In 1967, Harland and Lee made a startling discovery: in some humans, the colonic epithelium is covered with a “forest” of spirochetes (W. A. Harlan, and F. D. Lee, Br Med J 3:718–719, 1967, https://doi.org/10.1136/bmj.3.5567.718).
KEYWORDS: Brachyspira, Brachyspira aalborgi, Brachyspira pilosicoli, gastrointestinal infection, human infection, intestinal colonization
ABSTRACT
In 1967, Harland and Lee made a startling discovery: in some humans, the colonic epithelium is covered with a “forest” of spirochetes (W. A. Harlan, and F. D. Lee, Br Med J 3:718–719, 1967, https://doi.org/10.1136/bmj.3.5567.718). In this issue of Journal of Bacteriology, Thorell et al. present a systematic analysis of the prevalence and diversity of the spirochetes Brachyspira aalborgi and Brachyspira pilosicoli in the human colon. These and prior studies provide avenues toward resolving important questions: what bacterial and host parameters contribute to this extensive colonization, and what impact does it have on human health?
TEXT
The association of spirochetes with the gastrointestinal (GI) tracts of humans and other mammals has been known since the time of van Leeuwenhoek, who noted the presence of spiral organisms in human feces. Interest in these spirochetes grew rapidly with the explosion of microbiology during the late 1800s and early 1900s, leading to many publications dating back to 1884 (reviewed in references 1 and 2). However, attempts to culture these organisms during that time were either unsuccessful or not reproducible. Rejuvenation of the study of intestinal spirochetosis during the past 50 years was fueled by two discoveries. Harland and Lee (3) performed electron microscopy of a rectal biopsy specimen from a patient with chronic diarrhea and found that the epithelium was covered with a near-confluent “forest” of spirochetes firmly attached by one end to the surface of the host cells (Fig. 1). Additionally, spirochetes later identified as Brachyspira hyodysenteriae and Brachyspira pilosicoli, the latter meaning “short spiral-shaped organisms that make the colon look hairy,” were found to be frank pathogens in swine, causing dysentery and wasting in young pigs (4). Currently, nine Brachyspira species are recognized (B. aalborgi, B. alvinipulli, B. hampsonii, B. hyodysenteriae, B. innocens, B. intermedia, B. murdochii, B. pilosicoli, and B. suanatina), but to date only B. aalborgi and B. pilosicoli are known to colonize humans. While the natural host ranges of these organisms have not been studied thoroughly, B. aalborgi is reported to colonize humans, nonhuman primates, and opossums, whereas B. pilosicoli has been found in humans, swine, dogs, and birds (5, 6). A new species, “B. catarrhinii,” has been proposed; it encompasses a group of organisms (previously characterized as subset of B. aalborgi) that colonize monkeys (7).
FIG 1.
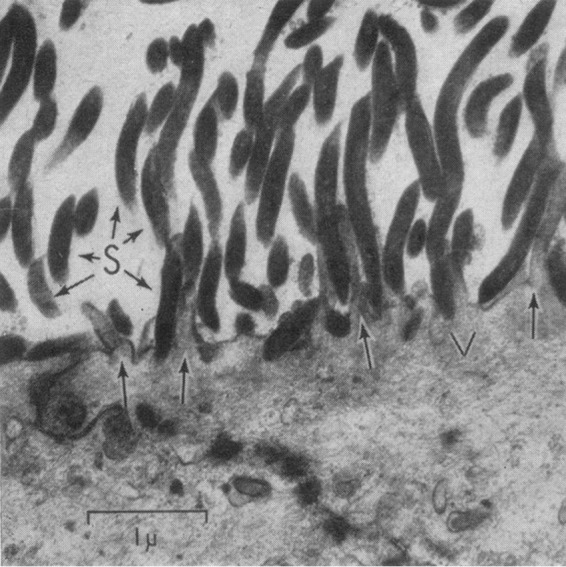
Human colonic spirochetosis. Spirochetes attach end-on to the colonic epithelium, as demonstrated by electron microscopy. This image is from the landmark 1967 publication by Harland and Lee (3) and is reprinted with permission from the publisher. Arrows indicate microvilli. S, spirochetes; V, reactive vacuole. Bar = 1 μm.
A number of studies have been carried out to determine the prevalence of Brachyspira colonization in humans and the potential association with disease manifestations. Some of these studies are summarized in Table 1; case reports or studies involving a small number of subjects (<5) are not included. Many of the early studies were published before the Brachyspira species were cultured or characterized, and methods such as light and electron microscopy cannot distinguish between different Brachyspira species. Therefore, information regarding the relative prevalence of B. aalborgi and B. pilosicoli was not available for several of the articles. Methods used for detection of colonic spirochetosis range from simple hematoxylin and eosin staining of tissue sections to culture, PCR, and fluorescent in situ hybridization (FISH). Culture from either fresh or frozen fecal samples or biopsy specimens collected during colonoscopy has been effective in isolation of Brachyspira organisms, yielding a large number of strains in some studies.
TABLE 1.
Selected studies examining the prevalence of human colonic spirochetosis
| Study | Reference | Detection method(s)a | Location(s) | Study population | Yr(s) of specimen collection | Specimen(s) | Total subjects | B. aalborgi positive | B. pilosicoli positive | Total HCS cases | Percent positive | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lee et al. (1971) | 2 | EM, HE, SS, PAS | Glasgow, Scotland, UK | Patients with diarrhea or suspected cancer | 1961 | Rectal biopsy specimen | 144 | —b | — | 10 | 6.9 | Includes the cases from Harland and Lee (3) |
| Lee et al. (1971) | 2 | EM, HE, SS, PAS | Glasgow, Scotland, UK | Appendectomy cases | 1963–1966 | Excised appendix | 790 | — | — | 62 | 7.8 | Included acute appendicitis (7/144 [4.4%]), “simulated” appendicitis (15/523 [9.8%]), incidental appendectomy (4/107 [4.4%]) |
| McMillan and Lee (1981) | 11 | HE | Glasgow, Scotland, UK | MSMc | NId | Colorectal biopsy specimen; FFPEe | 100 | — | — | 36 | 36.0 | |
| McMillan and Lee (1981) | 11 | HE | Glasgow, Scotland, UK | Heterosexual men | NI | Colorectal biopsy specimen; FFPE | 67 | — | — | 2 | 3.0 | |
| Mathan and Mathan (1985) | 26 | EM | Southern India | Healthy adults | NI | Rectal biopsy specimen | 14 | — | — | 9 | 64.3 | |
| Cooper et al. (1986) | 27 | EM | Southhampton, Hampshire, England, UK | MSM | NI | Rectal biopsy specimen | 8 | — | — | 5 | 62.5 | Reduction in microvillus density observed |
| Cooper et al. (1986) | 27 | EM | Southhampton, Hampshire, England, UK | Heterosexual men | NI | Rectal biopsy specimen | 5 | — | — | 0 | 0.0 | |
| Surawicz et al. (1986) | 28 | HE, AB, SS | Seattle, WA | MSM | NI | Rectal biopsy specimen | 100 | — | — | 28 | 28.0 | |
| Tompkins et al. (1986) | 29 | C | Great Britain, UK | Healthy adults | NI | Colorectal biopsy specimen | 1,527 | — | — | 23 | 1.5 | All positive specimens from either MSM or persons of Asian ethnicity |
| Barrett (1990) | 30 | C | Muskat Region, Oman | Healthy children and adults | 1988 | Feces | 292 | — | — | 78 | 26.7 | |
| Barrett (1990) | 30 | C | Muskat Region, Oman | Hospitalized patients | 1988 | Feces | 1,000 | — | — | 114 | 11.4 | |
| Lee and Hampson (1992) | 12 | C | Western Australia | Aboriginal children and adults | 1989–1991 | Feces | 181 | — | — | 59 | 32.6 | Isolates were shown subsequently to be B. pilosicoli (31) |
| Lee and Hampson (1992) | 12 | C | Western Australia, Northern Territory | Non-Aboriginal children and adults | 1989–1991 | Feces | 695 | — | — | 8 | 1.2 | |
| De Brito et al. (1996) | 32 | HE, SS, IHC, EM | Brazil | Patients with GIf symptoms | NI | Rectal and sigmoidal colonic biopsy specimens | 282 | — | — | 4 | 1.4 | |
| Trivett-Moore et al. (1998) | 13 | HE, EM, C | Sydney, Australia | MSM attending a sexual health clinic | NI | Rectal biopsy specimens | 41 | (0) | (13) | 22 | 53.7 | B. pilosicoli isolated from biopsy samples positive (11/22) and negative (2/19) for HCS by HE and EM |
| Brooke et al. (2001) | 10 | C | Western Australia | Aboriginal rural patients with GI complaints | 1998–1999 | Feces | 151 | 0 | 15 | 15 | 9.9 | High proportion of isolates from subjects aged 2 to 5 |
| Brooke et al. (2001) | 10 | C | Western Australia | Non-Aboriginal rural patients with GI complaints | 1998–1999 | Feces | 142 | 0 | 0 | 0 | 0 | |
| Brooke et al. (2001) | 10 | C | Australia | Entering migrants to Australia | 1998–1999 | Feces | 227 | 0 | 24 | 24 | 10.6 | Isolates/subjects for migrants from Asia (2/8), Eastern Europe (3/94), the Middle East (9/65), and Africa (10/50) |
| Margawani et al. (2004) | 33 | C | Bali, Indonesia | Adult and child residents | 1999 | Feces (August) | 500 | 0 | 59 | 59 | 11.8 | 375 subjects were sampled at both time points |
| Margawani et al. (2004) | 33 | C | Bali, Indonesia | Adult and child residents | 1999 | Feces (December) | 492 | 0 | 62 | 62 | 12.6 | 375 subjects were sampled at both time points |
| Esteve et al. (2006) | 16 | HE, SS, PAS, PCR | Barcelona, Spain | Patients with chronic watery diarrhea and control subjects | 1994–2004 | Colonic biopsy samples | 1,176 | (2) | (2) | 8 | 0.7 | Of 8 subjects positive for HCS by light microscopy, 2 were positive for B. pilosicoli and 2 for B. aalborgi by PCR |
| Calderaro et al. (2007) | 34 | PCR, C | Parma, Italy | Patients with suspected gastrointestinal infections | 2002–2006 | Feces, colonic biopsy samples, FFPE | 234 | 13 | 5 | 16 | 6.8 | Two patients were coinfected with B. aalborgi and B. pilosicoli |
| Tanahashi et al. (2008) | 35 | HE, SS, IHC, immuno-EM, PCR | Oita, Japan | Patients with colonoscopy or surgical resections | 2005–2006 | Colonic biopsy samples, FFPE | 2,556 | 20 | 3 | 20 | 0.8 | 11 cases identified by HE, SS, and IHC. 20 cases positive for B. aalborgi by PCR; 3 cases also positive for B. pilosicoli |
| Ichimata et al. (2017) | 36 | HE | Asahi, Matsumoto, Japan | Patients <20 yrs of age with gastrointestinal symptoms | NI | Biopsy specimens, surgical specimens | 479 | — | — | 1 | 0.2 | |
| Thorell et al. (2019) | 8 | HE, IHC, SS, C | Sweden | Adult population | 2000–2006 | Biopsy samples of terminal ileum and colon from cecum to rectum (5 sites) | 745 | 13 | 1 | 17 | 2.3 | HCS cases correspond to those described previously by Walker et al. (18); 3 subjects who were positive by HE, IHC, and SS positive were negative by culture |
| Mikosza et al. (2001) | 37 | PCR | Australia | HCS subjects (by HE) | NI | Colon, colorectal, cecum, and appendix biopsy samples; FFPEb | 28 | 24 | 4 | 26 | 92.9 | 2 subjects were positive for both B. aalborgi and B. pilosicoli; 2 subjects were negative by PCR |
| Mikosza et al. (2004) | 38 | PCR | Australia (20); USA (1); France (1); Norway (2) | HCS subjects (by HE) | NI | Colon, colorectal, cecum, and appendix biopsy samples; PET | 24 | 22 | 2 | 24 | 100 | Prescreened for intestinal spirochetosis by histology |
| Westerman et al. (2012) | 39 | Real-time PCR | The Netherlands | HCS subjects (by HE, IHC) | 2001–2011 | Colon biopsy samples; FFPE | 56 | 48 | 9 | 56 | 100 | Several genotypesg |
| Rojas et al. (2017) | 40 | FISH | Germany | HCS subjects (by HE) | NI | Intestinal biopsy samples (from ileum to rectum); PET | 91 | — | — | 77 | 84.6 | Prescreened for intestinal spirochetosis by histology; same specimens were analyzed by both FISH and PCR |
| Rojas et al. (2017) | 40 | PCR | Germany | HCS subjects (by HE) | NI | Intestinal biopsy samples (from ileum to rectum); PET | 91 | 53 | 23 | 75 | 82.4 | Prescreened for intestinal spirochetosis by histology; includes one subject with both B. aalborgi and B. pilosicoli |
In the studies described in references 37 to 40, specimens were prescreened for colonic spirochetosis. Abbreviations: AB, alcian blue-stained sections; C, culture; EM, transmission electron microscopy; FISH, fluorescent in situ hybridization; HE, hematoxylin and eosin-stained sections; IHC, immunohistochemistry; PAS, periodic acid-Schiff-stained sections; SS, silver-stained sections.
—, species not determined.
MSM, men who have sex with men.
NI, not indicated.
FFPE, formalin-fixed, paraffin-embedded tissue.
GI, gastrointestinal.
Includes 36 B. aalborgi cluster 1 organisms alone, 6 B. aalborgi cluster 2 (“B. hominis”) organisms alone, 6 B. pilosicoli organisms alone, 5 cluster 1 and cluster 2 organisms, 1 B. pilosicoli and cluster 2 organisms, and 2 B. pilosicoli, cluster 1, and cluster 2 organisms (triple positive).
In a comprehensive article featured in this issue of the Journal of Bacteriology, Thorell et al. (8) discuss the prevalence and properties of Brachyspira species in a group of 745 human subjects in Stockholm, Sweden. This study was part of a larger project called PopCol (9), which investigated the prevalence of endoscopic findings in randomly selected adults in the general population; it thus establishes a firm “baseline” of data useful in assessing the etiology of functional gastrointestinal disorders such as irritable bowel syndrome. Among the strengths of the article is the unbiased sampling of colonoscopy biopsy specimens from a representative cross section of the adult population. Random sampling was made at the terminal ileum (small intestine) and 4 different sites in the colon of each subject. The specimens were screened by light microscopy techniques for the presence of the “false brush border” appearance typical of human colonic spirochetosis (HCS). Seventeen subjects out of the 745 examined (2.3%) were determined to have HCS. Confirmatory studies and culture were performed on the samples from these 17 individuals, and 14 yielded positive cultures; these exhibited a predominance of B. aalborgi (13 positive individuals) relative to B. pilosicoli (1 positive individual), as reported in other European studies. Perhaps the most valuable information to come out of this study was the nearly complete genomic sequences of 16 B. aalborgi strains, including the type strain, 513A; these are the first available B. aalborgi genomic sequences. Finally, Thorell et al. determined that the primer sets commonly used for the amplification of 16S rRNA gene sequences for microbiome determinations are ineffective in amplifying Brachyspira 16S sequences. They also showed that Brachyspira organisms are likely underrepresented in prior human gut microbiome studies.
The article by Thorell et al. (8) thus represents a significant addition to the prior publications (Table 1). To an “outsider” who studies other spirochetes, the accumulating literature on the distribution and characteristics of HCS organisms is very impressive, given the relatively small number of groups contributing to these studies. What are some of the insights that have been derived by this cumulative work, and what questions still remain?
By all indications, HCS has a global distribution. Brachyspira species have been detected in humans on every continent (except Antarctica). However, the prevalence of HCS in healthy populations varies widely in different studies, from 0 to 64.8% (Table 1). Lower prevalences (0.2% to 3.2%) tend to be found among healthy individuals in temperate, highly urbanized areas (such as Western Europe and Japan), whereas higher levels (10.8% to 64.8%) have been observed in Indonesia, Oman, and India and in Australian Aboriginal populations. This trend does not appear to be associated strictly with rural versus urban environments; for example, Brooke et al. (10) found that Aboriginal and non-Aboriginal individuals with gastrointestinal disorders within the same rural region of Western Australia had HCS prevalences of 9.9% and 0%, respectively. The proportion of individuals with HCS is generally higher in subjects with gastrointestinal complaints. Men who have sex with men (MSM) populations also have relatively high proportions of colonic spirochete colonization, with prevalences ranging from 28 to 62.5% (Table 1). In one study in Scotland, McMillan and Lee (11) found that 36/100 MSM subjects (36%) were positive for colonic spirochetes, whereas a control group of male heterosexual subjects had an HCS prevalence of 2/67 (3%). As stated by Lee and Hampson (12), “there may either be ethnic or environmental influences predisposing to spirochaetal colonization of the intestine.”
Another variable aspect of HCS is the proportion of B. aalborgi versus B. pilosicoli colonization. Early studies utilized only light or electron microscopy to detect spirochete colonization, so distinction between Brachyspira species was not possible. As culture and PCR from colonic biopsy or stool specimens came into common use, most studies could utilize the sequences of 16S rRNA and NADH oxidase genes for species and subgroup identification. B. pilosicoli tends to be more common in populations with a high incidence of HCS, including Aboriginal Australian, Indonesian, and MSM groups (13). In comparison, B. aalborgi, which has not been associated with disease symptoms, is predominant in areas such as Europe, Japan, and urban Australia. The combination of low HCS prevalence (2.3%) and B. aalborgi dominance (14 of 16 isolates) is evident in the Swedish population studied by Thorell et al. (8). Genotyping by 16S rRNA gene sequencing further revealed that their B. aalborgi isolates fell into cluster 1 of the two major genotype clusters within this species (8).
Brachyspira species exhibit a spectrum of pathogenesis and host ranges, as reviewed previously (5, 14). B. hyodysenteriae is a frank pathogen in swine but does not colonize or cause disease in humans. B. pilosicoli has a broader host range, with swine, birds, humans, and nonhuman primates being among its known natural hosts. This organism is a cause of diarrheal disease and economic losses in farms raising swine and chickens. In humans, abdominal pain, diarrhea, and perirectal bleeding can be present in some B. pilosicoli-positive individuals, whereas many others are asymptomatic; B. pilosicoli also has been shown to cause spirochetemia in critically ill patients (15). Thus far, B. aalborgi has been detected only in humans, nonhuman primates, and opossums (5); some of the animal isolates may instead be the proposed species “B. catarrhinii” (7). Colonization of the colon by B. aalborgi is not significantly associated with gastrointestinal symptoms or histopathology, leading some investigators to conclude that it is essentially a commensal organism.
The association between Brachyspira colonization and gastrointestinal problems such as chronic diarrhea remains unclear. Thorell et al. (8) and Esteve et al. (16) noted that many individuals were positive for HCS at several regions of the colon yet were asymptomatic, indicating that an extensive “forest” of Brachyspira can be present throughout much of the colon without causing GI symptoms. Carr et al. (17) reviewed a series of HCS cases (113 colonic biopsy specimens and 16 appendixes) and concluded that there was a lack of inflammatory changes, except in cases with other causes of inflammation; they further stated that HCS in an inflamed biopsy specimen is “likely to be an incidental finding.” However, Walker et al. (18) noted that the occurrence of clusters of eosinophils in the subepithelial tissue of the colon was significantly associated with HCS. Additionally, clearance of HCS spontaneously or through treatment with metronidazole or other antimicrobial agents may result in resolution of symptoms in chronic diarrhea in some cases (6, 16, 19). Perhaps the occurrence of gastrointestinal symptoms may result from the combination of HCS and as yet undefined host factors.
A major contribution of the study by Thorell et al. was the addition of 18 new genomic sequences. The 16 B. aalborgi genome sequences revealed that the overall sizes (2.50 to 2.71 Mb) are smaller than those of B. hyodysenteriae (2.99 to 3.17 Mb) and lack the 36-kb plasmid thought to be important in pathogenesis (20). The B. aalborgi genomes have the characteristic low G+C content of Brachyspira (28.1 to 28.3%) and are otherwise similar to genomes of the 8 other Brachyspira species for which sequences are available (20, 21). Intraspecies comparisons of the B. aalborgi genomes revealed a heterogeneity greater than expected, although the organisms were clearly separated from other species. Black et al. (20) found that the genomes of B. hyodysenteriae strains had undergone significant rearrangements, and the heterogeneity in B. aalborgi may also be related in part to rearrangements. Availability of the B. aalborgi sequences will extend the comparative genomics possibilities within the Brachyspira genus, perhaps helping to reveal key differences important in host interactions, such as pathogenesis or host range (21).
The article by Thorell et al. (8) also revealed that the primer sets commonly used for amplifying 16S rRNA gene sequences for microbiome studies do not amplify Brachyspira sequences. As a result, analyses of the human gut microbiome lack representation from Brachyspira. An in silico reexamination of data from the Human Microbiome Project by Thorell et al. (8) found that only 1 individual out of 179 had Brachyspira 16S sequences, and the two samples from this individual contained only 0.03 to 0.04% Brachyspira sequences. This paucity of Brachyspira sequences is supported by a recent computational analysis of 11,850 human gut microbiomes, which yielded 92,143 metagenome-assembled genomes (MAGs) (22). Of these, only one MAG was identified as a Brachyspira species, and this genome assembly turned out to be from a bovine rumen specimen mistakenly included in the collection. Overall, these results are surprising given the extensive forests of Brachyspira found in some individuals and the ability to culture B. pilosicoli and B. aalborgi readily from both colonic biopsy and feces specimens. Perhaps the strong adherence of Brachyspira to the colonic epithelium limits the number present in feces, or the amplification of Brachyspira sequences in metagenomic studies is relatively inefficient because of its low G+C content.
The study by Thorell et al. and the cumulative work since the 1967 article by Harland and Lee have greatly increased our understanding of human colonic spirochetosis. However, many questions remain, as emphasized in several of the articles cited in this commentary. First and foremost, should B. pilosicoli (and perhaps also B. aalborgi) be considered a human pathogen? An “n of one” B. pilosicoli WesB self-inoculation experiment was conducted in the 1990s and resulted in colonization, abdominal discomfort, bloating, and headaches; the infection and symptoms resolved following metronidazole treatment (23). Should a more extensive, blinded, and controlled inoculation study in humans be performed, as has been done with norovirus and other human gastrointestinal pathogens? How is HCS transmitted: through direct human-to-human contact, through animal-to-human transmission (in the case of B. pilosicoli), via contaminated water or food supplies (6, 23, 24), or by other means? Do humans vary in susceptibility to colonization or disease manifestations, and to what extent is this aspect related to genetic factors, immune status, hygiene, overall health, sexual activity, socioeconomic influences, climate, and additional parameters? Does the immune response affect colonization or the progression of infection? What determines the host range and pathogenicity of Brachyspira species, and do certain strains within each species differ in their infectious and pathogenic properties? Can the expanding genomic and proteomic (25) information be effectively “mined” for this information? Specifically, what genes are involved in these processes, including the characteristic attachment mechanism and cytotoxicity (e.g., the strong and weak hemolysis by B. hyodysenteriae and B. pilosicoli, respectively)? Is genetic manipulation of Brachyspira possible, as has been accomplished to some extent with Borrelia, Treponema, and Leptospira organisms? Some of these questions may be addressed in part by studies of veterinary pathogens, e.g., those affecting the swine and poultry industries. It is hoped that special attention will be paid to the high incidence of B. pilosicoli in developing countries and its potential impact on human health.
ACKNOWLEDGMENTS
I gratefully acknowledge David J. Hampson (Murdoch University, Murdoch, Australia) and Bridget D. DeLay (McGovern Medical School at UTHealth, Houston, TX) for their careful reading of and insightful comments regarding the manuscript.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Footnotes
For the article discussed, see https://doi.org/10.1128/JB.00272-19.
REFERENCES
- 1.Parr LW. 1923. Intestinal spirochetes. J Infect Dis 33:369–383. doi: 10.1093/infdis/33.5.369. [DOI] [Google Scholar]
- 2.Lee FD, Kraszewski A, Gordon J, Howie JG, McSeveney D, Harland WA. 1971. Intestinal spirochaetosis. Gut 12:126–133. doi: 10.1136/gut.12.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harland WA, Lee FD. 1967. Intestinal spirochaetosis. Br Med J 3:718–719. doi: 10.1136/bmj.3.5567.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trott DJ, Stanton TB, Jensen NS, Duhamel GE, Johnson JL, Hampson DJ. 1996. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol 46:206–215. doi: 10.1099/00207713-46-1-206. [DOI] [PubMed] [Google Scholar]
- 5.Duhamel GE. 2001. Comparative pathology and pathogenesis of naturally acquired and experimentally induced colonic spirochetosis. Anim Health Res Rev 2:3–17. doi: 10.1079/AHRR200128. [DOI] [PubMed] [Google Scholar]
- 6.Hampson DJ. 2018. The spirochete Brachyspira pilosicoli, enteric pathogen of animals and humans. Clin Microbiol Rev 31:e00087-17. doi: 10.1128/CMR.00087-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips ND, La T, Hampson DJ. 2019. Brachyspira catarrhinii sp. nov., an anaerobic intestinal spirochaete isolated from vervet monkeys may have been misidentified as Brachyspira aalborgi in previous studies. Anaerobe 59:8–13. doi: 10.1016/j.anaerobe.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Thorell K, Inganäs L, Backhans A, Agréus L, Öst Å, Walker MM, Talley NJ, Kjellström L, Andreasson A, Engstrand L. 2019. Isolates from colonic spirochaetosis in humans show high genomic divergence and potential pathogenic features but are not detected using standard primers for the human microbiota. J Bacteriol 201:e00272-19. doi: 10.1128/JB.00272-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kjellström L, Molinder H, Agréus L, Nyhlin H, Talley NJ, Andreasson A. 2014. A randomly selected population sample undergoing colonoscopy: prevalence of the irritable bowel syndrome and the impact of selection factors. Eur J Gastroenterol Hepatol 26:268–275. doi: 10.1097/MEG.0000000000000024. [DOI] [PubMed] [Google Scholar]
- 10.Brooke CJ, Clair AN, Mikosza AS, Riley TV, Hampson DJ. 2001. Carriage of intestinal spirochaetes by humans: epidemiological data from Western Australia. Epidemiol Infect 127:369–374. doi: 10.1017/S095026880100588X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMillan A, Lee FD. 1981. Sigmoidoscopic and microscopic appearance of the rectal mucosa in homosexual men. Gut 22:1035–1041. doi: 10.1136/gut.22.12.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JI, Hampson DJ. 1992. Intestinal spirochaetes colonizing aborigines from communities in the remote north of Western Australia. Epidemiol Infect 109:133–141. [PMC free article] [PubMed] [Google Scholar]
- 13.Trivett-Moore NL, Gilbert GL, Law CL, Trott DJ, Hampson DJ. 1998. Isolation of Serpulina pilosicoli from rectal biopsy specimens showing evidence of intestinal spirochetosis. J Clin Microbiol 36:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikosza AS, Hampson DJ. 2001. Human intestinal spirochetosis: Brachyspira aalborgi and/or Brachyspira pilosicoli? Anim Health Res Rev 2:101–110. doi: 10.1079/AHRR200117. [DOI] [PubMed] [Google Scholar]
- 15.Trott DJ, Jensen NS, Saint Girons I, Oxberry SL, Stanton TB, Lindquist D, Hampson DJ. 1997. Identification and characterization of Serpulina pilosicoli isolates recovered from the blood of critically ill patients. J Clin Microbiol 35:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteve M, Salas A, Fernandez-Banares F, Lloreta J, Marine M, Gonzalez CI, Forne M, Casalots J, Santaolalla R, Espinos JC, Munshi MA, Hampson DJ, Viver JM. 2006. Intestinal spirochetosis and chronic watery diarrhea: clinical and histological response to treatment and long-term follow up. J Gastroenterol Hepatol 21:1326–1333. doi: 10.1111/j.1440-1746.2006.04150.x. [DOI] [PubMed] [Google Scholar]
- 17.Carr NJ, Mahajan H, Tan KL, Sharma R. 2010. The histological features of intestinal spirochetosis in a series of 113 patients. Int J Surg Pathol 18:144–148. doi: 10.1177/1066896908330203. [DOI] [PubMed] [Google Scholar]
- 18.Walker MM, Talley NJ, Inganas L, Engstrand L, Jones MP, Nyhlin H, Agreus L, Kjellström L, Ost A, Andreasson A. 2015. Colonic spirochetosis is associated with colonic eosinophilia and irritable bowel syndrome in a general population in Sweden. Hum Pathol 46:277–283. doi: 10.1016/j.humpath.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Carpentieri DF, Souza-Morones S, Gardetto JS, Ross HM, Downey K, Ingebo K, Siaw E. 2010. Intestinal spirochetosis in children: five new cases and a 20-year review of the literature. Pediatr Dev Pathol 13:471–475. doi: 10.2350/09-10-0725-CR.1. [DOI] [PubMed] [Google Scholar]
- 20.Black M, Moolhuijzen P, Barrero R, La T, Phillips N, Hampson D, Herbst W, Barth S, Bellgard M. 2015. Analysis of multiple Brachyspira hyodysenteriae genomes confirms that the species is relatively conserved but has potentially important strain variation. PLoS One 10:e0131050. doi: 10.1371/journal.pone.0131050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampson DJ, Wang P. 2018. Colonic spirochetes: what has genomics taught us? Curr Top Microbiol Immunol 415:273–294. doi: 10.1007/82_2017_48. [DOI] [PubMed] [Google Scholar]
- 22.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. 2019. A new genomic blueprint of the human gut microbiota. Nature 568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oxberry SL, Trott DJ, Hampson DJ. 1998. Serpulina pilosicoli, waterbirds and water: potential sources of infection for humans and other animals. Epidemiol Infect 121:219–225. doi: 10.1017/s0950268898008863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oxberry SL, Hampson DJ. 2003. Epidemiological studies of Brachyspira pilosicoli in two Australian piggeries. Vet Microbiol 93:109–120. doi: 10.1016/S0378-1135(03)00014-2. [DOI] [PubMed] [Google Scholar]
- 25.Casas V, Vadillo S, San Juan C, Carrascal M, Abian J. 2016. The exposed proteomes of Brachyspira hyodysenteriae and B. pilosicoli. Front Microbiol 7:1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathan MM, Mathan VI. 1985. Rectal mucosal morphologic abnormalities in normal subjects in southern India: a tropical colonopathy? Gut 26:710–717. doi: 10.1136/gut.26.7.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper C, Cotton DW, Hudson MJ, Kirkham N, Wilmott FE. 1986. Rectal spirochaetosis in homosexual men: characterisation of the organism and pathophysiology. Genitourin Med 62:47–52. doi: 10.1136/sti.62.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surawicz CM, Goodell SE, Quinn TC, Roberts PL, Corey L, Holmes KK, Schuffler MD, Stamm WE. 1986. Spectrum of rectal biopsy abnormalities in homosexual men with intestinal symptoms. Gastroenterology 91:651–659. doi: 10.1016/0016-5085(86)90635-9. [DOI] [PubMed] [Google Scholar]
- 29.Tompkins DS, Foulkes SJ, Godwin PG, West AP. 1986. Isolation and characterisation of intestinal spirochaetes. J Clin Pathol 39:535–541. doi: 10.1136/jcp.39.5.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett SP. 1990. Intestinal spirochaetes in a Gulf Arab population. Epidemiol Infect 104:261–266. doi: 10.1017/S0950268800059434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JI, Hampson DJ. 1994. Genetic characterisation of intestinal spirochaetes and their association with disease. J Med Microbiol 40:365–371. doi: 10.1099/00222615-40-5-365. [DOI] [PubMed] [Google Scholar]
- 32.De Brito T, Sandoval MP, Silva AG, Saad RC, Colaiacovo W. 1996. Intestinal spirochetosis: first cases reported in Brazil and the use of immunohistochemistry as an aid in histopathological diagnosis. Rev Inst Med Trop Sao Paulo 38:45–52. doi: 10.1590/s0036-46651996000100009. [DOI] [PubMed] [Google Scholar]
- 33.Margawani KR, Robertson ID, Brooke CJ, Hampson DJ. 2004. Prevalence, risk factors and molecular epidemiology of Brachyspira pilosicoli in humans on the island of Bali, Indonesia. J Med Microbiol 53:325–332. doi: 10.1099/jmm.0.05415-0. [DOI] [PubMed] [Google Scholar]
- 34.Calderaro A, Gorrini C, Peruzzi S, Piccolo G, Dettori G, Chezzi C. 2007. Occurrence of human intestinal spirochetosis in comparison with infections by other enteropathogenic agents in an area of the Northern Italy. Diagn Microbiol Infect Dis 59:157–163. doi: 10.1016/j.diagmicrobio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Tanahashi J, Daa T, Gamachi A, Kashima K, Kondoh Y, Yada N, Yokoyama S. 2008. Human intestinal spirochetosis in Japan; its incidence, clinicopathologic features, and genotypic identification. Mod Pathol 21:76–84. doi: 10.1038/modpathol.3800987. [DOI] [PubMed] [Google Scholar]
- 36.Ichimata S, Yoshizawa A, Kusakari M, Nakayama Y, Asaka S, Negishi T, Kasuga E, Matsumoto T, Honda T. 2017. Human intestinal spirochetosis in Japanese patients aged less than 20 years: histological analysis of colorectal biopsy and surgical specimens obtained from 479 patients. Pathol Int 67:302–305. doi: 10.1111/pin.12544. [DOI] [PubMed] [Google Scholar]
- 37.Mikosza AS, La T, de Boer WB, Hampson DJ. 2001. Comparative prevalences of Brachyspira aalborgi and Brachyspira (Serpulina) pilosicoli as etiologic agents of histologically identified intestinal spirochetosis in Australia. J Clin Microbiol 39:347–350. doi: 10.1128/JCM.39.1.347-350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikosza AS, Munshi MA, Hampson DJ. 2004. Analysis of genetic variation in Brachyspira aalborgi and related spirochaetes determined by partial sequencing of the 16S rRNA and NADH oxidase genes. J Med Microbiol 53:333–339. doi: 10.1099/jmm.0.05430-0. [DOI] [PubMed] [Google Scholar]
- 39.Westerman LJ, Stel HV, Schipper ME, Bakker LJ, Neefjes-Borst EA, van den Brande JH, Boel EC, Seldenrijk KA, Siersema PD, Bonten MJ, Kusters JG. 2012. Development of a real-time PCR for identification of Brachyspira species in human colonic biopsies. PLoS One 7:e52281. doi: 10.1371/journal.pone.0052281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rojas P, Petrich A, Schulze J, Wiessner A, Loddenkemper C, Epple HJ, Sterlacci W, Vieth M, Kikhney J, Moter A. 2017. Distribution and phylogeny of Brachyspira spp. in human intestinal spirochetosis revealed by FISH and 16S rRNA-gene analysis. Anaerobe 47:25–32. doi: 10.1016/j.anaerobe.2017.03.012. [DOI] [PubMed] [Google Scholar]


