Abstract
目的
探讨长链非编码RNA PTENP1在膀胱癌发生发展中的分子机制。
方法
通过逆转录实时定量PCR(qRT-PCR)分别检测PTENP1,PTEN,miR-17在膀胱癌中的基础表达并关联分析。利用Western blot检测膀胱癌细胞系T24与5637中超表达PTENP1后PTEN的表达变化。通过荧光素酶报告试验验证miR-17对PTENP1及PTEN的靶向作用。最后通过在膀胱癌细胞系T24和5637中共表达miR-17和PTENP1,建立稳定共表达细胞系进行增殖和迁移实验,探索长链非编码RNA PTENP1在膀胱癌发生发展中的分子机制。
结果
miR-17在膀胱癌中普遍呈现高表达趋势,PTENP1在膀胱癌中呈现普遍低表达趋势(P<0.05)。与此同时miR-17和PTENP1的基础表达为负相关,PTENP1与PTEN的基础表达为正相关。WB实验发现于膀胱癌细胞系T24和5637中过表达PTENP1后可以在翻译水平增加PTEN的表达,荧光素酶报道实验验证了miR-17可同时靶向PTENP1及PTEN,在膀胱癌中miR-17具有促癌功能,同时在膀胱癌细胞中我们发现miR-17可以部分回复PTENP1的抑癌功能。
结论
长链非编码RNA PTENP1在膀胱癌中发挥抑癌功能的分子机制可能是PTENP1结合miR-17作为竞争性内源RNA竞争,从而降低miR-17对抑癌基因PTEN的表达抑制。
Keywords: 膀胱癌, PTENP1, 分子机制, PTEN, miR-17, 竞争性内源RNA
Abstract
Objective
To explore the molecular mechanism underlying the biological function of lncRNA PTENP1 in bladder cancer.
Methods
Expressions of PTENP1, PTEN and miR-17 were examined by quantitative reverse transcriptase PCR (qRTPCR) in 12 bladder cancer tissues. The expression of PTEN was examined by Western blotting in bladder cancer cell lines T24 and 5637 overexpressing PTENP1. Luciferase reporter assay was performed to confirm the targeting of miR-17 to PTENP1 and PTEN. T24 and 5637 cell lines with stable overexpression of PTENP1 and mir-17 were used to investigate effect of PTNE and miR-17 on the function of PTENP1 in bladder cancer.
Results
The expression of miR-17 was up-regulated and PTENP1 and PTEN were down-regulated in bladder cancer tissues, where a positive correlation was found between PTENP1 and PTEN expressions and a negative correlation between PTENP1 and miR-17 (P < 0.05). Overexpression of PTENP1 in bladder cancer cell lines T24 and 5637 obviously enhanced the expression of PTEN protein. miR-17 was found to target both PTENP1 and PTEN and promote the growth of bladder cancer. miR-17 could partially restore the tumor-suppressing activity of PTENP1 in bladder cancer.
Conclusion
By binding with miR-17, lncRNA PTENP1 functions as a PTEN competing endogenous RNA (ceRNA) to suppress the progression of bladder cancer.
Keywords: bladder cancer, PTENP1, molecular mechanism, PTEN, miR-17, competing endogenous RNA
肿瘤是一组基因疾病,与遗传代谢和环境因素紧密相关,膀胱肿瘤作为我国人群中泌尿系统肿瘤中发病率第1位,随着经济医疗水平的提高和预期寿命的延长,其发病率还在不断升高,为公共卫生带来巨大压力,严重影响病患的生活质量[1]。在病理分型中主要为移行上皮癌,约占95%,其他类型占5%左右。临床工作中初诊的膀胱癌约有3/4为非肌层浸润性,剩下的为肌层浸润性膀胱肿瘤[2]。根据浸润与否其治疗方式完全不一样[3]。由于肌层浸润性膀胱癌愈合较差且膀胱癌的复发率高,目前治疗方法局限性日益突现,迫切需要新的治疗策略。多年来对膀胱肿瘤的研究提示膀胱肿瘤的发生涉及到一系列的分子生物学改变[4-6],但膀胱肿瘤的发生发展机制还远没有阐明,膀胱肿瘤的发生还有许多未知分子事件的参与。PTEN的肿瘤抑制作用已经在多种肿瘤中报道[7-9]。长链非编码RNA PTENP1作为PTEN的假基因,与PTEN高度同源,定位于9p13.3,参与了多种人类疾病的发生与发展,可以通过与小分子RNA相关作用影响PTEN的表达从而负向调节PI3-K/Akt/mTOR信号通路而发挥抑癌作用,同时有研究证明了PTENP1可以作为内源性竞争性RNA竞争靶向调节PTEN的小分子RNA调节肿瘤的发生发展[7, 10-14]。我们通过研究证明了膀胱肿瘤中PTENP1的低表达及其肿瘤抑制功能,探索在膀胱肿瘤中PTENP1是否可以作为内源性竞争性RNA参与膀胱肿瘤的发生发展,其研究意义在于在膀胱肿瘤中明确PTENP1表达的甲基化调控机制以及PTENP1的表达与临床特点之间的关系,明确PTENP1对膀胱癌细胞生长和转移的影响,继而探索PTENP1参与调节调剂膀胱癌发生和发展的分子机制。深入研究PTENP1在膀胱癌中的作用及其分子机制,不仅有利于我们更加深入地了解膀胱癌的发生发展过程中的关键的分子事件,同时有利于在目前的条件下寻找新的治疗靶点以及进一步丰富膀胱癌相关分子生物学理论。PTENP1在膀胱癌中作为内源性竞争性RNA的存在意义还知之甚少,并在临床研究方面缺乏细致数据,这是我们下一步的主要研究内容,明确PTENP1在膀胱癌诊疗中的价值。
1. 材料和方法
1.1. 细胞与实验试剂
膀胱癌细胞系购买自ATCC或者中科院上海细胞库,培养基分别为10%胎牛血清的1640,MEM和DMEM培养基于5% CO2,37 ℃培养箱中。胎牛血清、1640、MEM和DMEM培养基及流式检测试剂盒购自南京凯基生物有限公司;转染试剂Lipofectamine2000、逆转录试剂及荧光定量PCR试剂购自美国Invitrogen;miR-17模拟物(mimics)、抑制物(inhibitor)及miR-17荧光定量PCR检测引物由广州锐博生物科技公司设计并合成。CCK-8实验试剂及transwell小室购买自康宁公司,PTEN及抗GAPDH抗体购自美国CST。双荧光素酶报告基因试剂盒购自美国Promega。
1.2. 实验分组
分别将膀胱癌细胞T24及5637根据不同处理因素分别分为4组:感染Lenti-PTENP1组,感染Lenti-NC组,共转染Lenti-PTENP1及miR-17组,单独转染miR-17组。按照Lipofectamine2000试剂说明书常规转染,方法简述如下:将20 pmol的microRNA及6(T24)到8(5637)μL脂质体分别稀释于250 μL无血清培养基,室温静置5 min后轻柔混匀,再室温静置10 min后均与加入各组培养板中,使microRNA的终浓度为10 nmol/L,温箱培养6 h后换为完全培养基常规培养48 h行后续实验,病毒感染在感染72 h后进行后续实验。
1.3. 实验方法
1.3.1. 实时荧光定量PCR(Realtime-qPCR)
收集各组后的T24及5637细胞,Trizol一步法提取其中总RNA,常规逆转录合成cDNA,利用realtimeqPCR试剂盒PCR扩增,利用MX3000检测并分析细胞中miR-17,PTENP1的表达水平。
1.3.2. 双荧光素酶报告基因实验
将5637和T24细胞以1×105/孔密度接种于24孔板,第2天当细胞约70%融合时,利用Lipofectamine 2000共转染包含有PTEN的3'UTR的荧光素酶质粒和miR-17模拟物(mimics)或抑制物(inhibitor)或者Lenti-PTENP1,常规培养48 h后,按照双荧光素酶报告基因检测试剂盒(Promega)在多功能亮度计上测定并分析。
1.3.3. 蛋白免疫印迹(Western blot)实验
收集转染处理48 h或者72 h后的5637及T24细胞,加入适量NP40裂解液,获取细胞总蛋白液,加入适量SDS上样缓冲液100 ℃水浴解除蛋白交联。各组蛋白样品行聚丙烯酰胺凝胶电泳,PVDF膜转膜并用脱脂奶粉封闭2 h,然后依次孵育一抗4 ℃过夜及辣根过氧化物酶标记的二抗室温1 h后化学发光法显影,检测细胞中PTEN的表达情况,以GAPDH作为内参。
1.3.4. 细胞增殖实验及迁移侵袭实验
收集转染处理48 h或者72 h后的5637及T24细胞,严格按照实验设计分组消化获得细胞悬液后种于96孔板中,每孔1000个(T24)或者2000(5637)个,4个复孔,每隔24 h一个时间点应用CCK8试剂1: 10(无血清培养基)细胞培养箱孵育细胞2 h后酶标液检测并分析结果。迁移和侵袭实验中严格按照实验设计分组消化获得细胞悬液后种于24孔板transwell小室中,每孔1×104(T24)或者5×104(5637)于细胞培养箱培养16~18(T24)h后或者24 h后,弃细胞培养基后应用甲醇固定细胞10 min,之后应用0.5%的结晶紫染色细胞15 min后擦拭去未能迁移和侵袭的细胞,利用100倍显微镜观察并分析结果。
1.4. 统计学方法
用SPSS 18.0统计学软件进行统计学分析,符合正态分布的总体数据两组之间的比较采用t检验,3组数据之间的两两比较采用方差分析,P<0.05认为差异有统计学意义。
2. 结果
2.1. 膀胱癌中PTENP1和miR-17的表达检测及表达
在膀胱癌组织中运用实时定量PCR检测长链非编码RNA PTENP1的表达,发现在12对膀胱癌组织中有10对组织中肿瘤组织PTENP1呈现低表达(P<0.05,图 1A)。在膀胱癌组织中运用实时定量PCR检测miR-17的表达,发现在12对膀胱癌组织中有11对组织中肿瘤组织miR-17呈现高表达(P<0.05,图 1B)。将PTENP1的表达和miR-17在膀胱癌组织行表达相关分析,在膀胱癌组织中PTENP1的表达和miR-17的表达呈负相关(P<0.01,图 1C)。将PTEN的表达和miR-17在膀胱癌组织行表达相关分析,在膀胱癌组织中PTEN的表达和miR-17的表达呈负相关(P<0.01,图 1D)。将PTENP1的表达和PTEN在膀胱癌组织行表达相关分析,在膀胱癌组织中PTENP1的表达和PTEN的表达呈正相关(P<0.01,图 1E)。在膀胱癌细胞系T24和5637通过慢病毒感染稳定表达PTENP1后应用WB检测细胞中PTEN的表达变化,过表达PTENP1可以引起细胞中PTEN的表达增加(图 1F)。
1.

膀胱癌中PTENP1和miR-17的表达检测及表达
Expression of PTENP1 and miR-17 in bladder cancer. A: Expression of PTENP1 was down-regulated in bladder cancer tissues (P < 0.01); B: Expression of miR-17 was up-regulated in bladder cancer tissues (P < 0.01); C:Expressions of miR-17 and PTENP1 were negatively correlated in cancer tissues (P < 0.01); D: Expression of miR-17and PTEN were negatively correlated in cancer tissues (P < 0.01); E: Expressions of PTEN and PTENP1 were positively correlated in cancer tissues (P < 0.01); F: Expression of PTEN was up-regulated by overexpression of PTENP1 in bladder cancer cell lines (Western blotting).
2.1. 荧光素酶报告实验行靶标验证
为了验证PTENP1及PTEN是否为miR-17的直接靶标,我们构建了荧光素酶报告基因质粒系统,克隆了PTEN下游的野生型3'UTR及突变型3'UTR。通过在膀胱癌细胞系T24及5637细胞中共转染野生型3'UTR荧光素酶报告基因质粒系统和miR-17,孵育后,酶标仪检测发现miR-17可以减少含PTEN野生型3'UTR载体的荧光素酶的表达而对含PTEN突变型3'UTR的载体的荧光素酶的表达无影响;而在系统中用Inhibitor降低miR-17的表达水平能够增加含PTEN野生型3'UTR载体的荧光素酶的表达而对含PTEN突变型3'UTR的载体无影响(图 2)。通过膀胱癌两株细胞系均得到相同的结果。
2.
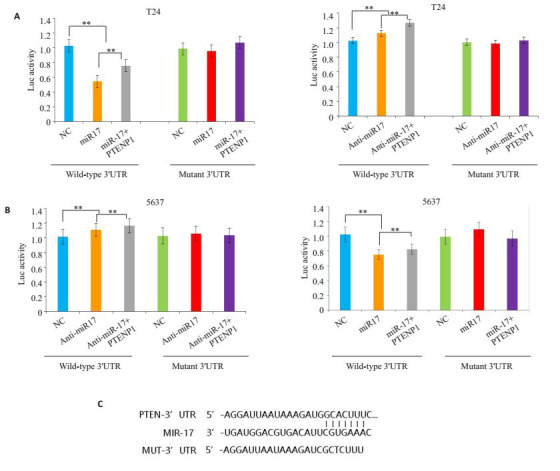
荧光素酶报告实验行靶标验证
Luciferase reporter system. A: luciferase signals of Wild-type or mutant PTEN reporters in bladder cancer cell line T24 transfected with miR-17 mimics, miR-17 inhibitors, miR-17 mimics with PTENP1, or miR-17 inhibitors with PTENP1 (P < 0.01); B: Luciferase signals of Wild-type or mutant PTEN reporters in bladder cancer cell line 5637transfected with miR-17 mimics, miR-17 inhibitors, miR-17 mimics with PTENP1, or miR-17 inhibitors with PTENP1(P < 0.01); C: Sequences of PTEN 3'UTR, miR-17, and mutant PTEN 3'UTR.
2.3. 在膀胱癌细胞系5637和T24细胞中共转染PTENP1及miR-17后细胞生物学行为的改变
在膀胱癌细胞系中进行相关功能实验(图 3A):在膀胱癌细胞系5637和T24中分4组行增殖实验,分别为:Lenti-PTENP1,Lenti-PTENP1+miR-17,Lenti-NC及miR-17。发现Lenti-PTENP1组增殖率最低,生长抑制最明显,抑制率接近60%,而miR-17实验组细胞增殖最快,相对于Lenti-NC,增值率达到120%左右,同时Lenti-PTENP1+miR-17的增值率介于Lenti-PTENP1组和Lenti-NC组,回复程度在50%左右。后续再次行细胞迁移和侵袭实验,选择的细胞不变,实验分组还是不变,可以发现:Lenti-PTENP1组迁移和侵袭最低,抑制最明显,抑制率接近65%左右,而miR-17实验组细胞迁移和侵袭最快,相对于Lenti-NC,迁移和侵袭能力是阴性对照组的1.2倍左右,同时Lenti-PTENP1+miR-17的迁移和侵袭能力介于Lenti-PTENP1组和Lenti-NC组,回复程度在50%左右(图 3B)。
3.
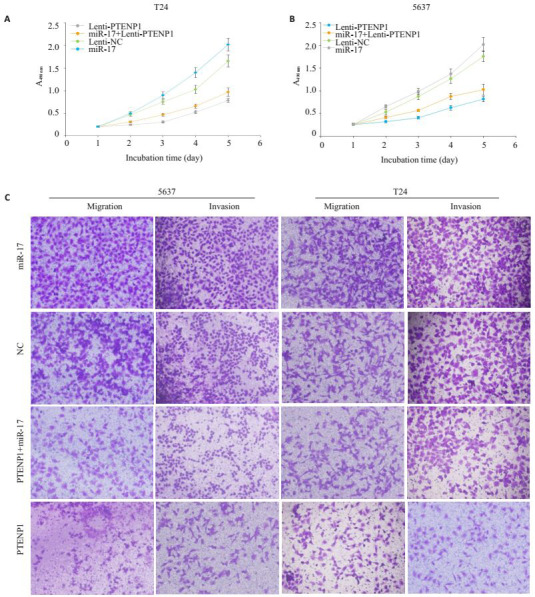
在膀胱癌细胞系5637和T24细胞中共转染PTENP1及miR-17后细胞生物学行为的改变
PTENP1 as a ceRNA in the regulation of tumor growth in bladder cancer. A, B: Proliferation of T24 and 5637 cells expressing control vector, PTENP1, miR-17, or miR-17 with PTENP1. Cell viability was quantified at each time point.PTENP1 reversed accelerated cell growth induced by miR-17; C: Migration and invasion assay of T24 and 5637 cells expressing control, miR-17, PTENP1, or miR-17 with PTENP1 (crystal violet staining, ×100).
3. 讨论
非编码RNA长期以来被认为转录垃圾,没有相关生物学功能,但是越来越多分子生物学事件无法解释,需要引入非编码RNA,且越来越多报道了非编码RNA在多种病理生理过程中发挥着重要的调节功能,非编码RNA根据其转录本长度可以分为长链非编码RNA和短链非编码RNA,短链非编码RNA中重要的一种小分子RNA已经有大量报道了其重要的生命过程调节功能,长链非编码RNA指的是转录本大于200 nt的RNA,PTENP1作为长链非编码RNA的一种,作为PTEN的假基因,与PTEN有高度同源性,在多种肿瘤中发挥重要的调节功能[15-16]。本实验中我们证明了PTENP1在膀胱肿瘤中的肿瘤抑制功能,并且探索了其作用机制,膀胱肿瘤的分子生物学理论进一步丰富。功能分子作用机制的研究,阐明其涉及的通路,明确通路中相关分子的位置及地位,不仅能够更好地解释生物现象,而且可以针对重要靶点设计相关药物,增强或阻断相关通路,从而产生相关影响,达到改变生物学过程的目的,同时可能可以发展出重要的疾病治疗策略[17-20]。长链非编码RNA的作用机制是多种多样的[21],本研究通过对PTENP1的作用机制进行研究发现了在膀胱肿瘤中PTENP1能够被同时靶向抑癌基因PTEN的miR-17靶向作用,PTENP1通过miR-17与PTEN相互作用,影响PTEN蛋白的表达而发挥作用。PTEN是经典的抑癌基因,在多种癌症中表达缺失,其能够编码一种磷酸酶,能够通过负向调节PI3-K/Akt/mTOR信号通路而发挥抑癌作用[8, 22-24]。RNA与RNA之间可以通过竞争结合共同小分子RNA而相互作用,通过小分子RNA桥接作用调节RNAs,非编码RNA及可编码RNA均可以被调控,故而可形成多量而复杂的CeRNA调节网络,调节多种人类肿瘤的发生和发展[22-29]。ceRNA作用使得mRNA不仅蛋白编码功能,还具备反式调节其他mRNA的作用,同时lncRNA也参与了这种RNA之间的作用,这在转录组学水平赋予了mRNAs、lncRNA新的生物学功能,扩大了基因组中的功能性遗传信息,是近年来的分子生物学上的一个重要进展[30]。本实验通过实验证明在膀胱癌PTENP1与PTEN存在某种相互作用,考虑到PTENP1位PTEN的假基因,具有高度同源性,我们有理由相信miR-17可能作为其中一个桥接分子参与PTENP1与PTEN的相互作用,初步判定长链非编码RNA PTENP1在膀胱癌中发挥抑制肿瘤增殖和迁移侵袭的分子机制可能是PTENP1结合miR-17可以作为竞争性内源RNA竞争,从而降低miR-17对抑癌基因PTEN的表达抑制。虽然已阐明PTENP1在膀胱肿瘤中CeRNA作用,但是并不代表PTENP1仅仅作为CeRNA调节膀胱肿瘤发生发展,其其他作用机制有待于进一步研究。综上所述,在膀胱肿瘤中PTENP1的功能作用及其与PTEN及miR-17的相互作用关系符合近年来提出的竞争性内源RNA的作用机制,通过共同的miRNA进行RNA-RNA相互作用调节膀胱肿瘤的发生发展,进一步丰富了膀胱肿瘤分子生物学理论。
Biography
余淦,博士,E-mail: cdyi1987@126.com
Funding Statement
澳门理工学院科研项目(RP/ESS-03/2015)
Contributor Information
余 淦 (Gan YU), Email: cdyi1987@126.com.
郎 斌 (Bin LANG), Email: blang@ipm.edu.mo.
References
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.v127:12. [Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008 [J]. Int J Cancer, 2010, 127(12): 2893-917.] [DOI] [PubMed] [Google Scholar]
- 2.Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. 2009;27(3):289–93. doi: 10.1007/s00345-009-0383-3. [Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world[J]. World J Urol, 2009, 27 (3): 289-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenzl A, Cowan NC, De Santis M, et al. Treatment of muscleinvasive and metastatic bladder cancer: update of the EAU guidelines. Actas Urol Esp. 2011;59(6):1009–18. doi: 10.1016/j.eururo.2011.03.023. [Stenzl A, Cowan NC, De Santis M, et al. Treatment of muscleinvasive and metastatic bladder cancer: update of the EAU guidelines [J]. Actas Urol Esp, 2011, 59(6): 1009-18.] [DOI] [PubMed] [Google Scholar]
- 4.Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. https://www.ncbi.nlm.nih.gov/pubmed/6206131. J Immunol. 1984;133(4):1710–5. [Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67 [J]. J Immunol, 1984, 133(4): 1710-5.] [PubMed] [Google Scholar]
- 5.Bertz S, Otto W, Denzinger S, et al. Combination of CK20 and Ki-67 immunostaining analysis predicts recurrence, progression, and cancer-specific survival in pT1 urothelial bladder cancer. Eur Urol. 2014;65(1):218–26. doi: 10.1016/j.eururo.2012.05.033. [Bertz S, Otto W, Denzinger S, et al. Combination of CK20 and Ki-67 immunostaining analysis predicts recurrence, progression, and cancer-specific survival in pT1 urothelial bladder cancer[J]. Eur Urol, 2014, 65(1): 218-26.] [DOI] [PubMed] [Google Scholar]
- 6.Brems-Eskildsen AS, Zieger K, Toldbod H, et al. Prediction and diagnosis of bladder cancer recurrence based on urinary content of hTERT, SENP1, PPP1CA, and MCM5 transcripts. http://d.wanfangdata.com.cn/OAPaper/oai_doaj-articles_9d87ce78fbdde9b3d0a462f7f389e832. BMC Cancer. 2010;10(2):646. doi: 10.1186/1471-2407-10-646. [Brems-Eskildsen AS, Zieger K, Toldbod H, et al. Prediction and diagnosis of bladder cancer recurrence based on urinary content of hTERT, SENP1, PPP1CA, and MCM5 transcripts [J]. BMC Cancer, 2010, 10(2): 646.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42(5):454–U136. doi: 10.1038/ng.556. [Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in Pten dose determine cancer susceptibility[J]. Nat Genet, 2010, 42 (5): 454-U136.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. http://d.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_1221969. Nat Rev Mol Cell Biol. 2012;13(5):283–96. doi: 10.1038/nrm3330. [Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor[J]. Nat Rev Mol Cell Biol, 2012, 13 (5): 283-96.] [DOI] [PubMed] [Google Scholar]
- 9.Bouali S, Chretien AS, Ramacci C, et al. PTEN expression controls cellular response to cetuximab by mediating PI3K/AKT and RAS/ RAF/MAPK downstream signaling in KRAS wild-type, hormone refractory prostate cancer cells. http://www.ncbi.nlm.nih.gov/pubmed/19212633. Oncol Rep. 2009;21(3):731–5. [Bouali S, Chretien AS, Ramacci C, et al. PTEN expression controls cellular response to cetuximab by mediating PI3K/AKT and RAS/ RAF/MAPK downstream signaling in KRAS wild-type, hormone refractory prostate cancer cells [J]. Oncol Rep, 2009, 21(3): 731-5.] [PubMed] [Google Scholar]
- 10.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133(3):403–14. doi: 10.1016/j.cell.2008.04.013. [Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression [J]. Cell, 2008, 133(3): 403-14.] [DOI] [PubMed] [Google Scholar]
- 11.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3206313/?cmd=HTOff&. Nature. 2010;465(731):1033–8. doi: 10.1038/nature09144. [Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology [J]. Nature, 2010, 465(731): 1033-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesson LB, Packham D, Pontzer E, et al. A reinvestigation of somatic hypermethylation at the PTEN CpG island in Cancer cell lines. Biol Proced Online. 2012;14(1):5. doi: 10.1186/1480-9222-14-5. [Hesson LB, Packham D, Pontzer E, et al. A reinvestigation of somatic hypermethylation at the PTEN CpG island in Cancer cell lines [J]. Biol Proced Online, 2012, 14(1): 5.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, Yao WM, Gumireddy K, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress Clear-Cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13(12):3086–97. doi: 10.1158/1535-7163.MCT-14-0245. [Yu G, Yao WM, Gumireddy K, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress Clear-Cell renal cell carcinoma progression [J]. Mol Cancer Ther, 2014, 13(12): 3086-97.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yndestad S, Austreid E, Knappskog SA, et al. High PTEN gene expression is a negative prognostic marker in human primary breast cancers with preserved p53 function. Breast Cancer Res Treat. 2017;163(1):177–90. doi: 10.1007/s10549-017-4160-5. [Yndestad S, Austreid E, Knappskog SA, et al. High PTEN gene expression is a negative prognostic marker in human primary breast cancers with preserved p53 function[J]. Breast Cancer Res Treat, 2017, 163(1): 177-90.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattick JS. Long noncoding RNAs in cell and developmental biology. Semin Cell Dev Biol. 2011;22(4):327. doi: 10.1016/j.semcdb.2011.05.002. [Mattick JS. Long noncoding RNAs in cell and developmental biology [J]. Semin Cell Dev Biol, 2011, 22(4): 327.] [DOI] [PubMed] [Google Scholar]
- 16.Tang JY, Lee JC, Chang YT, et al. Long noncoding RNAs-Related diseases, cancers, and drugs. http://www.ncbi.nlm.nih.gov/pubmed/23843741. ScientificWorldJournal. 2013;6:943539. doi: 10.1155/2013/943539. [Tang JY, Lee JC, Chang YT, et al. Long noncoding RNAs-Related diseases, cancers, and drugs[J]. ScientificWorldJournal, 2013, 6: 943539.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–68. doi: 10.1038/nbt.1685. [Portela A, Esteller M. Epigenetic modifications and human disease [J]. Nat Biotechnol, 2010, 28(10): 1057-68.] [DOI] [PubMed] [Google Scholar]
- 18.Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144(6):986–98. doi: 10.1016/j.cell.2011.02.016. [Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease [J]. Cell, 2011, 144(6): 986-98.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartwright BR, Goodman JM. Seipin: from human disease to molecular mechanism. J Lipid Res. 2012;53(6):1042–55. doi: 10.1194/jlr.R023754. [Cartwright BR, Goodman JM. Seipin: from human disease to molecular mechanism [J]. J Lipid Res, 2012, 53(6): 1042-55.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mcgonagle D, De Bari C, Arnold P, et al. Lessons from musculoskeletal stem cell research: the key to successful regenerative medicine development. Arthritis Rheum. 2007;56(3):714–21. doi: 10.1002/(ISSN)1529-0131. [Mcgonagle D, De Bari C, Arnold P, et al. Lessons from musculoskeletal stem cell research: the key to successful regenerative medicine development[J]. Arthritis Rheum, 2007, 56 (3): 714-21.] [DOI] [PubMed] [Google Scholar]
- 21.Guttman M, Rinn JL. Modular regulatory principles of large noncoding RNAs. Nature. 2012;482(7385):339–46. doi: 10.1038/nature10887. [Guttman M, Rinn JL. Modular regulatory principles of large noncoding RNAs [J]. Nature, 2012, 482(7385): 339-46.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan I, Kerwin J, Owen K, et al. Registered report: A codingindependent function of gene and pseudogene mRNAs regulates tumour biology. Elife. 2015;4 doi: 10.7554/eLife.08245. [Khan I, Kerwin J, Owen K, et al. Registered report: A codingindependent function of gene and pseudogene mRNAs regulates tumour biology [J]. Elife, 2015, 4. doi: 10.7554/eLife.08245.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo XQ, Deng L, Deng KY. etal/> Pseudogene PTENP1 suppresses gastric cancer progression by modulating PTEN. Anticancer Agents Med Chem. 2016;16(4):456–64. doi: 10.2174/1871520615666150507121407. [Guo XQ, Deng L, Deng KY, et al. Pseudogene PTENP1 suppresses gastric cancer progression by modulating PTEN[J]. Anticancer Agents Med Chem, 2016, 16(4): 456-64.] [DOI] [PubMed] [Google Scholar]
- 24.Dong L, Qi P, Xu MD, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137(5):1128–35. doi: 10.1002/ijc.v137.5. [Dong L, Qi P, Xu MD, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls [J]. Int J Cancer, 2015, 137(5): 1128-35.] [DOI] [PubMed] [Google Scholar]
- 25.Chen CL, Tseng YW, Wu JC, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [Chen CL, Tseng YW, Wu JC, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation[J]. Biomaterials, 2015, 44, (2): 71-81.] [DOI] [PubMed] [Google Scholar]
- 26.Tang JS, Ning RH, Zeng B, et al. Molecular evolution of PTEN pseudogenes in mammals. PLoS One. 2016;11(12):e0167851. doi: 10.1371/journal.pone.0167851. [Tang JS, Ning RH, Zeng B, et al. Molecular evolution of PTEN pseudogenes in mammals [J]. PLoS One, 2016, 11(12): e0167851.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Guo Y, Ma Z, et al. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. https://www.ncbi.nlm.nih.gov/pubmed/28212532/ Oncotarget. 2017;8(16):26079–89. doi: 10.18632/oncotarget.15317. [Zhang R, Guo Y, Ma Z, et al. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer[J]. Oncotarget, 2017, 8(16): 26079-89.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masunaga A, Omatsu M, Kunimura T, et al. Expression of PTEN and its pseudogene PTENP1, and promoter methylation of PTEN in non-tumourous thymus and thymic tumours. J Clin Pathol. 2017;70(8):690–6. doi: 10.1136/jclinpath-2016-204220. [Masunaga A, Omatsu M, Kunimura T, et al. Expression of PTEN and its pseudogene PTENP1, and promoter methylation of PTEN in non-tumourous thymus and thymic tumours [J]. J Clin Pathol, 2017, 70(8): 690-6.] [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Zhang N, Wang Z, et al. Pseudogene PTENP1 functions as a competing endogenous RNA (ceRNA) to regulate PTEN expression by sponging miR-499-5p. Biochemistry (Mosc) 2016;81(7):739–47. doi: 10.1134/S0006297916070105. [Wang L, Zhang N, Wang Z, et al. Pseudogene PTENP1 functions as a competing endogenous RNA (ceRNA) to regulate PTEN expression by sponging miR-499-5p[J]. Biochemistry (Mosc), 2016, 81(7): 739-47.] [DOI] [PubMed] [Google Scholar]
- 30.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition [J]. Nature, 2014, 505(7483): 344-52.] [DOI] [PMC free article] [PubMed] [Google Scholar]


