary Material
ACKNOWLEDGMENTS
This work received financial support from the Institute for Food Safety and Infection Biology at the Norwegian University of Life Sciences (NMBU), the Center for Food Safety (NMBU), and regional research funds in Norway (project no. 245881).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00451-19.
REFERENCES
- 1.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Y, Francke C, Abee T, Wells-Bennik MH. 2011. Clostridial spore germination versus bacilli: genome mining and current insights. Food Microbiol 28:266–274. doi: 10.1016/j.fm.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Abee T, Groot MN, Tempelaars M, Zwietering M, Moezelaar R, van der Voort M. 2011. Germination and outgrowth of spores of Bacillus cereus group members: diversity and role of germinant receptors. Food Microbiol 28:199–208. doi: 10.1016/j.fm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol 81:1061–1077. doi: 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen J, Pasman R, Manders EMM, Setlow P, Brul S. 2019. Visualization of germinosomes and the inner membrane in Bacillus subtilis spores. J Vis Exp 2019:59388. doi: 10.3791/59388. [DOI] [PubMed] [Google Scholar]
- 8.Moir A, Corfe BM, Behravan J. 2002. Spore germination. Cell Mol Life Sci 59:403–409. doi: 10.1007/s00018-002-8432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moir A, Smith DA. 1990. The genetics of bacterial spore germination. Annu Rev Microbiol 44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Jin K, Perez-Valdespino A, Federkiewicz K, Davis A, Maciejewski MW, Setlow P, Hao B. 2019. Structural and functional analyses of the N-terminal domain of the A subunit of a Bacillus megaterium spore germinant receptor. Proc Natl Acad Sci U S A 116:11470–11479. doi: 10.1073/pnas.1903675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jack DL, Paulsen IT, Saier MH. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146(Part 8):1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 12.Christie G, Lazarevska M, Lowe CR. 2008. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J Bacteriol 190:2014–2022. doi: 10.1128/JB.01687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie G, Lowe CR. 2008. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J Bacteriol 190:8009–8017. doi: 10.1128/JB.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol 188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie G, Lowe CR. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J Bacteriol 189:4375–4383. doi: 10.1128/JB.00110-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S, Ustok FI, Johnson CL, Bailey DM, Lowe CR, Christie G. 2013. Investigating the functional hierarchy of Bacillus megaterium PV361 spore germinant receptors. J Bacteriol 195:3045–3053. doi: 10.1128/JB.00325-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Setlow B, Setlow P, Hao B. 2010. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J Mol Biol 402:8–16. doi: 10.1016/j.jmb.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell RG, De Lacy KM. 1983. A note on the identity and properties of the spoilage microflora of chub-packed luncheon meat stored at ambient temperature. Can J Microbiol 29:1220–1223. doi: 10.1139/m83-187. [DOI] [PubMed] [Google Scholar]
- 19.Banyko J, Vyletelova M. 2009. Determining the source of Bacillus cereus and Bacillus licheniformis isolated from raw milk, pasteurized milk and yoghurt. Lett Appl Microbiol 48:318–323. doi: 10.1111/j.1472-765X.2008.02526.x. [DOI] [PubMed] [Google Scholar]
- 20.Crielly EM, Logan NA, Anderton A. 1994. Studies on the Bacillus flora of milk and milk products. J Appl Bacteriol 77:256–263. doi: 10.1111/j.1365-2672.1994.tb03072.x. [DOI] [PubMed] [Google Scholar]
- 21.Lucking G, Stoeckel M, Atamer Z, Hinrichs J, Ehling-Schulz M. 2013. Characterization of aerobic spore-forming bacteria associated with industrial dairy processing environments and product spoilage. Int J Food Microbiol 166:270–279. doi: 10.1016/j.ijfoodmicro.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Pavic S, Brett M, Petric I, Lastre D, Smoljanovic M, Atkinson M, Kovacic A, Cetinic E, Ropac D. 2005. An outbreak of food poisoning in a kindergarten caused by milk powder containing toxigenic Bacillus subtilis and Bacillus licheniformis. Arch Lebensmittelhyg 56:20–22. [Google Scholar]
- 23.Ruckert A, Ronimus RS, Morgan HW. 2004. A RAPD-based survey of thermophilic bacilli in milk powders from different countries. Int J Food Microbiol 96:263–272. doi: 10.1016/j.ijfoodmicro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JM, Waites WM, Dodd CER. 1998. Detection of rope spoilage in bread caused by Bacillus species. J Appl Microbiol 85:481–486. doi: 10.1046/j.1365-2672.1998.853512.x. [DOI] [Google Scholar]
- 25.Agerholm JS, Krogh HV, Jensen HE. 1995. A retrospective study of bovine abortions associated with Bacillus licheniformis. Zentralbl Veterinarmed B 42:225–234. [DOI] [PubMed] [Google Scholar]
- 26.Kramer JM, Gilbert JR. 1989. Bacillus cereus and other Bacillus species, p 58–61. In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, New York, NY. [Google Scholar]
- 27.Salkinoja-Salonen MS, Vuorio R, Andersson MA, Kampfer P, Andersson MC, Honkanen-Buzalski T, Scoging AC. 1999. Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl Environ Microbiol 65:4637–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haydushka IA, Markova N, Kirina V, Atanassova M. 2012. Recurrent sepsis due to Bacillus licheniformis. J Glob Infect Dis 4:82–83. doi: 10.4103/0974-777X.93768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugar AM, McCloskey RV. 1977. Bacillus licheniformis sepsis. JAMA 238:1180–1181. doi: 10.1001/jama.1977.03280120072022. [DOI] [PubMed] [Google Scholar]
- 30.Blue SR, Singh VR, Saubolle MA. 1995. Bacillus licheniformis bacteremia: five cases associated with indwelling central venous catheters. Clin Infect Dis 20:629–633. doi: 10.1093/clinids/20.3.629. [DOI] [PubMed] [Google Scholar]
- 31.Idelevich EA, Pogoda CA, Ballhausen B, Wüllenweber J, Eckardt L, Baumgartner H, Waltenberger J, Peters G, Becker K. 2013. Pacemaker lead infection and related bacteraemia caused by normal and small colony variant phenotypes of Bacillus licheniformis. J Med Microbiol 62:940–944. doi: 10.1099/jmm.0.051987-0. [DOI] [PubMed] [Google Scholar]
- 32.Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Baumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G. 2004. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7:204–211. doi: 10.1159/000079829. [DOI] [PubMed] [Google Scholar]
- 33.Rey MW, Ramaiya P, Nelson BA, Brody-Karpin SD, Zaretsky EJ, Tang M, Lopez de Leon A, Xiang H, Gusti V, Clausen IG, Olsen PB, Rasmussen MD, Andersen JT, Jørgensen PL, Larsen TS, Sorokin A, Bolotin A, Lapidus A, Galleron N, Ehrlich SD, Berka RM. 2004. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol 5:R77. doi: 10.1186/gb-2004-5-10-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borch-Pedersen K, Lindbäck T, Madslien EH, Kidd SW, O’Sullivan K, Granum PE, Aspholm M. 2016. The cooperative and interdependent roles of GerA, GerK, and Ynd in germination of Bacillus licheniformis spores. Appl Environ Microbiol 82:4279–4287. doi: 10.1128/AEM.00594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alzahrani OM, Moir A. 2014. Spore germination and germinant receptor genes in wild strains of Bacillus subtilis. J Appl Microbiol 117:741–749. doi: 10.1111/jam.12566. [DOI] [PubMed] [Google Scholar]
- 36.Cooper GR, Moir A. 2011. Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J Bacteriol 193:2261–2267. doi: 10.1128/JB.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Løvdal IS, From C, Madslien EH, Romundset KC, Klufterud E, Rosnes JT, Granum PE. 2012. Role of the gerA operon in L-alanine germination of Bacillus licheniformis spores. BMC Microbiol 12:34. doi: 10.1186/1471-2180-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christie G, Gotzke H, Lowe CR. 2010. Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J Bacteriol 192:4317–4326. doi: 10.1128/JB.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paredes-Sabja D, Setlow P, Sarker MR. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl Environ Microbiol 75:3813–3817. doi: 10.1128/AEM.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paredes-Sabja D, Torres JA, Setlow P, Sarker MR. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J Bacteriol 190:1190–1201. doi: 10.1128/JB.01748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banawas S, Paredes-Sabja D, Korza G, Li Y, Hao B, Setlow P, Sarker MR. 2013. The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination. J Bacteriol 195:5084–5091. doi: 10.1128/JB.00901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey R, Ter Beek A, Vischer NO, Smelt JP, Brul S, Manders EM. 2013. Live cell imaging of germination and outgrowth of individual Bacillus subtilis spores; the effect of heat stress quantitatively analyzed with SporeTracker. PLoS One 8:e58972. doi: 10.1371/journal.pone.0058972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Huang SS, Li YQ. 2006. Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal Chem 78:6936–6941. doi: 10.1021/ac061090e. [DOI] [PubMed] [Google Scholar]
- 44.Black EP, Koziol-Dube K, Guan D, Wei J, Setlow B, Cortezzo DE, Hoover DG, Setlow P. 2005. Factors influencing germination of Bacillus subtilis spores via activation of nutrient receptors by high pressure. Appl Environ Microbiol 71:5879–5887. doi: 10.1128/AEM.71.10.5879-5887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waschkau B, Waldeck J, Wieland S, Eichstadt R, Meinhardt F. 2008. Generation of readily transformable Bacillus licheniformis mutants. Appl Microbiol Biotechnol 78:181–188. doi: 10.1007/s00253-007-1278-0. [DOI] [PubMed] [Google Scholar]
- 46.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 47.Lefort V, Longueville J-E, Gascuel O. 2017. SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 49.Madslien EH, Granum PE, Blatny JM, Lindbäck T. 2014. l-Alanine-induced germination in Bacillus licheniformis—the impact of native gerA sequences. BMC Microbiol 14:101. doi: 10.1186/1471-2180-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janes BK, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovacs AT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ Microbiol 14:2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x. [DOI] [PubMed] [Google Scholar]
- 53.Mahillon J, Chungjatupornchai W, Decock J, Dierickx S, Michiels F, Peferoen M, Joos H. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett 60:205–210. doi: 10.1111/j.1574-6968.1989.tb03447.x. [DOI] [Google Scholar]
- 54.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 55.Baranyi J, Roberts TA. 1994. Special issue predictive modelling a dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.