Abstract
Characterized primarily by low BMI, anorexia nervosa is a complex and serious illness1, affecting 0.9-4% of women and 0.3% of men2-4, with twin-based heritability estimates of 50-60%5. Mortality rates are higher than other psychiatric disorders6, and outcomes are unacceptably poor7. Combining data from the Anorexia Nervosa Genetics Initiative (ANGI)8,9 and the Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC-ED), we conducted a genome-wide association study (GWAS) of 16,992 anorexia nervosa cases and 55,525 controls, identifying eight significant loci. The genetic architecture of anorexia nervosa mirrors its clinical presentation showing significant genetic correlations with psychiatric disorders, physical activity, metabolic (including glycemic), lipid, and anthropometric traits, independent of the effects of common variants associated with BMI. Results further encourage a reconceptualization of anorexia nervosa as a metabo-psychiatric disorder. Explicating the metabolic component is a critical direction, and attention to both psychiatric and metabolic components may be key to improving outcomes.
The first PGC-ED GWAS (3,495 cases, 10,982 controls) estimated the common genetic variant-based heritability of anorexia nervosa as ~20%, identified the first genome-wide significant locus, and reported significant genetic correlations (rg) between anorexia nervosa and psychiatric and metabolic/anthropometric phenotypes10. These rg pointed toward metabolic etiological factors, as they are robust to reverse causation although they could be mediated associations11 or reflect confounding processes12. To advance genomic discovery in anorexia nervosa and further explore genetic correlations, we combined samples from ANGI8,9, the Genetic Consortium for Anorexia Nervosa (GCAN)/Wellcome Trust Case Control Consortium-3 (WTCCC-3)13, and the UK Biobank14, quadrupling our sample size.
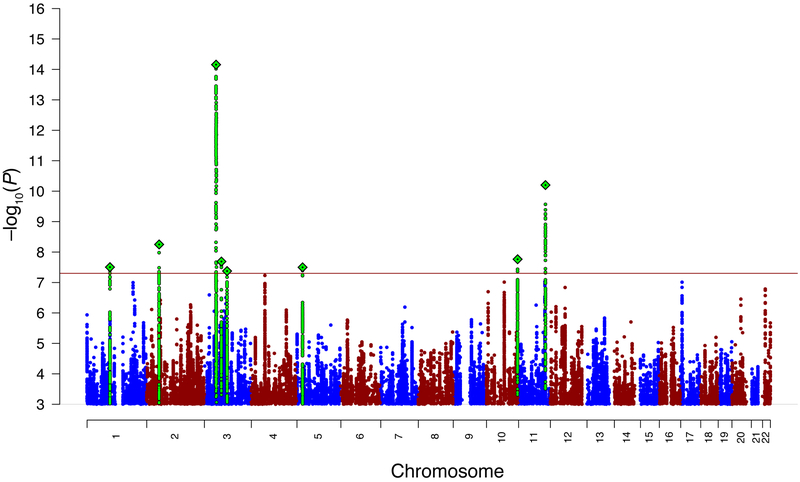
Our GWAS meta-analysis included 33 datasets comprising 16,992 cases and 55,525 controls of European ancestry from 17 countries (Supplementary Tables 1-4). We had 80% power to detect an odds ratio (OR) of 1.09-1.19 (additive model, 0.9% lifetime risk, α = 5 × 10−8, MAF 0.05–0.5). Typical of complex trait GWAS, we observed test statistic inflation (λ = 1.22) consistent with polygenicity, with no evidence of significant population stratification according to the LD intercept and attenuation ratio (Supplementary Results; Supplementary Fig. 1). Meta-analysis results were completed for autosomes and the X chromosome. We identified eight loci exceeding genome-wide significance (P < 5 × 10−8; Table 1 for loci; Fig. 1 for the Manhattan plot; Supplementary Figs. 2a-h and 3a-h for the forest and region plots). Many were near the threshold for significance, and no significant heterogeneity of SNP associations across cohorts was detected (P = 0.15-0.64; Supplementary Figs. 2a-h). Conditional and joint analysis (GCTA-COJO)15 confirmed independence of the lead SNPs within the significant loci (Supplementary Table 5). The eight loci were annotated to identify known protein-coding genes (Supplementary Table 6; Supplementary Table 7 reports a gene look-up restricted to the single-gene loci). The previously reported PGC-ED genome-wide significant variant (rs4622308)10 on 12q13.2 did not reach genome-wide significance (P = 7.02 × 10−5); however, between-cohort heterogeneity was apparent (I2 = 53.7; Supplementary Fig. 4 and Supplementary Results). The OR was in the same direction in 22 (67%) of the cohorts (z = 2.00, P = 0.05, 2-tailed).
Table 1.
Newly associated genome-wide significant loci for anorexia nervosa
| Locus | Chr | Basepair region | Lead SNP | BP | P | A1/A2 | OR | s.e. | Freq | Type | Number of genes |
Nearest gene | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| range left | range right | ||||||||||||
| 1 | 3 | 47588253 | 51368253 | rs9821797 | 48718253 | 6.99E-15 | A/T | 1.17 | 0.02 | 0.12 | multigenic | 111 | NCKIPSD |
| 2 | 11 | 114997256 | 115424956 | rs6589488 | 115096956 | 6.31E-11 | A/T | 1.14 | 0.02 | 0.13 | single-gene | 1 | CADM1 |
| 3 | 2 | 53881813 | 54362813 | rs2287348 | 54039813 | 5.62E-09 | T/C | 1.11 | 0.02 | 0.16 | multigenic | 13 | ASB3, ERLEC1 |
| 4 | 10 | 131269764 | 131463964 | rs2008387 | 131448764 | 1.73E-08 | A/G | 1.08 | 0.01 | 0.33 | single-gene | 2 | MGMT |
| 5 | 3 | 70670750 | 71074150 | rs9874207 | 71019750 | 2.05E-08 | C/T | 1.08 | 0.01 | 0.49 | single-gene | 2 | FOXP1 |
| 6 | 1 | 96699455 | 97284455 | rs10747478 | 96901455 | 3.13E-08 | T/G | 1.08 | 0.01 | 0.41 | single-gene | 2 | PTBP2 |
| 7 | 5 | 24945845 | 25372845 | rs370838138 | 25081845 | 3.17E-08 | G/C | 1.08 | 0.01 | 0.56 | intergenic | 0 | CDH10 |
| 8 | 3 | 93968107 | 95059107 | rs13100344 | 94605107 | 4.21E-08 | T/A | 1.08 | 0.01 | 0.54 | intergenic | 2 | NSUN3 |
Note. Shown are the results of the GWAS meta-analysis of anorexia nervosa (16,992 cases and 55,552 controls) which detected eight genome-wide significant loci. All of the eight loci are novel. Chr (chromosome) and Region (hg19) are shown for SNPs with P < 1e-05 and linkage-disequilibrium (LD) r2 > 0.1 with the most associated "lead" SNP, the location of which is given in BP (basepair). A1/A2 refers to Allele 1/Allele 2 and OR and s.e. are the odds ratio and standard error for the association between A1 and the phenotype. Freq is the frequency of A1 in controls. Number of genes was determined by genomic location, adult brain eQTL, regulatory chromatin interactions, and MAGMA gene-wise analysis (see Methods). Nearest gene is the nearest gene within the region of LD "friends" of the lead variant (LD-r2 > 0.6 +/− 500 Kb). The meta-analysis was restricted to variants with minor allele frequency (MAF) ≥ 0.01 and information quality (INFO) score ≥ 0.70. All loci were confirmed via forest plots based on consistent direction of effect in the majority of cohorts and via region plots whereby neighboring LD "friends" were required to show a similar effect. Chromosome X was analyzed but had no loci that reached genome-wide significance. Note that although lead variants are annotated to the nearest gene, this does not mean that the gene listed is a causal gene.
Figure 1. The Manhattan plot for the primary genome-wide association meta-analysis of anorexia nervosa with 33 case-control samples (16,992 cases and 55,525 controls of European descent).
The −log10(P) values for the association tests (two-tailed) are shown on the y-axis and the chromosomes are ordered on the x-axis. Eight genetic loci surpassed genome-wide significance (−log10(P) > 7.3). The lead variant is indicated by a diamond and green circles show the variants in linkage-disequilibrium. The blue and red colors differentiate adjacent chromosomes.
Although GWAS findings are informative genome-wide, identifying strong hypotheses about their connections to specific genes is not straightforward. We evaluated three ways to “connect” anorexia nervosa GWAS loci to genes: regulatory chromatin interactions; relationship to brain expression QTLs (eQTLs; using a superset of CommonMind16 and GTEx17) and the standard approach of gene location within a GWAS locus. The significant anorexia nervosa loci implicated 121 brain-expressed genes, 74% by location, 55% by adult brain eQTL, 93% by regulatory chromatin interaction, and 58 genes by all three methods. Supplementary Figs. 5a-h show the eight GWAS loci, GENCODE gene models, adult brain regulatory chromatin interactions, brain eQTLs, and functional genomic annotations.
Four single-gene loci were confirmed by eQTL, chromatin interaction, or both. These were the locus-intersecting genes CADM1 (locus 2 chr11:114.9-115.4 Mb, Supplementary Fig. 5b), MGMT (locus 4, chr10:131.2-131.4 Mb, Supplementary Fig. 5d), FOXP1 (locus 5, chr3:70.6-71.0 Mb, Supplementary Fig. 5e) and PTBP2 (locus 6, chr1:96.6-97.2 Mb, Supplementary Fig. 5f). For locus 5, eQTL data implicated a distal gene, GPR27. One intergenic locus (locus 7, chr5:24.9-25.3 Mb, Supplementary Fig. 5g) had no eQTL or chromatin interactions whereas the other intergenic locus (locus 8, chr3:93.9-95.0 Mb, Supplementary Fig. 5h) had eQTL connections to PROS1 and ARL13B. Two complex multigenic loci had many brain-expressed genes and dense chromatin and eQTL interactions that precluded identification of any single gene (locus 1, chr3:47.5-51.3 Mb; locus 3, chr2:53.8-54.3 Mb, Supplementary Figs. 5a and 5c). The clearest evidence and connections were for the single-gene loci intersecting CADM1, MGMT, FOXP1, and PTBP2 and we conclude these genes may play a role in anorexia nervosa etiology (Supplementary Results).
Supplementary Table 8 presents multi-trait analysis (GCTA-mtCOJO18 conditioning our genome-wide significant SNPs on associated variants in GWAS of BMI, type 2 diabetes, education years, HDL cholesterol, neuroticism, and schizophrenia. Seven loci appear to be independent. Locus 2 on chr11 may not be unique to anorexia nervosa and may be driven by genetic variation also associated with type 2 diabetes.
Liability-scale SNP heritability (SNP-h2) was estimated with LD score regression (LDSC)19,20. Assuming a lifetime prevalence of 0.9-4%2-4, SNP-h2 was 11-17% (s.e. = 1%), supporting the polygenic nature of anorexia nervosa. Polygenic risk score (PRS) analyses using a leave-one-out approach indicated that the PRS captures ~1.7% of the phenotypic variance on the liability scale for discovery P = 0.5. We did not observe differences in polygenic architecture between anorexia nervosa subtypes with binge eating (2,381 cases, 10,249 controls) or without (2,262 cases, 10,254 controls) or between males (447 cases, 20,347 controls) and females (14,898 cases, 27,545 controls) (Methods, Supplementary Results, Supplementary Fig. 6, Supplementary Table 9). Similar to females, males in the highest PRS decile had 4.13 (95% CI: 2.58-6.62) times the odds of anorexia nervosa than those in the lowest decile. Confirmation of these results requires larger samples.
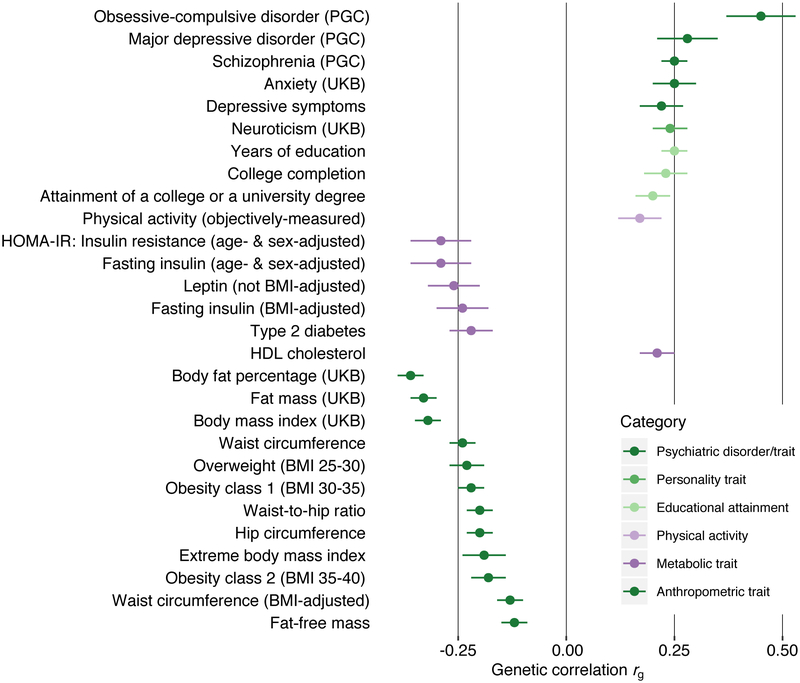
We tested SNP-based genetic correlations (SNP-rg) with external traits using bivariate LDSC19,20. Bonferroni-significant SNP-rg assorted into five trait categories: psychiatric and personality; physical activity; anthropometric; metabolic; and educational attainment (Supplementary Table 10). Fig. 2 presents Bonferroni-corrected positive SNP-rg with OCD (SNP-rg ± s.e. = 0.45 ± 0.08; P = 4.97 × 10−9), MDD (0.28 ± 0.07; P = 8.95 × 10−5), anxiety disorders (0.25 ± 0.05; P = 8.90 × 10−8), and schizophrenia (0.25 ± 0.03; P = 4.61 × 10−18). This pattern reflects observed comorbidities in clinical and epidemiological studies21,22. The newly-identified positive SNP-rg with physical activity (0.17 ± 0.05; P = 1.00 × 10−4) encourages further exploration of the refractory symptom of pathologically elevated activity in anorexia nervosa23. We note that the significant SNP-rg of anorexia nervosa with educational attainment (0.25 ± 0.03; P = 1.69 × 10−15) and related constructs was not seen for IQ24.
Figure. 2. Bonferroni-significant genetic correlations (SNP-rgs) and standard errors (error bars) between anorexia nervosa and other phenotypes as estimated by LD score regression.
Only traits with significant P values following Bonferroni correction are shown. Correlations with 447 phenotypes were tested (Bonferroni-corrected significance threshold P > 1.11 × 10−4). Complete results are shown in Table S10. PGC = Psychiatric Genomics Consortium, UKB = UK Biobank, HOMA-IR = Homeostatic model assessment - insulin resistance.
Expanding our previous observations10, we present a palette of metabolic and anthropometric rg with anorexia nervosa more pronounced than in other psychiatric disorders. We observed significant negative SNP-rg with fat mass (−0.33 ± 0.03; P = 7.23 × 10−25), fat-free mass (−0.12 ± 0.03; P = 4.65 × 10−5), BMI (−0.32 ± 0.03; P = 8.93 × 10−25), obesity (−0.22 ± 0.03; P = 2.96 × 10−11), type 2 diabetes (−0.22 ± 0.05; P = 3.82 × 10−5), fasting insulin (−0.24 ± 0.06; P = 2.31 × 10−5), insulin resistance (−0.29 ± 0.07; P = 2.83 × 10−5), and leptin (−0.26 ± 0.06; P = 4.98 × 10−5), and a significant positive SNP-rg with HDL cholesterol (0.21 ± 0.04; P = 3.08 × 10−7).
Systems biology analyses of our results revealed preliminarily interesting results (Methods, Supplementary Tables 11-13, Supplementary Figs. 7-15). Gene-wise analysis with MAGMA prioritized 79 Bonferroni-significant genes, most within the multigenic locus on chr3 (Supplementary Table 11). MAGMA indicated an association with NCAM1 (Supplementary Table 11) the expression of which increases in response to food restriction in a rodent activity-based anorexia nervosa model25. Partitioned heritability analysis showed, as with other GWAS26, considerable enrichment of SNP-h2 in conserved regions (fold enrichment = 24.97, s.e. = 3.29, P = 3.32 × 10−11; Supplementary Fig. 7)27. Cell type group-specific annotations revealed that the overall SNP-h2 is significantly enriched for CNS tissue (Supplementary Fig. 8). One biological pathway was significant: GO:positive_regulation_of_embryonic_development (32 genes, P = 1.39 × 10−7; Supplementary Table 12), which contains two Bonferroni-significant genes on chr3, CTNNB1 and DAG1. CTNNB1 encodes catenin beta-1, which is part of adherens junctions, and DAG1 encodes dystroglycan, a receptor which binds extracellular matrix proteins28. DAG1 falls within locus 1 (47.5-51.3 Mb). This pathway points to a potential role of developmental processes in the etiology of this complex phenotype (although this is currently speculative). Genes associated with anorexia nervosa were enriched for expression in most brain tissues, particularly the cerebellum, which has a notably high proportion of neurons29 (Supplementary Fig. 9). Among 24 brain cell types from mouse brain, significant enrichment was found for medium spiny neurons and pyramidal neurons from hippocampal CA1 (Supplementary Fig. 10). Both medium spiny and pyramidal neurons are linked to feeding behaviors including food motivation and reward30,31 (Supplementary Results). Using PrediXcan (Supplementary Methods), 36 genes were predicted to be differentially expressed in GTEx tissues or blood (Supplementary Table 13) with the expression of MGMT predicted to be downregulated in the caudate. We cautiously note that these results represent the first indications of specific pathways, tissues, and cell types that may mediate genetic risk for anorexia nervosa.
Because low BMI is pathognomonic of anorexia nervosa, we investigated the extent to which genetic variants associated with BMI accounted for genetic correlations with metabolic and anthropometric traits. First, covarying for the genetic associations of BMI (Methods) led to a mild but statistically non-significant attenuation of the SNP-rg between anorexia nervosa and fasting insulin, leptin, insulin resistance, type 2 diabetes, and HDL cholesterol (Supplementary Tables 14-15), suggesting that anorexia nervosa shares genetic variation with these metabolic phenotypes that may be independent of BMI. Second, we investigated bidirectional causality using generalized summary data-based Mendelian randomization18. GSMR analyses indicate a significant bidirectional causal relationship such that anorexia nervosa risk-increasing alleles may increase risk for low BMI and BMI-lowering alleles may increase the risk of anorexia nervosa (Supplementary Table 16). It is important to note that having only eight genome-wide significant loci for anorexia nervosa render this analysis marginally powered in the direction of anorexia nervosa to BMI, although this analysis is well powered in the direction of BMI to anorexia nervosa.
Replication is challenging with GWAS of low prevalence conditions like anorexia nervosa, as replication samples must be sufficiently powered to detect the initial findings. We included all available samples in our analysis to maximize chances of reaching the GWAS inflection point, after which there might be a linear increase in “hits”32. The PRS leave-one-out analyses provide evidence of replication by demonstrating a higher burden of anorexia nervosa common risk variants in cases, compared with controls, across all the cohorts (Supplementary Fig. 16).
In conclusion, we report multiple genetic loci alongside promising clinical and functional analyses and enrichments. The increased sample size in the present GWAS has allowed us to characterize more fully the metabolic contribution to anorexia nervosa than our previous report10 by revealing significant rg with metabolism related phenotypes including glycemic and anthropometric traits and by demonstrating that the effect is robust to correction for the effects of common variants significantly associated with BMI. Low BMI has traditionally been viewed as a consequence of the psychological features of anorexia nervosa (i.e., drive for thinness and body dissatisfaction). This perspective has failed to yield interventions that reliably lead to sustained weight gain and psychological recovery7. Fundamental metabolic dysregulation may contribute to the exceptional difficulty that individuals with anorexia nervosa have in maintaining a healthy BMI (even after therapeutic renourishment). Our results encourage consideration of both metabolic and psychological drivers of anorexia nervosa when exploring new avenues for treating this frequently lethal illness.
Methods
Samples and study design.
Thirty-three datasets with 16,992 anorexia nervosa cases and 55,525 controls were included in the primary GWAS. We included individuals from the Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC-ED) Freeze 110; newly collected samples from the Anorexia Nervosa Genetics Initiative (ANGI)8,9; archived samples from the Genetic Consortium for Anorexia Nervosa (GCAN)/Wellcome Trust Case Control Consortium-3 (WTCCC3)13; anorexia nervosa samples from UK Biobank14; and additional controls from Poland. Case definitions established a lifetime diagnosis of anorexia nervosa via hospital or register records, structured clinical interviews, or on-line questionnaires based on standardized criteria (DSM-III-R, DSM-IV, ICD-8, ICD-9, or ICD-10), whereas in the UK Biobank cases self-reported a diagnosis of anorexia nervosa. Controls were carefully matched for ancestry, and some, but not all control cohorts were screened for lifetime eating and/or some or all psychiatric disorders. Given the relative rarity of anorexia nervosa, large unscreened control cohorts were deemed appropriate for inclusion33.
The cohorts are detailed in the Supplement. Ethical approvals and consent forms were reviewed and archived for all participating cohorts (see Supplementary Methods ANGI-DK for Danish methods). Summary details about ascertainment (Supplementary Table 2), the genotyping platforms used (Supplementary Table 3), and genotype availability (Supplementary Table 4) can be accessed in the Supplement.
Statistical analysis.
Data processing and analysis were done on the Lisa Compute Cluster hosted by SURFsara (http://www.surfsara.nl) and the GenomeDK high-performance computing cluster (http://genome.au.dk).
Meta-analysis of genome-wide association data.
Quality control (QC), imputation, GWAS, and meta-analysis followed the standardized pipeline of the PGC, Ricopili (Rapid Imputation Consortium Pipeline). Ricopili versions used were 2017_Oct_11.002 and 2017_Nov_30.003. QC included SNP and sample QC, population stratification and ancestry outliers, and familial and cryptic relatedness. Further information about the Ricopili pipeline is available from the website (https://sites.google.com/a/broadinstitute.org/ricopili) and GitHub repository (https://github.com/Nealelab/ricopili/tree/master/rp_bin). Further details of the QC procedures can be found in the Supplementary Methods.
Imputation.
Imputation of SNPs and insertions-deletions was based on the 1000 Genomes Phase 3 (http://www.internationalgenome.org) data34.
GWAS.
GWASs were conducted separately for each cohort using imputed variant dosages and an additive model. Covariates nominally associated with the phenotype in univariate analysis (P < 0.05) and five ancestry PCs were included in GWAS (Supplementary Table 18). These analyses used the tests and methods programmed in the Ricopili pipeline. Genomic inflation factors (λ) of the final datasets indicated no evidence of inflation of the test statistics due to population stratification or other sources (Supplementary Table 1). The 33 cohorts were meta-analyzed with the Ricopili pipeline which uses an inverse-variance weighted fixed-effect model. We filtered our GWAS results with minor allele frequency (MAF) ≥ 0.01 and INFO score ≥ 0.70 (indicating “high-quality”).
Analysis of chrX.
Several cohorts in the primary GWAS did not have X chromosome variant data, specifically, some GCAN-based cohorts (fre1, ukd1, usa1, gns2) and were excluded. Imputation was performed separately from the autosome35. ChrX variants in the pseudoautosomal regions were excluded prior to imputation. SNPs exceeding MAF and INFO score thresholds of 0.01 and 0.70 were retained and analysis was performed with PLINK v1.9 (https://www.cog-genomics.org/plink2) and Ricopili.
Female-only GWAS.
A supplementary GWAS analysis was conducted on females only to determine the similarity of the results to the primary GWAS analysis which included both females and males. The cohorts that did not have chrX variants to verify sex could not be included (fre1, ukd1, usa1, gns2).
Distance- and LD-based clumping.
The GWAS results implicate genomic regions (“loci”). To define a locus, (1) SNPs that met the genome-wide significant threshold of P < 5 × 10−8 were identified; (2) clumping was used to convert significant SNPs to regions. The SNP with the smallest P value in a genomic window was kept as the index SNP and SNPs in high linkage disequilibrium (LD) with the index SNP defined the left and right end of the region (SNPs with P < 0.0001 and r2 > 0.1 within 3 Mb windows); (3) partially or wholly overlapping clumps within 50 Kb were identified and merged into one region; (4) only loci with additional evidence of association from variants in high LD as depicted by regional plots were retained; further, forest plots needed to confirm the associations based on the majority of cohorts; and (5) conditional analyses were conducted to identify SNPs with associations independent of the top SNP within the genomic chunk of interest.
Annotation.
Genome-wide significant loci were annotated with RegionAnnotator (https://github.com/ivankosmos/RegionAnnotator) to identify known protein-coding genes within loci (Supplementary Table 6).
Conditional and joint analysis.
Conditional and joint analysis was conducted using GCTA-COJO15. GCTA-COJO investigates every locus with a joint combination of independent markers via a genome-wide SNP selection procedure. It takes into account the LD correlations between SNPs and runs a conditional and joint analysis on the basis of conditional P values. After a model optimizing process, the joint effects of all selected SNPs are calculated. The largest subsample from our GWAS (sedk) was used to approximate the underlying LD structure of the investigated lead SNPs. The conditional regression was performed in a stepwise manner using the GCTA software36. We analyzed SNPs that had a P < 5 × 10−8 (Supplementary Table 5).
Multi-trait-based conditional and joint analysis.
To separate marginal effects from conditional effects (i.e., the effect of a risk factor on an outcome controlling for the effect of another risk factor), we performed a multi-trait-based conditional and joint analysis (GCTA-mtCOJO)18 using an extension of the GCTA software36 (Supplementary Table 8). This method uses summary-level data to perform the conditional analysis. We conditioned the results of our anorexia nervosa GWAS on GWAS results for education years37, type 2 diabetes38, HDL cholesterol39, BMI (Hübel, Gaspar, Coleman, Hanscombe, Purves…Breen, unpublished report), schizophrenia40, and neuroticism41. We again used the individual-level genotype data from our largest cohort (sedk) to approximate the underlying LD structure. As a first step, the method performs a generalized summary data-based Mendelian randomization (GSMR) analysis to test for causal association between the outcome (i.e., anorexia nervosa) and the risk factor (e.g., schizophrenia). We removed potentially pleiotropic SNPs from this analysis by the heterogeneity in dependent instruments (HEIDI) outlier method18. Pleiotropy is the phenomenon when a single locus directly affects several phenotypes. The power of the HEIDI-outlier method is dependent on sample size of the GWAS. Pleiotropic SNPs are defined as the SNPs that show an effect on the outcome that significantly diverges from that expected under a causal model. Second, the GCTA-mtCOJO method calculates the genetic correlation between the exposure and the outcome using linkage disequilibrium score regression (LDSC) to adjust for genetic overlap19,20. It also uses the intercept of the bivariate LDSC to account for potential sample overlap19,20. As a result, GCTA-mtCOJO calculates conditional betas, conditional standard errors, and conditional P values. Subsequently, we clumped the conditional GWAS results using the standard PLINK v1.942 algorithm (SNPs with P < 0.0001 and r2 > 0.1 within 3 Mb windows) to investigate if any of the genome-wide significant loci showed dependency on genetic variation associated with other phenotypes. As stated in Zhu et al.18, the GCTA-mtCOJO analysis requires the estimates of bxy of the covariate risk factors on the target risk factor and disease, rg of the covariate risk factors, heritability (h2snp) for the covariate risk factors, and the sampling covariance between SNP effects estimated from potentially overlapping samples.
eQTL and Hi-C interactions.
Although GWAS findings are informative genome-wide, identifying strong hypotheses about their connections to specific genes is not straightforward. The lack of direct connections to genes constrains subsequent experimental modeling and efforts to develop improved therapeutics. Genomic location is often used to connect significant SNPs to genes, but this is problematic because GWAS loci usually contain many correlated and highly significant SNP associations over hundreds of Kb. Moreover, the three-dimensional (3D) arrangement of chromosomes in cell nuclei enables regulatory interactions between genomic regions located far apart43. Chromosome conformation capture methods like Hi-C enable identification of 3D interactions in vivo44,45 and can clarify GWAS findings. For example, an intergenic region associated with multiple cancers was shown to be an enhancer for MYC via a long-range chromatin loop46,47, and intronic FTO variants are robustly associated with body mass but influence expression of distal genes via long-range interactions48. The Nature paper of Won et al49 used Hi-C to assess the 3D chromatin interactome in fetal brain, and asserted connections of some schizophrenia associations to specific genes.
To gain further understanding of 3D chromatin organization of the brain and to evaluate disease relevance, we applied “easy Hi-C”50 to postmortem samples (N = 3 adult temporal cortex). Library quality and yield from eHi-C are comparable to conventional Hi-C but requires much less starting material. Please refer to the following pre-print for details on methodology, data processing, quality control and statistical models used for these analyses51. We generated sufficient reads to enable a kilobase resolution map of the chromatin interactome from adult human brain. To our knowledge, these are the deepest Hi-C data on any human tissue (excluding cell lines) as they generated 22.5X as many cis-contacts as for the next largest datasets (DLPFC and hippocampus). We generated tissue RNA-seq, total-stranded RNA-seq, ChIP-seq (H3K27ac, H3K4me3, and CTCF), and open chromatin data (ATAC-seq) for adult brain to help interpret the eHi-C results. We also integrated brain expression and eQTL data from GTEx to aid these analyses. The Hi-C analysis is unbiased in that all chromatin interactions that pass a confidence threshold are considered when evaluating the associations between SNPs and genes (i.e., it is not a capture experiment where only “candidate” SNP-to-gene associations are evaluated).
Similar to the work by Won et al.49, we used Hi-C data generated from human adult brain to identify genes implicated by three-dimensional functional interactomics (Supplementary Figs. 5 a-h). These Hi-C data (N = 3, anterior temporal cortex) contain more than 103K high-confidence, regulatory chromatin interactions51. These interactions capture the physical proximity of two regions of the genome in brain nuclei (“anchors”, 10 Kb resolution) although they are separated by 20 Kb to 2 Mb in genomic distance. We focused on the regulatory subset of E-P or P-P (E = enhancer, P = promoter) chromatin interactions (with P defined by location of an open chromatin anchor near the transcription start site of an adult brain-expressed transcript and E defined by overlap with open chromatin in adult brain plus either H3K27ac or H3K4me3 histone marks). The presence of a regulatory chromatin interaction from a GWAS locus to a gene provides a strong hypothesis about SNP-to-gene regulatory functional interactions.
SNP-based heritability estimation.
LDSC software (https://github.com/bulik/ldsc) and method were used to estimate SNP-based heritabilities for each cohort and overall19,20. We used precomputed LD scores based on the 1000 Genomes Project European ancestry samples34 directly downloaded from https://github.com/bulik/ldsc. The liability scale estimate assumed a population prevalence of 0.9%-4% for anorexia nervosa2,3.
Within-trait prediction:
polygenic risk scoring. Polygenic leave-one-dataset-out analysis, using PRSice v2.1.352, was conducted in the first instance to identify any extreme outlying datasets. In addition, it enabled the evaluation of the association between anorexia nervosa polygenic risk score (PRS) and anorexia nervosa risk in an independent cohort as a means of replication of the GWAS results. We derived a PRS for anorexia nervosa from the meta-analysis of all datasets except for the target cohort, then applied the PRS to the target cohort to predict affected status (Supplementary Fig. 16). Logistic regression was performed, including as covariates the first five ancestry components and any other PCs significantly associated with the phenotype in the target cohort, and the target cohort was split into deciles based on anorexia nervosa PRS, with decile 1 comprised of those with the lowest anorexia nervosa PRS serving as the referent.
Anorexia nervosa subtype analysis.
PRS analyses were conducted with anorexia nervosa subgroups to investigate prediction of case status across the subtypes. For this, we split the anorexia nervosa cases to two groups based on whether binge eating was present. First, GWAS meta-analyses were conducted for (a) anorexia nervosa with binge eating vs controls (2,381 cases and 10,249 controls; k = 3 datasets: aunz, chop, usa2) and (b) anorexia nervosa with no binge eating vs controls (2,262 cases and 10,254 controls; k = 3 datasets: aunz, chop, usa2). Controls were randomly split between analyses to maintain independence (Supplementary Fig. 6). Genetic correlation analysis using LDSC19,20 was conducted to examine the potential genetic overlap of the two anorexia nervosa subtypes (Supplementary Table 9). Second, using PRSice52, we calculated PRS for each anorexia nervosa subtype separately in the three target cohorts for which anorexia nervosa subtype data were available. Finally, mean PRS scores were estimated for each subtype by cohort after accounting for covariates in R. Subtype phenotyping is described in the Supplementary Methods.
Males.
In order to assess whether sex-specific differences in anorexia nervosa genetic risk load exist, we calculated PRS, using PRSice52, from a GWAS meta-analysis performed on females only (14,898 cases and 27,545 controls) and applied it to a male-only target cohort (447 cases and 20,347 controls) to predict affected status.
Cross-trait analysis: genetic correlations.
Common variant-based genetic correlation (SNP-rg) measures the extent to which two traits or disorders share common genetic variation. SNP-rg between anorexia nervosa and 447 traits (422 from an internally curated dataset and 25 from LDHub53) were tested using GWAS summary statistics via an analytical extension of LDSC19,20. The sources of the summary statistics files (PMID, DOI, or unpublished results) used in the LDSC are provided in Supplementary Table 10. When there were multiple summary statistics files available for a trait, significant SNP-rg reported in the main text were chosen based on the largest sample size and/or matching ancestry with our sample (i.e., European ancestry).
Genetic correlations with anorexia nervosa corrected for BMI were carried out to investigate whether the observed genetic correlations between anorexia nervosa and metabolic phenotypes were attributable to BMI or partially independent. We used GCTA-mtCOJO18 to perform a GWAS analysis for anorexia nervosa conditioning on BMI using BMI summary data from our UK Biobank analysis (described in the next section) to derive anorexia nervosa GWAS summary statistics corrected for the common variants genetic component of BMI (Supplementary Tables 14 and 15).
GWAS of related traits in UK Biobank.
Several GWAS analyses were carried out for traits in UK Biobank to allow us to investigate body composition genetics in healthy individuals without a psychiatric disorder, a weight-altering disorder, or who were taking weight-altering medication. We also used UK Biobank to carry out GWAS of physical activity level, anxiety, and neuroticism. For details see the Supplementary Methods.
Generalized summary data-based Mendelian randomization (GSMR).
We performed two bidirectional GSMR analyses18 to test for the causal association between first, BMI and anorexia nervosa, and second, Type 2 diabetes and anorexia nervosa, using an extension of the GCTA software36 (Supplementary Table 16). We used the individual-level genotype data from our largest cohort (sedk) to approximate the underlying LD structure. We removed potentially pleiotropic SNPs from this analysis by the HEIDI outlier method18. Pleiotropic SNPs are defined as the SNPs which show an effect on the outcome that significantly diverges from the one expected under a causal model. The method uses the intercept of the bivariate LD score regression to account for potential sample overlap19,20. As a rule of thumb GSMR requires GWAS to have at least ten genome-wide significant hits. We lowered the threshold for this requirement to eight SNPs in our analyses of anorexia nervosa as an exposure and BMI or Type 2 diabetes as an outcome. Results, therefore, should be interpreted cautiously. We, furthermore, investigated bidirectional conditional effects between BMI or Type 2 diabetes and anorexia nervosa. We used GCTA-mtCOJO to perform a GWAS analysis for anorexia nervosa conditioning on (1) BMI using summary data from our UK Biobank analysis and (2) Type 2 diabetes using summary data38. Our anorexia nervosa GWAS and the BMI and Type 2 diabetes GWASs are based on independent samples. For BMI, we also re-ran the GSMR analysis using the BMI-adjusted anorexia nervosa GWAS summary data from the GCTA-mtCOJO analysis.
Gene-wise analysis.
MAGMA v1.0654 was used to perform a gene-wise test of association with anorexia nervosa based on GWAS summary statistics. MAGMA generates gene-based P values by combining SNP-based P values within a gene while accounting for LD. In order to include regulatory regions, SNPs are mapped to genes within a 35 kb upstream and 10 kb downstream window, and the gene P value is obtained using the “multi=snp-wise” model, which aggregates mean and top SNP association models. We tested 19,846 ENSEMBL genes, including the X chromosome (Supplementary Table 11). As reference panel for the underlying LD structure we used 1000 Genomes European data phase 334.
Pathway analysis.
MAGMA v1.0654 was used to perform a competitive pathway analysis, testing whether genes associated with anorexia nervosa were more enriched in a given pathway than all other pathways. The analysis included chrX. Biological pathways were defined using gene ontology pathways and canonical pathways from MSigDB v6.155, and psychiatric pathways mined from the literature. A total 7,268 pathways were tested (Supplementary Table 12).
Partitioned heritability.
Partitioned heritability was investigated using stratified LDSC26 which estimates the per-SNP contribution to overall SNP-heritability (SNP-h2) across various functional annotation categories of the genome (Supplementary Fig. 7). It accounts for linked markers and uses a ‘full baseline model’ of 24 annotations that are not specific to any cell type. We excluded the MHC region in our analysis. SNP-h2 can be partitioned in two different ways: a non-cell type-specific and a cell type-specific manner. Partitioned heritability analysis was used to test for cell type-specific enrichment in the GWAS of anorexia nervosa among 10 cell type groups; adrenal and pancreas, cardiovascular, central nervous system (CNS), connective and bone, gastrointestinal, immune and hematopoietic, kidney, liver, skeletal muscle, and other tissue, which includes adipose tissue (Supplementary Fig. 8).
Gene expression.
We conducted a series of gene expression analyses as detailed in the Supplementary Methods.
Reporting summary
Further information on research design is available in the Life Science Reporting Summary linked to this article.
Data availability
The Psychiatric Genomics Consortium’s (PGC) policy is to make genome-wide summary results public. Genome-wide summary statistics for the meta-analysis are freely downloadable from PGCs download website (http://www.med.unc.edu/pgc/results-and-downloads). Individual-level data are deposited in dbGaP (http://www.ncbi.nlm.nih.gov/gap) for ANGI-ANZ/SE/US (accession number phs001541.v1.p1) and CHOP/PFCG (accession number phs000679.v1.p1). ANGI-DK individual-level data are not available in dbGaP owing to Danish laws, but are available via collaboration with PIs. GCAN/WTCCC3 individual-level data are deposited in EGA (https://www.ebi.ac.uk/ega) (accession number EGAS00001000913) with the exception of Netherlands and US/Canada, which are available via collaboration with PIs. UK Biobank individual-level data can be applied for on the UK Biobank website (http://www.ukbiobank.ac.uk/register-apply).
Supplementary Material
Acknowledgements
Grant support for ANGI, the PGC-ED, and its component groups is shown in Supplementary Table 17. We thank all study volunteers, study coordinators, and research staff who enabled this study. ANGI: The Anorexia Nervosa Genetics Initiative was an initiative of the Klarman Family Foundation. Additional support was offered by the National Institute of Mental Health. We acknowledge support from the North Carolina Translational and Clinical Sciences Institute (NC TraCS), the Carolina Data Warehouse, and the Foundation of Hope, Raleigh, North Carolina. PGC: We are deeply indebted to the investigators who +comprise the PGC, and to the hundreds of thousands of individuals who have shared their life experiences with PGC investigators and the contributing studies. We are grateful to the Children’s Hospital of Philadelphia (CHOP), the Price Foundation Collaborative Group (PFCG), Genetic Consortium for Anorexia Nervosa (GCAN), Wellcome Trust Case-Control Consortium-3 (WTCCC-3), the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), the QSkin Sun and Health Study, Riksat (Swedish National Quality Register for Eating Disorders), the Stockholm Center for Eating Disorders (SCA), LifeGene, the UK Biobank, and all PGC-ED members for their support in providing individual samples used in this study. We thank SURFsara (http://www.surf.nl) for support in using the Lisa Compute Cluster. We thank M. Lam for Ricopili consultation. This study also represents independent research partly funded by the English National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the English Department of Health and Social Care. High performance computing facilities were funded with capital equipment grants from the GSTT Charity (TR130505) and Maudsley Charity (980). Research reported in this publication was supported by the National Institute of Mental Health of the US National Institutes of Health under Award Number U01MH109514. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institutes of Health.
Footnotes
Competing interests
The authors report the following potential competing interests. O.A.A. received a speaker’s honorarium from Lundbeck. G.B. received grant funding and consultancy fees in preclinical genetics from Eli Lilly, consultancy fees from Otsuka and has received honoraria from Illumina. C.M.B. is a grant recipient from Shire Pharmaceuticals and served on Shire Scientific Advisory Board; she receives author royalties from Pearson. D.D. served as a speaker and on advisory boards, and has received consultancy fees for participation in research from various pharmaceutical industry companies including: AstraZeneca, Boehringer, Bristol Myers Squibb, Eli Lilly, Genesis Pharma, GlaxoSmithKline, Janssen, Lundbeck, Organon, Sanofi, UniPharma, and Wyeth; he has received unrestricted grants from Lilly and AstraZeneca as director of the Sleep Research Unit of Eginition Hospital (National and Kapodistrian University of Athens, Greece). J.I.H. has received grant support from Shire and Sunovion, and has received consulting fees from DiaMentis, Shire, and Sunovion. A.S.K. is a member of the Shire Canadian BED Advisory Board and is on the steering committee for the Shire B/educated Educational Symposium: June 15-16, 2018. J.L.K. served as an unpaid member of the scientific advisory board of AssurexHealth Inc. M.L. declares that, over the past 36 months, he has received lecture honoraria from Lundbeck and served as scientific consultant for EPID Research Oy. No other equity ownership, profit-sharing agreements, royalties, or patent. P.F.S. is on the Lundbeck advisory committee and is a Lundbeck grant recipient; he has served on the scientific advisory board for Pfizer, has received a consultation fee from Element Genomics, and a speaker reimbursement fee from Roche. J.T. has received an honorarium for participation in an EAP meeting and has received royalties from several books from Routledge, Wiley, and Oxford University press. T.W. has acted as a lecturer and scientific advisor to H. Lundbeck A/S. All other authors have no conflicts of interest to disclose.
URLs. GCTA, http://cnsgenomics.com/software/gcta; GSMR, http://cnsgenomics.com/software/gsmr; LDSC, https://github.com/bulik/ldsc; MAGMA, http://ctg.cncr.nl/software/magma.
References
- 1.Schaumberg K et al. The science behind the Academy for Eating Disorders' nine truths about eating disorders. Eur. Eat. Disord. Rev 25, 432–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keski-Rahkonen A & Mustelin L Epidemiology of eating disorders in Europe: prevalence, incidence, comorbidity, course, consequences, and risk factors. Curr. Opin. Psychiatry 29, 340–345 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hudson JI, Hiripi E, Pope HG & Kessler RC The prevalence and correlates of eating disorders in the National Comorbidity Survey Replication. Biol. Psychiatry 61, 348–358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Micali N et al. Lifetime and 12-month prevalence of eating disorders amongst women in mid-life: a population-based study of diagnoses and risk factors. BMC Med. 15, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz Z, Hardaway JA & Bulik CM Genetics and epigenetics of eating disorders. Adv. Genomics Genet 5, 131–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arcelus J, Mitchell AJ, Wales J & Nielsen S Mortality rates in patients with anorexia nervosa and other eating disorders: a meta-analysis of 36 studies. Arch. Gen. Psychiatry 68, 724–731 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Watson H & Bulik C Update on the treatment of anorexia nervosa: review of clinical trials, practice guidelines and emerging interventions. Psychol. Med 43, 2477–2500 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Kirk KM et al. The Anorexia Nervosa Genetics Initiative: study description and sample characteristics of the Australian and New Zealand arm. Aust. N. Z. J. Psychiatry 51, 583–594 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Thornton L, Munn-Chernoff M, Baker J, Juréus A & et al. The Anorexia Nervosa Genetics Initiative (ANGI): Overview and methods. Contemp. Clin. Trials 74, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan L et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 173, 850–858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickrell JK et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet 48, 709–717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin J, Taylor MJ & Lichtenstein P Assessing the evidence for shared genetic risks across psychiatric disorders and traits. Psychol. Med 48, 1759–1774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boraska V et al. A genome-wide association study of anorexia nervosa. Mol. Psychiatry 19, 1085–1094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun 9, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cederlöf M et al. Etiological overlap between obsessive-compulsive disorder and anorexia nervosa: a longitudinal cohort, multigenerational family and twin study. World Psychiatry 14, 333–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye WH, Bulik CM, Thornton L, Barbarich N & Masters K Comorbidity of anxiety disorders with anorexia and bulimia nervosa. Am. J. Psychiatry 161, 2215–2221 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Dalle Grave R, Calugi S & Marchesini G Compulsive exercise to control shape or weight in eating disorders: prevalence, associated features, and treatment outcome. Compr. Psychiatry 49, 346–352 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Savage JE et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho EV, Klenotich SJ, McMurray MS & Dulawa SC Activity-based anorexia alters the expression of BDNF transcripts in the mesocorticolimbic reward circuit. PLoS One 11, e0166756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindblad-Toh K et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bello V et al. The dystroglycan: nestled in an adhesome during embryonic development. Dev. Biol 401, 132–142 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Azevedo FAC et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol 513, 532–541 (2009). [DOI] [PubMed] [Google Scholar]
- 30.O'Connor EC et al. Accumbal D1R neurons projecting to lateral hypothalamus authorize feeding. Neuron 88, 553–564 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Zhang X, Muralidhar S, LeBlanc SA & Tonegawa S Basolateral to central amygdala neural circuits for appetitive behaviors. Neuron 93, 1464–1479.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levinson DF et al. Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol. Psychiatry 76, 510–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moskvina V, Holmans P, Schmidt KM & Craddock N Design of case-controls studies with unscreened controls. Ann. Hum. Genet 69, 566–576 (2005). [DOI] [PubMed] [Google Scholar]
- 34.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang D et al. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One 9, e113684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris AP et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet 44, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hübel C et al. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am. J. Med. Genet. B Neuropsychiatr. Genet in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dekker J Mapping the 3D genome: aiming for consilience. Nat. Rev. Mol. Cell Biol 17, 741–742 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Dekker J Gene regulation in the third dimension. Science 319, 1793–1794 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ethier SD, Miura H & Dostie J Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta 1819, 401–410 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Pomerantz MM et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet 41, 882–884 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright JB, Brown SJ & Cole MD Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell Biol 30, 1411–1420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smemo S et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Won H et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu L, Liu X, Peng J, Li Y & Jin F Easy Hi-C: A simple efficient protocol for 3D genome mapping in small cell populations. bioRxiv, 245688 (2018). [Google Scholar]
- 51.Giusti-Rodriguez PM & Sullivan PF Schizophrenia and a high-resolution map of the three-dimensional chromatin interactome of adult and fetal cortex. bioRxiv, 406330 (2018). [Google Scholar]
- 52.Euesden J, Lewis CM & O'Reilly PF PRSice: Polygenic Risk Score software. Bioinformatics 31, 1466–1468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Psychiatric Genomics Consortium’s (PGC) policy is to make genome-wide summary results public. Genome-wide summary statistics for the meta-analysis are freely downloadable from PGCs download website (http://www.med.unc.edu/pgc/results-and-downloads). Individual-level data are deposited in dbGaP (http://www.ncbi.nlm.nih.gov/gap) for ANGI-ANZ/SE/US (accession number phs001541.v1.p1) and CHOP/PFCG (accession number phs000679.v1.p1). ANGI-DK individual-level data are not available in dbGaP owing to Danish laws, but are available via collaboration with PIs. GCAN/WTCCC3 individual-level data are deposited in EGA (https://www.ebi.ac.uk/ega) (accession number EGAS00001000913) with the exception of Netherlands and US/Canada, which are available via collaboration with PIs. UK Biobank individual-level data can be applied for on the UK Biobank website (http://www.ukbiobank.ac.uk/register-apply).