Abstract
Scope: Maternal obesity leads to glucose intolerance in the offspring. Changes in the gut microbiota are being increasingly implicated in the pathogenesis of diabetes. We hypothesized that inulin intervention during gestation and lactation improves glucose metabolism disorders in mouse offspring from high-fat diet (HD)-fed dams.
Procedures: Female C57BL mice were fed a control diet or HD for 4 weeks before mating. After mating, pregnant mice were randomly divided into three groups through gestation and lactation: control diet (CD) group, HD group, and HD treated with inulin (HD-inulin) group. At weaning, glucose metabolic status was assessed. Gut microbial DNA from offspring cecal contents was isolated and processed for metagenomic shotgun sequencing, and taxonomic and functional profiling were performed.
Results: Offspring from dams in the HD-inulin groups demonstrated reduced fasting blood glucose, decreased blood glucose area under the curve during the oral glucose tolerance test, and reduced fasting serum insulin and HOMA-IR compared to offspring from dams in the HD group. Nineteen differentially abundant bacterial species were identified between the HD-inulin and HD groups. The HD-inulin group displayed significantly greater abundances of Bacteroides_acidifaciens, Eubacterium_sp_CAG_786, Clostridium_sp_CAG_343, and Bifidobacterium_breve species and lower abundances of Oscillibacter_sp_1_3, Ruminococcus_gnavus_CAG_126, and Bacteroides_massiliensis species. Differentially abundant bacterial species among the three groups were involved in 38 metabolic pathways, including several glucose and lipid metabolism pathways.
Conclusion: Our results show that early inulin intervention in HD-fed mouse dams moderates offspring glucose metabolism and gut dysbiosis.
Keywords: maternal, offspring, inulin, glucose metabolism, gut microbiota
Introduction
Nutrition status during the intrauterine period has been reported to lead to the programming of metabolic disorders in the offspring throughout their whole life span (1). Maternal overnutrition can program an increased risk of diabetes in rodent offspring (2, 3). Improving maternal metabolism may help curb the burden of metabolic disease in offspring (4). However, the mechanism through which beneficial outcomes are programmed in offspring remains to be elucidated. Increasing evidence has shown that the gut microbiota greatly affects host metabolism (5–7). Type 2 diabetes patients have dysbiotic microbiota, which plays a central role in the process of diabetes and provides unique markers for its diagnosis (8). Transplantation of “diabetic microbiota” to germ-free mice results in insulin resistance (9). Moreover, the maternal gut microbiota has an important contribution to the colonization of the offspring gut (10) and impacts long-term metabolic health (11). These studies suggest that moderation of the maternal gut microbiota may be a potential target for the prevention of diabetes in offspring.
Dietary inulin-type prebiotic treatment represents a promising strategy for altering the gut microbiota and affecting host metabolism and physiology (12, 13). As one type of prebiotic, inulin is extracted from chicory roots. Inulin cannot be hydrolyzed by digestive enzymes in the human small intestine but is fermented by probiotics (12). Inulin intake has been proven to reduce blood glucose and moderate insulin resistance in diabetic rodent models (14, 15). In clinical trials, inulin supplementation was found to moderate glycemic status in diabetic patients (16). Furthermore, in high-fat diet (HD)-induced diabetic rats, inulin had a beneficial impact on gut microbiota profiles (17).
We hypothesized that inulin intervention during gestation and lactation improves glucose metabolism disorder in offspring from HD-fed dams. To identify this alteration in gut microbiota, we used a metagenomic shotgun sequencing approach to analyze the gut microbiota of offspring from inulin-supplemented HD-fed dams. In this study, we used metagenomic shotgun sequencing to sequence the whole set of genes present in the gut microbiome. This sequencing information can provide the relative abundance of genes not only in functional pathways but also at all taxonomical levels (18). The aim of this project was to provide a comprehensive understanding of the gut microbial mechanism in offspring of HD-fed dams receiving early inulin intervention.
Materials and Methods
Animal Treatments and Diets
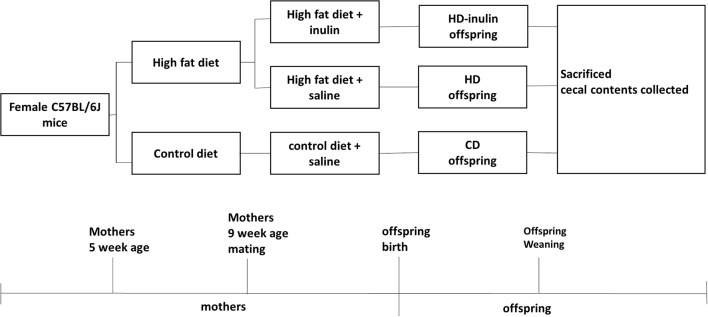
All animal experimental protocols were approved by the Animal Care Committee of Peking Union Medical Hospital (Permit Number: MC-07-6004). Five-week-old female C57BL6/J mice (body weight 13.03 ± 0.79 g) were given ad libitum access to control (kcal %: 10% fat, 20% protein, and 70% carbohydrate; 3.85 kcal/gm, n = 8) or high-fat diets (kcal %: 45% fat, 20% protein, and 35% carbohydrate; 4.73 kcal/gm; n = 16) for 4 weeks. At 9 weeks of age, females were bred with control male mice fed with a control diet. The vaginal plug was checked to confirm pregnancy. During the gestation and lactation period, female mice fed a control diet before gestation remained on a control diet (CD); female mice fed a high-fat diet before gestation went on either a high-fat diet (HD) only or a high-fat diet with 10% wt/wt inulin supplementation (HD-inulin, Vilof™ Soluble Dietary Fiber; BAHEAL Medical Inc., Qingdao, China and Fengning Ping'an High-tech Industrial Co., Ltd., Heber, China). Because of the differential sex phenotype following different maternal nutrition, this project researched only male offspring. At weaning, male offspring (n = 8 per group) were sacrificed. Mice were fasted for 10 h and anesthetized with chloral hydrate, and a blood sample was collected from the intraorbital retrobulbar plexus. The cecal contents were quickly removed, snap frozen on dry ice, and then stored at −80°C for further analysis. Figure 1 shows the experimental protocol.
Figure 1.
Timeline of animal protocol for different groups. CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
Measurement of Body Weight and Fasting Blood Glucose
The body weight of both the mother and pups was measured. Blood was collected from a tail bleed and analyzed to test glucose levels using a Contour TS glucometer (Bayer, Hamburg, Germany).
Oral Glucose Tolerance Test (OGTT)
At weaning, an OGTT was performed to assess glucose tolerance in pups. After 10 h of fasting, pups were given a glucose load (2.0 g/kg body weight) by gavage. Before (0 min) and at 30, 60, and 120 min after the gavage, the blood glucose levels were measured. The area under the glucose tolerance curve (AUC) of the OGTT was calculated as previously described (19).
Serum Insulin Assay and Homeostasis Model Assessment of Insulin Resistance (HOMA-IR)
At weaning, pups were fasted for 10 h to measure serum insulin by using an ELISA kit (Millipore, Billerica, MA, USA). Insulin sensitivity was assessed using HOMA-IR as previously described (19).
Microbial Sampling and DNA Isolation
Fifty micrograms of cecal contents was used for metagenomic DNA isolation using a TIANamp Stool DNA Kit (TIANGEN, Beijing, China). The quality and quantity of DNA was assessed using agarose gel electrophoresis and fluorometry (Qubit® dsDNA Assay Kit, Life Technologies, CA, USA).
Metagenomic Library Construction and Metagenomic Sequencing
One microgram of DNA was sheared to 350 bp fragments by sonication. After polishing and ligation with a full-length adaptor, DNA fragments were amplified using an NEBNext® Ultra™ DNA Library Prep Kit from Illumina (New England BioLabs Inc, Ipswich, MA, USA). Then, PCR amplification products were purified (AMPure XP System, Beckman Coulter, Woerden, Netherlands). DNA libraries underwent size distribution via an Agilent 2100 Bioanalyzer. Finally, DNA libraries were sequenced on the Illumina HiSeq 2000 Platform (Beijing Compass Biotechnology Company, China). Paired-end reads were generated for further processing.
Metagenome Preprocessing and Assembly
First, we removed the adaptors and low quality reads. Then, reads were filtered to exclude the host DNA genome based on the Mus musculus reference genome using Bowtie 2.2.4 software (20). Then, the clean data were assembled into contigs and analyzed by SOAPdenovo software v2.04 (21).
Gene Prediction, Taxonomy, and Functional Profiling
After assembling contigs, open reading frames (ORFs) were predicted by MetaGeneMark software (22). DIAMOND software was used to blast the Unigenes to the sequences of bacteria that were extracted from NCBI (23). For beta diversity, principal component analysis (PCA) plots were constructed. LEfSe analysis was conducted by using LEfSe software (24). DIAMOND software was used to blast the Unigenes to a KEGG functional database (25).
Statistics
The data are expressed as the mean ± SD. Differences among the groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. For sequencing data, statistical analyses were performed using R software (v. 2.15.3). For determination of the abundance of genes, taxonomies, and KEGG ontologies, the Mann-Whitney test was used for statistics between two groups, and the Kruskal-Wallis test was used for comparisons among three groups. Statistical analyses were performed in GraphPad Prism 6 (GraphPad Software Inc., CA, USA). Statistical significance was defined as P < 0.05.
Results
Maternal Body Weight and Changes in Fasting Blood Glucose
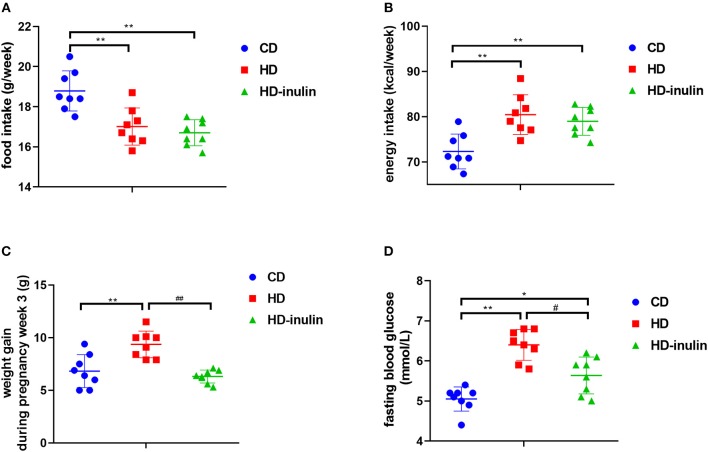
Before consuming a HD, dam body weight between groups did not differ (P > 0.05). After eating a HD for 4 weeks, the HD group had a higher body weight than the CD group (19.77 ± 1.46 g vs. 17.28 ± 1.16 g, P < 0.01). Despite decreased food intake in the HD group compared with the CD group (Figure 2A, P < 0.01), the energy intake of the HD group was higher than that of the CD group during the pregnancy period (Figure 2B, P < 0.01). Accordingly, the body weight change in HD mice was 139% of that in CD mice (Figure 2C, P < 0.01). Although no significant difference in food intake and energy intake was observed between the HD and HD-inulin groups, the body weight change in HD-inulin dams was 33.9% lower than that in the HD mice during the pregnancy period (Figure 2C, P < 0.01). Similarly, maternal HD feeding increased fasting blood glucose by 24.8% compared with CD mice (Figure 2D, P < 0.01). Interestingly, fasting blood glucose was reduced by 11.3% in HD-inulin dams compared with HD mice (Figure 2B, P < 0.05).
Figure 2.
Maternal food intake, energy intake, body weight and fasting blood glucose in different groups at weaning time. (A) Maternal weekly food intake during gestation and lactation period. (B) Maternal weekly energy intake during gestation and lactation period. (C) Maternal body weight. (D) Maternal fasting blood glucose. The data were shown as mean ± SD. n = 8 in each group. *P < 0.05, **P < 0.01 vs. CD group; #P < 0.05, ##P < 0.01 vs. HD group. CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
Offspring Body Weight
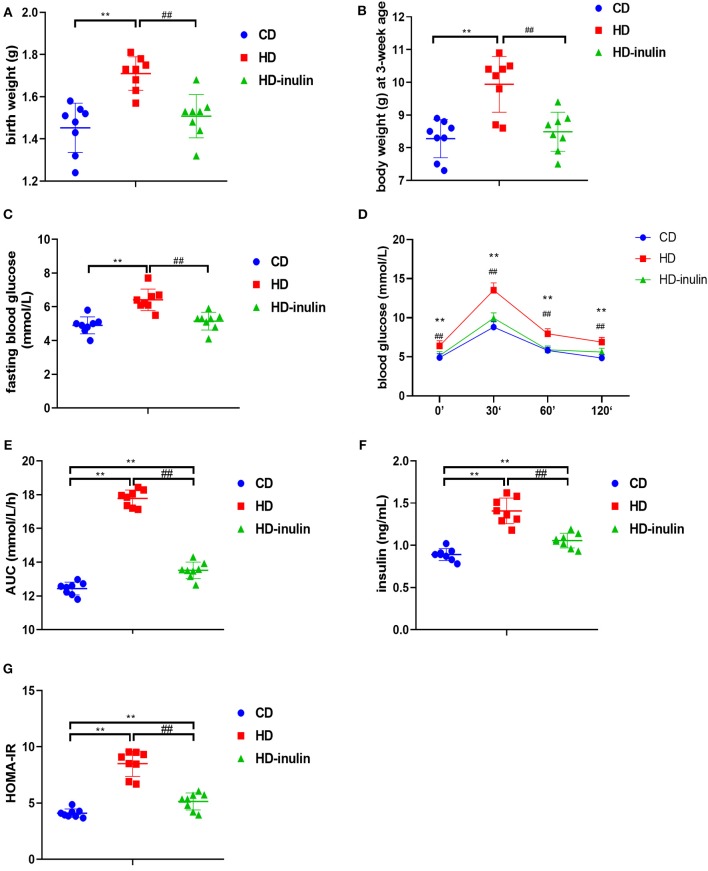
Litters from HD dams weighed more than litters from CD dams both at birth and at weaning time (P < 0.01, Figures 3A,B). However, the pups in the HD-inulin group had reduced body weight compared with pups in the HD group both at birth and at weaning (P < 0.01, Figures 3A,B).
Figure 3.
Metabolic variables of male mice offspring in different groups. (A) birth weight; (B) body weight at weaning; (C) fasting blood glucose; (D) oral glucose tolerance test (OGTT); (E) area under curve (AUC) in OGTT; (F) serum insulin; (G) HOMA-IR. The data were shown as mean ± SD. n = 8 in each group. **P < 0.01 vs. CD group; ##P < 0.01 vs. HD group. CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
Offspring Glucose Metabolic Index, Serum Insulin, and HOMA-IR
Compared with pups from CD dams, pups from dams fed a HD displayed increased fasting blood glucose, blood glucose during OGTT, and AUC during OGTT (P < 0.01, Figures 3C–E). Offspring from HD-inulin dams had lower fasting blood glucose, and blood glucose and AUC during OGTT (P < 0.01, Figures 3C–E). Fasting insulin and HOMA-IR were significantly higher in pups from HD dams than in pups from CD controls (P < 0.01, Figures 3F,G). Maternal inulin supplementation reduced serum fasting insulin and HOMA-IR in male pups at weaning (P < 0.01, Figures 3F,G).
Characterization of Gut Microbiota
Cecal contents from 15 mice (n = 5 per group) were used to perform whole-metagenome shotgun sequencing to understand the gut microbial composition. After quality control, we acquired a total of 113.5 Gbp of high-quality metagenomic data (7.56 ± 0.62 Gbp per sample) for further analysis. The sequence data generated in this study were submitted to the NCBI Sequence Read Archive database (accession number PRJNA552163). After de novo assembly and gene data calling, we constructed a non-redundant gene catalog of all cecal contents containing 1,048,576 genes. This gene catalog was qualified for further gut microbial analysis.
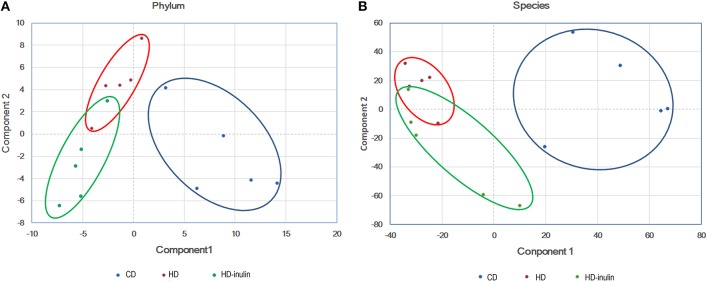
Gut Microbial Beta Diversity
According to the principal component analysis of the bacterial abundance, the gut microbial communities were significantly different among the three groups (Figure 4). Offspring gut microbial composition was affected by both maternal HD and maternal inulin supplementation.
Figure 4.
Principal component analysis plot at phylum level (A) and species level (B). CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
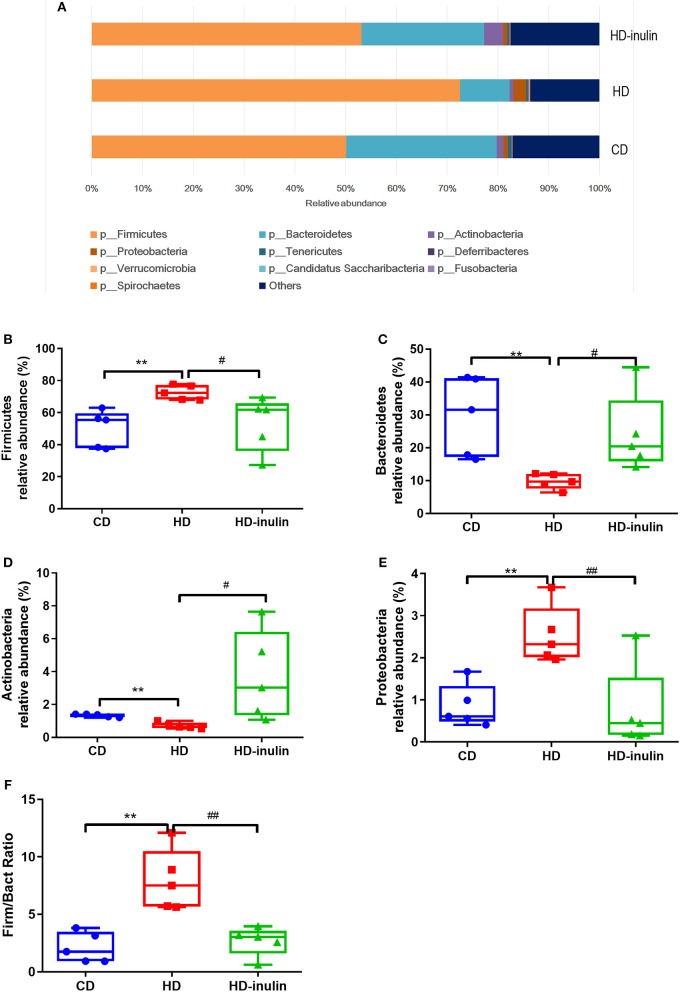
Relative Abundance of Gut Microbiota at the Phyla Level
The distribution and relative abundance of bacterial taxa at the phyla level are shown in Figures 5A–F. Offspring gut microbiota were distributed across 10 bacterial phyla, including Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Tenericutes, Deferribacteres, Verrucomicrobia, Candidatus Saccharibacteria, Fusobacteria, and Spirochaetes. The relative abundance analysis revealed a significant increase in the abundance of Firmicutes and Proteobacteria in pups from HD dams (P < 0.05, Figures 5B,D). On the other hand, the abundance of Bacterodetes and Actinobacteria was reduced in pups from HD dams (P < 0.05, Figures 5C,E). Inulin supplementation moderated the disturbance of the abundance of Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria (P < 0.05, Figures 5B–E). The ratio of Firmicutes and Bacteroidetes in pups from HD dams was greatly increased (P < 0.05, Figure 5F). Inulin supplementation moderated this ratio (P < 0.05, Figure 5F).
Figure 5.
The relative abundance of bacterial population at phylum level (A). The abundance of Firmicutes (B), Bacteroidetes (C), Actinobacteria (D), Proteobacteria (E), and Firmicutes/Bacteroidetes (Firm/Bact) ratios (F) in pups from CD, HD, and HD-inulin dams. Kruskal-Wallis non-parametric test, followed by the Wilcoxon tests. The data were shown as median (min-max). n = 5 in each group. **P < 0.01 vs. CD group; #P < 0.05, ##P < 0.01 vs. HD group. CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
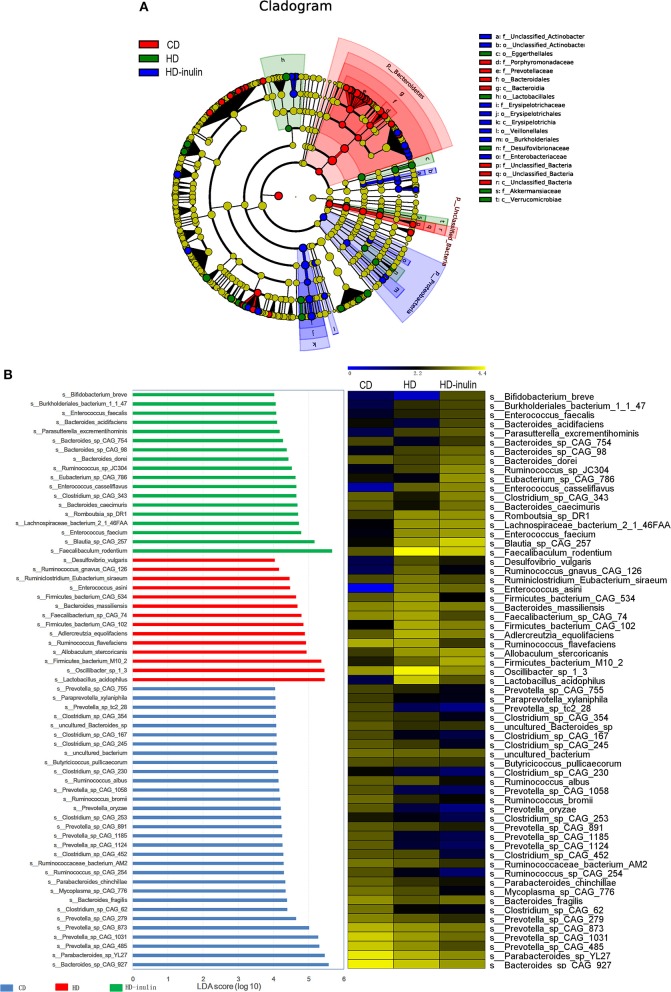
LEfSe Analysis in Different Groups at the Species Level
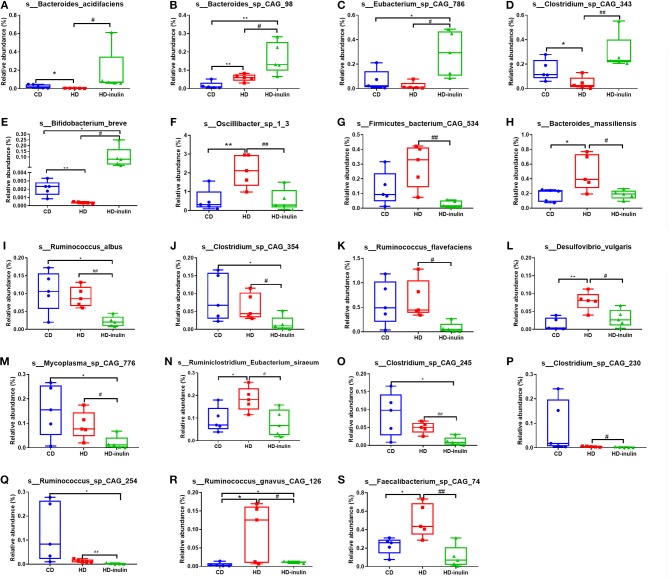
We further compared changes in the gut microbiota among the three groups by using LEfSe analysis. The histogram of the LDA scores further revealed a clear difference among pups from the CD, HD, and HD-inulin dams at the species level (Figure 6). Sixty-three bacterial species changed significantly among the pups from the CD, HD, and HD-inulin dams. Among these bacteria species, 19 species showed a significant difference between the pups from HD-inulin dams and HD dams (P < 0.05). Bacteroides_acidifaciens (P < 0.05), Bacteroides_sp_CAG_98 (P < 0.05), Eubacterium_sp_CAG_786 (P < 0.05), Clostridium_sp_CAG_343 (P < 0.01), and Bifidobacterium_breve (P < 0.05) were significantly elevated in pups from HD-inulin dams vs. pups from HD dams. However, Oscillibacter_sp_1_3 (P < 0.01), Firmicutes_bacterium_CAG_534 (P < 0.01), Bacteroides_massiliensis (P < 0.05), Ruminococcus_albus (P < 0.05), Clostridium_sp_CAG_354 (P < 0.05), Ruminococcus_flavefaciens (P < 0.05), Desulfovibrio_vulgaris (P < 0.05), Mycoplasma_sp_CAG_776 (P < 0.05), Ruminiclostridium_Eubacterium_siraeum (P < 0.05), Clostridium_sp_CAG_245 (P < 0.01), Clostridium_sp_CAG_230 (P < 0.05), Ruminococcus_sp_CAG_254 (P < 0.01), Ruminococcus_gnavus_CAG_126 (P < 0.05), and Faecalibacterium_sp_CAG_74 (P < 0.01) were reduced significantly in pups from HD-inulin dams vs. HD dams (Figure 7).
Figure 6.
The LEfSe analysis of the gut microbiota in pups from HD-inulin dams differed significantly from that in pups from CD and HD dams. (A) Differences are represented in the color of the most abundant group. (B) The left histogram shows the lineages with LDA values of 4.0 or higher as determined by LEfSe in species level. The right heap map shows the relative abundance (log10 transformation). CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
Figure 7.
The relative abundance of bacterial population at species level. Bacteroides_acidifaciens (A), Bacteroides_sp_CAG_98 (B), Eubacterium_sp_CAG_786 (C), Clostridium_sp_CAG_343 (D), and Bifidobacterium_breve (E), Oscillibacter_sp_1_3 (F), Firmicutes_bacterium_CAG_534 (G), Bacteroides_massiliensis (H), Ruminococcus_albus (I), Clostridium_sp_CAG_354 (J), Ruminococcus_flavefaciens (K), Desulfovibrio_vulgaris (L), Mycoplasma_sp_CAG_776 (M), Ruminiclostridium_Eubacterium_siraeum (N), Clostridium_sp_CAG_245 (O), Clostridium_sp_CAG_230 (P), Ruminococcus_sp_CAG_254 (Q), Ruminococcus_gnavus_CAG_126 (R), and Faecalibacterium_sp_CAG_74 (S) in pups from CD, HD, and HD-inulin dams. Kruskal-Wallis non-parametric test, followed by the Wilcoxon tests. The data were shown as median (min–max). n = 5 in each group. *P < 0.05, **P < 0.01 vs. CD group; #P < 0.05, ##P < 0.01 vs. HD group. CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
KEGG Metabolic Pathways
We used the KEGG pathway database to analyze functional metagenomic profiles. The comparison among groups indicated significant differences in 38 metabolic functions (P < 0.05, Table 1). Most of these features revealed a similar abundance between pups from CD and HD-inulin dams. A series of glucose and lipid metabolism pathways were modified in pups from HD-inulin dams compared with pups from HD dams, such as glycolysis/gluconeogenesis, citrate cycle (TCA cycle), fatty acid biosynthesis, adipocytokine signaling pathway, glucagon signaling pathway, type II diabetes mellitus, and carbohydrate digestion and absorption.
Table 1.
The relative abundance (%) of bacterial groups that showed statistical significance of KEGG pathway categories among different groups.
| KEGG function level 3 | CD | HD | HD-inulin | P-value | |
|---|---|---|---|---|---|
| HD vs. CD | HD-inulin vs. HD | ||||
| Glycolysis/Gluconeogenesis | 0.6148(0.5570–0.6355) | 0.7049(0.6043–0.7745)* | 0.57650(0.53656–0.66263)# | 0.03260 | 0.04934 |
| Citrate cycle (TCA cycle) | 0.2828(0.2680–0.3155) | 0.2597(0.2323–0.2822)* | 0.31907(0.25165–0.36493)# | 0.03499 | 0.03844 |
| Fatty acid biosynthesis | 0.2342(0.2135–0.2573) | 0.1920(0.1875–0.2256)* | 0.22575(0.20333–0.24197)# | 0.01960 | 0.04073 |
| Geraniol degradation | 0.0001(0.0000–0.0002) | 0.0010(0.0004–0.0025)* | 0.00011(0.00007–0.00038)# | 0.03166 | 0.03742 |
| Lysine biosynthesis | 0.2983(0.2841–0.3296) | 0.3294(0.3021–0.3332)* | 0.28247(0.25319–0.33476)# | 0.04331 | 0.04813 |
| Lysine degradation | 0.0304(0.0257–0.0359) | 0.0481(0.0466–0.0529)** | 0.03409(0.02443–0.04769)# | 0.00179 | 0.03139 |
| Furfural degradation | 0.0000(0.0000–0.0001) | 0.0001(0.0001–0.0001)* | 0.00000(0.00000–0.00005)# | 0.01573 | 0.03019 |
| Tryptophan metabolism | 0.0218(0.0162–0.0222) | 0.0353(0.0310–0.0393)** | 0.02619(0.01823–0.03353)# | 0.00390 | 0.04139 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.3522(0.3181–0.3744) | 0.3180(0.2895–0.3444)* | 0.34770(0.32533–0.39867)# | 0.03330 | 0.02792 |
| D-Alanine metabolism | 0.0517(0.0479–0.0564) | 0.0612(0.0482–0.0720)* | 0.04862(0.04265–0.05741)# | 0.04520 | 0.03154 |
| Other glycan degradation | 0.1488(0.1202–0.2094) | 0.1025(0.0970–0.1262)* | 0.17478(0.11970–0.26930)# | 0.03166 | 0.03805 |
| Amino sugar and nucleotide sugar metabolism | 0.7181(0.6680–0.7482) | 0.8406(0.7493–0.8665)* | 0.67503(0.60146–0.80699)# | 0.01718 | 0.03934 |
| Peptidoglycan biosynthesis | 0.4221(0.4010–0.4480) | 0.4537(0.4386–0.4615)* | 0.36959(0.34915–0.45002)# | 0.02066 | 0.02034 |
| Sphingolipid metabolism | 0.1130(0.1059–0.1476) | 0.0937(0.0879–0.1058)* | 0.12882(0.09625–0.17644)# | 0.03166 | 0.02805 |
| Pyruvate metabolism | 0.4972(0.4639–0.5164) | 0.5507(0.4991–0.5585)* | 0.45720(0.43866–0.54814)# | 0.01683 | 0.02934 |
| Toluene degradation | 0.0002(0.0001–0.0003) | 0.0015(0.0002–0.0022)* | 0.00049(0.00018–0.00081)*# | 0.01625 | 0.04934 |
| Vitamin B6 metabolism | 0.0891(0.0814–0.0979) | 0.0756(0.0667–0.0801)* | 0.08564(0.07998–0.10059)# | 0.01620 | 0.02803 |
| Folate biosynthesis | 0.1733(0.1254–0.1937) | 0.1198(0.0987–0.1432)* | 0.00158(0.00130–0.00181)# | 0.03425 | 0.03128 |
| Limonene and pinene degradation | 0.0074(0.0057–0.0111) | 0.0148(0.0119–0.0189)* | 0.00005(0.00002–0.00014)# | 0.01300 | 0.00934 |
| Caprolactam degradation | 0.0019(0.0002–0.0074) | 0.0028(0.0026–0.0029) | 0.00002(0.00001–0.00002)# | 0.30751 | 0.02752 |
| Flavone and flavonol biosynthesis | 0.0046(0.0028–0.0064) | 0.0025(0.0019–0.0035) | 0.00004(0.00003–0.00004)# | 0.04153 | 0.03792 |
| ABC transporters | 0.9351(0.7441–1.5006) | 1.5091(1.4626–1.6103)* | 0.00187(0.00101–0.01708)# | 0.01960 | 0.04584 |
| Ribosome biogenesis in eukaryotes | 0.0193(0.0176–0.0201) | 0.0246(0.0237–0.0252)** | 0.00020(0.00016–0.00025)# | 0.00159 | 0.03139 |
| Ribosome | 1.1228(1.0636–1.3193) | 1.3400(1.3128–1.3650)* | 0.01147(0.01099–0.01296)# | 0.02666 | 0.00302 |
| RNA polymerase | 0.1164(0.0976–0.1221) | 0.1253(0.1240–0.1342)* | 0.00105(0.00099–0.00122)# | 0.02489 | 0.00317 |
| Bacterial secretion system | 0.3061(0.2976–0.3245) | 0.3300(0.3252–0.3336)* | 0.00307(0.00237–0.00321)# | 0.01300 | 0.03128 |
| PPAR signaling pathway | 0.0591(0.0479–0.0702) | 0.0476(0.0419–0.0549)* | 0.00066(0.00046–0.00077)# | 0.04254 | 0.04936 |
| Base excision repair | 0.2119(0.2023–0.2278) | 0.2394(0.2347–0.2505)* | 0.21334(0.18743–0.23365)# | 0.01048 | 0.00726 |
| HIF-1 signaling pathway | 0.0494(0.0434–0.0553) | 0.0615(0.0520–0.0642)* | 0.05271(0.05079–0.05575)# | 0.01960 | 0.01655 |
| Sulfur relay system | 0.0934(0.0889–0.1164) | 0.1249(0.1200–0.1327)* | 0.08055(0.01371–0.13565)# | 0.01145 | 0.03893 |
| Apoptosis | 0.0149(0.0068–0.0158) | 0.0030(0.0013–0.0043)* | 0.01686(0.00387–0.05312)# | 0.01300 | 0.04234 |
| Adipocytokine signaling pathway | 0.0346(0.0337–0.0574) | 0.0266(0.0194–0.0329)* | 0.03450(0.02599–0.05028)# | 0.02666 | 0.04397 |
| Glucagon signaling pathway | 0.0892(0.0816–0.0971) | 0.1270(0.1009–0.1336)* | 0.70773(0.09824–0.73901)*# | 0.01573 | 0.04893 |
| Type II diabetes mellitus | 0.0226(0.0222–0.0239) | 0.0264(0.0258–0.0274)** | 0.02215(0.02147–0.02677)# | 0.00381 | 0.02674 |
| Carbohydrate digestion and absorption | 0.0155(0.0097–0.0323) | 0.0071(0.0052–0.0132)* | 0.03536(0.00509–0.04880)# | 0.04024 | 0.02193 |
| Protein digestion and absorption | 0.0160(0.0069–0.0175) | 0.0042(0.0016–0.0058)* | 0.02259(0.00643–0.06965)# | 0.01573 | 0.04139 |
| Proteoglycans in cancer | 0.0191(0.0175–0.0199) | 0.0245(0.0236–0.0247)** | 0.02156(0.01852–0.02434)*# | 0.00119 | 0.01655 |
| Renal cell carcinoma | 0.0030(0.0022–0.0047) | 0.0074(0.0048–0.0083) | 0.00434(0.00282–0.00604)# | 0.01620 | 0.01655 |
CD, control diet; HD, high fat diet; HD-inulin, high fat diet with inulin supplement.
P < 0.05,
P < 0.01 vs. CD group;
P < 0.05, ##P < 0.01 vs. HD group.
Discussion
In this study, female mice fed a HD exhibited higher energy intake and weight gain during the pregnancy period. In spite of the change in energy intake, inulin treatment reduced weight gain during pregnancy and fasting blood glucose in female mice. In rodent experiments, several groups found that inulin-type fructans dramatically reduced epididymal, inguinal, and visceral adipose tissue fat masses by 30–40% (26–30). Previous studies showed that inulin-type fructans effectively reduced fasting blood glucose in both T2D patients (31) and diabetic rats (15, 17, 32). Meta-analysis also revealed that dietary prebiotic consumption improved satiety and reduced postprandial blood glucose and insulin levels (33).
Maternal HD feeding led to an increase in blood glucose and insulin resistance in male offspring at weaning. This result is consistent with previous literature (34–36). Interestingly, our results showed that early maternal inulin intervention reduces fasting blood glucose and moderates glucose intolerance and insulin resistance in offspring from HD dams. Other studies also prove that prebiotics can reduce glycemia in offspring in rats (37–39).
The gut microbiome plays an important role in the pathogenesis of various health disorders, such as diabetes (40) and obesity (41). In the current study, the β-diversity of the gut microbiota showed a significant difference among offspring from CD, HD, and HD-inulin dams.
Our results showed that offspring from HD-inulin dams exhibited marked changes in microbial abundance at the phylum level compared with offspring from HD dams. First, the ratio of Firmicutes to Bacteroidetes in pups from HD dams was greatly increased, and inulin supplementation moderated this ratio. Previous studies have shown that the abundance of Firmicutes is associated with metabolic disease in mice (9, 42). Second, offspring from HD dams had a lower Actinobacteria abundance than those from CD dams. Maternal inulin supplementation moderated Actinobacteria abundance. Although Actinobacteria make up a tiny proportion, they are critical for the maintenance of gut homeostasis (43). As an important probiotic, Bifidobacterium, all under the classes of Actinobacteria, has favorable effects on human health (43). Type 1 diabetic children have a lower abundance of Actinobacteria compared with healthy children (44). Moreover, there was a significant increase in the abundance of Proteobacteria in offspring from HD dams compared to CD dams. A previous study showed that dietary inulin affects the gut microbiota of healthy suckling piglets fed a normal diet (45). In a rodent study, prebiotic intervention may change gut microbiota richness and diversity in mice fed either a normal diet and or a high-fat diet (45, 46). In our research, maternal inulin supplementation reduced the abundance of Proteobacteria. An increased abundance of Proteobacteria is an indicator of an unhealthy gut microbial community (47). Type 2 diabetic mice and humans have high levels of Proteobacteria (48, 49). Collectively, our study confirms that maternal inulin supplementation moderates dysbiosis in offspring of HD dams.
At the species level, the abundance of Bifidobacterium_breve was decreased in offspring of HD dams compared to those of CD dams. HD-inulin group pups had a higher Bifidobacterium_breve relative abundance. Bifidobacterium is one of the most abundant probiotic species present in the mammalian gut (50). Previous studies indicate that a lower abundance of Bifidobacterium is a hallmark of a type 2 diabetic gut microbiota and accelerates the progression of type 2 diabetes (51, 52). Type 2 diabetic, overweight and obese patients have a lower abundance of Bifidobacterium than healthy adults and children (53–55). Bifidobacterium modulates glucose metabolism, improves insulin resistance, and gut barrier function, and stimulates the host immune system (56, 57). Paul et al. found that oligofructose treatment during gestation increased the maternal relative abundance of Bifidobacterium compared to rats fed a high-fat/sucrose diet (37). Interestingly, the increased abundance of Bifidobacterium is similar between mothers and pups. According to previous literature (58), the increased abundance of Bifidobacterium in offspring affected by maternal inulin supplementation may also be linked to the observed improvement in HOMA-IR scores. Some data raised the role of gut microbiota in metabolic programming (59). Gut microbiota is one of the most important environmental factors that will affect offspring metabolism (60). Although we cannot exclude the transmission of microbial components from mothers to pups during vaginal delivery and breastfeeding (61), this study could provide some evidence that inulin intervention during gestation could increase maternal Bifidobacterium abundance, and may have colonized the pup gut.
Our findings indicate that maternal inulin supplementation induced a marked increase in the abundance of Bacteroides_acidifaciens, Eubacterium_sp_CAG_786, and Clostridium_sp_CAG_343 in offspring. Bacteroides, Eubacterium, and Clostridium are butyrate-producing bacteria (62, 63). Butyrate promotes intestinal barrier function to maintain colonic health through increased fatty acid oxidation (64, 65). Type 2 diabetic patients have reduced levels of butyrate-producing bacteria (66). A previous study showed that Bacteroides_acidifaciens improves insulin sensitivity in mice (67). Oral administration of Eubacterium improved insulin sensitivity in db/db mice (68). Clostridium improved fasting glucose, glucose tolerance and insulin tolerance in STZ induced HD-fed diabetic C57BL/6J mice (69). Butyrate enhances the secretion of the gut satiety hormones glucagon-like peptide-1 (GLP-1) and peptide YY (PYY) (70). GLP-1 and PYY played an important role in body weight reduction. A previous study showed that maternal oligofructose treatment increased plasma GLP-1 and PYY levels both in rat mothers and pups (37). Therefore, another explanation for the effect of maternal inulin treatment on glucose metabolism in pups might be the transfer of gut satiety hormones from mothers to pups through the placenta.
Ruminococcus is considered a butyrate-producing acid bacteria (71). However, in this study, the relative abundance of several Ruminococcus was lower in the HD-inulin group than in the HD group, such as Ruminococcus_albus, Ruminococcus_flavefaciens, Ruminococcus_sp_CAG_254, and Ruminococcus_gnavus_CAG_126. Researchers found that Ruminococcus was highly abundant in adults with T2DM (72). Random blood glucose was positively correlated with Ruminococcus in rats (73). A number of Ruminococcus species are thought to be related to metabolic diseases. Ruminococcus also acts as transient blooms of proinflammation (74). Ruminococcus_gnavus has been found to secrete a unique L-rhamnose oligosaccharide that induces tumor necrosis factor alpha (TNFa), a major pro-inflammatory cytokine (75).
The abundance of Oscillibacter_sp_1_3 and Bacteroides_massiliensis in offspring from HD-inulin dams was significantly reduced compared to HD dams. Bacteroides_massiliensis was significantly enriched in obese children (76) and in German adults who were unsuccessful with weight loss (77). It is also positively correlated with body mass index (BMI) z-scores (76). Oscillibacter was strongly associated with the diabetic phenotype (78) and also positively correlated with gut permeability (79).
Metagenomic function prediction is based on the relative expression abundance of each bacterial taxonomy, which belongs to each metabolic pathway. KEGG pathway analysis seems to show differences in 38 metabolic functions among offspring from CD, HD, and HD-inulin dams. Specifically, maternal inulin supplementation may alter gut microbiota involved in glucose and lipid metabolism, such as glycolysis/gluconeogenesis, citrate cycle (TCA cycle), fatty acid biosynthesis, adipocytokine signaling pathway, glucagon signaling pathway, type II diabetes mellitus, and carbohydrate digestion and absorption. Previous studies indicated the beneficial effect of prebiotics on metabolic disorders, such as obesity and type 2 diabetes (80, 81). Given that metagenomic function prediction is based on the relative expression abundance of gut microbiota, more experiments about the metabolic function of gut microbiota need to be performed.
Conclusion
In conclusion, maternal inulin supplementation has beneficial effects on glucose metabolism in offspring, including improvements in glucose intolerance and insulin resistance. Importantly, maternal inulin supplementation increased the abundance of Bifidobacterium and several butyrate-producing bacteria. Thus, maternal inulin supplementation is promising for the prevention of metabolic disorders in the offspring.
Data Availability Statement
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to The datasets supporting the conclusions of this manuscript are available from the corresponding author (xiaoxh2014@vip.163.com) on reasonable request.
Ethics Statement
All animal experimental protocols were approved by the Animal Care Committee of Peking Union Medical Hospital (Permit Number: MC-07-6004).
Author Contributions
XX conceived and designed the experiments. QZ, JZ, TW, and XW performed the experiments. MY, ML, and FP analyzed the data. XX contributed reagents, materials, and analysis tools. QZ wrote the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the grants from National Key R&D Program of China (2017YFC1309603), National Key Research and Development Program of China (2016YFA0101002), Medical Epigenetics Research Center, Chinese Academy of Medical Sciences (2017PT31036,2018PT31021), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2017PT32020, No. 2018PT32001), National Natural Science Foundation of China (No. 81170736, 81570715, 81870579, 81870545), National Natural Science Foundation for Young Scholars of China (No. 81300649), China Scholarship Council foundation (201308110443), PUMC Youth Fund (33320140022) and Fundamental Research Funds for the Central Universities, and Scientific Activities Foundation for Selected Returned Overseas Professionals of Human Resources and Social Security Ministry. We are very grateful to Beijing Compass Biotechnology Company for excellent technical assistance with the sequencing experiments.
References
- 1.Barella LF, de Oliveira JC, Mathias PC. Pancreatic islets and their roles in metabolic programming. Nutrition. (2014) 30:373–9. 10.1016/j.nut.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 2.Zheng J, Zhang Q, Mul JD, Yu M, Xu J, Qi C, et al. Maternal high-calorie diet is associated with altered hepatic microRNA expression and impaired metabolic health in offspring at weaning age. Endocrine. (2016) 54:70–80. 10.1007/s12020-016-0959-9 [DOI] [PubMed] [Google Scholar]
- 3.Ribaroff GA, Wastnedge E, Drake AJ, Sharpe RM, Chambers TJG. Animal models of maternal high fat diet exposure and effects on metabolism in offspring: a meta-regression analysis. Obes Rev. (2017) 18:673–86. 10.1111/obr.12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantham P, Aye IL, Powell TL. Inflammation in maternal obesity and gestational diabetes mellitus. Placenta. (2015) 36:709–15. 10.1016/j.placenta.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greiner TU, Hyotylainen T, Knip M, Backhed F, Oresic M. The gut microbiota modulates glycaemic control and serum metabolite profiles in non-obese diabetic mice. PLoS ONE. (2014) 9:e110359. 10.1371/journal.pone.0110359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai N, Wong FS, Wen L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev Endocr Metab Disord. (2015) 16:55–65. 10.1007/s11154-015-9309-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. (2016) 15:108. 10.1186/s12944-016-0278-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 10.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 11.Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. The first thousand days - intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol. (2014) 25:428–38. 10.1111/pai.12232 [DOI] [PubMed] [Google Scholar]
- 12.Alles MS, Hautvast JG, Nagengast FM, Hartemink R, Van Laere KM, Jansen JB. Fate of fructo-oligosaccharides in the human intestine. Br J Nutr. (1996) 76:211–21. 10.1079/BJN19960026 [DOI] [PubMed] [Google Scholar]
- 13.Shoaib M, Shehzad A, Omar M, Rakha A, Raza H, Sharif HR, et al. Inulin: properties, health benefits and food applications. Carbohydr Polym. (2016) 147:444–54. 10.1016/j.carbpol.2016.04.020 [DOI] [PubMed] [Google Scholar]
- 14.Busserolles J, Gueux E, Rock E, Demigne C, Mazur A, Rayssiguier Y. Oligofructose protects against the hypertriglyceridemic and pro-oxidative effects of a high fructose diet in rats. J Nutr. (2003) 133:1903–8. 10.1093/jn/133.6.1903 [DOI] [PubMed] [Google Scholar]
- 15.Cani PD, Knauf C, Iglesias MA, Drucker DJ, Delzenne NM, Burcelin R. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes. (2006) 55:1484–90. 10.2337/db05-1360 [DOI] [PubMed] [Google Scholar]
- 16.Roshanravan N, Mahdavi R, Alizadeh E, Jafarabadi MA, Hedayati M, Ghavami A, et al. Effect of butyrate and inulin supplementation on glycemic status, lipid profile and glucagon-like peptide 1 level in patients with type 2 diabetes: a randomized double-blind, placebo-controlled trial. Horm Metab Res. (2017) 49:886–91. 10.1055/s-0043-119089 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Yu H, Xiao X, Hu L, Xin F, Yu X. Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ. (2018) 6:e4446. 10.7717/peerj.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. (2016) 14:508–22. 10.1038/nrmicro.2016.83 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Sun X, Xiao X, Zheng J, Li M, Yu M, et al. Maternal chromium restriction induces insulin resistance in adult mice offspring through miRNA. Int J Mol Med. (2018) 41:1547–59. 10.3892/ijmm.2017.3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. (2012) 9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. (2009) 25:1966–7. 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- 22.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. (2010) 38:e132. 10.1093/nar/gkq275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. (2015) 12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 24.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. (2014) 42:560–6. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kok NN, Morgan LM, Williams CM, Roberfroid MB, Thissen JP, Delzenne NM. Insulin, glucagon-like peptide 1, glucose-dependent insulinotropic polypeptide and insulin-like growth factor I as putative mediators of the hypolipidemic effect of oligofructose in rats. J Nutr. (1998) 128:1099–103. 10.1093/jn/128.7.1099 [DOI] [PubMed] [Google Scholar]
- 27.Daubioul CA, Taper HS, De Wispelaere LD, Delzenne NM. Dietary oligofructose lessens hepatic steatosis, but does not prevent hypertriglyceridemia in obese zucker rats. J Nutr. (2000) 130:1314–9. 10.1093/jn/130.5.1314 [DOI] [PubMed] [Google Scholar]
- 28.Cani PD, Dewever C, Delzenne NM. Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. Br J Nutr. (2004) 92:521–6. 10.1079/bjn20041225 [DOI] [PubMed] [Google Scholar]
- 29.Cani PD, Neyrinck AM, Maton N, Delzenne NM. Oligofructose promotes satiety in rats fed a high-fat diet: involvement of glucagon-like Peptide-1. Obes Res. (2005) 13:1000–7. 10.1038/oby.2005.117 [DOI] [PubMed] [Google Scholar]
- 30.Delzenne NM, Cani PD. A place for dietary fibre in the management of the metabolic syndrome. Curr Opin Clin Nutr Metab Care. (2005) 8:636–40. 10.1097/01.mco.0000171124.06408.71 [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Prabhakar M, Ju J, Long H, Zhou HW. Effect of inulin-type fructans on blood lipid profile and glucose level: a systematic review and meta-analysis of randomized controlled trials. Eur J Clin Nutr. (2017) 71:9–20. 10.1038/ejcn.2016.156 [DOI] [PubMed] [Google Scholar]
- 32.Cani PD, Daubioul CA, Reusens B, Remacle C, Catillon G, Delzenne NM. Involvement of endogenous glucagon-like peptide-1(7-36) amide on glycaemia-lowering effect of oligofructose in streptozotocin-treated rats. J Endocrinol. (2005) 185:457–65. 10.1677/joe.1.06100 [DOI] [PubMed] [Google Scholar]
- 33.Kellow NJ, Coughlan MT, Reid CM. Metabolic benefits of dietary prebiotics in human subjects: a systematic review of randomised controlled trials. Br J Nutr. (2014) 111:1147–61. 10.1017/s0007114513003607 [DOI] [PubMed] [Google Scholar]
- 34.Zheng J, Xiao X, Zhang Q, Yu M, Xu J, Wang Z. Maternal high-fat diet modulates hepatic glucose, lipid homeostasis and gene expression in the PPAR pathway in the early life of offspring. Int J Mol Sci. (2014) 15:14967–83. 10.3390/ijms150914967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Xiao X, Zhang Q, Yu M, Xu J, Wang Z, et al. Maternal and post-weaning high-fat, high-sucrose diet modulates glucose homeostasis and hypothalamic POMC promoter methylation in mouse offspring. Metab Brain Dis. (2015) 30:1129–37. 10.1007/s11011-015-9678-9 [DOI] [PubMed] [Google Scholar]
- 36.Schellong K, Melchior K, Ziska T, Ott R, Henrich W, Rancourt RC, et al. Hypothalamic insulin receptor expression and DNA promoter methylation are sex-specifically altered in adult offspring of high-fat diet (HFD)-overfed mother rats. J Nutr Biochem. (2019) 67:28–35. 10.1016/j.jnutbio.2019.01.014 [DOI] [PubMed] [Google Scholar]
- 37.Paul HA, Bomhof MR, Vogel HJ, Reimer RA. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Sci Rep. (2016) 6:20683. 10.1038/srep20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dennison CA, Eslinger AJ, Reimer RA. Preconception prebiotic and sitagliptin treatment in obese rats affects pregnancy outcomes and offspring microbiota, adiposity, and glycemia. Front Endocrinol. (2017) 8:301. 10.3389/fendo.2017.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul HA, Collins KH, Nicolucci AC, Urbanski SJ, Hart DA, Vogel HJ, et al. Maternal prebiotic supplementation reduces fatty liver development in offspring through altered microbial and metabolomic profiles in rats. Faseb J. (2019) 33:5153–67. 10.1096/fj.201801551R [DOI] [PubMed] [Google Scholar]
- 40.Soverini M, Turroni S, Biagi E, Quercia S, Brigidi P, Candela M, et al. Variation of carbohydrate-active enzyme patterns in the gut microbiota of italian healthy subjects and type 2 diabetes patients. Front Microbiol. (2017) 8:2079. 10.3389/fmicb.2017.02079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. (2017) 22:589–99. 10.1016/j.chom.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 42.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. (2016) 8:42. 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binda C, Lopetuso LR, Rizzatti G, Gibiino G, Cennamo V, Gasbarrini A. Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig Liver Dis. (2018) 50:421–8. 10.1016/j.dld.2018.02.012 [DOI] [PubMed] [Google Scholar]
- 44.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. (2013) 11:46. 10.1186/1741-7015-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passlack N, Vahjen W, Zentek J. Dietary inulin affects the intestinal microbiota in sows and their suckling piglets. BMC Vet Res. (2015) 11:51. 10.1186/s12917-015-0351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everard A, Lazarevic V, Gaia N, Johansson M, Stahlman M, Backhed F, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. (2014) 8:2116–30. 10.1038/ismej.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. (2015) 33:496–503. 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 48.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. (2010) 5:e9085. 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. (2011) 2:149. 10.3389/fmicb.2011.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. (2015) 5:15782. 10.1038/srep15782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuno MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. (2014) 5:190. 10.3389/fmicb.2014.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Upadhyaya S, Banerjee G. Type 2 diabetes and gut microbiome: at the intersection of known and unknown. Gut Microbes. (2015) 6:85–92. 10.1080/19490976.2015.1024918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. (2010) 18:190–5. 10.1038/oby.2009.167 [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol. (2010) 61:69–78. 10.1007/s00284-010-9582-9 [DOI] [PubMed] [Google Scholar]
- 55.Munyaka PM, Khafipour E, Ghia JE. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr. (2014) 2:109. 10.3389/fped.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An HM, Park SY, Lee DK, Kim JR, Cha MK, Lee SW, et al. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. (2011) 10:116. 10.1186/1476-511x-10-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Philippe D, Favre L, Foata F, Adolfsson O, Perruisseau-Carrier G, Vidal K, et al. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J Gastroenterol. (2011) 17:459–69. 10.3748/wjg.v17.i4.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. (2007) 50:2374–83. 10.1007/s00125-007-0791-0 [DOI] [PubMed] [Google Scholar]
- 59.Luoto R, Collado MC, Salminen S, Isolauri E. Reshaping the gut microbiota at an early age: functional impact on obesity risk? Ann Nutr Metab. (2013) 63 (Suppl. 2):17–26. 10.1159/000354896 [DOI] [PubMed] [Google Scholar]
- 60.Garcia-Mantrana I, Collado MC. Obesity and overweight: Impact on maternal and milk microbiome and their role for infant health and nutrition. Mol Nutr Food Res. (2016) 60:1865–75. 10.1002/mnfr.201501018 [DOI] [PubMed] [Google Scholar]
- 61.Jenmalm MC. The mother-offspring dyad: microbial transmission, immune interactions and allergy development. J Intern Med. (2017) 282:484–95. 10.1111/joim.12652 [DOI] [PubMed] [Google Scholar]
- 62.Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. (2016) 7:979. 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park S, Yasin M, Jeong J, Cha M, Kang H, Jang N, et al. Acetate-assisted increase of butyrate production by Eubacterium limosum KIST612 during carbon monoxide fermentation. Bioresour Technol. (2017) 245(Pt A):560-566. 10.1016/j.biortech.2017.08.132 [DOI] [PubMed] [Google Scholar]
- 64.Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res. (2007) 61:37–41. 10.1203/01.pdr.0000250014.92242.f3 [DOI] [PubMed] [Google Scholar]
- 65.Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. (2008) 100:297–305. 10.1017/S0007114508888733 [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE. (2013) 8:e71108. 10.1371/journal.pone.0071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang JY, Lee YS, Kim Y, Lee SH, Ryu S, Fukuda S, et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. (2017) 10:104–16. 10.1038/mi.2016.42 [DOI] [PubMed] [Google Scholar]
- 68.Udayappan S, Manneras-Holm L, Chaplin-Scott A, Belzer C, Herrema H, Dallinga-Thie GM, et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes. (2016) 2:16009. 10.1038/npjbiofilms.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia L, Li D, Feng N, Shamoon M, Sun Z, Ding L, et al. Anti-diabetic effects of Clostridium butyricum CGMCC0313.1 through promoting the growth of gut butyrate-producing bacteria in type 2 diabetic mice. Sci Rep. (2017) 7:7046. 10.1038/s41598-017-07335-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parnell JA, Reimer RA. Prebiotic fibres dose-dependently increase satiety hormones and alter Bacteroidetes and Firmicutes in lean and obese JCR:LA-cp rats. Br J Nutr. (2012) 107:601–13. 10.1017/S0007114511003163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin M, Kalainy S, Baskota N, Chiang D, Deehan EC, McDougall C, et al. Faecal microbiota from patients with cirrhosis has a low capacity to ferment non-digestible carbohydrates into short-chain fatty acids. Liver Int. (2019) 39:1437–47. 10.1111/liv.14106 [DOI] [PubMed] [Google Scholar]
- 72.Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol. (2013) 27:73–83. 10.1016/j.bpg.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 73.Zhou W, Xu H, Zhan L, Lu X, Zhang L. Dynamic development of fecal microbiome during the progression of diabetes mellitus in zucker diabetic fatty rats. Front Microbiol. (2019) 10:232. 10.3389/fmicb.2019.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. (2017) 9:103. 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci USA. (2019) 116:12672–7. 10.1073/pnas.1904099116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hollister EB, Foster BA, Dahdouli M, Ramirez J, Lai Z. Characterization of the stool microbiome in hispanic preschool children by weight status and time. Child Obes. (2018) 14:122–30. 10.1089/chi.2017.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PLoS ONE. (2016) 11:e0149564. 10.1371/journal.pone.0149564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jung MJ, Lee J, Shin NR, Kim MS, Hyun DW, Yun JH, et al. Chronic repression of mTOR complex 2 induces changes in the gut microbiota of diet-induced obese mice. Sci Rep. (2016) 6:30887. 10.1038/srep30887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE. (2012) 7:e34233. 10.1371/journal.pone.0034233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delzenne NM, Cani PD. Gut microbiota and the pathogenesis of insulin resistance. Curr Diab Rep. (2011) 11:154–9. 10.1007/s11892-011-0191-1 [DOI] [PubMed] [Google Scholar]
- 81.Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. MBio. (2014) 5:e01011–01014. 10.1128/mBio.01011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to The datasets supporting the conclusions of this manuscript are available from the corresponding author (xiaoxh2014@vip.163.com) on reasonable request.