Abstract
The evolution of brain function in the regulation of physiology may depend in part upon the numbers and locations of neurons. Wild populations of rodents contain natural genetic variation in the inhibition of reproduction by winter-like short photoperiod, and it has been hypothesized that this functional variation might be due in part to heritable variation in the numbers or location of gonadotropin releasing hormone (GnRH) neurons. A naturally variable wild-source population of white-footed mice was used to develop lines artificially selected for or against mature gonads in short, winter-like photoperiods. We compared a selection line that is reproductively inhibited in short photoperiod (Responsive) to a line that is weakly inhibited by short photoperiod (Nonresponsive) for differences in counts of neurons identified using in situ hybridization for GnRH mRNA. There was no effect of photoperiod, but there were 60% more GnRH neurons in total in the Nonresponsive selection line than the Responsive selection line. The lines differed specifically in numbers of GnRH neurons in more anterior regions, whereas numbers of GnRH neurons in posterior areas were not statistically different between lines. We compare these results to those of an earlier study that used immunohistochemical labeling for GnRH neurons. The results are consistent with the hypothesis that the selection lines and natural source population contain significant genetic variation in the number and location of GnRH neurons. The variation in GnRH neurons may contribute to functional variation in fertility that occurs in short photoperiods in the laboratory and in the wild source population in winter.
The evolution of brain function in the regulation of physiology may depend partially upon the numbers, locations, and interconnections of neurons (Williams and Herrup, ‘88; Bittner and Friedman, 2000). Neurons that release gonadotropin releasing hormone (GnRH) are essential for fertility (Ebling, 2005). Even in non-inhibitory laboratory conditions, GnRH neurons must be present in sufficient numbers in order to maintain fertility (Herbison et al., 2008; Hoffmann et al., 2014; Quaynor et al., 2015). Reproduction can be inhibited in rodents by inhibitory environments that may include restricted food, stress, and short photoperiod (Bronson, ‘89). Within species, these inhibitory factors differentially affect specific genetic strains or individuals; genetic variation exists both among laboratory strains (Heideman and Sylvester, ‘97; Lorincz et al., 2001; Ross et al., 2009) and within wild-source populations (Prendergast et al., 2001; Mintz et al., 2007; Heideman and Pittman, 2009). Genetic variation in the neurons and endocrine cells that regulate fertility is a potential source of natural genetic variation in sensitivity to reproductive inhibition in humans and other mammals. Genetic variation in the number or location of GnRH neurons could contribute to functional variation in sensitivity to reproductive inhibition, and in natural populations could contribute both to the potential for microevolutionary change and to individual variation in sensitivity to environmental inhibitors of reproduction (Heideman, 2014).
The response of the reproductive system to winter-like short photoperiods is a naturally variable neuroendocrine trait within multiple natural populations (Prendergast et al., 2001; Mintz et al., 2007; Heideman and Pittman, 2009). In laboratory populations derived from wild populations in the genus Peromyscus, variation in reproduction is associated with neuroendocrine variation in the system that transduces photoperiodic information and/or in the hypothalamic-pituitary-gonadal (HPG) axis regulating fertility (Blank, ‘92; Heideman et al., ‘99a; Mintz et al., 2007; Heideman and Pittman, 2009). In two wild-source populations, there is evidence for heritable variation in the hypothalamic neurons that release GnRH (Korytko et al., ‘95, ‘97, ‘98; Avigdor et al., 2005; Heideman et al., 2007; Mintz et al., 2007). One hypothesis is that the total number and location of GnRH neurons is related to sensitivity to reproductive inhibition by short photoperiod. Alternatively, a null hypothesis is that other sources of neuroendocrine variation cause this variation.
Although no method unambiguously identifies a neuron to type (Underwood, 2015), estimates for the number or location of GnRH neurons are commonly made using immunohistochemistry (IHC) for the GnRH peptide (e.g., Heideman et al., ‘99a; Heideman, 2014) or by in situ hybridization (ISH) for mRNA from the GnRH gene (reviewed by Stevenson et al. (2012)). IHC-based counts have the potential to vary due to differences in peptide content among neurons (e.g., Korytko et al., ‘95; Kriegsfeld et al., 2000); reviewed by Stevenson et al. (2012)). Neurons that have low levels of GnRH may be too lightly stained to be detectable, causing underestimation of numbers. ISH-based counts remove variability due to differential content of GnRH peptide, although GnRH gene expression might also be too low for detection in some neurons (Stevenson et al., 2012). If GnRH neurons are differentially detectable in conditions that are reproductively stimulatory versus inhibitory, then counts in two conditions, stimulatory long photoperiods versus inhibitory short photoperiods, may reveal differences due to differential content of peptide or of mRNA. Together, use of both IHC and ISH conducted on individuals in long and short photo-period may provide the best estimates for total numbers and locations of GnRH neurons.
Here, we test for heritable variation in GnRH neurons using ISH on two selection lines (Heideman et al., ‘99a; Heideman, 2014) of white-footed mice (Peromyscus leucopus) derived from a natural population. One selection line resists suppression of reproduction in short, winter-like photoperiods (photoperiod nonresponsive, or NR selection line), and the other selection line strongly suppresses reproduction in short, winter-like photoperiods (photoperiod responsive or R selection line) (Heideman et al., ‘99a; Heideman and Pittman, 2009). The number of IHC-GnRH neurons is significantly heritable in an unselected control line from this population of white-footed mice (Heideman et al., 2007). The two selection lines have been found to differ in the number of GnRH neurons assessed by IHC (Avigdor et al., 2005), but it is plausible that the difference is due to differences in detectability of GnRH neurons rather than a difference in absolute numbers. For this study, therefore, we tested whether the two selection lines differed similarly in numbers of neurons identified by in situ hybridization in two environmental treatments, long photoperiods that are stimulatory to the reproductive axis, and short photoperiods that can have strong inhibitory effects on the reproductive axis. We assessed whether the numbers and location of GnRH neurons identified using ISH provide estimates that are similar or dissimilar to those from IHC (Avigdor et al., 2005). We use the results to discuss (i) the likely amount of natural variation in the number and location of GnRH neurons and (ii) how fertility might be affected by variation in the numbers and location of GnRH neurons.
METHODS
Animals and Selection Lines
Two selection lines of Peromyscus leucopus were initiated in 1995 from a wild population near Williamsburg, VA (latitude 37 × 3°N, longitude 76 × 7°W) (Heideman et al., ‘99a). A parental laboratory generation was used to found three lines: one control line not bred for a photoperiod response and two lines bred for either a photoperiod response (responsive, R) or the lack of a photoperiod response (nonresponsive, NR) (Heideman et al., ‘99a). Mice from a parental generation were raised in short day (SD, 8L:16D), examined at 70 ± 3 days of age and assessed for reproductive phenotype based on the size of gonads. For males, a testis size measure (length × width of testis) was used to classify males as eitherR (<24 mm2) or NR (>32mm2). R mice were paired with R mates and NR mice were paired with NR mates in long day (LD, 16L:8D) to find the R and NR lines, respectively. Selective breeding was continued for a subsequent 10 generations, after which selection was relaxed. Mice from the parental generation were chosen and paired randomly to form the unselected control line, which was maintained without selective breeding (Heide- man and Pittman, 2009). Additional details about the selection lines are available elsewhere (Heideman et al., ‘99a; Heideman, 2014).
Animal use procedures conform to NIH guidelines and were approved by the Institutional Animal Care and Use Committee as IACUC-2009–10-19–6267-pdheid.
Primer Design
Primers for P. leucopus GnRH mRNA were created from known GnRH mRNA sequences from the following species: Mus musculus (NM_008145.1), Rattus norvegicus (NM_012767.2), Homo sapiens (NM_000825.3), Mesocricetus auratus (U91938.1), Cavia porcellus (NM_001172957.1), and Xenopus tropicalis (NM_001113693.1). The primers are as follows: forward (5′-TGT GTT TGG AAG GCT GCT C-3′) and reverse (5′-ACT TTA TTA TGA AAT CTA CGC TGC T-3′).
RT-PCR and Cloning
Hypothalamus tissue was extracted from R and NR males and flash-frozen in liquid nitrogen. Tissue was stored at −80°C. Total RNA was extracted from the tissue using the RNeasy Mini Kit (Qiagen, Venlo, Netherlands). cDNA was synthesized using the iScript cDNA Synthesis kit. Subsequent PCR was performed using the Supertaq DNA Polymerase Kit (Ambion, Waltham, MA). PCR products were cloned into StrataClone pSC-A-amp/kan PCR cloning vector and transformed into StrataClone competent Escherichia coli cells (Stratagene, San Diego, CA). Cells that had incorporated the plasmid with insert were selected using X-gal. White colonies were grown in 3 ml LB medium containing 100 μg/ml ampicillin overnight at 37°C with shaking.
Plasmid DNA was isolated from liquid bacterial cultures using the Wizard Plus SV Miniprep kit (Promega Corp., Fitchburg, WI). A restriction digest was performed using EcoRl to assess whether the insert was of appropriate size. Plasmid DNA was sequenced using the ABI Big Dye Terminator v3.1 sequencing reaction and Applied Biosystems 3100-Avant sequencer. Midi preps were carried out using Bio-Rad Midiprep kits.
Linearization and Probe Synthesis
Midi prep DNA was linearized using the restriction enzymes HindIII (sense probe) and BamHI (antisense probe). Sense and antisense digoxigenin UTP-labeled probes were created using T3 RNA polymerase and T7 RNA polymerase, respectively, with linearized midi prep DNA as a template. Probes were purified using RNeasy MinElute Cleanup kit and stored at −20°C (Qiagen).
Perfusions and Sectioning
Mice were euthanized with an overdose of isoflurane (Abbot Laboratories) followed by vascular perfusion. Once the mice entered respiratory arrest, they were perfused through the left ventricle at ~4 ml/min using a perfusion pump and bled via the right atrium. Perfusion for 5 min with 5 ml of 0.1 M PBS at a pH of 7.4 was followed by perfusion for 15–20 min with 4% paraformaldehyde in PBS. Brains were removed and postfixed in 4% paraformaldehyde in PBS for 5–7 hr, and then transferred to 0.1 M PBS until sectioning. Following perfusion, testes length and width were recorded. Within 36 hr of perfusion, brains were sectioned on a vibratome at a thickness of 100 μm. Sections were dehydrated in serial washes to 100% methanol and stored at −20°C until in situ hybridization.
In Situ Hybridization
An in situ hybridization protocol was devised by combining and modifying several existing protocols (Correia and Conlon, 2001; Lewis et al., 2009). Sections from each brain were divided into four 35 mm Petri dishes. Tissue was rehydrated from 100% methanol to 1 × PTw (PBS + 0.1% Tween 20), treated with 10 μg/ml proteinase K in PTw at 37°C for 10–30 min, and postfixed in 4% paraformaldehyde in PTw for 20 min. Tissue was washed with PTw and incubated with hybridization buffer (50% formamide, 1.3 × SSC pH 7, 5mM EDTA pH 8, 50 μg/ml yeast RNA, 0.2% Tween 20,0.5% CHAPS, and 100 μg/ml heparin in sdd H2O) at 60°C for 4–8 hr. Tissue was then incubated overnight at 60°C in hybridization buffer containing 1 ug/ml of digoxigenin-labeled probe. After removal from the probe solution, the tissue was washed with hybridization buffer and washing solution (50% formamide, 1× SSC, and 0.1% Tween 20 diluted to 50 ml with sdd H2O). Tissue was then treated with RNase A in RNase buffer (0.5 M NaCl, 10 mM PIPES pH 7.2, and 0.1% Tween 20 diluted to 50 ml with sdd H2O) for 1 hr. Tissue underwent one 30 min wash in 1:1 washing solution with MABT (100 mM maleic acid, 150 mM NaCl, and 0.1% Tween 20 pH adjusted to 7.5 with solid NaOH), followed by blocking in MABT + 2% BMB for 1 hr and MABT + 2% BMB + 20% goat serum for 1–2 hr. Tissue was incubated overnight at 4°C with a 1:2,000 dilution of anti-digoxigenin alkaline phosphatase antibody in MABT + 2% BMB + 20% heat-inactivated goat serum. Tissue was washed for at least 5 hr in MABT and then incubated in the dark at room temperature for 2–3 days in NTMT (1 ml of 5 M NaCl, 5 ml of 1 M Tris-HCl pH 9.5, 2.5 ml 1 M MgCl2, and 0.5 ml 100% Tween 20 diluted to 50 ml with sdd H2O) containing 4.5 mL NBT stock and 3.5 μL BCIP stock per ml of solution. Once the color reaction was complete, tissue was postfixed overnight at 4°C in 4% paraformaldehyde in PBS, then washed with 1× PBS and stored in 1 × PBS at 4°C. Following ISH, sections were mounted in 1 × PBS on albumin-treated slides and allowed to dry, then immersed in xylene for 8–10 min and coverslipped with Permount (Fisher Scientific, Waltham, MA).
Neuron Counts
ISH-GnRH neurons (Fig. 1) were counted blind with respect to treatment by KEK using an Olympus CH2 compound light microscope. Counting accuracy was assessed by independent counts of randomly selected sections by other investigators. The regions in which GnRH neurons were identified with ISH were compared with those in a stereotaxic coordinate atlas for a closely related species, Peromyscus maniculatus (Eleftheriou and Zolovick, ‘65) and a rat brain atlas with finer resolution (Paxinos and Watson, ‘86). To assess regional variation in counts of IHC-GnRH neurons (Avigdor et al., 2005), we separated counts into more anterior and more posterior regions. The anterior region in this study included rostral areas that held GnRH neurons, the diagonal band of Broca, the septal nuclei, the preoptic area, and regions of the anterior and lateral hypothalamus anterior to the tuber cinereum (plates 8 through 24 in Paxinos and Watson (‘86)). The posterior region extended posteriorly through the lateral hypothalamus to the ventromedial nucleus (plates 25 through 40 in Paxinos and Watson (‘86)). Because P. leucopus have comparatively large eyes, the rat brain in the region of the optic tract is structurally more similar to that of P. leucopus than either is to laboratory mice, but brain areas in the rat and P. leucopus are not necessarily homologous.
Figure 1.
Representative coronal sections at a position equivalent to Plate 19 in Paxinos and Watson (‘86) showing GnRH neurons identified by in situ hybridization in the nonresponse and responsive selection line. The lower panel shows a negative sense-probe control with a lack of specific staining. Abbreviations: MPOA, medial preoptic area; OC, optic chiasm; 3V, third ventricle. Scale bar: 300μm.
In order to obtain approximate total counts for GnRH neurons comparable with other studies, we applied a correction factor (Abercrombie, ‘46) to adjust for overcounting of single cells that are visible in two adjacent sections. The correction factor is an approximation with potential biases (Guillery and Herrup, ‘97), but allows approximate comparisons of numbers of neurons obtained in different studies. We used the formula N = n(T/T + L), where n is the raw count of neurons, T is the section thickness in micrometers, and L is the average diameter of a GnRH neuron in micrometers. The maximum reported diameter of typical GnRH neurons is 10–20 μm (Silverman et al., ‘94), a range consistent with our measurements, and we used 15 μm as an approximation for L. This adjustment can be biased (Guillery and Herrup, ‘97), and, therefore, the adjusted totals may be biased relative to the true number of neurons.
Sampling and Statistical Analysis
Young male mice were used as available from the colony photoperiod treatments, with a final sample size of 28 individuals (NR: 15, R: 13). One additional individual raised in LD was infertile despite being raised in the stimulatory LD photoperiod, a photoperiod treatment in which all normal males become reproductively mature by age 70 days and, therefore, was removed from the study. Sample sizes were unevenly distributed among LD and SD groups (NRLD: 11, NR SD: 4, RLD: 5, R SD: 8). With ANOVA, low sample sizes and unbalanced sampling among groups produces unreliable estimates of probabilities (Hector et al., 2010; Landsheer and van den Wittenboer, 2015). Therefore, we used a statistical method that has been demonstrated to provide more reliable P-values than ANOVA when samples are unbalanced and some cell sizes are small (Xu et al., 2013a, 2013b): the parametric bootstrap (Xu et al., 2013a, 2013b) with restricted maximum likelihood mixed model analysis (Faraway, 2006). Because mice were obtained as available from the two photoperiods, we treated photoperiod as a random effect (Faraway, 2006), but results of analyses were qualitatively identical if photoperiod was considered as a fixed effect. In a sequential procedure, we performed initial tests for significant effects of line and of photoperiod. Next, if one or both effects were significant, we followed by testing whether a model with both factors was statistically significant. For purposes of comparison, comparable statistical analyses were conducted on these data using conventional Type III ANOVA; comparable analyses using ANOVA gave qualitatively identical results. Probability values given are those obtained using the parametric bootstrap for line and for photoperiod, and, if one of these was statistically significant, for the combination of both terms.
In order to compare the results from this study to a previous result on variation in IR-GnRH neurons in these same selection lines (Avigdor et al., 2005), we evaluated the hypothesis that selection line alone accounted best for variation in numbers of neurons (Avigdor et al., 2005). For this test, we used general linear models (GLM) evaluated using Akaike Information Criteria (AIC) (Anderson, 2008; Mundry, 2011; Symonds and Moussalli, 2011) with correction for small sample bias (QAICc) (Richards, 2008; Symonds and Moussalli, 2011). Models were compared according to AICc model probabilities, relative likelihood, and evidence ratios (Anderson, 2008; Mundry, 2011).
All statistical analyses were carried out using RStudio 0.95.258.
RESULTS
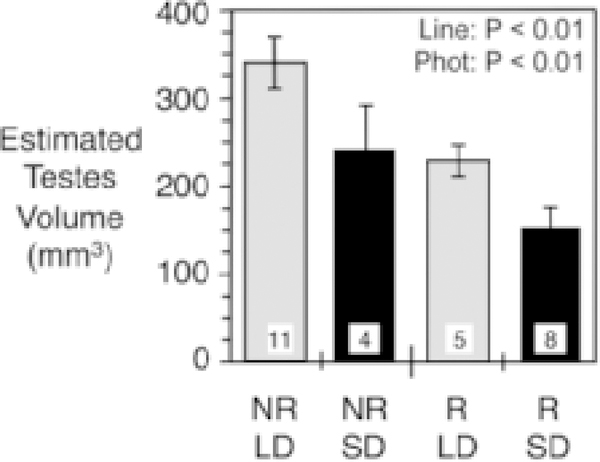
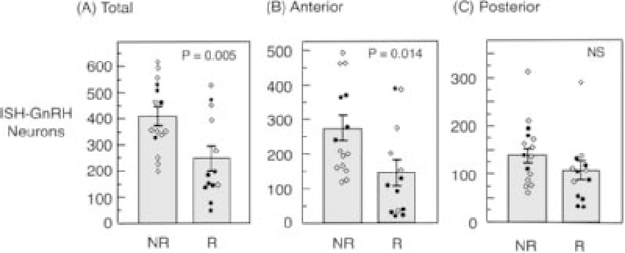
As in previous studies (reviewed by (Heideman, 2014)), testis size was greater in LD than in SD (P = 0.009), and greater in the NR line than in the R line (P = 0.002) (Fig. 2). After accounting for selection line, testis size was unrelated to counts of total GnRH neurons (P = 0.867). Young adult males from the NR selection line had more ISH-GnRH neurons (Fig. 1) in total than young adult males from the R line (P = 0.005; Fig. 3A). In anterior regions of the hypothalamus, there was a significant difference in the number of GnRH neurons between selection lines (P = 0.014; Fig. 3B), with approximately twice the number of GnRH neurons in the NR line relative to the R line. In more posterior regions of the hypothalamus, the difference in the number of GnRH neurons between selection lines was not statistically significant (P = 0.16), with approximately equivalent numbers of neurons in the NR and R lines (Fig. 3C). Photoperiod had no significant effects on the total number of ISH-GnRH neurons (P = 0.08), on the number of ISH-GnRH neurons in anterior regions of the hypothalamus (P = 0.57), or on the number of ISH-GnRH neurons in posterior regions of the hypothalamus (P = 0.07) (Table 1).
Figure 2.
Estimated paired testes volume in males raised in long photoperiod (LD) or short photoperiod (SD) from lines artificially selected to be photoperiod nonresponsive (NR) or responsive (R). Sample sizes are shown in bars at the base of each bar. Statistical significance is indicated for selection line (Line) and photoperiod (Phot) (Mean +/− SEM).
Figure 3.
Estimated numbers of GnRH neurons, after adjustment for potential over-counting (see Methods section) labeled using in situ hybridization (ISH) in lines artificially selected to be photoperiod nonresponsive (NR) or responsive (R) to short photoperiod: (A) total number of neurons, (B) neurons in more anterior regions of the brain, and (C) neurons in more posterior regions of the brain (see Methods section for region boundaries). Statistical significance levels are indicated for differences between selection lines; there were no statistically significant effects of photoperiod. Individual values are plotted as either open diamonds (long photoperiod) or closed circles (short photoperiod) (Mean +/− SEM).
Table 1.
Means and 95% confidence intervals for counts of neurons adjusted for overcounting (see Methods section).
| ISH-GnRH neurons | R | 95%CI | NR | 95%CI |
|---|---|---|---|---|
| Anterior | 145 | (73, 217) | 274 | (209, 340) |
| Posterior | 102 | (62, 138) | 138 | (106, 165) |
| Total | 247 | (160, 334) | 412 | (344, 479) |
Using GLM evaluated with QAICc, we found that the model including only selection line for variation in total counts of ISH- GnRH neurons had the best support (Table 2). Similarly, for anterior neurons analyzed separately, the model with selection line alone had the best support (Table 3). The probabilities of models that included photoperiod alone or photoperiod with line (Tables 2 and 3) were in the range of 0.1–0.19, raising the possibility that photoperiod may have a slight effect on counts of ISH-GnRH neurons. For posterior ISH-GnRH neurons, photoperiod provided a better fit than other models, but no models were a good fit, all having a low adjusted R-squared. Overall, the results indicated that variation in ISH-GnRH neurons was best accounted for by selection line alone.
Table 2.
QAICc summary for a priori models for variation in the total number of ISH-GnRH neurons.
| Model | K | Log-likelihood | QAICc | Delta-i | Model probability (w-sub-i) | Evidence ratio | Adjusted R2 |
|---|---|---|---|---|---|---|---|
| Line | 3 | −182.2156 | 133.8504 | 0 | 0.4416 | 0.2253 | |
| Phot | 3 | −184.7018 | 135.5438 | 1.6934 | 0.1894 | 2.33 | 0.0748 |
| Line + phot | 4 | −181.7044 | 136.4903 | 2.6399 | 0.1180 | 3.74 | 0.2232 |
| Line + phot + line: phot | 5 | −179.0851 | 137.979 | 4.1286 | 0.0560 | 7.88 | 0.3289 |
| ETV | 3 | −184.6171 | 135.4861 | 1.6357 | 0.1949 | 2.27 | 0.0804 |
K, number of terms estimated in the model; QAICc, Quasi-Akaike Information Criteria with second order bias correction; Delta-i, difference between best (lowest) QAICc and QAICc for the model in that row; Phot, photoperiod; ETV, estimated testis volume.
Table 3.
QAICc summary for a priori models for variation in the number of anterior ISH-GnRH neurons.
| Model | K | Log-likelihood | QAICc | Delta-i | Model probability (w-sub-i) | Evidence ratio | Adjusted R2 |
|---|---|---|---|---|---|---|---|
| Line | 3 | −178.9518 | 128.9301 | 0 | 0.4346 | 0.1785 | |
| Phot | 3 | −181.5426 | 130.6558 | 1.7257 | 0.1834 | 2.37 | 0.0115 |
| Line + phot | 4 | −178.8818 | 131.8717 | 2.9416 | 0.0998 | 4.35 | 0.1499 |
| Line + phot + line: phot | 5 | −177.3226 | 134.1059 | 5.1758 | 0.0327 | 13.30 | 0.2078 |
| ETV | 3 | −180.6169 | 130.0392 | 1.1091 | 0.2496 | 1.74 | 0.0748 |
K, number of terms estimated in the model; QAICc, Quasi-Akaike Information Criteria with second order bias correction; Delta-i, difference between best (lowest) QAICc and QAICc for the model in that row; Phot, photoperiod; ETV, estimated testis volume.
For the data from Avigdor et al. (2005), the best models included selection line alone and accounted for 38–40% of the variation in both total counts (Supplemental Table S1) and anterior counts of IHC-GnRH neurons (Supplemental Table S2). The best model for posterior IHC-GnRH neurons also included only selection line, but accounted for only 6% of the variation (Supplemental Table S3). In no model did photoperiod account for more than 2% of the variation, and in no case did the addition of photoperiod or an interaction between line and photoperiod improve the fit of the model.
Based on the results from IHC, we tested the counts of ISH-GnRH neurons using an a priori model including only selection line against four biologically reasonable alternative models (Tables 2–4). The results were consistent with predictions: for total ISH-GnRH neurons and more anterior ISH-GnRH neurons, the best model included only selection line, accounting for 18–23% of variation in counts. For more posterior ISH-GnRH neurons, no model accounted for much of the variation (Table 4). We cannot eliminate the possibility that photoperiod had a weak effect on identifiable ISH-GnRH neurons, but in the analyses of ISH-GnRH neurons, photoperiod alone had less support than selection line in the models (Tables 2 and 3: evidence ratio >2 and model probabilities for photoperiod 0.18–0.19).
Table 4.
QAICc summary for a priori models for variation in the number of posterior ISH-GnRH neurons.
| Model | K | Log-likelihood | QAICc | Delta-i | Model probability (w-sub-i) | Evidence ratio | Adjusted R2 |
|---|---|---|---|---|---|---|---|
| Phot | 3 | −158.2879 | 140.6501 | 0 | 0.3785 | 0.0838 | |
| Line | 3 | −158.9505 | 141.1981 | 0.548 | 0.2878 | 1.32 | 0.0394 |
| Line + phot | 4 | −157.8563 | 143.2813 | 2.6312 | 0.1015 | 3.73 | 0.0761 |
| Line + phot + line: phot | 5 | −156.7523 | 145.641 | 4.9909 | 0.0312 | 12.13 | 0.1106 |
| ETV | 3 | −159.8179 | 141.9155 | 1.2654 | 0.2010 | 1.88 | −0.0220 |
K, number of terms estimated in the model; QAICc, Quasi-Akaike Information Criteria with second order bias correction; Delta-i, difference between best (lowest) QAICc and QAICc for the model in that row; Phot, photoperiod; ETV, estimated testis volume.
The mean numbers of neurons identified in the selection lines in this study by in situ hybridization were similar to the numbers reported previously for the NR and R selection lines using antibodies for mature GnRH peptide. When identified using ISH, after adjustment for overcounting, the R line averaged approximately 250 neurons and the NR line approximately 410 neurons (Table 1). When identified using IHC (Avigdor et al., 2005), after adjustment for overcounting, the R line averaged approximately 290 neurons and the NR line approximately 460 neurons. For both selection lines, the means from Avigdor et al. (2005) fall within the confidence intervals for counts of GnRH neurons identified by ISH (Table 1). In both studies, the NR line had an average of approximately 60% more GnRH neurons than the R line.
DISCUSSION
Two artificial selection lines that develop (NR) or fail to develop (R) large, fertile gonads in winter-like short photoperiod differed by 60% in counts of ISH-GnRH neurons (Fig. 3A). This difference was regionally heterogeneous, with large and significant differences in more anterior regions of the hypothalamus (Fig. 3B), but smaller and statistically non-significant differences in more posterior regions of the hypothalamus (Fig. 3C). This regional variation indicates a genetic difference not solely in the number of GnRH neurons but also developmental differences in either regional migration or regional retention.
The results provide independent confirmation of similar genetic differences identified previously for these lines using a different assessment method for GnRH neurons, IHC for mature GnRH peptide (Avigdor et al., 2005). In birds, but not mammals, it has been proposed that a subset of GnRH I neurons in more rostral areas selectively respond to changes in photoperiod, whereas more caudal GnRH I neurons do not (Stevenson and Ball, 2009; Stevenson et al., 2009, 2012). In mammals, a subset of more rostrally located neurons may have roles specifically during the ovulatory LH surge (Stevenson et al., 2012). We hypothesize that in this population, posterior GnRH neurons support fertility in the absence of inhibitory factors, whereas the number of GnRH neurons in more anterior regions may affect the likelihood of fertility in short photoperiod. In previous studies, photoperiod has been found to have regional differences on more anterior versus more posterior GnRH neurons of photoperiod-responsive in comparison to photoperiod-nonresponsive individuals (Korytko et al., ‘95, ‘98; Kriegsfeld and Nelson, ‘99).
Although photoperiod had a statistically non-significant effect on the number of ISH-GnRH neurons, the effect of photoperiod approached significance both for the total and for posterior ISH-GnRH neurons. To further assess the effect of photoperiod and additional factors, we first conducted an a posteriori analysis of previous data on IHC-GnRH neurons (Avigdor et al., 2005) in an information-theoretic analysis using AIC. Using data from Avigdor et al. (2005), the best a posteriori models for variation in IHC-GnRH neurons included only selection line and accounted for 38–40% of the variation in total and anterior IHC-GnRH neurons. In contrast, for posterior IHC-GnRH neurons, no model accounted for more than 6% of the variation. The results of the analysis of IHC-generated data were considered a priori models for new tests of the effect of selection line against the alternative, biologically plausible models assessed using ISH. As with IHC-GnRH neurons, selection line was the sole factor in the best fitting models for total and anterior ISH-GnRH neuron counts. The fit was not improved by adding photoperiod (Tables 2 and 3), indicating that with our staining protocols, counts taken when males are either fertile or infertile may identify most or all GnRH neurons.
With both IHC and ISH and in either photoperiod, we obtained estimates of 60% more GnRH neurons in the Nonresponsive line (~430 neurons) than in the Responsive line (~270 neurons) (Table 1 and see Results section). Interestingly, as with results using IHC (Avigdor et al., 2005), the number of ISH-GnRH neurons was not significantly correlated with the mass of testes. This suggests that the number of GnRH neurons may affect the likelihood of fertility in short photoperiod, but once a male becomes fertile, factors independent of the number of GnRH neurons affect the abundance of gametes. Together, these studies provide evidence for a heritable difference in the number of GnRH neurons in the two selection lines.
Differences in the number of GnRH neurons may have functional consequences in the two selection lines. In young prepubertal males, odor cues from soiled bedding of mature females increased reproductive development in the NR line, but not the R line, suggesting a greater capacity in the NR than R line to respond to an environmental cue that can stimulate reproductive development (Sharp et al., 2015). Young males in the NR line had greater binding of iodomelatonin in the hypothalamus than young males from the R line (Heideman et al., ‘99b), suggesting the potential for differential modulation of responses of GnRH neurons to photoperiodic cues. In SD, males in the NR line were more likely than R males to mate and inseminate females, indicating a potential behavioral correlate in SD. Females in the NR line had higher concentrations of circulating LH than the R line (Heideman et al., 2010), potentially because of differences in GnRH release.
Reducing the numbers of GnRH neurons through experimental manipulations or natural mutations have been shown in laboratory mice and other species to affect the function of the HPG axis and the likelihood of infertility (Herbison et al., 2008; Wierman et al., 2011). Genetic manipulation of the number of GnRH neurons in laboratory mice from a normal average of600 (in counts unadjusted for overcounting) to 20% or less of the average has been found to reduce or block fertility (Herbison et al., 2008; Diaczok et al., 2011). In our wild-derived population in which natural variation was manipulated using artificial selection, only one mouse from 83 in the two studies (this study and Avigdor et al. (2005)) had fewer than 20% of the population average (combined mean of adjusted totals for NR and R using ISH ~330 and using IHC ~375 neurons). If the results from laboratory mice generalize to other rodents, including our population, this observation suggests that few if any individuals in our laboratory population would suffer impaired fertility solely due to insufficient numbers of GnRH neurons. However, assessments of fertility in the laboratory have been conducted under benign laboratory conditions of abundant food, water, and temperature, and in the absence of natural stressors, parasites, and serious disease. As challenges from reduced food or cold, stress, parasites, or disease begin to inhibit the reproductive axis, higher numbers of GnRH neurons may be necessary to maintain fertility.
The NR line is less sensitive to inhibitory factors than the R line, but the 60% greater number of GnRH neurons in the NR line does not protect males from environmentally induced infertility (Reilly et al., 2006). In the NR line, although neither SD alone nor slight restriction of access to food suppresses fertility, the two factors combine to suppress fertility (Reilly et al., 2006). Although the R line is more sensitive to inhibitory factors than the NR line, males in LD in the R line and NR line are similar in sexual behavior and insemination of females (Sharp et al., 2015). In our unselected control line, different families varied more than threefold in the number IR-GnRH neurons (Heideman et al., 2007). We speculate that in the wild, the number of GnRH neurons may contribute to normal variation in fertility. Field studies on the wild source population of our selection lines have identified individual variation in fertility in both winter and summer seasons (Terman, ‘93; Heideman et al., ‘99a), and we suggest that this variation in fertility may be related to numbers of GnRH neurons.
A major hypothesis in evolutionary physiology is that selection on complex physiological pathways may be able to remove most functional variation, producing physiological systems that function optimally (Lindstedt and Jones, ‘87; Weibel et al., ‘98). The alternative hypothesis is that selection is weak and variable when there are many genes that affect a pathway, permitting the maintenance of high levels of functional genetic variation (Bartholemew, ‘87; Lindstedt and Jones, ‘87). The results of this study suggest that substantial variation exists in the number and location of GnRH neurons in this population. This neuronal variation could allow rapid microevolutionary change in response to selection (Heideman and Pittman, 2009). If similar variation is widespread in GnRH neurons or other classes of neurons in mammals, then standing natural genetic variation would provide a substrate for substantial differences in neuroendocrine function among populations as well as a source of variation for macroevolutionary neuronal change.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support from NIH R15NS067566 (to MSS), R15HD068962 (to PDH), and a Howard Hughes Medical Institute Undergraduate Science Education grant to the College of William and Mary. J. Upperman assisted with in situ hybridization, and G. Loiseau and G. Smith assisted by recounting to assess reliability of counts, assisting in data analysis, and reading earlier drafts of the manuscript.
Grant sponsor: NIH; grant number: R15NS067566; grant sponsor: NIH; grant number: R15HD068962; grant sponsor: Howard Hughes Medical Institute Undergraduate Science Education grant to the College of William and Mary.
Footnotes
Conflicts of interest: None.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
LITERATURE CITED
- Abercrombie M 1946. Estimation of nuclear population from microtome sections. Anat Rec 94:239–247. [DOI] [PubMed] [Google Scholar]
- Anderson DR. 2008. Model based inference in the life sciences: a primer on evidence. New York: Springer. [Google Scholar]
- Avigdor M, Sullivan SD, Heideman PD. 2005. Response to selection for photoperiod responsiveness on the density and location of mature GnRH-releasing neurons. Am J Physiol Regul Integr Comp Physiol 288:1226–1236. [DOI] [PubMed] [Google Scholar]
- Bartholemew GA. 1987. Interspecific comparison as a tool for ecological physiologists In: Feder ME, Bennett AF, Burggren WW, Huey RB, editors. New directions in ecological physiology. Cambridge, UK: Cambridge University Press; p 11–35. [Google Scholar]
- Bittner GD, Friedman BX. 2000. Evolution of brain structures and adaptive behaviors in humans and other animals: role of polymorphic genetic variations. Neuroscientist 6:241–251. [Google Scholar]
- Blank JL. 1992. Phenotypic variation in physiological response to seasonal environments In: Tomasi TE, Horton T, editors. Mammalian energetics: interdisciplinary views of metabolism and reproduction. Ithaca: Comstock Publishing Associates; p 186–212. [Google Scholar]
- Bronson FH. 1989. Mammalian reproductive biology. Chicago: Univ. Chicago Press. [Google Scholar]
- Correia KM, Conlon RA. 2001. Whole-mount in situ hybridization to mouse embryos. Methods 23:335–338. [DOI] [PubMed] [Google Scholar]
- Diaczok D, DiVall S, Matsuo I, et al. 2011. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol 25:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebling FJP. 2005. The neuroendocrine timing of puberty. Reproduction 129:675–683. [DOI] [PubMed] [Google Scholar]
- Eleftheriou BE, Zolovick AJ. 1965. The forebrain of the deermouse in stereotaxic coordinates. Kansas State University of Agriculture and Applied Science Technical Bulletin 146:A7b. [Google Scholar]
- Faraway JJ. 2006. Extending the linear model with R: generalized linear, mixed effects and nonparametric regression models. Boca Raton, FL: Chapman and Hall/CRC. [Google Scholar]
- Guillery RW, Herrup K. 1997. Quantification without pontification: choosing a method for counting objects in sectioned tissues. J Comp Neurol 386:2–7. [DOI] [PubMed] [Google Scholar]
- Hector A, Von Felten S, Schmid B. 2010. Analysis of variance with unbalanced data: an update for ecology & evolution. J Anim Ecol 79:308–316. [DOI] [PubMed] [Google Scholar]
- Heideman PD, Pittman JT. 2009. Microevolution of neuroendocrine mechanisms regulating reproductive timing in Peromyscus leucopus. Int Comp Biol 49:550–562. [DOI] [PubMed] [Google Scholar]
- Heideman PD, Sylvester CJ. 1997. Reproductive photoresponsiveness in unmanipulated Fischer 344 laboratory rats. Biol Reprod 57:134–138. [DOI] [PubMed] [Google Scholar]
- Heideman PD, Bruno TA, Singley JW, Smedley JV. 1999a. Genetic variation in photoperiodism in Peromyscus leucopus: geographic variation in an alternative life-history strategy. J Mamm 80: 1232–1242. [Google Scholar]
- Heideman PD, Kane SL, Goodnight AL. 1999b. Differences in hypothalamic 2-[125I]iodomelatonin binding in photoresponsive and non-photoresponsive white-footed mice, Peromyscus leuco- pus. Brain Res 840:56–64. [DOI] [PubMed] [Google Scholar]
- Heideman PD, Broussard DR, Tate JA, Avigdor M. 2007. Number of immunoreactive GnRH-containing neurons is heritable in a wild-derived population of white-footed mice (Peromyscus leucopus). Physiol Biochem Zool 80:534–541. [DOI] [PubMed] [Google Scholar]
- Heideman PD, Pittman JT, Schubert KA, et al. 2010. Variation in levels of luteinizing hormone and reproductive photoresponsiveness in a population of white-footed mice (Peromyscus leucopus). Am J Physiol Regul Integr Comp Physiol 298:R1543–R1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman PD. 2014. Circannual clocks in tropical bats and heritable variation in seasonal reproductive timing in temperate zone mice In: Numata H, Helm B, editors. Annual, lunar, and tidal clocks: patterns and mechanisms of nature’s enigmatic rhythms. Japan: Springer; p 309–331. [Google Scholar]
- Herbison AE, Porteous R, Pape J-R, Mora JM, Hurst PR. 2008. Gonadotropin-releasing hormone neuron requirements for puberty, ovulation, and fertility. Endocrinology 149:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM,Tamrazian A, Xie H, et al. 2014. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology 155:4043–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korytko AI, Marcelino J, Blank JL. 1995. Differential testicular responses to short daylength in deer mice are reflected by regional and morphological differences in the GnRH neuronal system. Brain Res 685:135–142. [DOI] [PubMed] [Google Scholar]
- Korytko AI, Dluzen DE, Blank JL. 1997. Photoperiod and steroid- dependent adjustments in hypothalamic gonadotropic hormonereleasing hormone, dopamine, and norepinephrine content in male deer mice. Biol Reprod 56:617–624. [DOI] [PubMed] [Google Scholar]
- Korytko AI, Vessey SH, Blank JL. 1998. Phenotypic differences in the GnRH neuronal system of deer mice Peromyscus maniculatus under a natural short photoperiod. J Reprod Fertil 114:231–235. [DOI] [PubMed] [Google Scholar]
- Kriegsfeld LJ, Nelson RJ. 1999. Photoperiod affects the gonadotropin-releasing hormone neuronal system of male prairie voles (Microtus ochrogaster). Neuroendocrinology 69:238–244. [DOI] [PubMed] [Google Scholar]
- Kriegsfeld LJ, Ranalli NJ, Bober MA, Nelson RJ. 2000. Photoperiod and temperature interact to affect the GnRH neuronal system of male prairie voles (Microtus ochrogaster). J Biol Rhythms 15: 306–316. [DOI] [PubMed] [Google Scholar]
- Landsheer JA, van den Wittenboer G. 2015. Unbalanced 2×2 factorial designs and the interaction effect: a troublesome combination. PLoS ONE 10:e0121412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BB, Wester MR, Miller LE, et al. 2009. Cloning and characterization ofvoltage-gated calcium channel alpha1 subunits in Xenopus laevis during development. Dev Dyn 238:2891–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt SL, Jones JH. 1987. Symmorphosis: the concept of optimal design In: Feder ME, Bennett AF, Burggren WW, Huey RB, editors. New directions in ecological physiology. Cambridge: Cambridge University Press; p 289–309. [Google Scholar]
- Lorincz AM, Shoemaker MB, Heideman PD. 2001. Genetic variation in photoperiodism among naturally photoperiodic rat strains. Am J Physiol Regul Integr Comp Physiol 281:R1817–R1824. [DOI] [PubMed] [Google Scholar]
- Mintz EM, Lavenburg KR, Blank JL. 2007. Short photoperiod and testosterone-induced modification of GnRH release from the hypothalamus of Peromyscus maniculatus. Brain Res 1180:20–28. [DOI] [PubMed] [Google Scholar]
- Mundry R 2011. Issues in information theory-based statistical inference—a commentary from a frequentist’s perspective. Behav Ecol Sociobiol 65:57–68. [Google Scholar]
- Paxinos G, Watson C. 1986. The rat brain in stereotaxic coordinates. San Diego, California: Academic Press. [Google Scholar]
- Prendergast BJ, Kriegsfeld LJ, Nelson RJ. 2001. Photoperiodic polyphenisms in rodents: neuroendocrine mechanisms, costs and functions. Quart Rev Biol 76:293–325. [DOI] [PubMed] [Google Scholar]
- Quaynor SD, Ko EK, Chorich LP, et al. 2015. NELF knockout is associated with impaired pubertal development and subfertility. Mol Cell Endocrinol 407:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SJ, Oum R, Heideman PD. 2006. Phenotypic plasticity of reproduction in response to timed food access and photoperiod in artificially selected white-footed mice (Peromyscus leucopus). Oecologia 150:373–382. [DOI] [PubMed] [Google Scholar]
- Richards SA. 2008. Dealing with overdispersed count data in applied ecology. J Appl Ecol 45:218–227. [Google Scholar]
- Ross A, Johnson CE, Bell L, et al. 2009. Divergent regulation of hypothalamic neuropeptide Y and agouti-related protein by photoperiod in F344 rats with differential food intake and growth. J Neuroendocrinol 21:610–619. [DOI] [PubMed] [Google Scholar]
- Sharp K, Bucci D, Zelensky PK, et al. 2015. Genetic variation in male sexual behaviour in a population of white-footed mice in relation to photoperiod. Anim Behav 104:203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman AJ, Livne I, Witkin JW. 1994. The gonadotropin-releasing hormone (GnRH), neuronal systems: immunocytochemistry and in situ hybridization In: Knobil E, Neill JD, editors. The physiology of reproduction. New York: Raven Press; p 1683–1709. [Google Scholar]
- Stevenson TJ, Ball GF. 2009. Anatomical localization of the effects of reproductive state, castration, and social milieu on cells immunoreactive for gonadotropin-releasing hormone-I in male European starlings (Sturnus vulgaris). J Comp Neurol 517: 146–155. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, Bernard DJ, Ball GF. 2009. Photoperiodic condition is associated with region-specific expression of GNRH1 mRNA in the preoptic area of the male starling (Sturnus vulgaris). Biol Reprod 81:674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Hahn TP, MacDougall-Shackleton SA, Ball GF. 2012. Gonadotropin-releasing hormone plasticity: a comparative perspective. Front Neuroendocrinol 33:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds ME, Moussalli A. 2011. A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike’s information criterion. Behav Ecol Sociobiol 65: 13–21. [Google Scholar]
- Terman CR. 1993. Studies of natural populations of white-footed mice: reduction of reproduction at varying densities. J Mamm 74:678–687. [Google Scholar]
- Underwood E 2015. The brain’s identity crisis. Science 349:575–577. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Taylor CR, Bolis L, editors. 1998. Principles of animal design: the optimization and symmorphosis debate. New York: Cambridge University Press. [Google Scholar]
- Wierman ME, Kiseljak-Vassiliades K, Tobet S. 2011. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol 32:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Herrup K. 1988. The control of neuron number. Ann Rev Neurosci 11:423–453. [DOI] [PubMed] [Google Scholar]
- Xu L-W, Mei B, Chen R-R, Guo H-X, Wang J-J. 2013a. Parametric bootstrap tests for unbalanced nested designs under hetero- scedasticity. J Stat Comput Simulation 84:2059–2070. [Google Scholar]
- Xu L-W, Yang F-Q, Abula AE, Qin S. 2013b. A parametric bootstrap approach for two-way ANOVA in presence of possible interactions with unequal variances. J Multivariate Anal 115:172–180. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.