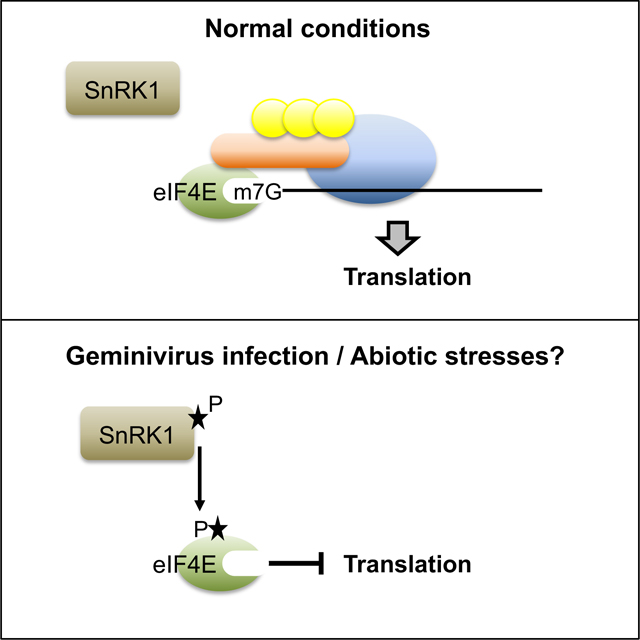
Graphical Abstract
In unstressed conditions, the cap-binding protein, eIF4E, binds the 5’ cap structure (m7G) on mRNA and recruits other translation initiation factors to bring the ribosome to the mRNA. Bruns et al show that, in plants, phosphorylation of eIF4E by the master metabolic regulatory protein, sucrose non-fermenting-related kinase 1 (SnRK1), reduces translation globally. This negative regulation of translation inhibits geminivirus infection and is speculated to be a response to various abiotic stresses.
The process of protein synthesis is well-conserved among eukaryotes [1]. However, regulation of translation differs between plants and animals [2]. One means of translational control involves eukaryotic initiation factor 4E (eIF4E), which binds the 5’ cap structure of mRNA, and its partner eIF4G to recruit other initiation factors and ultimately the ribosome to the mRNA. Plants also contain an isoform, eIFiso4E, which pairs with eIFiso4G. The different biological roles of eIF4E/4G vs eIFiso4E/iso4G are unclear, but Arabidopsis thaliana (At) plants lacking eIFiso4E or eIFiso4G show developmental abnormalities [3]. In mammals, under low nutrient or various stress conditions, eIF4E-binding protein (4E-BP) inhibits translation initiation by preventing eIF4E from binding eIF4G and thus from recruiting the other factors to the mRNA (Figure 1) [4]. Excessive translation initiation due to aberrant lack of inhibition of eIF4E by 4E-BP can lead to many types of cancer and metabolic disorders [5]. While in plants and animals eIF4E is structurally similar [6], plants lack an ortholog to the mammalian 4E-BP [7]. However, Patrick et al. [8] identified a different eIF4E binding protein (CBE1) in Arabidopsis that negatively regulates accumulation of mRNAs that control the cell cycle and mitosis, via a mechanism that is currently still unknown.
Figure 1. Negative regulation of translation in mammals and plants.
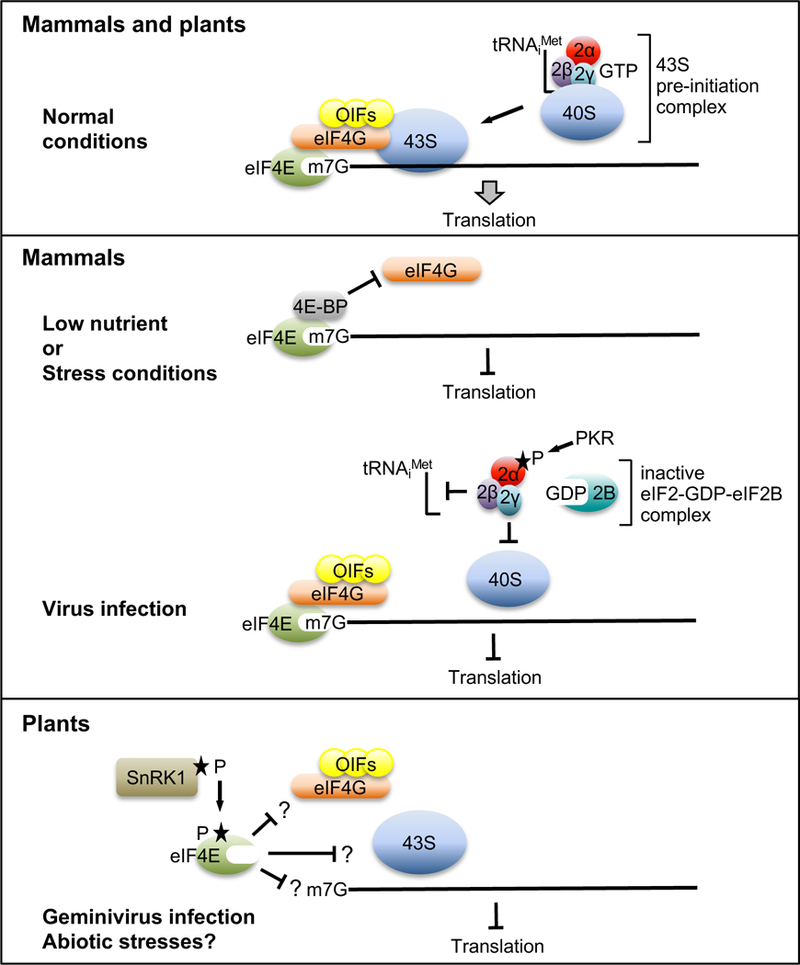
Top: In normal, unstressed conditions, cap-binding protein eIF4E simultaneously binds the m7GpppN cap structure on the mRNA and scaffolding protein eIF4G, which binds other initiation factors (OIFs) to recruit the 43S complex consisting of the 40S ribosomal subunit bound to the ternary complex of eIF2•GTP•tRNAiMet. Middle: In mammals, under low nutrient or stress conditions, eIF4E binding protein (4E-BP) binds eIF4E to prevent eIF4G binding, thus blocking translation initiation. In response to many virus infections, PKR phosphorylates eIF2α, preventing exchange of GTP with GDP in eIF2B, which, in turn, prevents formation of the ternary complex. Bottom: In plants, Bruns et al provide evidence that phosphorylation of eIF4E by SnRK1 attenuates translation initiation, but the interactions disrupted by this modification are unknown (question marks).
In vertebrates, translation is often attenuated greatly during virus infection. This inhibits virus gene expression because viruses depend on the host’s translation machinery. During virus infection, protein kinase R (PKR) becomes activated by binding to double-stranded RNA, which is generated during RNA virus replication, and it then phosphorylates eIF2α [9]. This prevents the exchange of GDP for GTP required for regeneration of the ternary complex eIF2•GTP•tRNAiMet, necessary for initiation of translation (Figure 1). Plants lack a PKR homolog, and do not appear to use this mechanism as an antiviral response [10]. Given that translation attenuation is important for rapid response to changes in the environment or during virus infection, it is likely that such regulation takes place in plants by mechanisms that have yet to be discovered.
Indeed, a new mechanism of translation attenuation, apparently unique to plants, has been reported by the Bisaro lab in a recent issue of The FEBS Journal [11]. They provide evidence for translation attenuation mediated by phosphorylation of eIF4E and eIFiso4E by sucrose non-fermenting (SNF)-related kinase 1 (SnRK1). SnRK1 is an ortholog of yeast SNF1 and mammalian AMP-activated kinase (AMPK) that functions as a master regulator of energy homeostasis [12]. During biotic or abiotic stress, SNF1/AMPK1/SnRK1 activates energy-generating catabolic pathways and inhibits energy-consuming anabolic pathways, at the transcriptional level and post-translationally, by phosphorylation of key enzymes [13]. Here, Bruns et al [11] identified two consensus SnRK1 target sites in Arabidopsis eIF4E and eIFiso4E that are absent in mammalian eIF4E. The purified SnRK1 kinase domain (SnRK1-KD), but not a catalytically inactive mutant (SnRK1-KDKR), phosphorylates the predicted target sites in At eIFiso4E. Consistent with this, SnRK1 interacts with At eIF4E and At eIFiso4E in a yeast two-hybrid assay and in bimolecular fluorescent complementation (BiFC) assays in plant cells. Furthermore, SnRK1 phosphorylates At eIFiso4E when both are transiently co-expressed in plant cells.
To determine the effect of SnRK1-mediated phosphorylation on At eIF4E/iso4E activity, the authors expressed SnRK1 under a copper inducible promoter in a yeast strain that lacks endogenous eIF4E (cdc33Δ) but expresses At eIF4E, At eIFiso4E or human eIF4E (Hs eIF4E). Interestingly, induction of SnRK1 increased doubling time of yeast that depended on At eIF4E or At eIFiso4E but not Hs eIF4E. This reduced growth of yeast is likely due to phosphorylation of At eIF4E/iso4E by SnRK1 because Hs eIF4E lacks the SnRK1 phosphorylation site present on plant eIF4E/iso4E. In the SnRK1-growth-inhibited yeast, there was a substantial reduction in translation as measured by a 50% reduction in the ratio of polysomes (ribosome-studded, actively translating mRNAs) to non-polysomes (non-translating ribosomes). However, as predicted, no such SnRK1-mediated reduction in translation was observed in yeast complemented with Hs eIF4E or non-phosphorylatable mutant At eIF4E‐T67V/T91A (called At eIF4E-VA). A similar reduction in polysome/non-polysome ratio was also observed in Nicotiana benthamiana plants engineered to overexpress SnRK1. Finally, transient overexpression of At eIF4E or At eIF4E-VA in SnRK1-overexpressing plants restored the polysome/non-polysome ratio to that found in wild type plants, presumably because the amount of overexpressed eIF4E was sufficient to allow full translation. As expected, overexpression of the phosphomimic mutant, At eIF4E-T67D, had no effect on the reduced polysome/non-polysome ratio, because it behaves as a phosphorylated (inactivated) eIF4E under all conditions.
The results described in Bruns et al. [11] provide compelling evidence for translation attenuation due to SnRK1-mediated phosphorylation of eIF4E and eIFiso4E. However, many questions remain. How does phosphorylation inhibit eIF4E/iso4E activity (Figure 1)? Does it affect binding to eIF4G or to the 5’ cap on the mRNA or other components of the translation machinery? Does SnRK1-mediated phosphorylation of eIF4E functionally replace the role that 4E-BP plays in mammals? If it does, then the phosphorylation state of eIF4E should increase under nutrient stress/low energy conditions in plants. Does SnRK1-mediated translation inhibition play an antiviral role? Previously, Bisaro’s group showed that SnRK1 overexpression increases resistance to a geminivirus, while SnRK1 knockdown makes plants more susceptible to the virus [14]. Therefore, translation attenuation would inhibit geminivirus gene expression because its mRNAs, generated by host transcription machinery from the viral DNA, are translated by normal cellular mechanisms, unlike many RNA viruses that are translated cap-independently [15] and thus may not be subject to eIF4E regulation. How is translation of individual mRNAs affected by eIF4E phosphorylation? It’s unlikely that all are affected equally. One would predict that mRNAs needed during low energy conditions would translate more efficiently than those that are not. Ribosome profiling [16] could be one approach to answer this question.
In summary, Bruns et al. have provided one answer to the complex question of how plants negatively control translation, identifying a novel regulatory mechanism that is unknown in mammals. How SnRK1-mediated eIF4E phosphorylation is regulated, and the biological pathways controlled by this mechanism remain to be determined.
Acknowledgement.
The authors thank Karen Browning for advice. Research on viral RNA-eIF4E interactions in WAM’s laboratory has been supported by the US National Institutes of Health (R01 GM067104). Research on host-virus interactions in Dinesh-Kumar laboratory is supported by US National Science Foundation (IOS-1339185) and National Institute of Health (R01 GM132582)
Abbreviations
- eIF4E
eukaryotic initiation factor 4E
- eIFiso4E
eukaryotic initiation factor iso4E
- 4E-BP
eIF4E-binding protein
- PKR
protein kinase R
- SNF1
sucrose non-fermenting 1
- SnRK1
SNF1-related kinase 1
- AMPK
AMP-activated kinase
- SnRK1-KD
SnRK1 kinase domain
- SnRK1-KDKR
SnRK1 kinase domain inactive
- BiFC
bimolecular fluorescent complementation
Footnotes
Conflict of interest statement. The authors declare no conflicts of interest.
References
- 1.Jackson RJ, Hellen CU & Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation, Nat Rev Mol Cell Biol. 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning KS & Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation, The Arabidopsis book / American Society of Plant Biologists. 13, e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lellis AD, Allen ML, Aertker AW, Tran JK, Hillis DM, Harbin CR, Caldwell C, Gallie DR & Browning KS (2010) Deletion of the eIFiso4G subunit of the Arabidopsis eIFiso4F translation initiation complex impairs health and viability, Plant Mol Biol. 74, 249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter JD & Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins, Nature. 433, 477–80. [DOI] [PubMed] [Google Scholar]
- 5.Musa J, Orth MF, Dallmayer M, Baldauf M, Pardo C, Rotblat B, Kirchner T, Leprivier G & Grunewald TG (2016) Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): a master regulator of mRNA translation involved in tumorigenesis, Oncogene. 35, 4675–88. [DOI] [PubMed] [Google Scholar]
- 6.Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD & Browning KS (2007) The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond, Plant Physiol. 143, 1504–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning KS (2004) Plant translation initiation factors: it is not easy to be green, Biochem Soc Trans. 32, 589–591. [DOI] [PubMed] [Google Scholar]
- 8.Patrick RM, Lee JCH, Teetsel JRJ, Yang SH, Choy GS & Browning KS (2018) Discovery and characterization of conserved binding of eIF4E 1 (CBE1), a eukaryotic translation initiation factor 4E-binding plant protein, J Biol Chem. 293, 17240–17247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou-Nader C, Gordon JM, Henderson FE & Zhang J (2019) The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators, RNA. 25, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilgin DD, Liu Y, Schiff M & Dinesh-Kumar SP (2003) P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis, Dev Cell. 4, 651–661. [DOI] [PubMed] [Google Scholar]
- 11.Bruns AN, Li S, Mohannath G & Bisaro DM (2019) Phosphorylation of Arabidopsis eIF4E and eIFiso4E by SnRK1 inhibits translation, FEBS J. 10.1111/febs.14935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emanuelle S, Doblin MS, Stapleton DI, Bacic A & Gooley PR (2016) Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1, Trends Plant Sci. 21, 341–353. [DOI] [PubMed] [Google Scholar]
- 13.Hulsmans S, Rodriguez M, De Coninck B & Rolland F (2016) The SnRK1 Energy Sensor in Plant Biotic Interactions, Trends Plant Sci. 21, 648–661. [DOI] [PubMed] [Google Scholar]
- 14.Hao L, Wang H, Sunter G & Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase, Plant Cell. 15, 1034–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miras M, Miller WA, Truniger V & Aranda MA (2017) Non-canonical Translation in Plant RNA Viruses, Front. Plant Sci. 8, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U & Benfey PN (2016) Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis, Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]


