Abstract
Cerebral palsy (CP) and epilepsy are two interactive neurological diseases, and their clinical treatment can cause severe side-effects in children's development, especially when it involves long-term administration of antiepileptic drugs. Accumulating studies on the gut-brain axis indicated that the gut microbiota (GM), which participates in various neurological diseases, would provide a harmless therapeutic target for the treatment of CP and epilepsy. To explore the GM characteristics in children with both CP and epilepsy (CPE), we collected fecal samples from 25 CPE patients (CPE group) and 21 healthy children (Healthy group) for 16S rDNA sequencing. In this study, we discovered significantly higher microbial diversity in the CPE group compared to healthy group (P < 0.001). After selecting the top 15 most abundant genera in each group, we found significantly enriched Bifidobacterium, Streptococcus, Akkermansia, Enterococcus, Prevotella, Veillonella, Rothia, and Clostridium IV in the CPE group, and noticeably reduced Bacteroides, Faecalibacterium, Blautia, Ruminococcus, Roseburia, Anaerostipes, and Parasutterella. A GM co-occurrence network was also constructed, and negative correlations were discovered between Bacteroides and Lactobacillus (r = −0.768, P < 0.001, FDR < 0.001), as well as Intestinibacter and Bifidobacterium (r = −0.726, P < 0.001, FDR < 0.001). After KEGG annotation and functional enrichment, 24 functional categories exhibited different enrichment levels between the CPE and Healthy groups. The functions, associated with xenobiotics metabolism, immune system diseases, and neurodegenerative diseases, were enriched in the CPE group. Conversely, the functional categories related to the biosynthesis of secondary metabolites were reduced. Furthermore, the neurodegenerative diseases were mainly attributed to Streptococcus, while an increased risk of immune system diseases was associated with enriched Akkermansia in the CPE patients. Generally, this study characterized the GM in CPE patients, illustrated the microbial co-occurrence relationships, and detected the functional distributions of the bacteria.
Keywords: cerebral palsy, epilepsy, gut microbiota, co-occurrence network, KEGG functional category
Introduction
Cerebral palsy (CP) refers to the non-progressive brain injuries that occur in the fetus or infant, and is a life-long physical disease with a childhood-onset (1, 2). Although it is defined by central dyskinesia and abnormal motor function (1, 2), CP is often accompanied with other complications, including epilepsy, orthopedic disorder, speech impairment, and neurobehavioral disorders (3). As one of the common complications, epilepsy occurs in 25–45% of CP patients (4), which is higher than the prevalence of 0.1–0.3% in all neonates (5). Epilepsy aggravates the brain injury in patients with CP (3, 4, 6). The current clinical therapy for CP patients with epilepsy is long-term physical rehabilitation training (2) and antiepileptic drug administration (or ketogenic diet) (7, 8). However, severe developmental side-effects have been reported in children following these clinical treatments (9). The rapid progress in gut microbiota (GM) research raise the possibility of detecting harmless therapies for these neurologic diseases through GM intervention (10, 11).
Increasing studies have suggested a close relationship between GM and various neurologic diseases, such as Autism Spectrum Disorder (ASD) (12), Parkinson's Disease (PD) (13), and Alzheimer's Disease (AD) (14). By secreting secondary metabolites into blood circulation, GM could stimulate the central nervous system (CNS), and affect the stress, cognition, and mood of hosts (10, 11). For example, De Angelis et al. reported increased Clostridia species in ASD patients (12), which might repress the activation of enterochromaffin cells and the secretion of serotonin (15). Additionally, decreased Prevotellaceae and increased Lactobacilliaceae, which relate to neuroinflammation, were discovered in neurodegenerative diseases, such as PD (13). In Gan Xie et al.'s study, increased Proteobacteria and Cronbacteria were found in patients with epilepsy, and the role of GM in seizure controlling was hinted (16). Although GM has been investigated in various single neurologic diseases, its characteristics in patients with both CP and epilepsy (CPE) remain unexplored.
We recruited 25 CPE patients and 21 healthy children aged 3–18 years old to investigate the GM features in CPE. In addition to exploring the GM differences between the CPE and Healthy groups, we also aimed to: (I) evaluate the microbial correlations in children; (II) elucidate the alteration of GM functions in CPE patients and their corresponding intestinal commensals. We hope this study will enhance our understanding of the roles of GM in the pathogenesis of CPE, and provide a solid theoretical basis for microbial intervention in CPE patients.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Committee of The Hospital of Maternal and Child Health (Longgang, China) under registration number LGFY2017005. As the guardian of the CPE patients, the Longgang District Social Welfare Center provided written informed consent and volunteered their children for investigation for scientific research, as well as the parents of the healthy children.
Participant Recruitment
The CPE patients were recruited from the Longgang District Social Welfare Center, and diagnosed by the Department of Neurology, The Hospital of Maternal and Child Health (Longgang, China) with the following inclusion criteria: (I) older than 3 years of age and younger than 18 years of age; (II) the patients were diagnosed with clear clinical manifestations of both CP and epilepsy in accordance with the diagnosis guidelines (17, 18). Healthy children were selected from those who passed physical examinations by the Department of Physical Examination, The Hospital of Maternal and Child Health (Longgang, China) with the following standards: (I) older than 3 years of age and younger than 18 years; (II) no allergic history (e.g., food allergy, AD, and asthma); (III) no hereditary diseases (e.g., thalassemia, hereditary deafness, and phenylketonuria); (IV) no metabolic diseases (e.g., obesity, diabetes, and rheumatoid arthritis). Moreover, the subjects were excluded from the study if they have been exposed to antibiotics, probiotics, or proton pump inhibitors 2 weeks before fecal sample collection. Finally, 25 CPE patients and 21 healthy children were enrolled between Aug. 2017 and Dec. 2017 (Supplementary File 1).
Fecal Sample Collection
Fresh stool samples were collected from CPE and healthy subjects during the clinical examination using sample swabs [iClean, Huachenyang (Shenzhen) Technology Co., LTD, China], and these were stored in sterilized tubes (62-558-201, SARSTEDT AG & Co. KG, Germany). Then the fecal samples were transferred to a −80°C freezer for long-term storage within 30 min.
DNA Extraction, Library Construction, and Sequencing
In compliance with the protocols of E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross GA, U.S.A.), bacterial DNA was isolated from fecal samples. Using a PCR kit (AP221-02, TransGen Biotech, China), the hyper-variable V3-V4 regions of 16S rRNA were amplified with 338F and 806R primers. Then, the PCR products were prepared for sequencing library construction (TruSeq DNA PCR-Free kit, Illumina, San Diego CA, U.S.A.), and sequenced by MiSeq platform (Illumina, San Diego CA, U.S.A.) as 300 (nt) paired-end reads. Raw reads were uploaded to the NCBI Sequence Read Archive (SRA) Database (BioProject ID: PRJNA530084). After sequencing, the remaining DNA and fecal samples were stored in a −80°C refrigerator, and will be eliminated after 2 years, according to the regulations of standard biosecurity and institutional safety in The Hospital of Maternal and Child Health (Longgang, China).
Taxonomical Annotation
The raw data was filtered as previously described (19). Based on at least 50 bases overlapping, the filtered reads were connected into tags, and the tags were clustered into operational taxonomic units (OTUs) with 97% similarity by USEARCH (v7.0.1090) (20). The taxonomical positions of OTUs were obtained by aligning their sequences to the RDP 16S rRNA database (trainset 16/release 11.5) (21) (Supplementary File 2).
PERMANOVA to Evaluate the Influence of Physical Indices
PERMANOVA was carried out on GM composition of all samples to assess the impacts of physical indices, which are listed in Table 1 (22). The vegan package in R was applied with 9,999 permutations and Euclidean distances.
Table 1.
Background information of the 46 study subjects.
| Healthy (n = 21) | CPE patients (n = 25) | P-value (PERMANOVA) | FDR | |
|---|---|---|---|---|
| Gender (male) | 12 | 13 | 0.460 | 0.575 |
| Age (month) | 70.43 ± 20.93 | 108.13 ± 42.83 | 0.035 | 0.088 |
| BMI | 17.37 ± 4.60 | 12.35 ± 2.29 | 0.843 | 0.843 |
| Anti-seizure drug usage | 0 | 15 | 0.378 | 0.575 |
Functional Prediction and Enrichment
With 16S rRNA OTUs profiling, the functional distributions of GM were acquired by PICRUSt with the default settings (23). For each sample, the abundance of KEGG Orthology (KO) was calculated, and the enriched functional categories on level III and level II of the KEGG database were detected (Supplementary File 3).
Statistics
All statistical analysis was performed in R (version 3.4.1). The differentially enriched genera and functional categories between CPE patients and healthy children were detected by Wilcoxon rank-sum test (P < 0.05, using “wilcox.test” in R). The genera that were enriched in the CPE and Healthy groups were selected, and their associations were evaluated by Spearman correlations (using “cor” in R). The co-occurrence networks were visualized by Cytoscape software (v2.2.0) (24). Statistical results from the multiple tests were adjusted with Benjamini and Hochberg method (FDR < 0.05) using “p.adjust,” and were plotted using package “ggplot2” in R.
Results
Sample Characteristics and Data Output
A total of 25 CPE patients (CPE group) and 21 healthy children (Healthy group) were enrolled for fecal sample collection in this study (Table 1, Supplementary File 1). The high-quality reads from 16S rRNA sequencing were connected into 2,293,673 tags, and the number of OTUs was significantly higher in the CPE group than that in the Healthy group (P < 0.001): it ranged from 157 to 1,333 in the CPE group, and 109 to 242 in the Healthy group. After RDP database alignment, 341 genera of 21 phyla were identified in all samples (Supplementary File 2), and the number of genera was remarkably higher in the CPE group as compared to the Healthy group (P < 0.001): the averaged number was 152 ± 47 and 53 ± 8 in CPE and Healthy groups, respectively. Other than age, gender, body mass index (BMI), or anti-seizure drug exposure, health condition exhibited a significant impact on the GM differences between the CPE and Healthy groups (P < 0.001, FDR < 0.001, PERMANOVA analysis, Table 1).
CPE Patients Held Discrepant GM Structure
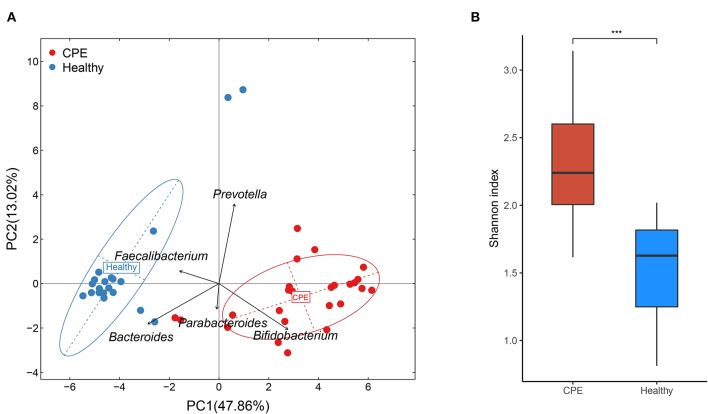
Principal component analysis (PCA) indicated that microbial samples from the CPE group were separated from those in the Healthy group, and the microbial samples were dominated by Bacteroides, Bifidobacterium, Prevotella, Faecalibacterium, and Parabacteroides (Figure 1A). Moreover, higher microbial diversity was identified in the CPE group (P < 0.001): the Shannon indexes were 2.33 ± 0.43 and 1.49 ± 0.36 in CPE and Healthy groups, respectively (Figure 1B).
Figure 1.
PCA and bacterial diversity analysis on fecal samples from CPE and healthy children. (A) PCA analysis indicated that samples from the Healthy group were clustered together, and they were apart from those of the CPE group. The GM of the participants was mainly dominated by Bacteroides, Bifidobacterium, Prevotella, Faecalibacterium, and Parabacteroides. (B) The microbial diversity was significantly higher in the CPE group (2.33 ± 0.43) than that in the Healthy group (1.49 ± 0.36). ***stands for P-value smaller than 0.001.
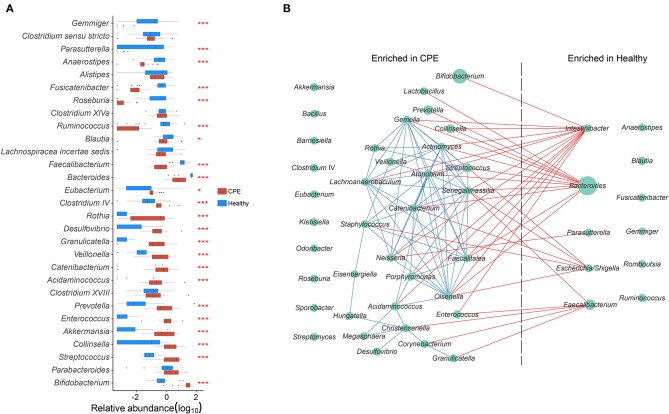
The top 15 most abundant genera from the CPE and Healthy groups were selected, and 23 of them were differentially enriched between these two groups (Figure 2A). Compared to the Healthy group, the CPE group contained higher proportions of Bifidobacterium (29.59 ± 15.07%, P < 0.001, FDR < 0.001), 0.001), Streptococcus (4.70 ± 3.61%, P < 0.001, FDR < 0.001), Akkermansia P < 0.001, FDR < 0.001), Enterococcus (1.88 ± 2.49%, P < 0.001, FDR < 0.001), FDR < 0.001), Prevotella (1.86 ± 2.52%, P < 0.001, FDR < 0.001), Veillonella (0.98 ± 1.21%, P < 0.001, FDR < 0.001), Rothia (0.62 ± 0.82%, P < 0.001, P < 0.001, FDR < 0.001), and Clostridium IV (0.60 ± 1.35%, P < 0.001, FDR < FDR < 0.001) (Figure 2A). In contrast, the relative abundances of Bacteroides (10.94 ± P < 0.001, FDR < 0.001), Faecalibacterium (0.78 ± 0.82%, P < 0.001, FDR < 0.001), Blautia (1.44 ± 2.68%, P = 0.022, FDR = 0.037), Ruminococcus (0.01 ± 0.02%, P < 0.001, FDR < 0.001), Roseburia (0.00 ± 0.00%, P < 0.001, FDR < 0.001), Anaerostipes (0.04 ± 0.06%, P < 0.001, FDR < 0.001), and Parasutterella (0.00 ± 0.00%, P < 0.001, FDR < 0.001) were notably decreased in CPE patients (Figure 2A).
Figure 2.
GM differences between CPE and Healthy groups, and bacterial co-occurrence network in all children. (A) Top 15 abundant genera were selected from CPE and Healthy groups, and their abundances were compared between these two groups. * and *** stand for the P-value smaller than 0.05 and 0.001. Healthy and CPE groups were represented with blue and red boxes, respectively. (B) The genera enriched in CPE and Healthy groups were selected, and their co-occurrence network was constructed. The blue and red edges suggest the positive and negative correlations, respectively, while the diameter of the circle is proportional to the relative abundance of the genera.
A GM co-occurrence network was constructed based on the differentially enriched bacteria between the CPE and Healthy groups (Figure 2B). Bacteroides was negatively correlated with Lactobacillus (r = −0.768, P < 0.001, FDR < 0.001) and Prevotella (r = −0.722, P < 0.001, FDR < 0.001), while Intestinibacter was negatively correlated with Bifidobacterium (r = −0.726, P < 0.001, FDR <0.001) and Enterococcus (r = −0.707, P < 0.001, FDR = 0.002) (Figure 2B). On the other hand, complex positive correlations were unveiled among bacteria that were enriched in CPE patients. Positive relationships were discovered between Streptococcus and Actinomyces (r = 0.833, P < 0.001, FDR < 0.001), Actinomyces and Veillonella (r = 0.811, P < 0.001, FDR < 0.001), Veillonella and Staphylococcus (r = 0.682, P < 0.001, FDR = 0.009), and Staphylococcus and Catenibacterium (r = 0.790, P < 0.001, FDR < 0.001).
Altered GM Function in the CPE Patients Was Correlated With Their Dysbiotic GM
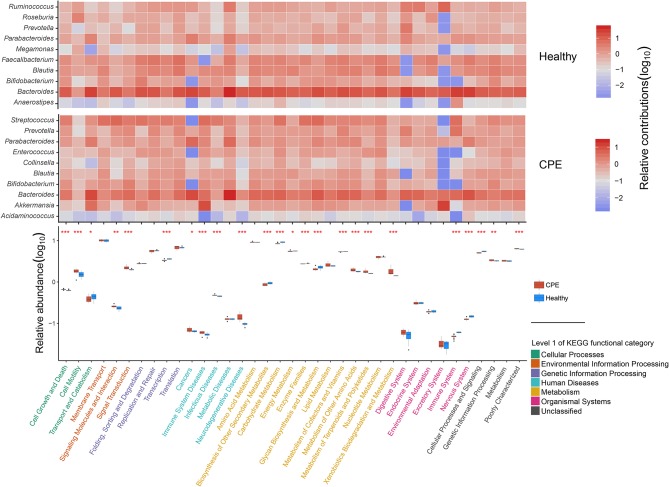
Using PICRUSt software and the KEGG database, we obtained 6,909 KEGG orthologous groups (KOs) from all the samples (Supplementary File 3). All pathways collapsed into 37 functional categories at KEGG level II, and 24 of them were differentially enriched between the Healthy and CPE groups (Figure 3). The enriched functional categories in the CPE group included “Signal transduction” (2.22 ± 0.20%, P < 0.001, FDR < 0.001) and “Xenobiotics biodegradation and metabolism” (1.78 ± 0.30%, P < 0.001, FDR < 0.001) (Figure 3). By contrast, the proportions of microbial genes involved in “Biosynthesis of in “Biosynthesis of other secondary metabolites” (0.86 ± 0.06%, P < 0.001, system” (0.04 ± 0.00%, P < 0.001, FDR < 0.001), and “Nervous system” (0.12 ± 0.00%, P < 0.001, FDR < 0.001) were decreased in CPE patients, which corresponded with the increased risks increased risks of “Immune system diseases” (0.06 ± 0.00%, P < 0.001, FDR < 0.001) and “Neurodegenerative diseases” (0.14 ± 0.02%, P < 0.001, FDR < 0.001).
Figure 3.
Distribution of functional categories for GM in CPE and Healthy groups. In the heatmap, the contributions of the top 10 genera on 37 KEGG level II functional categories were detected in CPE and Healthy groups, respectively. The deeper red square means the genera contribute to the functional category importantly, while the deeper blue square means the functional category obtained less contribution from the genera. In the box plot, the enriched pathways were compared between CPE and Healthy groups, and their P-values were indicated by asterisks. *, **, and *** stand for the P-value smaller than 0.05, 0.01, and 0.001, respectively. Functional classifications of KEGG level I were suggested by different colors, while the blue and red boxes represented Healthy and CPE groups, respectively.
The increased risk of “Neurodegenerative diseases” in CPE patients was probably attributed to Streptococcus, Parabacteroides, and Bacteroides (Figure 3), among which Streptococcus was significantly enriched in the patients (P < 0.001, FDR < 0.001, Figure 2A). In healthy children, both Faecalibacterium and Blautia interacted with the “Nervous system” closely (Figure 3), but they decreased in CPE patients (Figure 2A). Moreover, the enriched Akkermansia and Streptococcus (Figure 2A) in the CPE group were the major contributors for the increased risk of “Immune system diseases” (Figure 3). Besides regulating the “Immune system,” Bacteroides participated in the metabolism of most nutrition in its host (Figure 3).
Discussion
Though they are two different neurologic diseases, CP and epilepsy interact with each other closely: over 25% of CP patients are diagnosed with epilepsy (1), and epilepsy aggravates the motor and mental impairment of CP patients (3, 4, 6). An increasing numbers of studies on the gut-brain axis have suggested the potential impact of GM on various neurologic diseases (10, 11), though little is known regarding patients with CP and epilepsy. In this study, we characterized the GM features in CPE patients and their potential roles in the pathogenesis of CPE.
Compared with healthy children, the CPE patients exhibited lower proportions of Anaerostipes, Faecalibacterium, and Bacteroides. Jamie Joseph et al. have reported that Anaerostipes and Faecalibacterium spp. could produce butyrate with acetate (25), and butyrate could stimulate the differentiation of regulatory T (Treg) cells and relieve the neuroinflammation burdens (26). However, large amounts of acetate would accumulate in CPE patients with low abundances of Anaerostipes and Faecalibacterium spp., which could activate the parasympathetic nervous system (27) and trigger a seizure. In addition, the decreased Bacteroides would also reduce butyrate secretion and attenuate its neuroprotective effect in CPE patients (26). On the other hand, higher abundances of Enterococcus, Bifidobacterium, Clostridium IV, and Akkermansia were discovered in the CPE patients. Enterococcus could stimulate the production of serotonin in chromaffin cells and improve autistic behaviors through neuromodulation (28), whereas, excessive serotonin, most likely caused by the multiple drug treatments in the CPE patients, was harmful to the central nervous system (29). Although Bifidobacterium has been regarded as a beneficial microbe due to its ability to enforce the epithelial barrier, its higher abundance also indicated the immature microbiota in the patients (30). Furthermore, a higher abundance of Clostridium would aggravate microbial dysbiosis in CPE patients as a potentially pathogenic bacterium (31). With increasing non-dominated bacteria, dysbiotic GM in CPE patients exhibited higher microbial diversity than healthy children. Since complex competitive and synergistic relationships exist inside the GM (10), we further constructed the bacterial co-occurrence network to deepen our understanding of the GM features in CPE patients.
The co-occurrence network exhibited the negative correlations between bacteria enriched in healthy children (such as Bacteroides, Escherichia/Shigella, and Intestinibacter) and those enriched in CPE patients (such as Bifidobacterium, Lactobacillus, and Enterococcus). These antagonistic relationships indicated that the dominance of Bacteroides would repress the overgrowth of Bifidobacterium and Lactobacillus in individuals with mature GM (32). Probably due to its high diversity and abundance (33), Bacteroides participated in multiple metabolic pathways, including amino acid metabolism, energy metabolism, and lipid metabolism. Its abundance also reflected that Bacteroides could adapt to the diversified diet in adults, and provide energy and substrates to the host and other gut commensals.
Further analysis of microbial functions revealed the increased risks of immune systemic diseases and neurodegenerative diseases in CPE patients, and that neuroinflammation probably played a key role in the pathology of CPE (34). The elevated risk of immune system diseases was mainly attributed to the higher proportion of Akkermnsia in CPE patients. According to a previous report, the overgrowth of Akkermnsia would degrade mucin in mucous layers and increase the mucosal permeability (35), which enables more bacterial antigens to be exposed to the host immune system and trigger systematic immunoreactions in CPE patients. Moreover, the increased growth of Streptococcus would raise the levels of IL-6 and TNF-α (36), and induce neurodegenerative diseases by inducing neuroinflammation. Combined with the co-occurrence relationships among bacteria, we speculated that the mature GM was crucial for the development of the nervous system in children, and the clinical symptoms would possibly be relieved by GM interventions in CPE patients.
A limitation of the current research is the lack of a large number of patients with epilepsy. The comparison between CPE and epilepsy patients would better illustrate the roles of GM in the pathogenesis of CP and epilepsy, which would improve our understanding of the gut-brain axis. Additional work, as listed here, is also imperative in further studies: (I) performing large-cohort studies to verify the current findings; (II) exploring the alteration of intestinal metabolites in CPE patients, and their associations with microbiota; (III) recording the clinical improvement after treatment in CPE patients, and detecting their GM alterations.
In summary, this study presented the GM characteristics in CPE patients, illustrated the bacterial relationships inside the GM, and detected the functional distributions of GM in CPE patients. These findings suggest the roles of GM in the pathogenesis of CPE, and provide references for the bacterial adjuvant interventions in the treatment of CPE patients.
Data Availability Statement
The datasets generated for this study can be found in NCBI Sequence Read Archive (SRA) Database, BioProject ID: PRJNA530084.
Ethics Statement
This study was approved by the Ethics Committee of The Hospital of Maternal and Child Health (Longgang, China) under registration number LGFY2017005. As the guardian of the CPE patients, the Longgang District Social Welfare Center provided written informed consent and volunteered to receive investigation on their children for scientific research, as well as the parents of the healthy children.
Author Contributions
YP, SL, KZ, and XX managed the project. CH, XL, QO, GW, and PW performed the fecal sampling and information collection. CH and QO prepared the DNA. YL, DL, and XF performed the bioinformatics analysis in this work. CH, YL, and DL interpreted the analysis results and wrote the paper. XF, WD, and QZ optimized the graphs. KZ, XX, SL, and YP guided statistical analysis and polished the article. All authors reviewed this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all participants for their support in completing this work. We thank the doctors and nurses at The Hospital of Maternal and Child Health (Longgang, China), who assisted the study team with clinical examination and fecal sample collection. We also thank Mr. Xiaofeng (Lawrence) Lin from EasyPub for polishing the manuscript.
Footnotes
Funding. The work was supported by Longgang District Science and Technology Innovation Bureau (No. p020180720388406844161) and Shenzhen Science and Technology Project (No. JCYJ20170816170527583).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2019.00394/full#supplementary-material
Background information for enrolled subjects.
Distribution of GM in all subjects at the phylum and genus levels.
Distribution of bacterial functions in all subjects with KEGG database.
References
- 1.Rosenbaum P, Paneth N, Leviton A, Goldstein M, Bax M, Damiano D, et al. A report: the definition and classification of cerebral palsy April 2006. Dev Med Child Neurol Suppl. (2007) 109:8–14. 10.1111/j.1469-8749.2007.tb12610.x [DOI] [PubMed] [Google Scholar]
- 2.Graham HK, Rosenbaum P, Paneth N, Dan B, Lin JP, Damiano DL, et al. Cerebral palsy. Nat Rev Dis Primers. (2016) 2:15082 10.1038/nrdp.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajewska E, Sobieska M, Samborski W. Associations between manual abilities, gross motor function, epilepsy, and mental capacity in children with cerebral palsy. Iran J Child Neurol. (2014) 8:45–52. [PMC free article] [PubMed] [Google Scholar]
- 4.Sellier E, Uldall P, Calado E, Sigurdardottir S, Torrioli MG, Platt MJ, et al. Epilepsy and cerebral palsy: characteristics and trends in children born in 1976-1998. Eur J Paediatr Neurol. (2012) 16:48–55. 10.1016/j.ejpn.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 5.Sundrum R, Logan S, Wallace A, Spencer N. Cerebral palsy and socioeconomic status: a retrospective cohort study. Arch Dis Child. (2005) 90:15–8. 10.1136/adc.2002.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peduzzi M, Defontaine E, Misson JP. Epilepsy in children with cerebral palsy. Rev Med Liege. (2006) 61:237–9. 10.1055/s-0033-1337759 [DOI] [PubMed] [Google Scholar]
- 7.Vigevano F, Arzimanoglou A, Plouin P, Specchio N. Therapeutic approach to epileptic encephalopathies. Epilepsia. (2013) 54 (Suppl. 8):45–50. 10.1111/epi.12423 [DOI] [PubMed] [Google Scholar]
- 8.van der Louw E, van den Hurk D, Neal E, Leiendecker B, Fitzsimmon G, Dority L, et al. Ketogenic diet guidelines for infants with refractory epilepsy. Eur J Paediatr Neurol. (2016) 20, 798–809. 10.1016/j.ejpn.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 9.Ijff DM, Aldenkamp AP. Cognitive side-effects of antiepileptic drugs in children. Handb. Clin. Neurol. (2013) 111:707–18. 10.1016/B978-0-444-52891-9.00073-7 [DOI] [PubMed] [Google Scholar]
- 10.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK. The central nervous system and the gut microbiome. Cell. (2016) 167:915–32. 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinan TG, Cryan JF. The microbiome-gut-brain axis in health and disease. Gastroenterol Clin North Am. (2017) 46:77–89. 10.1016/j.gtc.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 12.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE. (2013) 8:e76993 10.1371/journal.pone.0076993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa S, Goto S, Tsuji H, Okuno T, Asahara T, Nomoto K, et al. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson's disease. PLoS ONE. (2015) 10:e0142164. 10.1371/journal.pone.0142164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Li G, Huang P, Liu Z, Zhao B. The gut microbiota and Alzheimer's disease. J Alzheimers Dis. (2017) 58:1–15. 10.3233/JAD-161141 [DOI] [PubMed] [Google Scholar]
- 15.O'Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. (2015) 277:32–48. 10.1016/j.bbr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 16.Xie G, Zhou Q, Qiu CZ, Dai WK, Wang HP, Li YH, et al. Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J Gastroenterol. (2017) 23:6164–71. 10.3748/wjg.v23.i33.6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skiold B, Eriksson C, Eliasson AC, Aden U, Vollmer B. General movements and magnetic resonance imaging in the prediction of neuromotor outcome in children born extremely preterm. Early Hum Dev. (2013) 89:467–72. 10.1016/j.earlhumdev.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 18.Parisi P, Paolino MC, Raucci U, Ferretti A, Villa MP, Trenite DK. “Atypical forms” of benign epilepsy with centrotemporal spikes (BECTS): How to diagnose and guide these children. A practical/scientific approach. Epilepsy Behav. (2017) 75:165–9. 10.1016/j.yebeh.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 19.Guo C, Li Y, Wang P, Li Y, Qiu C, Li M, et al. Alterations of gut microbiota in cholestatic infants and their correlation with hepatic function. Front Microbiol. (2018) 9:2682. 10.3389/fmicb.2018.02682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 21.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. (2014) 42:D633–42. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang ZZ, Chen G, Alekseyenko AV. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics. (2016) 32:2618–25. 10.1093/bioinformatics/btw311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. (2013) 31:814–21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph J, Depp C, Shih PB, Cadenhead KS, Schmid-Schonbein G. Modified Mediterranean diet for enrichment of short chain fatty acids: potential adjunctive therapeutic to target immune and metabolic dysfunction in schizophrenia? Front Neurosci. (2017) 11:155. 10.3389/fnins.2017.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamawaki Y, Yoshioka N, Nozaki K, Ito H, Oda K, Harada K, et al. Sodium butyrate abolishes lipopolysaccharide-induced depression-like behaviors and hippocampal microglial activation in mice. Brain Res. (2018) 1680:13–38. 10.1016/j.brainres.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 27.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. (2016) 534:213–7. 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. (2015) 161:264–76. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett D. Drug-induced serotonin syndrome. Crit Care Nurse. (2017) 37:49–54. 10.4037/ccn2017169 [DOI] [PubMed] [Google Scholar]
- 30.Subramanian S, Blanton LV, Frese SA, Charbonneau M, Mills DA, Gordon JI. Cultivating healthy growth and nutrition through the gut microbiota. Cell. (2015) 161:36–48. 10.1016/j.cell.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortier LC. The contribution of bacteriophages to the biology and virulence of pathogenic clostridia. Adv Appl Microbiol. (2017) 101:169–200. 10.1016/bs.aambs.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 32.Shimizu H, Benno Y. Membrane filter method to study the effects of Lactobacillus acidophilus and Bifidobacterium longum on fecal microbiota. Microbiol Immunol. (2015) 59:643–52. 10.1111/1348-0421.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. (2016) 4:15. 10.1186/s40168-016-0160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paouri E, Georgopoulos S. Systemic and CNS inflammation crosstalk: implications for Alzheimer's disease. Curr Alzheimer Res. (2019) 16:559–74. 10.2174/1567205016666190321154618 [DOI] [PubMed] [Google Scholar]
- 35.Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr. (2018) 63:33–5. 10.3164/jcbn.18-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. (2015) 5:8096. 10.1038/srep08096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Background information for enrolled subjects.
Distribution of GM in all subjects at the phylum and genus levels.
Distribution of bacterial functions in all subjects with KEGG database.
Data Availability Statement
The datasets generated for this study can be found in NCBI Sequence Read Archive (SRA) Database, BioProject ID: PRJNA530084.