Abstract
Mechanical forces are well known for modulating heart valve developmental programs. Yet, it is still unclear how genetic programs and mechanosensation interact during heart valve development. Here, we assessed the mechanosensitive pathways involved during zebrafish outflow tract (OFT) valve development in vivo. Our results show that the hippo effector Yap1, Klf2, and the Notch signaling pathway are all essential for OFT valve morphogenesis in response to mechanical forces, albeit active in different cell layers. Furthermore, we show that Piezo and TRP mechanosensitive channels are important factors modulating these pathways. In addition, live reporters reveal that Piezo controls Klf2 and Notch activity in the endothelium and Yap1 localization in the smooth muscle progenitors to coordinate OFT valve morphogenesis. Together, this work identifies a unique morphogenetic program during OFT valve formation and places Piezo as a central modulator of the cell response to forces in this process.
Research organism: Zebrafish
Introduction
Heart pumping and shaping take place concomitantly during embryonic development. These two processes require a tight and dynamic coordination between mechanical forces and tissue morphogenesis. Heart valve development is a great model for studying these interactions when considering that heart valve defects are common congenital cardiac malformation in human (Hoffman and Kaplan, 2002; Øyen et al., 2009). The four-chambered heart contains two different sets of valves (Lin et al., 2012):
- Tricuspide valves including the two semilunar (SL) valves, the aortic valve and the pulmonary valve, as well as the tricuspide valve located between the right atrium and the right ventricle.
- A bicuspide valve called the mitral valve separating the left atrium from the left ventricle. Abnormalities of the arterial valve leaflets are the most-common congenital malformations, in particular bicuspid aortic valve (Hoffman and Kaplan, 2002). Aortic valves mainly derive from endocardial cushion progenitors with a potential contribution from other cellular sources (epicardial cells and neural crest) (Wu et al., 2017). Generally, valve formation depends on two main events: cell proliferation, which is mainly mediated by the vegf-nfat1 pathway, matrix deposition and an endothelial-to-mesenchymal transformation (endMT) under the control of Gata transcription factors (Laforest et al., 2011; Stefanovic et al., 2014), Notch signaling (Luxán et al., 2016), Smad/tgf-beta/Bmp, and Wnt-beta catenin signals (Combs and Yutzey, 2009). Nevertheless, recent evidence suggests that arterial valves develop differently from atrioventricular valves by differentiating directly from progenitors in the outflow wall independently from endMT in mouse (Eley et al., 2018).
Congenital valve defects may originate from developmental origins and/or abnormal haemodynamic forces between these two sets of valves, and it remains unclear how general these developmental programs are. Aortic valves are located in areas of high flow velocity and mechanical forces have a great impact on valve morphogenesis (Butcher et al., 2008). Abnormal blood circulation is widely recognized as a cardiovascular risk factor and abnormal mechanotransduction has been associated with valvulopathies (Bäck et al., 2013). Congenital heart valve malformations are usually associated with genetic mutations in genes essential for heart valve development, such as signaling factors (Notch1, TGFβ) for the aortic valves (Bäck et al., 2013), and actin-binding proteins (FilaminA) for the mitral valves (Sauls et al., 2012). The reoccurring discovery of genetic mutations linking valve defects with genes involved in controlling developmental programs (e.g., in NOTCH1, TBX5, GATA4, TBX20, LMCD1, TNS1, and DCHS1) (PROMESA investigators et al., 2015; Durst et al., 2015; Garg et al., 2005; Richards and Garg, 2010), has spurred interest in valve morphogenesis. A key issue is to further define the genetic or environmental causes of valve malformation.
The zebrafish constitutes a powerful model to study cardiac valve development and the role of mechanical forces at the cellular scale. Zebrafish heart is two chambered and contains three sets of valves (the outflow tract (OFT), atrioventricular (AVC) and the inflow tract (IFT) valve [Figure 1A]) that are all bicuspid (Beis et al., 2005; Hsu et al., 2019; Tessadori et al., 2012). While the developmental programs driving mitral valve development in response to mechanical forces start to be well established in zebrafish, less is known about OFT and IFT valves (Paolini and Abdelilah-Seyfried, 2018; Steed et al., 2016a). The cellular processes leading to valve formation are dynamic and are particularly challenging to address in vivo. Zebrafish heart valves originate from progenitors located in the ventricle and atrium that generate the valve leaflets through a coordinated set of endocardial tissue movements (Boselli et al., 2017; Pestel et al., 2016; Steed et al., 2016a; Steed et al., 2016b; Vermot et al., 2009). The sequence of cellular events leading to AVC valve formation in zebrafish embryonic hearts is initiated through cell shape changes that lead to EC convergence towards the AVC (Boselli et al., 2017) and cellular rearrangements that will form a multilayered tissue (Beis et al., 2005; Pestel et al., 2016; Scherz et al., 2008; Steed et al., 2016b). In the zebrafish AVC, blood flow and Klf2a control notch1b and bmp4 expression, both of which are necessary for valve formation (Vermot et al., 2009). Klf2a regulates the deposition of matrix protein (in particular Fibronectin1) in the valve forming area (Steed et al., 2016b), as well as Wnt signaling by controlling wnt9b expression (Goddard et al., 2017). The latter is consistent with the fact that canonical Wnt signals arise specifically in sub-endocardial, abluminal cells and that these Wnt signals are dependent upon hemodynamic forces in zebrafish (Pestel et al., 2016). In addition, Notch signaling is essential for aortic valve formation (Garg, 2016) and OFT development (MacGrogan et al., 2016; Wang et al., 2017). The role of mechanical forces during OFT valve development at the cellular and molecular scale, however, is largely unknown.
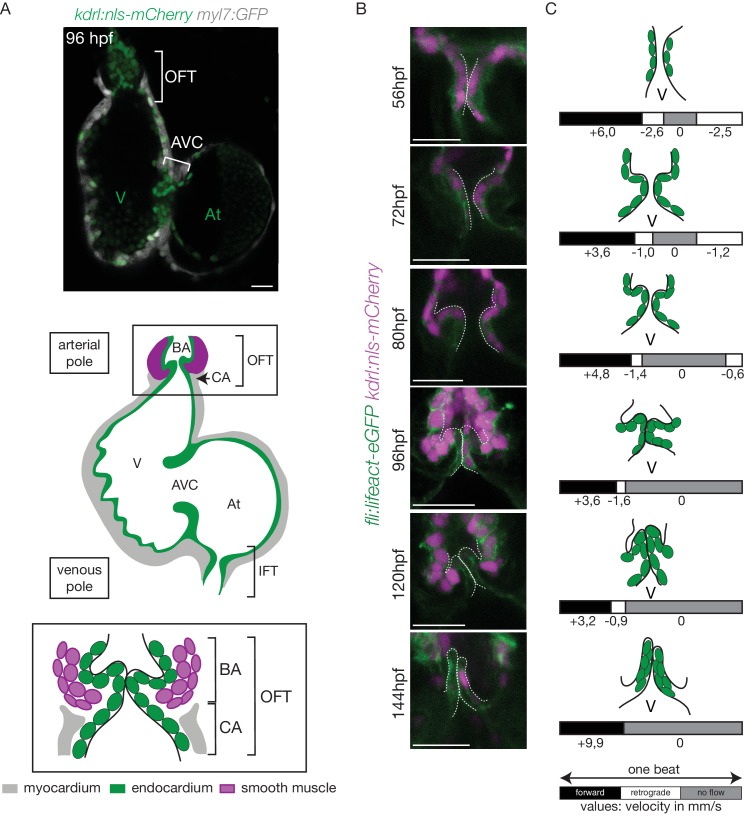
Figure 1. The OFT develops from 56 hpf to form functional valves at 144 hpf.
(A) Top: Z-section of the double transgenic line Tg(kdrl:nls-mCherry; myl7:GFP) showing the overall structure of the heart. Bottom: Scheme of the zebrafish heart with the endocardium, myocardium and smooth muscles and zoom on the OFT structure. The OFT includes the CA and the BA. The CA is the zone of the myocardial connection of the ventricle to the BA. The BA is after the ventricle and is surrounded by smooth muscles. Scheme adapted from Felker et al. (2018). OFT: outflow tract, IFT: inflow tract, AVC: atrioventricular canal, At: atrium, V: ventricle, BA: bulbus arteriosus, CA: conus arteriosus. Scale bar: 20 µm. (B) Z-sections of the double transgenic line Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) at different time-points showing the endocardial OFT structure. Scale bar: 25 µm. (C) Schematic representation summarizing the formation of the valve leaflets over time and flow profile in the OFT during development (from 56 hpf to 144 hpf) showing the forward flow (black), retrograde flow (white) and no flow (grey) fractions with the velocity of the red blood cells (in mm/s) using the double transgenic line Tg(gata1:ds-red; kdrl:EGFP). V: ventricle.
Fluid shear stress is an important environmental cue that governs vascular physiology and pathology (Baeyens et al., 2016), but the molecular mechanisms that mediate endocardial responses to flow are only partially understood. In zebrafish, the mechanosensitive channels Trpv4 and Trpp2 modulate endocardial calcium signaling and klf2a expression is necessary for valve morphogenesis and downstream pathway activation (Heckel et al., 2015; Steed et al., 2016b). Notch signaling is tightly involved in cellular mechanosensitive responses in human aortic valves (Godby et al., 2014) and Notch1 is a potent mechanosensor in adult arteries (Mack et al., 2017). More recently, it has been shown that stretch-sensitive channels from the Piezo family (Murthy et al., 2017) are important for vascular development (Li et al., 2014; Ranade et al., 2014) and lymphatic valve formation (Nonomura et al., 2018). In the embryo, Piezo channels exert essential roles during cell differentiation (He et al., 2018) and can affect lineage choice by modulating the nuclear localization of the mechanoreactive transcription coactivator Yap (Pathak et al., 2014). Nevertheless, the role of Piezo-mediated mechanotransduction during cardiac development and its potential targets remain unclear.
In the developing cardiovascular system, biomechanics is key for modulating flow propagation (Anton et al., 2013). In the teleost heart, the OFT constitutes a specialized organ comprising the conus arteriosus (CA) and the bulbus arteriosus (BA) (Figure 1A). The BA dampens the pressure wave down the arterial tree (Braun et al., 2003b). To perform its function, the BA expresses elastic fiber genes that are thought to provide the mechanical properties necessary for its physiological function (Braun et al., 2003a; Braun et al., 2003b; Keith et al., 1977). The BA is separated from the ventricle by the OFT valve and is composed of smooth muscle. The extracellular matrix (ECM) gene, elastin b, contributes to the development of the BA by regulating cell fate determination of cardiac precursor cells into smooth muscle via a process that involves the mechanotransducer Yap1 (Moriyama et al., 2016). How these factors contribute to OFT valve development and interact with other mechanosensitive pathways remains unclear.
In this study, we investigated the signaling events taking place during OFT valve formation and addressed their regulation by the mechanosensitive channels Piezo and transient potential channels (Trp) as well as the flow-responsive transcription factor Klf2a. We show that OFT valve formation proceeds via an initial stage of endothelial cell folding, which is associated with the generation of a cluster of smooth muscle cell progenitors surrounding the endothelial layer. Subsequent global tissue remodeling events result in the appearance of functional leaflets, which defines a unique process of valvulogenesis. Using live reporters to highlight the signaling changes accompanying these temporally coordinated cell-movement events and genetics, we identified Notch and Klf2 as key flow-dependent factors as well as Yap1 as necessary factors for the correct coordination of OFT valvulogenesis. We show that Piezo and Trp channels are key regulators of klf2 activity in the endothelium and that piezo modulates Yap1 localization in the smooth muscle cells, providing a molecular link between mechanosensitivity and cell signaling in the multilayered valve structure. These data describe the cell responses that are coordinated by the mechanical environment and mechanotransduction via mechanosensitive channels in the endothelium.
Results
Outflow tract valve morphogenesis is unique
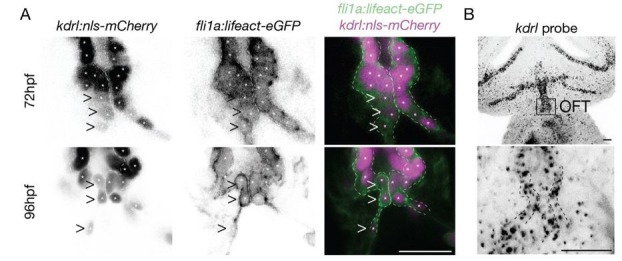
In order to better understand the roles played by blood flow during outflow tract (OFT) valve development, we have developed imaging techniques to capture cardiac motion and analyze blood flow in the OFT. Live imaging of the double transgenic line Tg(gata1:ds-red; kdrl:EGFP) to follow red blood cells and endothelial cell wall movements reveal dramatic changes in intracardiac blood flow patterns during OFT valve development: as the heart matures, blood flow in the OFT is bidirectional until functional valve leaflets emerge in the OFT at 144 hpf (Figure 1A–C). Throughout development, the periods within the cardiac cycle in which reversing flow can be observed decrease in length until 144 hpf, the stage at which we could not observe reversing flow anymore (Figure 1C). Using the Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) line, which labels endothelial cells, we found that these flow profile modifications are linked to changes in OFT tissue geometry and the state of OFT maturation (Figure 1B). At 56 hpf, the endothelium resembles a tube (Figure 1B,C), maturing into cushions by 72 hpf, into premature leaflets by 96 hpf and finally into elongated, thin leaflets by 144 hpf (Figure 1B,C).
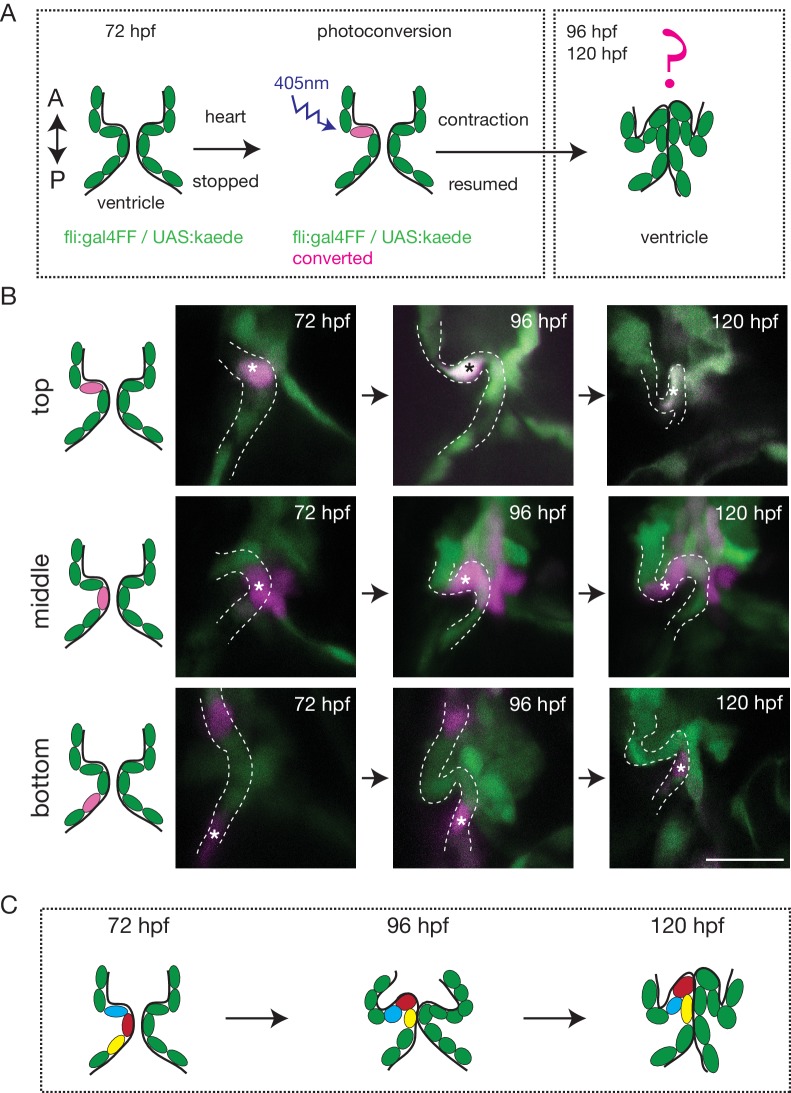
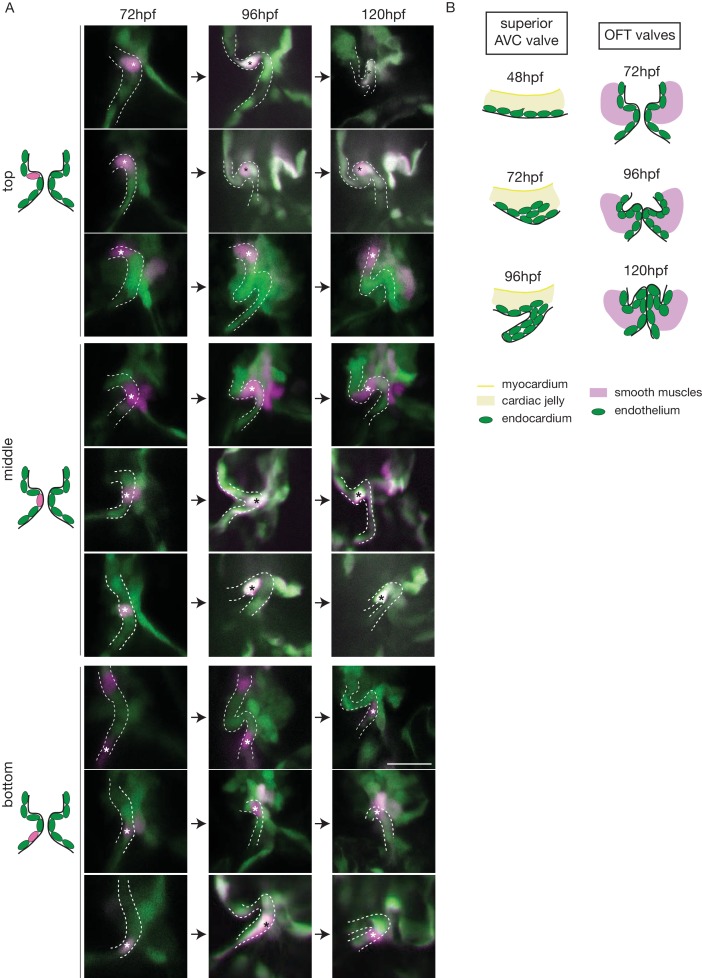
To better characterize how the endothelium changes shape over time and how cells reorganize to form OFT valves, we performed photoconversion experiments using the Tg (fli:gal4FF;UAS:kaede) (Figure 2A). We photoconverted Kaede from green to red in the cells located in the anterior, middle or posterior part of the valve at 72 hpf and assessed their position at 96 hpf and 120 hpf (Figure 2A,B). The results suggest that the endothelium folds to form the valve without multilayering (Figure 2B,C and Figure 2—figure supplement 1A). Indeed, we could observe that the photoconverted cells remain attached to each other and do not show signs of delamination as observed in the AVC (Figure 2B,C and Figure 2—figure supplement 1B).
Figure 2. The endothelium contribution to emerging OFT valve leaflets.
(A) Experimental set-up for the photoconversion studies. Heart was stopped at 72 hpf, the region of interest exposed to 405 nm light to convert kaede from green to red (shown in magenta) fluorescent form and heart contraction was resumed until 120 hpf. Beating hearts were imaged at 96 hpf and 120 hpf by spinning disk microscopy. A. Anterior, P. Posterior. (B) Z-sections of the Tg(fli:gal4FF; UAS:Kaede) line just after photoconversion (72 hpf), and at 96 hpf and 120 hpf. The star highlights the photoconverted cell in the top (n = 6), middle (n = 5) and bottom (n = 4) part of the OFT valve. The other photoconverted cell anteriorly goes out of the frame. Scale bar: 20 µm. Results obtained from three independent experiments. (C) Schematic representation of the results of the photoconversion studies showing the folding of the endothelium in the OFT.
Figure 2—figure supplement 1. The endothelium contribution to emerging OFT versus AVC valves.
Together, these results suggest that OFT valves form by a folding process that might involve the adjacent tissue.
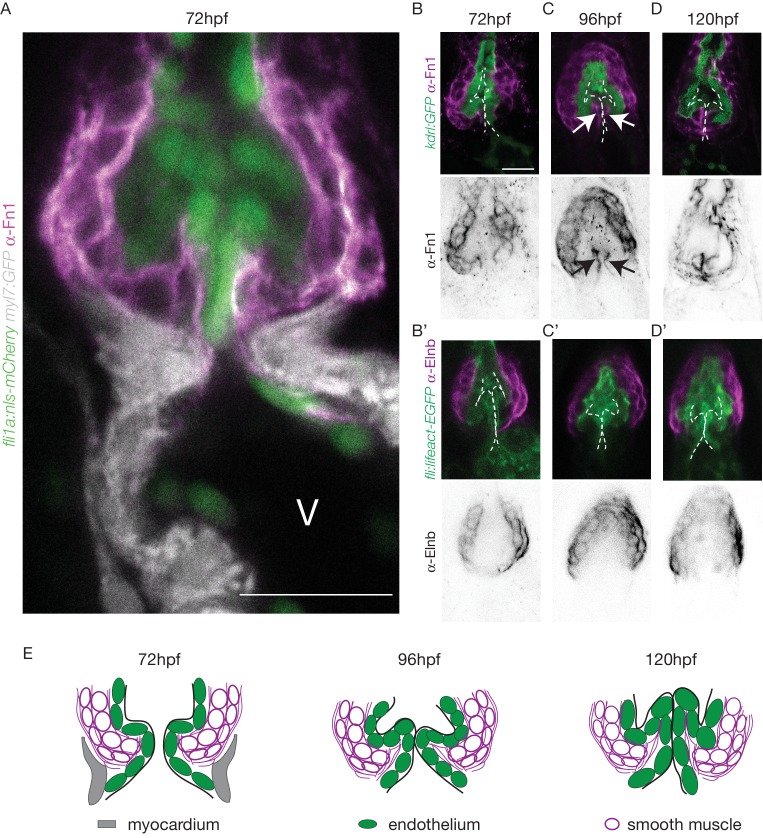
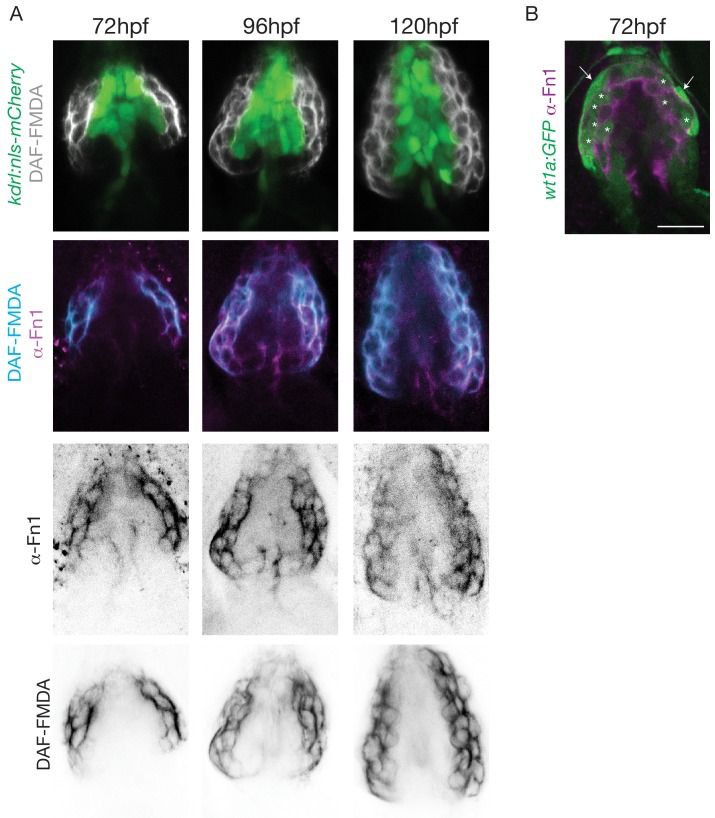
The cellular contribution of the OFT valve led us to the hypothesis that the surrounding tissue could contribute to valve morphogenesis. We analyzed Fibronectin1 (Fn1) expression in the OFT by counterstaining the Tg(kdrl:EGFP) with the Fn1 antibody (Figure 3A–D). We found that Fn1 deposition in the OFT is different from that in the AVC (Steed et al., 2016a). At 72 hpf, cushions appear and Fn1 is observed in the cells around the endothelial cells, that are themselves surrounded at their base by myocardium in the posterior part (Figure 3A,B,C,D,E). Fn1 expression level increases at 96 hpf and delineates a group of cells surrounding the OFT as well as in the basal side of a few endothelial cells that form the cushions (Figure 3C, arrows, E). In the developed leaflets at 120 hpf, Fn1 expression is maintained within the leaflets (Figure 3D,E). We found that most of the cells surrounding the endothelium expressing Fn1 also express Elastin b (Elnb, Eln2 or Tropoelastin), a marker of smooth muscle cells (Grimes et al., 2006; Miao et al., 2007; Paffett-Lugassy et al., 2017) (Figure 3B’, C’, D’). We could confirm that the cells surrounding the Fn1 cells are not myocardial cells as the myocardium stops just before the BA region (Figure 3A,E). To better characterize the smooth muscle identity and their activity, we performed a DAF-FMDA assay and counterstained with Fn1 at 72 hpf, 96 hpf and 120 hpf (Figure 3—figure supplement 1A). These results suggest that all cells expressing Fn1 are also DAF-FMDA-positive (Figure 3—figure supplement 1A). Some of these cells also express Tg(wt1a:GFP) (asterisks in Figure 3—figure supplement 1B) a marker of epicardial cells (Peralta et al., 2013). One hypothesis could therefore be that they might have originated from epicardial precursors. Together, these results suggest a developmental sequence of OFT morphogenesis in vivo where endothelial cell reorganization is associated with changes in gene expression in the surrounding smooth muscle cell progenitors. This indicates that the OFT morphogenesis involves remodeling of not just the endothelium, but also of a group of smooth muscle cells that express Fn1 and Elnb, and are functionally active.
Figure 3. The OFT endothelium is surrounded by smooth muscle cell progenitors expressing fibronectin and elastin.
(A) Staining of Fibronectin1 (magenta) on Tg(myl7:GFP; fli1a:nls-mCherry), highlighting the myocardium (white) and the endothelium (green) at 72 hpf. Scale bar: 20 μm. V: ventricle. Fibronectin1 (anti-Fn1, magenta) counterstaining on Tg(kdrl:GFP) and Elastinb (anti-Elnb, magenta) counterstaining on Tg(fli:lifeact-EGFP) showing their expressions in the OFT at 72 hpf (B), (B’) respectively) at 96 hpf (C), (C’) respectively) and at 120 hpf (D, D’ respectively). Scale bar: 20 µm. Arrows show the Fn1 localisation within the valve leaflets. Results obtained from three independent experiments. (E) Scheme of the three layers shown in A’, B, C and D (magenta, smooth muscles; green, endothelium; grey, myocardium; Fibronectin1, magenta lines) at 72hpf, 96hpf and 120hpf.
Figure 3—figure supplement 1. Smooth muscle identity is revealed by NO.
We conclude that the tissue remodeling occurring during OFT valve development is significantly different from AVC valve development where the endocardium is the main remodeling tissue (Beis et al., 2005; Pestel et al., 2016; Steed et al., 2016b).
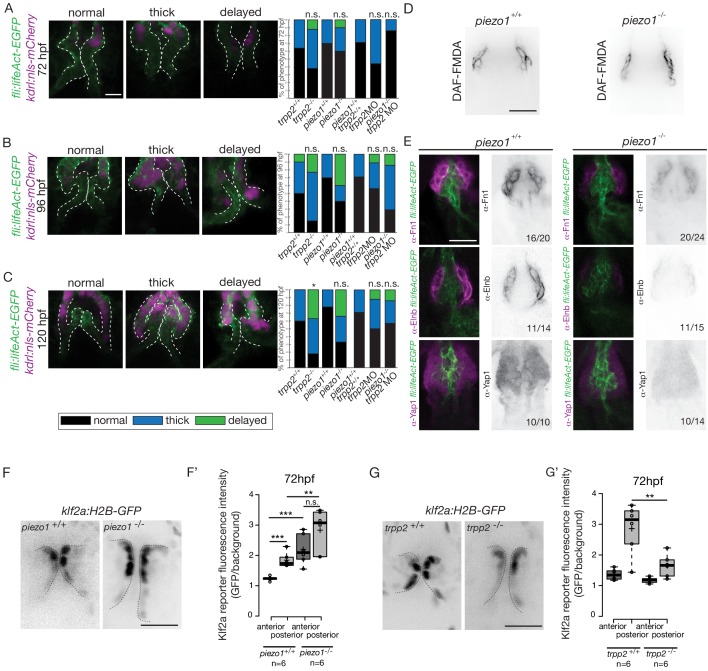
Klf2, Notch signaling, and Hippo pathways are active in the OFT in different cell layers and are all necessary for proper OFT valve development
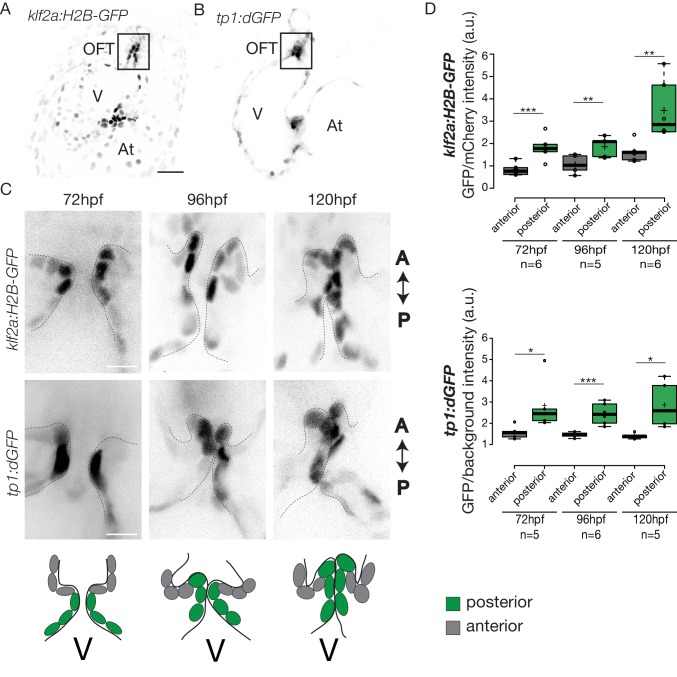
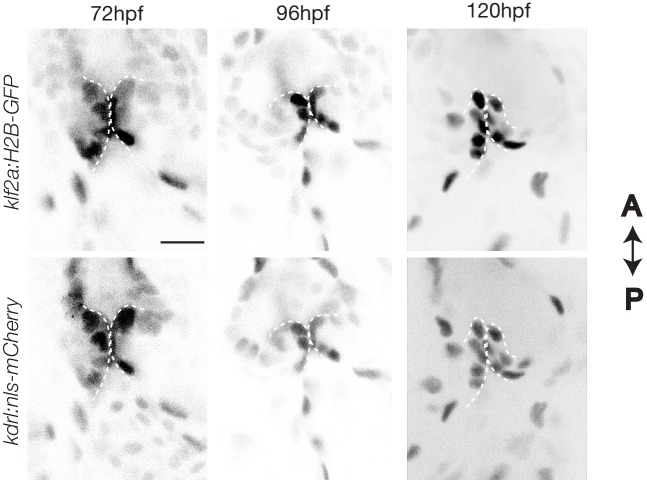
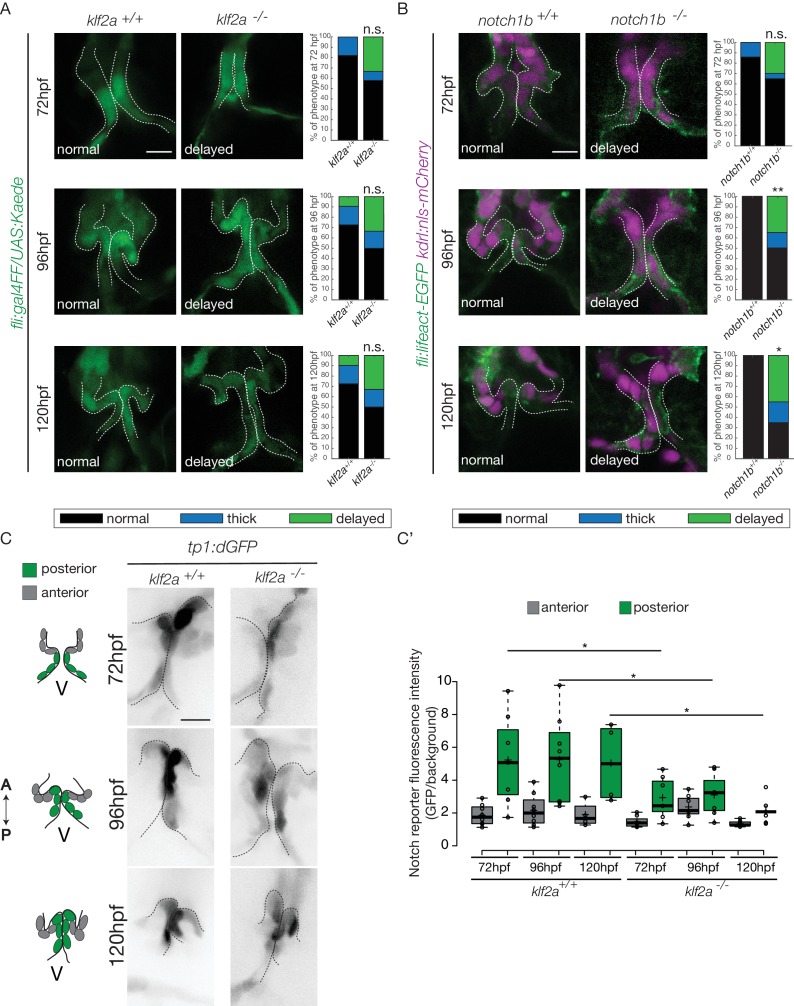
To elucidate how these early events are regulated, we sought to determine the mechanosensitive signaling pathways activated at these early stages of OFT valve formation. We first assessed the activity of a Klf2a reporter line (Tg(klf2a:H2B-GFP)) (Figure 4A), which is a well described flow responsive reporter (Heckel et al., 2015; Steed et al., 2016b) and the Notch reporter Tg(tp1:dGFP) (Figure 4B) which is well active in the progenitors of the AVC cardiac valves (Pestel et al., 2016). We could observe a specific activation of the Klf2a reporter in the OFT endothelium (Figure 4A,C and Figure 4—figure supplement 1), in particular in the ventricular part of the valve (posterior) from 72 hpf to 120 hpf (Figure 4C,D and Figure 4—figure supplement 1). Similarly, the Notch reporter Tg(tp1:dGFP) is specifically expressed in the OFT endothelium (Figure 4B,C), in the ventricular part of the valve from 72 hpf to 120 hpf (Figure 4C,D). Interestingly, the spatial activation of the reporter varies within the valve forming area - the transgene activation is stronger in the posterior part of the valve than the anterior part of the valve throughout the process of valve maturation (Figure 4C,D). We made similar observations for the Notch reporter (Figure 4C,D). These results suggest that Klf2a and Notch signaling are activated specifically in the part of the valve corresponding to where the OFT has the smallest diameter and where shear stress is expected to be the highest, consistent with the hypothesis that Klf2a and Notch signaling are flow-responsive.
Figure 4. Klf2a and Notch reporters are activated in the OFT endothelium Confocal z-section of the Tg(klf2a:H2B-GFP; fli:nls-mCherry).
(A) and Tg(tp1:dGFP) (B) at 72hpf. OFT: outflow tract, At: atrium, V: ventricle. Scale bar: 20 µm. (C) Confocal z-section of OFT valves expressing the Klf2a reporter and Notch reporter at 72 hpf, 96 hpf, and 120 hpf. Schemes explaining the considered anterior (grey) and posterior (green) parts of the valve at 72 hpf, 96 hpf, and 120 hpf. A: anterior, P: posterior. V: ventricle. Scale bar: 10 µm. (D) Quantification of the fluorescent intensity of the Klf2a (GFP over mCherry) and Notch (GFP over background) reporters in the anterior versus posterior part of the valves at 72 hpf (n = 6 embryos, p=0001 and n = 5 embryos, p=0,02 respectively), 96 hpf (n = 5 embryos, p=0005 and n = 6 embryos, p=0,0007 respectively) and 120 hpf (n = 6 embryos, p=0,01 and n = 5 embryos, p=0,01) in wild-type embryos using the student’s t-test. Boxplots: Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. Results obtained from three independent experiments.
Figure 4—figure supplement 1. Expression of the klf2a reporter Z-section of the Tg(klf2a:H2B-EGFP; kdrl:nls-mCherry) at 72 hpf, 96 hpf and 120 hpf used to quantify the reporter expression.
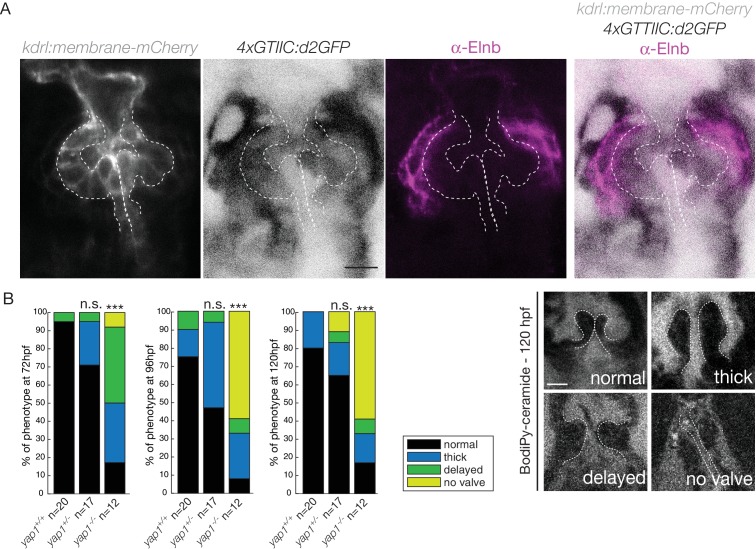
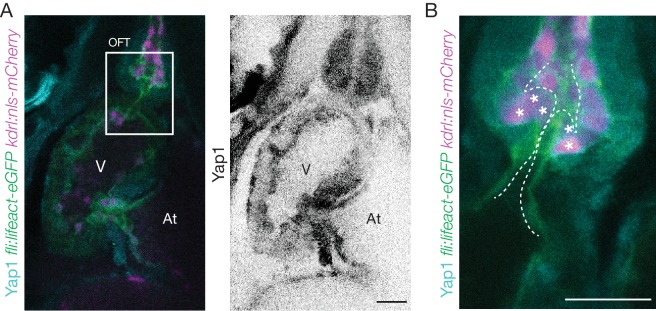
We next investigated the expression of the Hippo effector Yap1 and a reporter of the Hippo pathway in vivo. We found that Yap1 is localized in the heart at 72 hpf, in particular in the OFT smooth muscle cells (Figure 5—figure supplement 1A,B). To better characterize its tissue-specific expression, we used the double transgenic line Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) and could show that the smooth muscle cells surrounding the OFT, but also some OFT endothelial cells express Yap1 (asterisks in Figure 5—figure supplement 1B). To assess the activity of the Hippo pathway in the heart at 72 hpf, we made use of the Tg(4xGTIIC:d2GFP) and could see that the Yap/Wwtr1-Tead reporter was activated in the OFT endothelial cells (labelled using the kdrl:membrane-mCherry line) as well as the smooth muscle cells surrounding the OFT (labelled by the Elnb staining) (Figure 5A). To assess the implication of Yap1 during OFT valve development, we followed the same embryos over time and looked at the valve phenotype in yap1 mutant embryos, yap embryos and yap1 control embryos (Agarwala et al., 2015) (Figure 5B). When analyzed from 72 hpf until 120 hpf, a significant fraction of yap1-/- embryos displayed abnormal valves (17% at 72 hpf, 8% at 96 hpf, 17% at 120 hpf, n = 12) and an increasing fraction of yap1 embryos did not have recognizable OFT valves (58% at 120hpf, n = 12). Thus, these results suggest that yap1 is involved during OFT valve morphogenesis (Figure 5B) and that smooth muscle cell progenitors are likely to play a role in the process.
Figure 5. Hippo pathway effector Yap1 is active and Yap1 is essential for valve formation in the OFT.
(A) Confocal z-sections of the double transgenic line 4xGTIIC:d2GFP; kdrl:membrane-mCherry counterstained with the Elnb antibody and focused on the OFT. Scale bar: 20 µm. (B) Example of the valve phenotypes (normal, thick, delayed and no valve) and quantification of the phenotypes in the yap1+/+ controls embryos, yap1+/- and in yap1-/- mutant embryos. Chi-square test. n.s.: non significant, ***: p<10−3. Scale bar: 10 µm. Results obtained from two independent experiments.
Figure 5—figure supplement 1. Yap1 is expressed in the OFT Yap1 antibody staining on Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) (A) and zoom on the OFT (B) at 72 hpf. .
Together, these results show that Klf2a, Notch signaling, and Hippo pathways are active in the OFT during valve morphogenesis.
Klf2a and notch reporter activity is flow-dependent in the OFT
As blood flow is an important regulator of klf2a expression and cardiac valve formation, we next wanted to assess whether changes in flow properties impact Klf2a and the Notch reporter activity in the OFT.
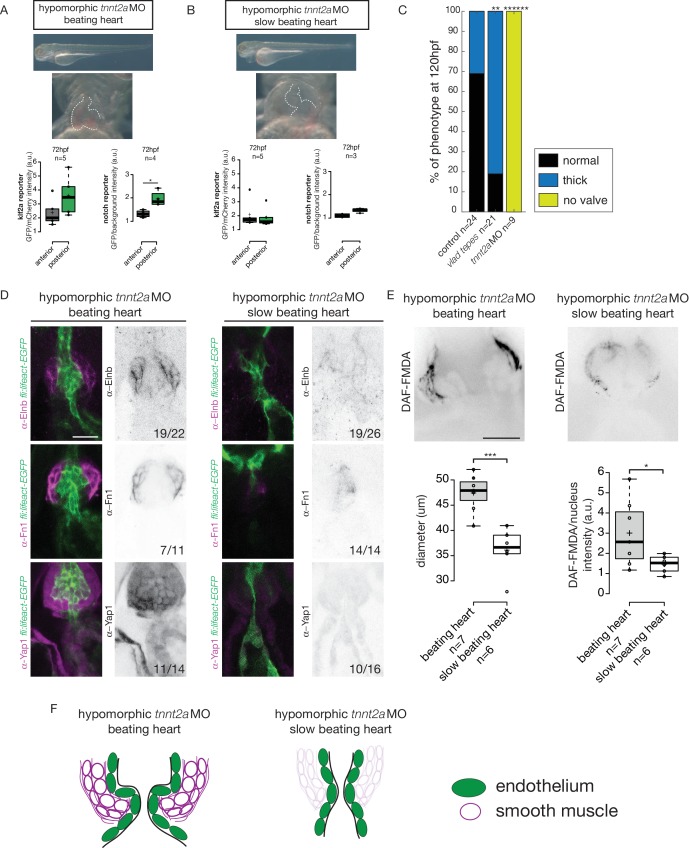
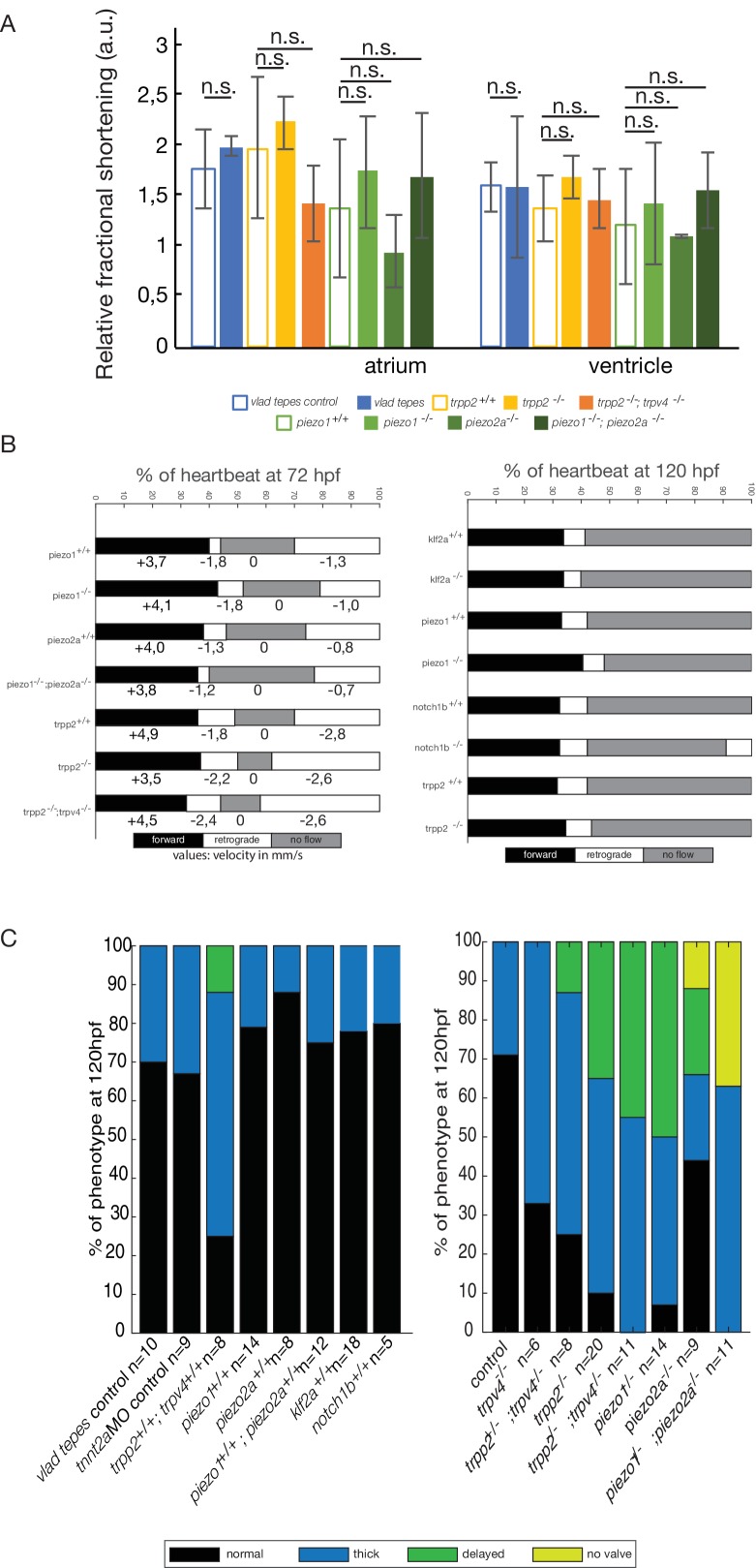
We analyzed the reporters activity following injection of a morpholino specific for troponin T2a (tnnt2a), which is necessary for heart contraction and reliably mimics the sih mutants (Sehnert et al., 2002), to determine whether these signaling pathways were impacted when heart contraction is abnormal and/or are activated upon shear stress forces. As the absence of heart contraction can impact heart morphogenesis, we injected highly diluted tnnt2a morpholino (hypomorphic condition) into these two reporter lines (Figure 6A,B). Such treatment allows us to decrease heart function and flow without dramatically altering heart shape (Figure 6A,B, Videos 1 and 2). Depending on the knockdown efficiency in single embryos, this treatment leads to the generation of two groups of embryos: group1 where the heart beats ‘normally' (normal heart rate at 2–3 Hz and function) and group2 where the heartbeat is slower. In the group of ‘beating heart’ embryos (group1, Video 1), we still observe stronger GFP expression in the posterior part of the valve for both reporters at every stage analyzed (Figure 6A). In the group of embryos where the heart is still beating but at an abnormal slow rate (less than 2 Hz, group2, Video 2), we observe no difference between the anterior and posterior part of the valve (p=0.99) for the Klf2a reporter at 72 hpf (Figure 6B). For the Notch reporter, we observed no difference in fluorescence intensity at 72 hpf (p=0.1) (Figure 6B). In addition, we analyzed the localization of Fn1, Elnb and Yap1 by immunostaining in both ‘beating heart’ (n = 4, n = 7 and n = 7, respectively) and ‘slow beating heart’ (n = 6, n = 7 and n = 7 respectively) groups and could observe that Fn1, Elnb, and Yap1 are down-regulated in the smooth muscle cells of the ‘slow beating heart’ embryos (Figure 6D,F). Moreover, we assessed the BA diameter and the activity of the smooth muscle using the DAF-FMDA assay (Figure 6E). The results suggest that the ‘slow beating heart’ group has a smaller BA (p<0,001) and the smooth muscle are much less active (p=0,05) (Figure 6E,F). We next assessed valve morphology in which blood flow is altered due to slow heart contraction and selected the fish with almost no contraction (group2). All the tnnt2aMO-injected embryos have no valve (n = 9/9, p<10−6) (Figure 6C). To confirm the role of flow in the process, we analyzed the effect of altered blood viscosity and shear stress by lowering red blood cell content in the gata1 mutants (Vlad Tepes) as previously described (Steed et al., 2016b; Vermot et al., 2009). In vlad tepes mutant embryos (n = 21, p<10−2), more than 80% of the mutants displayed abnormal OFT valves (Figure 6C).
Figure 6. Klf2a and notch response, as well as the smooth muscle cell identity, are flow-dependent.
Quantification of the Klf2a and Notch reporter expressions in tnnt2a-morpholino injected embryos showing a ‘beating heart’ (p=0,27 and p=0,01 respectively). (A) and a ‘slow beating heart’ (p=0,7 and p=0,1 respectively) (B) at 72 hpf. N = 2 independent experiments. (C) Quantification of the phenotypes in the control (n = 24), vlad tepes mutant (n = 21 embryos from two independent experiments), tnnt2aMO-injected embryos (n = 9 embryos from two independent experiments). Chi-square test. **: p<10−2, ******: p<10−6. (D) Z-sections of the Tg(fli:lifeact-eGFP) counterstained with either Fibronectin1 (Fn1), elastin (Elnb) or Yap1 in tnnt2a-morpholino injected embryos (slow beating and beating heart). Scale bar: 20 µm. N = 2 independent experiments. (E) Z-section and quantification of the BA diameter and DAF-FMDA intensity in tnnt2a-morpholino injected embryos (p=0,0005 and p=0,05 respectively). Student’s t-test. Boxplots: Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. (F) Scheme summarizing the down-regulation of the smooth muscle markers in ‘slow beating heart’ embryos compared to ‘beating heart”embryos.
Video 1. Bright field videomicroscopy of the typical heart function of a tnnt2a MO group1 embryo where the heart is still beating normally.
Video 2. Bright field videomicroscopy of the typical heart function of a tnnt2a MO group2 embryo where the heart is still beating but at an abnormal slow rate (less than 2 Hz).
Together, these results suggest that the expression of both Klf2a and Notch reporters is flow-dependent and that mechanical forces associated with heart activity are necessary for valve development and smooth muscle cell identity in the OFT.
Klf2a regulates OFT valve morphogenesis via notch signaling activation and is necessary for the smooth muscle cells differentiation
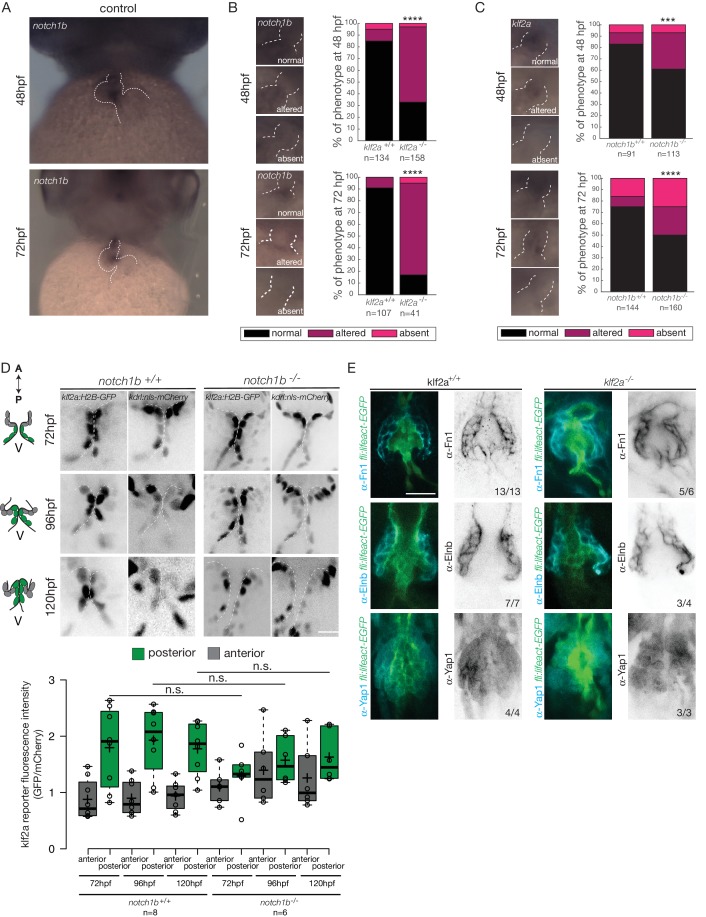
As notch1b and klf2a are expressed in the OFT endothelium in response to flow forces, we hypothesized that they have a role during OFT valve morphogenesis. Therefore, we imaged the Tg(fli:gal4/UAS:kaede) and Tg(flli:lifeact-EGFP; kdrl:nls-mCherry) transgenic lines (Figure 7A,B). We looked at the phenotype of OFT valve endothelium in the klf2a-/- and notch1b-/- embryos at 72 hpf, 96 hpf and 120 hpf (Figure 7A,B). We found that most of the klf2a-/- embryos display proper valves at 72 hpf (n = 7/12), 96 hpf (n = 6/12) and 120 hpf (n = 6/12). However, 33% of the klf2a-/- embryos have abnormal ‘delayed phenotype’ at 72 hpf (n = 4/12), at 96 hpf (n = 4/12) and 120 hpf (n = 4/12) (Figure 7A). In this case, the OFT valves are larger, leading to big cushions instead of thin valve leaflets (Figure 7A). In addition, notch1b is also necessary for proper valve formation since 30% (n = 6/20) of the notch1b-/- embryos have a ‘delayed phenotype’ at 72 hpf, 35% (n = 7/20) at 96 hpf and 45% (n = 9/20) at 120 hpf (Figure 7B) while almost all control embryos have proper valves at 72 hpf (n = 6/7), 96 hpf (n = 7/7) and 120 hpf (n = 7/7) (Figure 7B). Next, we wondered whether a cross-regulation between notch1b and klf2a exists since they are both necessary for proper valve formation. First, we compared notch1b expression between klf2a-/- embryos and klf2a+/+ control embryos (Figure 7—figure supplement 1A). In controls, 85% and 91% of the embryos have notch1b expression in the OFT at 48 hpf and 72 hpf, respectively (Figure 7—figure supplement 1A–C). However, in klf2a-/- embryos, notch1b expression is altered with no clear expression defining the OFT region at 48 hpf (64%) and at 72 hpf (78%) (Figure 7—figure supplement 1A,B). However, in the reverse experiment, most of the embryos show proper klf2a expression in notch1b -/- embryos (61% at 48 hpf and 51% at 72 hpf) (Figure 7—figure supplement 1C).
Figure 7. Klf2a and notch are necessary for valve formation.
Quantification of the valve phenotypes at 72 hpf, 96 hpf and 120 hpf (normal, thick, delayed) in klf2a+/+(n = 11), and klf2a-/- (n = 12) using the Tg(fli:gal4FF/UAS:Kaede). (A) and notch1b+/+ (n = 7) and notch1b-/- (n = 20) using Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) embryos. (B) Scale bar: 10 µm. N = 3 independent experiments. (C) Confocal z-sections of the Tg(tp1:dGFP) in klf2a+/+ and klf2a-/- embryos at 72 hpf, 96 hpf and 120 hpf. V: ventricle. Scale bar: 10 µm. (C’) Quantification of the fluorescent intensity of the Notch reporter (GFP over background) in the anterior versus posterior parts of the valves in in klf2a+/+ (n = 5) and klf2a-/- (n = 4) embryos. Statistical test were performed to compare the posterior intensities in klf2a+/+versus klf2a-/- at 72 hpf (p=0,05), 96 hpf (p=0,03) and 120 hpf (p=0,04). Student’s t-test. Boxplot: Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots. Results obtained from three independent experiments.
Figure 7—figure supplement 1. Klf2a regulates notch1b expression but Notch1b does not regulate klf2a expression.
In order to assess the regulation of the Notch pathway activity by Klf2a in the endothelium, we analyzed the Notch reporter activity in the posterior and anterior parts of the valve in klf2a+/+ (n = 5) versus klf2a-/- (n = 4) embryos. As for Notch reporter expression in wild-type (Figure 4C,D), the Notch reporter is significantly more expressed in the posterior part compared to the anterior part of the valves at 72 hpf (p<10−2), 96 hpf (p<10−2) and 120 hpf (p<10−1) (Figure 7C,C’) in the klf2a+/+ embryos. Interestingly, the stronger posterior expression is lost in the klf2a-/- embryos and the fluorescent intensity in the posterior part of the valve is significantly reduced in the klf2a-/- compared to klf2a+/+ at 72 hpf (p<10–1), 96 hpf (p<10−1) and 120 hpf (p<10−1) (Figure 7C,C’). In comparison, the expression in the posterior part of the valve in the notch1b-/- embryos compared to the notch1b+/+ control embryos is not significantly different at any time point analyzed (Figure 7—figure supplement 1D). To further assess whether Klf2 has an effect on the OFT formation, we performed Fn1, Elnb and Yap1 stainings on klf2a-/- and controls at 72 hpf. We could observe that Fn1, Elnb, and Yap1 are properly localized in controls (n = 13/13, n = 7/7 and n = 4/4, respectively), klf2a-/- (n = 5/6, n = 3/4, and n = 3/3) (Figure 7—figure supplement 1E).
These results suggest that klf2a and notch1b are involved for proper valve morphogenesis and that Klf2a modulates notch expression in the process. Moreover, klf2a does not seem necessary for the smooth muscle cell differentiation surrounding the endothelium.
Piezo channels regulate both the endothelial and smooth muscle cell markers expression
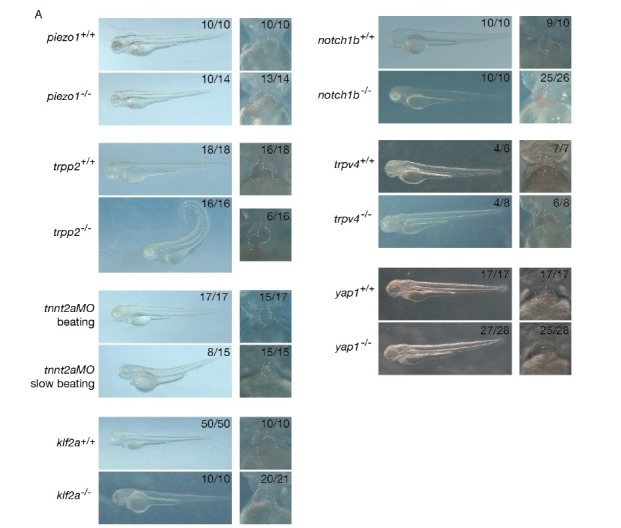
In order to decipher whether flow and mechanosensitive channels could be involved in the regulation the OFT valve formation, we proceeded to analyze the phenotype of different potent mechanosensitive channel mutants. We focused on the non-selective ion channels Trpp2, Trpv4, Piezo1 and Piezo2a (Figure 8—figure supplement 1A,B,C) that are known for their mechanosensitive properties (Coste et al., 2010; Köttgen et al., 2008; Li et al., 2014; Sharif-Naeini et al., 2009; Thodeti et al., 2009). First, we evaluated the relative fractional shortening (RFS) at 72hpf in the atrium and in the ventricle of each mutant and their corresponding controls (Figure 8—figure supplement 1A). We did not observe any significant difference in the RFS between mutants and their respective controls neither in the atrium nor in the ventricle, suggesting that heart function in these mutants is normal. As another readout of flow forces and heart function, we quantified the retrograde flow fraction (RFF) at 72hpf and 120hpf in these mutants and the time windows for forward, reverse or no flow. We found that they are equivalent in all mutants when compared to their respective controls (Figure 8—figure supplement 1B), confirming that heart function is not different in these mutants. We looked at valve morphology at 120 hpf when leaflets are normally fully formed. In control embryos, the valves are extending into the lumen (Figure 8—figure supplement 1C). We found that 33% of the trpv4 mutant embryos (n = 7) have normal valves and mainly display thick valves (67%). In the trpp2 mutant embryos (n = 20), a stronger phenotype is observed with only 10% of normal valves. We observed that some embryos are delayed with respect to valve phenotype, meaning that the valve forming area still displays cushions at 120 hpf instead of having leaflets (35%). The trpv4-/-; trpp2+/- (n = 8) has an intermediate phenotype with 25% of the embryos having normal valves. Finally, the trpp2-/-; trpv4-/- embryos (n = 11) have a stronger phenotype, with none of the embryos showing proper OFT valve development and displaying mainly a delayed valve phenotype (45%). Interestingly, the piezo1 mutant embryos (n = 14) show mainly a delayed valve formation (50%), similarly to the trpv4-/-; trpp2-/- embryos. The piezo2a-/- (n = 9) has a less stringent phenotype with mainly normal valves (44%) but nevertheless 11% of piezo2a-/- fish do not have valves at all. This phenotype is even more prevalent in piezo1-/-; piezo2a-/- embryos (n = 11), where none of the fish display proper valve development and most of them do not form any valves (36%).
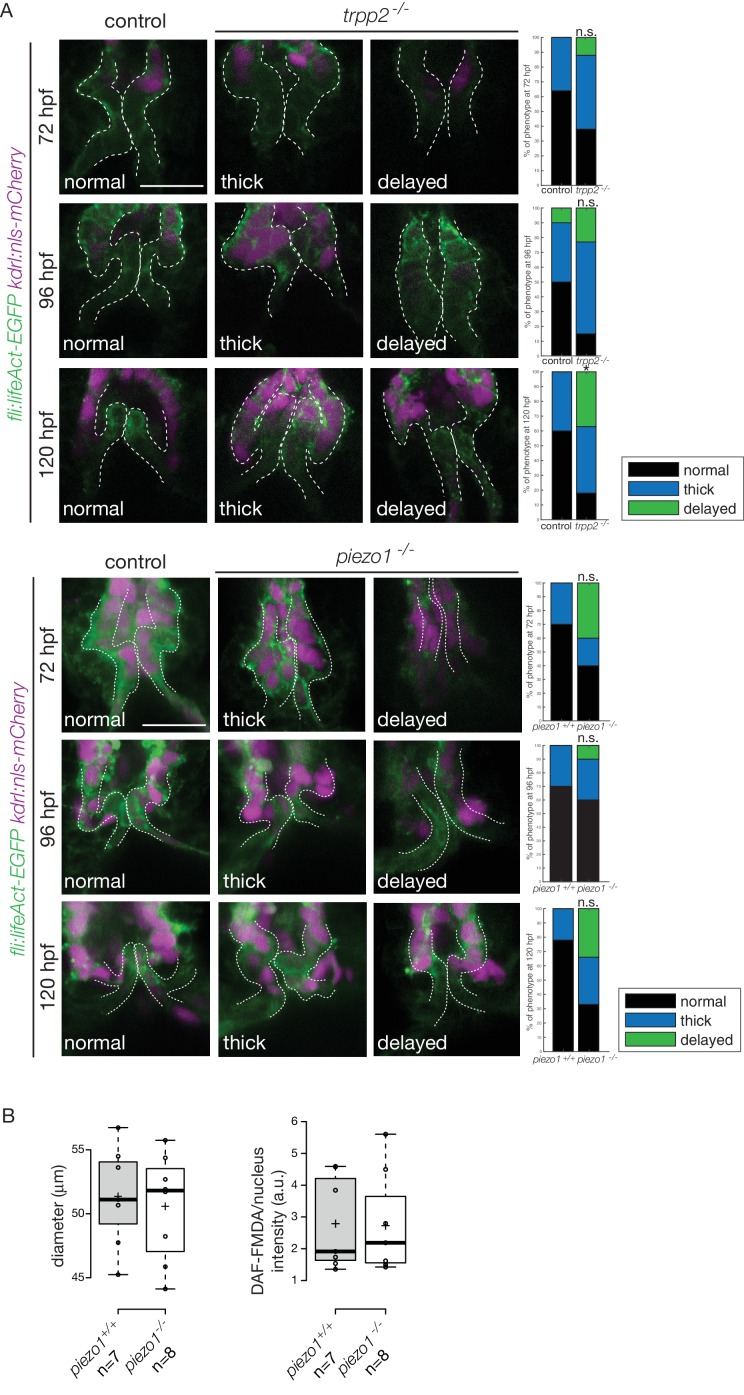
To better characterize the valve phenotype in trpp2-/- and piezo1-/- embryos, we made use of the Tg(fli:lifeact-EGFP; kdrl:nls-mCherry) transgenic line and assessed the shape of the endothelium at 72 hpf (Figure 8A and Figure 8—figure supplement 2A), 96 hpf (Figure 8B and Figure 8—figure supplement 2A) and 120 hpf (Figure 8C and Figure 8—figure supplement 2A). trpp2-/- embryos display mainly thick valves at all time points (n = 4/8 at 72 hpf, n = 8/13 at 96 hpf and n = 5/11 at 120 hpf) (Figure 8A–C and Figure 8—figure supplement 2A). Indeed, the endothelial layer is not a single cell layer anymore (Figure 8B) which leads to a thicker leaflet at 120 hpf (Figure 8C). Although piezo1-/- embryos have mainly normal valves at 72 hpf (Figure 8A and Figure 8—figure supplement 2A) (n = 6/10), they display a delayed phenotype (still cushions) at 96 hpf (Figure 8B and Figure 8—figure supplement 2) (n = 4/10) and thick or delayed phenotype at 120 hpf (Figure 8C and Figure 8—figure supplement 2A) (n = 3/10 and n = 3/10 respectively). To assess the redundancy between trpp2 and piezo1, we performed the same experiment in piezo1-/-; trpp2-morpholino injected embryos at 72 hpf (Figure 8A), 96hpf (Figure 8B) and 120 hpf (Figure 8C). Although the trpp2-MOrpholino injected embryos do not show a phenotype as strong as the trpp2-/- mutant embryos at 96 hpf and 120 hpf (possibly due to less effective morpholinos at these stages), the double piezo1-/-; trpp2-MO do not clearly show a stronger phenotype than the single trpp2-/- or piezo1-/- embryos (normal valves in n = 6/7, n = 2/7 n = 4/7 and at 72 hpf, 96 hpf and 120 hpf respectively). These results suggest that Trp and Piezo channels are necessary for the proper folding of the endothelium from 72 hpf to 120 hpf.
Figure 8. Flow and mechanosensitive channels are necessary for proper OFT valve formation.
(A) Z-sections and quantifications of the valves phenotypes (normal, thick, delayed) of the Tg(fli:lifeaAct-EGFP; kdrl:nls-mCherry) at 72 hpf (B), 96 hpf (C) and 120 hpf (D) in trpp2+/+(n = 11, n = 10, n = 10), trpp2-/- (n = 8, n = 13, n = 13 from three independent experiments), piezo1+/+ (n = 10, n = 10, n = 9), piezo1-/-(n = 10, n = 10, n = 9 from two independent experiments), piezo1+/+; trpp2+/+ (n = 7), trpp2-morpholino injected embryos (n = 9) and piezo1-/-; trpp2-morpholino injected embryos (n = 7). Scale bar: 10 µm. (D) Z-section of the OFT stained with DAF-FMDA in piezo1+/+ and piezo1-/-. Scale bar: 20 µm. (E) Fibronectin1 (Fn1), elastin (Elnb) and Yap1 staining on Tg(fli:lifeact-eGFP) in piezo1+/+ (n = 12, n = 4 and n = 10 respectively) and piezo1-/- (n = 12, n = 4 and n = 10 respectively from three independent experiments). Scale bar: 20 µm. Z-sections (F) and quantification (F’) of the klf2a reporter (GFP over background) in the anterior and posterior parts of the valves in piezo1+/+ (n = 6) and piezo1-/- (n = 6) obtained from two independent experiments). Scale bar: 20 µm. Z-sections (G) and quantification (G’) of the klf2a reporter (GFP over background) in the anterior and posterior parts of the valves in trpp2+/+ (n = 6) and trpp2-/- (n = 6) (obtained from two independent experiments). Scale bar: 20 µm. Student’s t-test. Boxplot: Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots.
Figure 8—figure supplement 1. Embryo phenotypes in controls and mutants.
Figure 8—figure supplement 2. Valve phenotypes in trpp2-/- and in piezo1-/-.
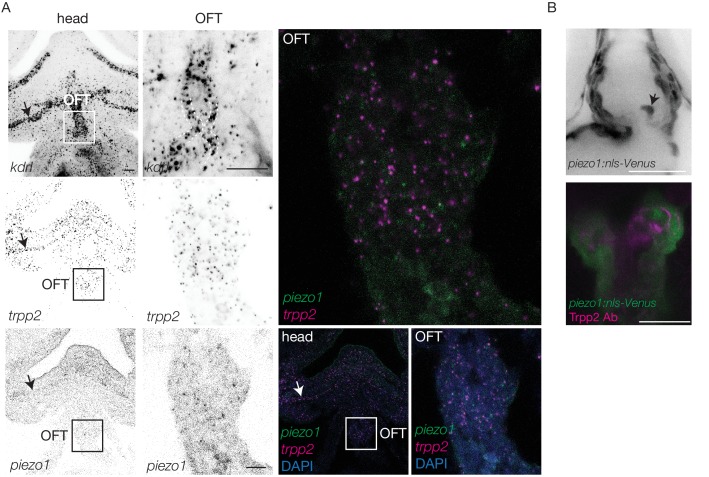
Figure 8—figure supplement 3. Trpp2 and Piezo1 expression in the OFT.
To better characterize the cell layer affected by piezo1 loss of function, we performed immunohistochemistry against Yap1 and two smooth muscle identity markers (Fn1 and Elnb) as well as their active functionality in piezo1+/+ controls and piezo1-/- mutants at 72 hpf (Figure 8D and Figure 8—figure supplement 2B). A bit more than two thirds of piezo1-/- embryos (n = 7/10) display reduced Yap1 expression as well as a down-regulation of Elnb (n = 9/12) and Fn1 (n = 3/4) (Figure 8E). However, the BA diameter as well as the DAF-FMDA fluorescence intensity are not affected in the piezo1-/- compared to piezo1+/+ (Figure 8D and Figure 8—figure supplement 2B) demonstrating that smooth muscle cells are still present in the piezo mutants. These results suggest a selective role of piezo1 in the regulation of the smooth muscle cell maturation and proper Yap1 localization in the OFT. We next assessed Klf2a reporter expression in piezo1-/- and trpp2-/-. We found that Klf2a expression was misregulated in piezo1-/- mutant embryos with stronger Klf2a reporter activation in the anterior and posterior part of the valve endothelium when compared to controls (p<10−3 for the anterior part and p<10−2 for the posterior part of the valves) (Figure 8F,F'). By contrast, we found that the posterior part of the valve has a decreased expression of GFP (p<10−2), highlighting a down-regulation of klf2a in the trpp2-/- embryos (Figure 8G,G’). These results suggest that Piezo1 inhibits klf2a overall the valve endothelium (Figure 8F,F’) while Trpp2 is required for klf2a activation in the posterior part of the OFT valve endothelium (Figure 8G,G’). To assess if the localization of piezo1 and trpp2 mRNA could explain the differential function of these channels, we performed RNAscope assay at 72hpf (Figure 8—figure supplement 3A). We found that trpp2 is ubiquitously expressed in the embryo, including in the different layers composing the OFT. Similarly, we found that piezo1 is expressed in both endothelium and smooth muscles, albeit at a lower level than trpp2 (Figure 8—figure supplement 3A). To confirm these results, we generated a transgenic reporter line with 3 kb of the piezo1 promoter upstream of the start codon (piezo1:nls-Venus). We observed the expression of the reporter line mostly in smooth muscles at 72hpf (Figure 8—figure supplement 3B) and cells of the endothelium of the OFT valve. Trpp2 immunohistochemistry showed that Trpp2 is expressed in the endothelium and the smooth muscles confirming that trpp2 is ubiquitously expressed in the OFT (arrow in Figure 8—figure supplement 3B). We conclude that Piezo1 plays a dual role in the OFT: it modulates klf2a expression in the endothelium and it is necessary for proper smooth muscle cell maturation around the OFT endothelium.
Discussion
Using cardiac live imaging and functional studies combined with in vivo reporter analysis, we uncover key mechanosensitive signaling pathways involved in OFT valve morphogenesis (Figure 9). We identify two tissue layers sensitive to mechanical forces in the OFT: (1) The endothelial cells where klf2a expression is modulated both by Piezo and Trp channels (2) The smooth muscles surrounding the endothelium, where Piezo channels regulate Yap1 localization and smooth muscle cell specific marker expression (Figure 9). These observations enable us to confirm the universal role of mechanical forces in cardiac valve morphogenesis and suggest a specific mechanism for OFT valve morphogenesis in which the origins of the valve progenitors, the implication of particular groups of cells, the mechanosensors involved and the impact of the mechanotransduction cascade are identified.
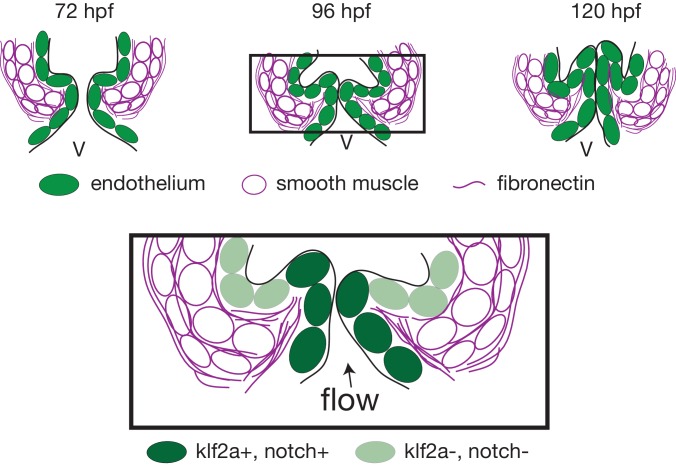
Figure 9. Working model summarizing OFT valve morphogenesis in response to mechanical forces.
Two cell layers forming the OFT respond to piezo1 activity: the endothelium (green) and smooth muscle cells (magenta). Endothelial klf2a expression is repressed by Piezo1. In the smooth muscle cells, the expression of elastin (Elnb), fibronectin (Fn1) and Yap1 is modulated by Piezo1. Fibronectin is localized in the smooth muscle cell layer and within the forming valve (magenta lines). Future work will help to decipher how the two cell layers interact with each other to modulate OFT valve formation. V, ventricle.
Klf2a modulates notch signaling specifically in the OFT endothelium
Valve morphogenesis occurs in complex mechanical environments. In the AVC, endocardial cells experience both shearing forces and mechanical deformation due to the contraction of the heart and its associated blood flow. The situation is slightly different in the OFT because endothelial valvular progenitors are not surrounded by contractile cardiomyocytes but passive smooth muscle cells that can provide a counter force to flow and pressure. Here, we show that the main mechanosensitive pathways involved in AVC valve development are required for OFT valve development, even though the mechanical stimuli vary greatly between these two sets of valves. Previous studies have proposed that klf2a and notch1b are important during the formation of functional cardiac valve leaflets in zebrafish. Both are specifically expressed in the AVC (Beis et al., 2005; Pestel et al., 2016; Steed et al., 2016b; Vermot et al., 2009) and are transcriptionally misregulated in models of Cerebral cavernous malformation (CCM) where cardiac valve development is altered (Donat et al., 2018). Here, we show that both Notch signaling and klf2a are active in the endothelial cells of the OFT between 48 hpf and 120 hpf. During the process in which zebrafish cardiac AVC cushion remodel into valve leaflets, endocardial Klf2a expression and Notch activity are high on the luminal side of the developing valve leaflet, which is exposed to blood-flow, whereas their expression is lower on the abluminal side of the leaflet (Pestel et al., 2016; Steed et al., 2016b). In the OFT, Klf2a expression and Notch activation follow a different pattern because we could not clearly identify abluminal cells in the OFT. This might reflect a complete lack of endothelial to mesenchymal transition in the OFT by comparison to the AVC. Nevertheless, the impact of both pathways on valve morphogenesis remains the same as we found that klf2a-/- and notch1b-/- embryos show similar valve phenotypes and Notch signaling pathway is significantly decreased in klf2a-/- mutants. Even though the downstream players of the notch pathway remain to be established in the OFT, these results validate previous observations, suggesting that klf2a acts upstream of Notch signaling in the endocardium (Donat et al., 2018; Samsa et al., 2015; Vermot et al., 2009). Considering that Notch has been proposed to be mechanosensitive in blood vessels (Mack et al., 2017), an attractive hypothesis is that the Klf2-Notch axis could be a general mechanosensitive cascade in endothelial cells. In that case, it would be interesting to address notch activity in a flow related context, similarly to what is currently being done with Nf-kb, Klf2a, and other flow responsive pathways (Feaver et al., 2013). This assumption is particularly interesting in the context of sprouting angiogenesis and other aspects of angiogenesis where Notch is broadly required (Choi et al., 2017; Hasan et al., 2017; Pitulescu et al., 2017; Tammela et al., 2011). We further show that the expression of Fibronectin1 and Elastinb in the smooth muscle cells surrounding the OFT is not altered in the klf2a-/-embryos. Considering that Piezo is emerging as an important contributor to human diseases related to blood cells (Ma et al., 2018; Zarychanski et al., 2012) and lymphatic diseases in humans (Lukacs et al., 2015), our work should motivate the search of potential involvement of Piezo in cardiac pathologies such as valvulopathies.
Piezo is necessary for proper OFT smooth muscle cell identity and endothelial klf2a expression in the OFT
Cardiac valve development is highly dependent on endothelial/endocardial cell mechanosentivity. Previous work identified the membrane-bound mechanosensitive channels (Trpp2 and Trpv4) and a calcium-activated intracellular signaling cascade leading to Klf2a expression and valve morphogenesis as key elements of the endocardial mechanodetection-signaling pathway (Heckel et al., 2015 ) Consistently, we found that these Trp channels are also required for endothelial Klf2a expression and OFT valve formation. In addition, we identify another type of mechanosensitive channel belonging to the Piezo family. Piezo channel mutants show valve dysgenesis phenotype, suggesting their requirement during OFT valve morphogenesis. Interestingly, Piezo channels are important both for modulating Klf2a expression in the endothelium and for smooth muscle cell-specific expression of Elnb and Fn1. Piezo function in these two cell layers is consistent with the fact that Piezo1 is expressed in smooth muscle cells and is important for tissue remodeling in mouse arteries (Retailleau et al., 2015) as well as in endothelial cells in mouse vasculature (Li et al., 2014; Ranade et al., 2014) and lymphatic valves (Nonomura et al., 2018). These studies along with our results suggest that Piezo1 might be necessary for both layers to regulate distinguishable functions: activation of signaling cascade upon shear stress in the OFT endothelium and proper cell identity acquisition in smooth muscles. Both tissue layers might be sensing different stimuli: shear plus strain for the endothelial cells and strain for the smooth muscle cells. This hypothesis is consistent with the fact that Piezo is sensitive to stretch (Ranade et al., 2015), compression (Lee et al., 2014; Qi et al., 2015), and rhythmic mechanical stimuli (Lewis et al., 2017). Importantly, Piezo-dependent mechanisms can transduce forces at the cell-cell or cell-matrix interface (Eisenhoffer et al., 2012; Poole et al., 2014). Thus, Piezo can have different mechanosensitive roles both in smooth muscle cells and endothelium to coordinate the expression of Klf2 and ECM proteins. An interesting hypothesis is that the smooth muscle cell layer participates in the shaping up of the OFT valve and that both tissue layers establish paracrine interactions to fine tune the morphogenetic process. Further work will be needed to identify if this is the case and how it affects OFT valve morphogenesis at the cellular scale.
Yap1 and hippo pathways are regulators of the OFT valve formation in the endothelium and smooth muscles
Our work shows that Yap1 is specifically localized in the OFT in both endothelial cells and smooth muscle cells and is required for valvulogenesis. Accordingly, the Hippo pathway is active in smooth muscles and in the endothelium. Our work suggests that Piezo constitutes a plausible regulator of Yap1 localization as Piezo1 seems important for its localization in smooth muscle cells. Interestingly, Yap1 has been shown to translocate less in the nucleus in Piezo1 mutant mice neural stem cells (Pathak et al., 2014) and to induce proliferation in smooth muscles during cardiovascular development in mouse (Wang et al., 2014). Even though we were not able to assess Yap1 subcellular localization in vivo, the regulation of Yap1 activity and ECM assembly might be a general feature of Piezo function. The connection between Piezo and Yap1 is particularly interesting in the context of OFT development and function. In teleost, the OFT has an important biomechanical role for the control of flow propagation within the vascular networks by contributing to the dampening of the pressure wave down the arterial tree (Braun et al., 2003b). In zebrafish, Yap1 is involved in the determination of cardiac precursor cells into smooth muscle cell fate via a process that involves the regulation of Elastinb expression (Moriyama et al., 2016). Besides the control of cell identity, ECM contributes to biomechanical properties of tissues (Dzamba and DeSimone, 2018). It is thus possible that Piezo acts as a regulator of tissue mechanical properties by regulating Yap1 expression both in the OFT and in the vascular system where Yap1 expression is flow inducible (Nakajima et al., 2017).
In summary, this study reveals a novel function for mechanosensitive Piezo and Trp channels in modulating OFT valve development as well OFT smooth muscle cell maturation. It will be important to further investigate endothelial-smooth muscle cells interactions during OFT cardiac wall maturation.
Materials and methods
Zebrafish strains, husbandry, embryo treatments, and morpholinos
Animal experiments were approved by the Animal Experimentation Committee of the Institutional Review Board of the IGBMC (reference numbers MIN APAFIS#4669–2016032411093030 v4 and MIN 4669–2016032411093030 v4-detail of entry 1). Zebrafish lines used in this study were Tg(fli1a:lifeact-EGFP) (Phng et al., 2013), Tg(kdrl:nls-mCherry) (Nicenboim et al., 2015), Tg(kdrl:EGFP) (Jin et al., 2005), Tg(fli1a:nls-mCherry) (Heckel et al., 2015), Tg(myl7:egfp) (Huang et al., 2003), Tg(−26.5Hsa.WT1-gata2:eGFP)cn12 (Sánchez-Iranzo et al., 2018), Tg(klf2a(6 kb):H2B-eGFP) (Heckel et al., 2015), Tg(tp1:dGFP) (Ninov et al., 2012), Tg(4xGTIIC:d2GFP) (Miesfeld and Link, 2014), vlad tepesm651 (Lyons et al., 2002), cuptc321 (Schottenfeld et al., 2007), piezo1sa12608 (EZRC), piezo2asa12414 (ZIRC), trpv4sa1671 (ZIRC), notch1bsa11236 (ZIRC), klf2aig4 (Steed et al., 2016b), yap1fu48 (Agarwala et al., 2015) and wild-type AB. Cup mutant embryos were phenotyped based on the curled tail phenotype. The Tg(piezo1:nls-Venus) was generated by injection of the piezo1:nls-Venus plasmid and the mRNA of the Tol2 transposase. The plasmid was generated by cloning of 3 kb of the zebrafish piezo1 promoter upstream of the ATG start site and 3xnls-Venus into a pTol2-GAGGS vector. All animals were incubated at 28.5°C for 5 hr before treatment with 1-phenyl- 2-thiourea (PTU) (Sigma Aldrich) to prevent pigment formation. Morpholino specific for tnnt2a (Sehnert et al., 2002) (5’-CATGTTTGCTCTGATCTGACACGCA-3’) were obtained from GeneTools. It was injected into the yolk at the one-cell stage at a concentration of 5,8 ng to stop the heart. It was diluted 40 times in order to get fish with either a decreased heartbeat (‘slow beating heart’ group in this study) or close to the non-injected fish heartbeat (‘beating heart’ group in this study).
Immunofluorescence
Embryos were fixed at the desired stage in 4% paraformaldehyde overnight at 4°C. After washing in 1X PBST (PBS-0.1% Tween-20), embryos were permeabilized in 1X PBST containing 1% Triton-X 100 for 30 min at room temperature or overnight at 4°C. For Fibronectin1 staining, the pericardial cavity was then carefully pierced with the tip of a needle to facilitate antibody penetration before blocking in permeabilization buffer supplemented with 5% BSA. Embryos were incubated in blocking solution containing 5% BSA and 15% NGS (α-Fn1), 1% BSA, 2% NGS (anti-Elnb) and 2% BSA, 2% MgCl2 (1M), 5% NGS supplemented by 1,5% Tween-20 (anti-Yap1) for 2 hr at room temperature or overnight at 4°C. Primary antibodies were added to the relevant blocking solution and incubated 2 overnights at 4°C. Secondary antibodies were added in blocking solution after thorough washing in PBST and incubated for 2 days at 4°C. Embryos were thoroughly washed in PBST and mounted for imaging on a Leica SP8 confocal. Antibodies were used as follows: rabbit α-Fn1 (F3648, Sigma) 1:100, rabbit anti-Elnb (Miao et al., 2007; kind gift from Burns lab; Paffett-Lugassy et al., 2017) 1:1000, rabbit anti-Yap1 (generated by the Lecaudey lab; Agarwala et al., 2015) 1:300, rabbit anti-Trpp2 (kind gift from Drummond lab) 1:100 and Alexa Fluor 647 goat anti-rabbit IgG secondary antibody (A21245, Life Technologies) were used at 1:500.
In situ hybridization
In situ hybridization was performed as in Thisse and Thisse (2008). Anti-sense probes for notch1b and klf2a were generated from a plasmid containing the cDNA of zebrafish notch1b in pCR-script SK+ (provided by the Bakkers lab, The Netherlands) and zebrafish klf2a in IRBOp991B0734D (provided by RPDZ, Berlin; Vermot et al., 2009) and subsequently transcribed using the T3 polymerase and T7 polymerase, respectively. After ISH, embryos were incubated subsequently in 45% and 90% D-fructose (Sigma F0127) containing 0.5% of 1-Thioglycerol (Sigma M6145) for 20 min. Imaging of ISH was done using a Leica M165 macroscope with a TrueChrome Metrics (Tucsen) with a Leica 1.0X objective (10450028).
RNAscope
72 hpf wildtype zebrafish embryos were fixed in 4% PFA overnight at 4°C. The fixed embryos were dehydrated to 100% ethanol gradually. Embryos were stained using the RNAscope Fluorescent Multiplex kit (Advanced Cell Diagnostics).
Confocal imaging
For live imaging, zebrafish embryos were staged, anaesthetized with 0.02% tricaine solution or 50 mM BDM, to stop the heart when necessary, and mounted in 0.7% low melting-point agarose (Sigma Aldrich). Confocal imaging was performed either on a Leica SP8 confocal microscope (experiments with BODIPY-ceramide or fixed samples) or a Leica spinning disk (valve structure, flow profile, reporter experiments). Fast confocal imaging to image valve leaflets from 72 hpf to 120 hpf stained with BODIPY-ceramide was performed using the resonant scanner mode of the Sp8 microscope. Images were acquired with a low-magnification water immersion objective (Leica HCX IRAPO L, 25X, N.A. 0.95). The optical plane was moved 2 µm between the z-sections until the whole OFT was acquired. 2-colored fast confocal imaging was used to image valve structure, red blood cells, and reporter activities from 56 hpf to 144 hpf was performed using a Leica DMi8 combined with a CSU-X1 (Yokogawa) spinning at 10 000 rpm, two simultaneous cameras (TuCam Flash4.0, Hamamatsu) and a water immersion objective (Leica 20X, N.A. 0.75 or Leica 40X, N.A. 1.1). 1 ms exposure was used for red blood cells imaging and 20 ms exposure for valve structure and reporter activity experiments. 50% of 488 laser power and 40% of 561 laser power were used for reporter activity experiments.
BODIPY-ceramide imaging
Embryos were incubated with 4 mM BODIPY-ceramide (Molecular Probes) overnight and then processed as in Heckel et al. (2015) and Vermot et al. (2009) to visualize the valve shape.
DAF-FMDA labelling
To reveal the presence of NO, embryos were incubated in zebrafish medium containing 5 µM DAF-FM DA (Life Technologies, D23842) for 30 min in the dark at 28°C. Fluorescence intensities of the smooth muscles were measured using ImageJ software.
Photoconversion
Photoconversion was performed using the FRAP module on a SP8 confocal microscope and a Leica HCX IRAPO L, 25X, NA0.95 water immersion objective. Tg(fli1a:Gal4FF; UAS:Kaede) embryos were mounted in 0.7% low melting-point agarose supplemented with 50 mM BDM to inhibit heart contraction for the duration of the procedure. A region of interest corresponding to the anterior, middle or posterior part of the valve was selected and exposed to 405 nm light (25% laser power). One pre-bleach frame was acquired, followed by 3–6 bleach pulses (3–5 ms each) without acquisition to achieve conversion of the kaede protein to its red form. A z-stack of the photoconverted heart was then acquired in the standard confocal mode to record the starting point of each experiment. Embryos were then carefully dissected from the agarose, placed in fish water for 5–10 min until heart contraction resumed and then put at 28.5°C to develop individually under standard conditions until the next time point of interest.
Fractional shortening
Imaging was performed on a Leica DMIRBE inverted microscope using a Photron SA3 high-speed CMOS camera (Photron, San Diego, CA) and water immersion objective (Leica 20X, NA 0.7). Image sequences were acquired at a frame rate of 1000 frames per second. FS% = (Dm diastole- Dm systole)/ (Dm diastole) and where Dm is the diameter of the chamber of interest (atrium or ventricle).
Flow analysis
Red blood cells were manually tracked through the OFT and their velocity calculated from image sequences of the Tg(gata1:ds-red; kdrl:EGFP) beating heart, acquired at 1000 frames per second as described previously. Red blood cells transiting through the OFT were tracked manually on Imaris and their velocity calculated. The tracks of multiple cells in at least four embryos per stage were assembled to obtain an estimate of the flow velocity over multiple cardiac cycles (typically 3). Velocities estimated at the same time point by tracking different cells were averaged.
Image analysis
For fluorescence intensity analysis of the Klf2a reporter, the Tg(klf2a:H2B-EGFP) reporter line was crossed with the Tg(fli1a:nls-mCherry) line and the mCherry signal was used for normalization. The maximum intensity of each channel on a single Z-section through the valves was quantified and the EGFP over mCherry ratio generated. For fluorescence intensity analysis of the Notch reporter, the ratio of the maximum intensity of the GFP signal from the Tg(tp1:dGFP) reporter line on a single Z-section through the valves over the maximum intensity of the background was generated.
These ratios were then averaged for three cells in the anterior part and three cells in the posterior part of both valves in the OFT of individual embryos. Finally, the averages of the anterior and posterior parts were compared.
Statistical analyses
We did not compute or predict the number of samples necessary for statistical differences because the standard deviation of our study’s population was not known before starting our analysis. Biological replicate corresponds to the analysis of different embryos of the same stage. Technical replicate corresponds to the analysis of the same embryo imaged the same way. The sample size (biological replicate and number) to use was as defined by our ability to generate our datasets. For analyses between two groups of embryos, differences were considered statistically significant when the p-value<0.05, as determined using a two-tailed and paired Student’s t-test (klf2a and notch reporter expression). For boxplots, center lines show the medians, crosses show the means, box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, data points are represented as circles.
Acknowledgements
We thank the Vermot laboratory for discussion and H Fukui and R Chow for thoughtful comments on the manuscript. We thank C Burns, V Lecaudey and I Drummond for providing antibodies, mutants and protocols for immunohistochemistry. We thank J-M Garnier for the cloning of the piezo1:nls-Venus construct. We thank the IGBMC fish facility (S Pajot and C Moebs) and the IGBMC imaging center, in particular B Gurchenkov, D Hentsch, E Guiot and E Grandgirard. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme: GA N°682939, the Fondation pour la Recherche Médicale: DEQ29553, Agence Nationale de la Recherche: ANR-15-CE13-0015-01, ANR-10-IDEX-0002–02, ANR-12-ISV2-0001-01 and ANR-10-LABX-0030-INRT and the European Molecular Biology Organization Young Investigator Program. HV was supported by the IGBMC International PhD program: ANR-10-LABX-0030-INRT. ALD was supported by a post doctoral fellowship from the Lefoulon-Delalande Foundation.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Julien Vermot, Email: julien@igbmc.fr.
Holger Gerhardt, Max Delbrück Center for Molecular Medicine, Germany.
Didier Y Stainier, Max Planck Institute for Heart and Lung Research, Germany.
Funding Information
This paper was supported by the following grants:
H2020 European Research Council 682938 - EVALVE to Julien Vermot.
Fondation pour la Recherche Médicale DEQ29553 to Julien Vermot.
Agence Nationale de la Recherche ANR-15-CE13-0015-01 to Anne-Laure Duchemin, Hélène Vignes, Julien Vermot.
European Molecular Biology Organization Young Investigator Program to Julien Vermot.
Fondation Lefoulon Delalande to Anne-Laure Duchemin.
Agence Nationale de la Recherche ANR-10-IDEX-0002-02 to Anne-Laure Duchemin, Hélène Vignes, Julien Vermot.
Agence Nationale de la Recherche ANR-12-ISV2-0001-01 to Anne-Laure Duchemin, Hélène Vignes, Julien Vermot.
Agence Nationale de la Recherche ANR-10-LABX-0030-INRT to Anne-Laure Duchemin, Hélène Vignes, Julien Vermot.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Methodology, Writing—original draft.
Data curation.
Conceptualization, Supervision, Funding acquisition, Writing—original draft, Project administration.
Ethics
Animal experimentation: Animal experiments were approved by the Animal Experimentation Committee of the Institutional Review Board of the IGBMC.(reference numbers MIN APAFIS#4669-2016032411093030 v4 and MIN 4669-2016032411093030 v4-detail of entry 1).
Additional files
Data availability
Source data for the figures has been uploaded.
References
- Agarwala S, Duquesne S, Liu K, Boehm A, Grimm L, Link S, König S, Eimer S, Ronneberger O, Lecaudey V. Amotl2a interacts with the hippo effector Yap1 and the wnt/β-catenin effector Lef1 to control tissue size in zebrafish. eLife. 2015;4:e08201. doi: 10.7554/eLife.08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton H, Harlepp S, Ramspacher C, Wu D, Monduc F, Bhat S, Liebling M, Paoletti C, Charvin G, Freund JB, Vermot J. Pulse propagation by a capacitive mechanism drives embryonic blood flow. Development. 2013;140:4426–4434. doi: 10.1242/dev.096768. [DOI] [PubMed] [Google Scholar]
- Bäck M, Gasser TC, Michel JB, Caligiuri G. Biomechanical factors in the biology of aortic wall and aortic valve diseases. Cardiovascular Research. 2013;99:232–241. doi: 10.1093/cvr/cvt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA. Endothelial fluid shear stress sensing in vascular health and disease. Journal of Clinical Investigation. 2016;126:821–828. doi: 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Boselli F, Steed E, Freund JB, Vermot J. Anisotropic shear stress patterns predict the orientation of convergent tissue movements in the embryonic heart. Development. 2017;144:4322–4327. doi: 10.1242/dev.152124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun MH, Brill RW, Gosline JM, Jones DR. Form and function of the bulbus arteriosus in yellowfin tuna (Thunnus albacares), bigeye tuna (Thunnus obesus) and blue marlin (Makaira nigricans): static properties. Journal of Experimental Biology. 2003a;206:3311–3326. doi: 10.1242/jeb.00575. [DOI] [PubMed] [Google Scholar]
- Braun MH, Brill RW, Gosline JM, Jones DR. Form and function of the bulbus arteriosus in yellowfin tuna (Thunnus albacares): dynamic properties. Journal of Experimental Biology. 2003b;206:3327–3335. doi: 10.1242/jeb.00576. [DOI] [PubMed] [Google Scholar]
- Butcher JT, Simmons CA, Warnock JN. Mechanobiology of the aortic heart valve. The Journal of Heart Valve Disease. 2008;17:62–73. [PubMed] [Google Scholar]
- Choi D, Park E, Jung E, Seong YJ, Yoo J, Lee E, Hong M, Lee S, Ishida H, Burford J, Peti-Peterdi J, Adams RH, Srikanth S, Gwack Y, Chen CS, Vogel HJ, Koh CJ, Wong AK, Hong YK. Laminar flow downregulates notch activity to promote lymphatic sprouting. Journal of Clinical Investigation. 2017;127:1225–1240. doi: 10.1172/JCI87442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development. Circulation Research. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat S, Lourenço M, Paolini A, Otten C, Renz M, Abdelilah-Seyfried S. Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. eLife. 2018;7:e28939. doi: 10.7554/eLife.28939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst R, Sauls K, Peal DS, deVlaming A, Toomer K, Leyne M, Salani M, Talkowski ME, Brand H, Perrocheau M, Simpson C, Jett C, Stone MR, Charles F, Chiang C, Lynch SN, Bouatia-Naji N, Delling FN, Freed LA, Tribouilloy C, Le Tourneau T, LeMarec H, Fernandez-Friera L, Solis J, Trujillano D, Ossowski S, Estivill X, Dina C, Bruneval P, Chester A, Schott JJ, Irvine KD, Mao Y, Wessels A, Motiwala T, Puceat M, Tsukasaki Y, Menick DR, Kasiganesan H, Nie X, Broome AM, Williams K, Johnson A, Markwald RR, Jeunemaitre X, Hagege A, Levine RA, Milan DJ, Norris RA, Slaugenhaupt SA. Mutations in DCHS1 cause mitral valve prolapse. Nature. 2015;525:109–113. doi: 10.1038/nature14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamba BJ, DeSimone DW. Extracellular matrix (ECM) and the sculpting of embryonic tissues. Current Topics in Developmental Biology. 2018;130:245–274. doi: 10.1016/bs.ctdb.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA, Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley L, Alqahtani AM, MacGrogan D, Richardson RV, Murphy L, Salguero-Jimenez A, Sintes Rodriguez San Pedro M, Tiurma S, McCutcheon L, Gilmore A, de La Pompa JL, Chaudhry B, Henderson DJ. A novel source of arterial valve cells linked to bicuspid aortic valve without raphe in mice. eLife. 2018;7:e34110. doi: 10.7554/eLife.34110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver RE, Gelfand BD, Blackman BR. Human haemodynamic frequency harmonics regulate the inflammatory phenotype of vascular endothelial cells. Nature Communications. 2013;4:1525. doi: 10.1038/ncomms2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felker A, Prummel KD, Merks AM, Mickoleit M, Brombacher EC, Huisken J, Panáková D, Mosimann C. Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nature Communications. 2018;9:2001. doi: 10.1038/s41467-018-04402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Garg V. Notch Signaling in Aortic Valve Development and Disease. In: Nakanishi T, Markwald R. R, Baldwin H. S, Keller B. B, Srivastava D, Yamagishi H, editors. Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. Springer; 2016. pp. 371–376. [PubMed] [Google Scholar]
- Godby R, Munjal C, Opoka A, Smith J, Yutzey K, Narmoneva D, Hinton R. Cross talk between NOTCH signaling and biomechanics in human aortic valve disease pathogenesis. Journal of Cardiovascular Development and Disease. 2014;1:237–256. doi: 10.3390/jcdd1030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard LM, Duchemin AL, Ramalingan H, Wu B, Chen M, Bamezai S, Yang J, Li L, Morley MP, Wang T, Scherrer-Crosbie M, Frank DB, Engleka KA, Jameson SC, Morrisey EE, Carroll TJ, Zhou B, Vermot J, Kahn ML. Hemodynamic forces sculpt developing heart valves through a KLF2-WNT9B paracrine signaling Axis. Developmental Cell. 2017;43:274–289. doi: 10.1016/j.devcel.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Developmental Biology. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Hasan SS, Tsaryk R, Lange M, Wisniewski L, Moore JC, Lawson ND, Wojciechowska K, Schnittler H, Siekmann AF. Endothelial notch signalling limits angiogenesis via control of artery formation. Nature Cell Biology. 2017;19:928–940. doi: 10.1038/ncb3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Si G, Huang J, Samuel ADT, Perrimon N. Mechanical regulation of stem-cell differentiation by the stretch-activated piezo channel. Nature. 2018;555:103–106. doi: 10.1038/nature25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel E, Boselli F, Roth S, Krudewig A, Belting HG, Charvin G, Vermot J. Oscillatory flow modulates mechanosensitive klf2a expression through trpv4 and trpp2 during heart valve development. Current Biology. 2015;25:1354–1361. doi: 10.1016/j.cub.2015.03.038. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39:1890–1900. doi: 10.1016/S0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Hsu JJ, Vedula V, Baek KI, Chen C, Chen J, Chou MI, Lam J, Subhedar S, Wang J, Ding Y, Chang C-C, Lee J, Demer LL, Tintut Y, Marsden AL, Hsiai TK. Contractile and hemodynamic forces coordinate Notch1b-mediated outflow tract valve formation. JCI Insight. 2019;4:124460. doi: 10.1172/jci.insight.124460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Developmental Dynamics. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Keith DA, Paz A, Gallop PM, Glimcher MJ. Histologic and biochemical identification and characterization of an elastin in cartilage. Journal of Histochemistry & Cytochemistry. 1977;25:1154–1162. doi: 10.1177/25.10.72098. [DOI] [PubMed] [Google Scholar]
- Köttgen M, Buchholz B, Garcia-Gonzalez MA, Kotsis F, Fu X, Doerken M, Boehlke C, Steffl D, Tauber R, Wegierski T, Nitschke R, Suzuki M, Kramer-Zucker A, Germino GG, Watnick T, Prenen J, Nilius B, Kuehn EW, Walz G. TRPP2 and TRPV4 form a polymodal sensory channel complex. The Journal of Cell Biology. 2008;182:437–447. doi: 10.1083/jcb.200805124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest B, Andelfinger G, Nemer M. Loss of Gata5 in mice leads to bicuspid aortic valve. Journal of Clinical Investigation. 2011;121:2876–2887. doi: 10.1172/JCI44555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Leddy HA, Chen Y, Lee SH, Zelenski NA, McNulty AL, Wu J, Beicker KN, Coles J, Zauscher S, Grandl J, Sachs F, Guilak F, Liedtke WB. Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. PNAS. 2014;111:E5114–E5122. doi: 10.1073/pnas.1414298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AH, Cui AF, McDonald MF, Grandl J. Transduction of repetitive mechanical stimuli by Piezo1 and Piezo2 ion channels. Cell Reports. 2017;19:2572–2585. doi: 10.1016/j.celrep.2017.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, Yuldasheva NY, Majeed Y, Wilson LA, Rode B, Bailey MA, Kim HR, Fu Z, Carter DA, Bilton J, Imrie H, Ajuh P, Dear TN, Cubbon RM, Kearney MT, Prasad RK, Evans PC, Ainscough JF, Beech DJ. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515:279–282. doi: 10.1038/nature13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-J, Lin C-Y, Chen C-H, Zhou B, Chang C-P. Partitioning the heart: mechanisms of cardiac septation and valve development. Development. 2012;139:3277–3299. doi: 10.1242/dev.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs V, Mathur J, Mao R, Bayrak-Toydemir P, Procter M, Cahalan SM, Kim HJ, Bandell M, Longo N, Day RW, Stevenson DA, Patapoutian A, Krock BL. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nature Communications. 2015;6:8329. doi: 10.1038/ncomms9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxán G, D'Amato G, MacGrogan D, de la Pompa JL. Endocardial notch signaling in cardiac development and disease. Circulation Research. 2016;118:e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. PNAS. 2002;99:5454–5459. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Cahalan S, LaMonte G, Grubaugh ND, Zeng W, Murthy SE, Paytas E, Gamini R, Lukacs V, Whitwam T, Loud M, Lohia R, Berry L, Khan SM, Janse CJ, Bandell M, Schmedt C, Wengelnik K, Su AI, Honore E, Winzeler EA, Andersen KG, Patapoutian A. Common PIEZO1 allele in african populations causes RBC dehydration and attenuates plasmodium infection. Cell. 2018;173:443–455. doi: 10.1016/j.cell.2018.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrogan D, D'Amato G, Travisano S, Martinez-Poveda B, Luxán G, Del Monte-Nieto G, Papoutsi T, Sbroggio M, Bou V, Gomez-Del Arco P, Gómez MJ, Zhou B, Redondo JM, Jiménez-Borreguero LJ, de la Pompa JL. Sequential Ligand-Dependent notch signaling activation regulates valve primordium formation and morphogenesis. Circulation Research. 2016;118:1480–1497. doi: 10.1161/CIRCRESAHA.115.308077. [DOI] [PubMed] [Google Scholar]
- Mack JJ, Mosqueiro TS, Archer BJ, Jones WM, Sunshine H, Faas GC, Briot A, Aragón RL, Su T, Romay MC, McDonald AI, Kuo CH, Lizama CO, Lane TF, Zovein AC, Fang Y, Tarling EJ, de Aguiar Vallim TQ, Navab M, Fogelman AM, Bouchard LS, Iruela-Arispe ML. NOTCH1 is a mechanosensor in adult arteries. Nature Communications. 2017;8:1620. doi: 10.1038/s41467-017-01741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Bruce AE, Bhanji T, Davis EC, Keeley FW. Differential expression of two tropoelastin genes in zebrafish. Matrix Biology. 2007;26:115–124. doi: 10.1016/j.matbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Miesfeld JB, Link BA. Establishment of transgenic lines to monitor and manipulate Yap/Taz-Tead activity in zebrafish reveals both evolutionarily conserved and divergent functions of the Hippo pathway. Mechanisms of Development. 2014;133:177–188. doi: 10.1016/j.mod.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama Y, Ito F, Takeda H, Yano T, Okabe M, Kuraku S, Keeley FW, Koshiba-Takeuchi K. Evolution of the fish heart by sub/neofunctionalization of an elastin gene. Nature Communications. 2016;7:10397. doi: 10.1038/ncomms10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SE, Dubin AE, Patapoutian A. Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nature Reviews Molecular Cell Biology. 2017;18:771–783. doi: 10.1038/nrm.2017.92. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Yamamoto K, Agarwala S, Terai K, Fukui H, Fukuhara S, Ando K, Miyazaki T, Yokota Y, Schmelzer E, Belting H-G, Affolter M, Lecaudey V, Mochizuki N. Flow-Dependent endothelial YAP regulation contributes to vessel maintenance. Developmental Cell. 2017;40:523–536. doi: 10.1016/j.devcel.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Nicenboim J, Malkinson G, Lupo T, Asaf L, Sela Y, Mayseless O, Gibbs-Bar L, Senderovich N, Hashimshony T, Shin M, Jerafi-Vider A, Avraham-Davidi I, Krupalnik V, Hofi R, Almog G, Astin JW, Golani O, Ben-Dor S, Crosier PS, Herzog W, Lawson ND, Hanna JH, Yanai I, Yaniv K. Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature. 2015;522:56–61. doi: 10.1038/nature14425. [DOI] [PubMed] [Google Scholar]
- Ninov N, Borius M, Stainier DY. Different levels of notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Lukacs V, Sweet DT, Goddard LM, Kanie A, Whitwam T, Ranade SS, Fujimori T, Kahn ML, Patapoutian A. Mechanically activated ion channel PIEZO1 is required for lymphatic valve formation. PNAS. 2018;115:12817–12822. doi: 10.1073/pnas.1817070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- Paffett-Lugassy N, Novikov N, Jeffrey S, Abrial M, Guner-Ataman B, Sakthivel S, Burns CE, Burns CG. Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development. 2017;144:4616–4624. doi: 10.1242/dev.153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini A, Abdelilah-Seyfried S. The mechanobiology of zebrafish cardiac valve leaflet formation. Current Opinion in Cell Biology. 2018;55:52–58. doi: 10.1016/j.ceb.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DT, Bernardis E, Flanagan LA, Tombola F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. PNAS. 2014;111:16148–16153. doi: 10.1073/pnas.1409802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta M, Steed E, Harlepp S, González-Rosa JM, Monduc F, Ariza-Cosano A, Cortés A, Rayón T, Gómez-Skarmeta JL, Zapata A, Vermot J, Mercader N. Heartbeat-driven pericardiac fluid forces contribute to epicardium morphogenesis. Current Biology. 2013;23:1726–1735. doi: 10.1016/j.cub.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DY. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143:2217–2227. doi: 10.1242/dev.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng LK, Stanchi F, Gerhardt H. Filopodia are dispensable for endothelial tip cell guidance. Development. 2013;140:4031–4040. doi: 10.1242/dev.097352. [DOI] [PubMed] [Google Scholar]
- Pitulescu ME, Schmidt I, Giaimo BD, Antoine T, Berkenfeld F, Ferrante F, Park H, Ehling M, Biljes D, Rocha SF, Langen UH, Stehling M, Nagasawa T, Ferrara N, Borggrefe T, Adams RH. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nature Cell Biology. 2017;19:915–927. doi: 10.1038/ncb3555. [DOI] [PubMed] [Google Scholar]
- Poole K, Herget R, Lapatsina L, Ngo HD, Lewin GR. Tuning piezo ion channels to detect molecular-scale movements relevant for fine touch. Nature Communications. 2014;5:3520. doi: 10.1038/ncomms4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROMESA investigators. MVP-France. Leducq Transatlantic MITRAL Network. Dina C, Bouatia-Naji N, Tucker N, Delling FN, Toomer K, Durst R, Perrocheau M, Fernandez-Friera L, Solis J, Le Tourneau T, Chen MH, Probst V, Bosse Y, Pibarot P, Zelenika D, Lathrop M, Hercberg S, Roussel R, Benjamin EJ, Bonnet F, Lo SH, Dolmatova E, Simonet F, Lecointe S, Kyndt F, Redon R, Le Marec H, Froguel P, Ellinor PT, Vasan RS, Bruneval P, Markwald RR, Norris RA, Milan DJ, Slaugenhaupt SA, Levine RA, Schott JJ, Hagege AA, Jeunemaitre X. Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nature Genetics. 2015;47:1206–1211. doi: 10.1038/ng.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Andolfi L, Frattini F, Mayer F, Lazzarino M, Hu J. Membrane stiffening by STOML3 facilitates mechanosensation in sensory neurons. Nature Communications. 2015;6:8512. doi: 10.1038/ncomms9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy SE, Cahalan SM, Xu J, Mathur J, Bandell M, Coste B, Li YS, Chien S, Patapoutian A. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. PNAS. 2014;111:10347–10352. doi: 10.1073/pnas.1409233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Syeda R, Patapoutian A. Mechanically Activated Ion Channels. Neuron. 2015;87:1162–1179. doi: 10.1016/j.neuron.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retailleau K, Duprat F, Arhatte M, Ranade SS, Peyronnet R, Martins JR, Jodar M, Moro C, Offermanns S, Feng Y, Demolombe S, Patel A, Honoré E. Piezo1 in smooth muscle cells is involved in Hypertension-Dependent arterial remodeling. Cell Reports. 2015;13:1161–1171. doi: 10.1016/j.celrep.2015.09.072. [DOI] [PubMed] [Google Scholar]
- Richards AA, Garg V. Genetics of congenital heart disease. Current Cardiology Reviews. 2010;6:91–97. doi: 10.2174/157340310791162703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa LA, Givens C, Tzima E, Stainier DY, Qian L, Liu J. Cardiac contraction activates endocardial notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142:4080–4091. doi: 10.1242/dev.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Iranzo H, Galardi-Castilla M, Minguillón C, Sanz-Morejón A, González-Rosa JM, Felker A, Ernst A, Guzmán-Martínez G, Mosimann C, Mercader N. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nature Communications. 2018;9:428. doi: 10.1038/s41467-017-02650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauls K, de Vlaming A, Harris BS, Williams K, Wessels A, Levine RA, Slaugenhaupt SA, Goodwin RL, Pavone LM, Merot J, Schott J-J, Le Tourneau T, Dix T, Jesinkey S, Feng Y, Walsh C, Zhou B, Baldwin S, Markwald RR, Norris RA. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovascular Research. 2012;96:109–119. doi: 10.1093/cvr/cvs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- Schottenfeld J, Sullivan-Brown J, Burdine RD. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development. 2007;134:1605–1615. doi: 10.1242/dev.02827. [DOI] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nature Genetics. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Sharif-Naeini R, Folgering JHA, Bichet D, Duprat F, Lauritzen I, Arhatte M, Jodar M, Dedman A, Chatelain FC, Schulte U, Retailleau K, Loufrani L, Patel A, Sachs F, Delmas P, Peters DJ, Honoré E. Polycystin-1 and -2 dosage regulates pressure sensing. Cell. 2009;139:587–596. doi: 10.1016/j.cell.2009.08.045. [DOI] [PubMed] [Google Scholar]
- Steed E, Boselli F, Vermot J. Hemodynamics driven cardiac valve morphogenesis. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2016a;1863:1760–1766. doi: 10.1016/j.bbamcr.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Steed E, Faggianelli N, Roth S, Ramspacher C, Concordet JP, Vermot J. klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nature Communications. 2016b;7:11646. doi: 10.1038/ncomms11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Barnett P, van Duijvenboden K, Weber D, Gessler M, Christoffels VM. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nature Communications. 2014;5:3680. doi: 10.1038/ncomms4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T, Zarkada G, Nurmi H, Jakobsson L, Heinolainen K, Tvorogov D, Zheng W, Franco CA, Murtomäki A, Aranda E, Miura N, Ylä-Herttuala S, Fruttiger M, Mäkinen T, Eichmann A, Pollard JW, Gerhardt H, Alitalo K. VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing notch signalling. Nature Cell Biology. 2011;13:1202–1213. doi: 10.1038/ncb2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, van Weerd JH, Burkhard SB, Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM, Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLOS ONE. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Thodeti CK, Matthews B, Ravi A, Mammoto A, Ghosh K, Bracha AL, Ingber DE. TRPV4 channels mediate cyclic strain-induced endothelial cell reorientation through integrin-to-integrin signaling. Circulation Research. 2009;104:1123–1130. doi: 10.1161/CIRCRESAHA.108.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Forouhar AS, Liebling M, Wu D, Plummer D, Gharib M, Fraser SE. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLOS Biology. 2009;7:e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu B, Farrar E, Lui W, Lu P, Zhang D, Alfieri CM, Mao K, Chu M, Yang D, Xu D, Rauchman M, Taylor V, Conway SJ, Yutzey KE, Butcher JT, Zhou B. Notch-Tnf signalling is required for development and homeostasis of arterial valves. European Heart Journal. 2017;38:675–686. doi: 10.1093/eurheartj/ehv520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Wang Y, Xiao F, Butcher JT, Yutzey KE, Zhou B. Developmental mechanisms of aortic valve malformation and disease. Annual Review of Physiology. 2017;79:21–41. doi: 10.1146/annurev-physiol-022516-034001. [DOI] [PubMed] [Google Scholar]
- Zarychanski R, Schulz VP, Houston BL, Maksimova Y, Houston DS, Smith B, Rinehart J, Gallagher PG. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood. 2012;120:1908–1915. doi: 10.1182/blood-2012-04-422253. [DOI] [PMC free article] [PubMed] [Google Scholar]