Abstract
Mechanical force regulates periodontal ligament cell (PDL) behavior. However, different force types lead to distinct PDL responses. Here, we report that pretreatment with an intermittent compressive force (ICF), but not a continuous compressive force (CCF), promoted human PDL (hPDL) osteogenic differentiation as determined by osteogenic marker gene expression and mineral deposition in vitro. ICF-induced osterix (OSX) expression was inhibited by cycloheximide and monensin. Although CCF and ICF significantly increased extracellular adenosine triphosphate (ATP) levels, pretreatment with exogenous ATP did not affect hPDL osteogenic differentiation. Gene-expression profiling of hPDLs subjected to CCF or ICF revealed that extracellular matrix (ECM)-receptor interaction, focal adhesion, and transforming growth factor beta (TGF-β) signaling pathway genes were commonly upregulated, while calcium signaling pathway genes were downregulated in both CCF- and ICF-treated hPDLs. The TGFB1 mRNA level was significantly increased, while those of TGFB2 and TGFB3 were decreased by ICF treatment. In contrast, CCF did not modify TGFB1 expression. Inhibiting TGF-β receptor type I or adding a TGF-β1 neutralizing antibody attenuated the ICF-induced OSX expression. Exogenous TGF-β1 pretreatment promoted hPDL osteogenic marker gene expression and mineral deposition. Additionally, pretreatment with ICF in the presence of TGF-β receptor type I inhibitor attenuated the ICF-induced mineralization. In conclusion, this study reveals the effects of ICF on osteogenic differentiation in hPDLs and implicates TGF-β signaling as one of its regulatory mechanisms.
Subject terms: Cell signalling, Osteoblasts
Introduction
The periodontal ligament (PDL) is a connective tissue that links the tooth root to alveolar bone1,2. Fibroblasts are the main cell type residing in the PDL3. The PDL’s mechanoreceptors play an important role in the reflexes that prevent damage to the tooth and periodontium4,5. The PDL functions to resist occlusal forces, to transmit forces from the teeth to alveolar bone, to secure the teeth in the alveolar socket, and as a protective scaffold for cells, vessels, and nerves6,7. The PDL is exposed to various mechanical stimuli in both physiological and pathological conditions. During normal mastication, the PDL is subjected to various force types. The PDL fibers are arranged in several orientations, generating resistance to chewing forces from various directions. Compressive and tensile stresses are observed in different locations of the PDL as demonstrated by finite element analysis of simulated parafunctional and traumatically loaded teeth8. During orthodontic tooth movement (OTM), the PDL normally receives the force in a directional manner, resulting in a change of tooth position. Thus, mechanical force clearly participates in the regulation of PDL homeostasis.
Many publications demonstrate that mechanical force controls the biological activities and responses of cells isolated from human PDL tissue9–11. Cyclic stretch exposure results in differential expression of genes related to the ECM, cell adhesion, and ECM proteases in human PDL cells (hPDLs)9. Continuous compressive force (CCF) treatment significantly increases interleukin 6 (IL6), but decreases alkaline phosphatase (ALP), mRNA expression10. Hence, these results indicate the influence of different force types on hPDL behavior. Studies illustrate that CCF and intermittent compressive force (ICF) differentially regulate hPDL behavior11,12. CCF and ICF both significantly increased SOST, TGFB1, and HEY1 mRNA expression in hPDLs11. However, the upregulation was less in the CCF-treated group compared with the ICF-treated group11. Furthermore, CCF treatment did not significantly alter the expression of HES1 mRNA, while a significant upregulation of HES1 was observed after ICF treatment11. Moreover, in an in vivo OTM model, CCF generated more intermediate root resorption than that of ICF application12. Correspondingly, histomorphometrical analysis illustrated that the percentage of osteoclast length per bone length is lower after ICF treatment compared with CCF treatment in rat molars13. A clinical study demonstrated that CCF application during canine retraction resulted in gradual bodily movement, while ICF led to rapid tipping of the retracted canine14. Both force types influenced anchorage loss in a similar manner14. These data imply that different mechanical forces have distinct roles in regulating hPDL responses.
Mechanical forces have been shown to regulate osteogenic differentiation in osteoblasts. However, the effect of compressive force on osteogenic differentiation potency of hPDLs remains unclear. The aim of the present study was to evaluate the effect of CCF and ICF stimuli on hPDL osteogenic differentiation. In addition, the regulatory mechanisms of ICF-pretreatment on the osteogenic differentiation of hPDLs were examined.
Materials and methods
Cell isolation and culture
The cell isolation protocol was approved by the Human Research Ethics Committee, Faculty of Dentistry, Chulalongkorn University (Approval number 008/2018). The inclusion criteria for the donors were: (1) age between 18‒35 years old, (2) normal healthy teeth without infection or inflammation, (3) the teeth were treatment planned for extraction, and (4) orthodontically untreated patients. Cell isolation was performed as previously described15. The periodontal tissues were gently scraped from the middle third of the root surface and the tissues were placed on 35-mm tissue culture dishes (cat. No. 430165, Corning, Oneonta, NY, USA) for cell explant. The explanted cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM cat. No. 11960, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (cat. No. 10270, Gibco), 1% l-glutamine (GlutaMAXTM−1, cat. No. 35050, Gibco), and 1% Antibiotic-Antimycotic (penicillin, streptomycin, amphotericin B, cat. No. 15240, Gibco). The cells were incubated at 37 °C in a humidified 5% carbon dioxide atmosphere. The culture medium was changed every 48 h. After reaching confluence, the cells were subcultured at a 1:3 ratio using a trypsin/EDTA solution (cat. No. 25200, Gibco).
Compressive force treatment
The compressive force treatment was performed using a computer-controlled apparatus11,16. Cells were seeded in 6-well tissue culture plates (cat. No. 430166, Corning) at a density of 37,500 cells/cm2 for 24 h and the cells were then serum starved for 8 h prior to compressive force treatment. The compressive force application parameters were as previously described11,16. Briefly, an ICF was applied on the cells with a loading frequency of 0.23 Hz at a 1.5 g/cm2 force. The CCF was applied using continuous loading with a 1.5 g/cm2 force. The mechanical stimulation was performed in serum-free culture medium. In some conditions, the cells were pretreated with TGF-β receptor type 1 inhibitor (SB431542 4 μM, cat. No. S4317, Sigma-Aldrich, St. Louis, MO, USA), TGF-β1 neutralizing antibody (5 μg/ml, cat. No. MAB240, R&D Systems, Minneapolis, MN, USA), suramin (15 μM, cat. No. 574625, Calbiochem®, La Jolla, CA, USA), JNK inhibitor (40 nM, cat. No. 420119, Calbiochem®), p38 MAP kinase inhibitor (35 nM, cat. No. 506126, Calbiochem®), cycloheximide (1 μg/ml, cat. No. C-0934, Sigma-Aldrich), Rho-kinase inhibitor (12.7 nM, cat. No. 555550, Calbiochem®), or monensin (1 μM, cat. No. M5273, Sigma-Aldrich) for 30 min prior to mechanical force treatment.
Osteogenic differentiation
Cells were seeded on 6-well tissue culture plates at a density of 37,500 cells/cm2. The cells were subjected to CCF or ICF stimulation in serum-free medium for 24 h. Subsequently, the culture medium was changed to osteogenic medium, which was normal growth medium supplemented with β-glycerophosphate (5 mM, cat. No. G9422, Sigma-Aldrich), l-ascorbic acid (50 μg/mL, cat. No. A-4034, Sigma-Aldrich), and dexamethasone (250 nM, cat. No. D8893, Sigma-Aldrich). The medium was changed every 48 h.
In other experiments, cells were seeded on 48-well tissue culture plates (cat. No. 3548, Costar®, Corning) at a density of 37,500 cells/cm2 and allowed to attach for 24 h. The cells were then starved in serum-free medium for 8 h and subsequently exposed to adenosine 5′-triphosphate disodium salt hydrate (ATP, cat. No. A6419, Sigma-Aldrich) or recombinant human TGF-β1 (cat. No. 616455, Calbiochem®) for 24 h in serum-free culture medium. The cells were then maintained in osteogenic medium.
Alizarin Red S staining
Mineral deposition was determined using alizarin red s staining. Briefly, the samples were fixed with ice-cold methanol (cat. No. 100230, Honeywell, Ulsan, Korea) for 10 min. The mineral deposits were stained with 1% alizarin red s (cat. No. A5533, Sigma-Aldrich) solution for 3 min at room temperature. The samples were gently rinsed with deionized water between procedures. The stained mineral deposits were solubilized in 10% cetylpyridinium chloride solution and the optical density was measured at 570 nm. The relative absorbance was calculated by normalizing the results to the corresponding controls.
ATP assay
The amount of extracellular ATP after mechanical stimuli was determined using an ENLITEN® ATP assay system bioluminescence detection kit (cat. No. FF2000, Promega, Madison, WI, USA). Briefly, aliquots of culture media (50 μL) were added to the microplate. Subsequently 50 μL of Enliten Luciferase/Luciferin solution was added and the signal was immediately measured using a microplate reader (Synergy H1, Biotek multi-mode reader, Winooski, VT). The amount of extracellular ATP was quantified using a standard curve and the value was normalized to the control of each donor.
Immunofluorescence staining
The cells were fixed with 4% buffered formalin (cat. No. F-1268, Sigma-Aldrich) for 10 min and permeabilized with 0.1% Triton-X100 (cat. No. 22686, USB corporation, Cleveland, OH, USA) for 5 min. Nonspecific binding was blocked by incubating the cells with 2% horse serum (cat. No. SH30074, Hyclone, South Logan, UT, USA) for 30 min. The specimens were then incubated with primary antibody at 4 °C overnight. The primary antibodies used in the present study were rabbit anti-osterix (OSX) antibody (1000× dilution, cat. No. ab22552, Abcam, Cambridge, UK) and mouse anti-TGF-β1 antibody (10x dilution, cat. No. MAB240, R&D Systems). A biotinylated goat-anti rabbit antibody (1000× dilution, cat. No. sc-2040, Santa Cruz Biotechnology, Dallas, TX, USA) or a biotinylated goat-anti mouse antibody (1000× dilution, cat. No. B2763, LifetechnologiesTM, Eugene, OR, USA) were employed as secondary antibodies. Streptavidin-FITC (cat. No. S3762, Sigma-Aldrich) was used for fluorescence labeling. The specimens were incubated with secondary antibodies and Strep-FITC for 40 min each. The specimens were gently washed with PBS between each step. DAPI (cat. No. 5748, TOCRIS bioscience, Bristol, UK) was used to counterstain the nuclei. The specimens were then observed under an Apotome.2 (Carl Zeiss, Jena, Germany) fluorescence microscope.
RNA sequencing
After compressive force treatment for 24 h, total RNA was isolated using an RNeasy Plus Mini Kit (cat. No. 74134, Qiagen, Germantown, MD, USA). RNA sequencing was performed at the Omics Science and Bioinformatics Center, Faculty of Science, Chulalongkorn University. Briefly, total RNA quality was examined using an Agilent 2100 BioAnalyzer (Agilent Technologies, CA, USA). Sequencing libraries were constructed using a TrueSeq mRNA stranded library prep kit (Illumina, CA, USA). Subsequently, the library quality was determined using an Agilent 2100 BioAnalyzer and Qubit 3.0 fluorometer (Thermo Fisher Scientic, MA, USA). RNA sequencing was performed using a NextSeq 500 (Illumina). The reads quality was checked, trimmed, and filtered by FastQC and Trimmomatic17,18. Read mapping was performed using HISAT2 against Homo sapiens UCSC hg3819. Transcript quantification was performed using HTseq count20. For data filtering, the genes with the lowest 15% variance based on inter-quartile range and the genes with less than 4 counts in total were removed. Differential gene expression was determined using EdgeR21,22. Differentially expressed genes that exhibited an at least twofold up- or downregulation were included. A significant difference was defined as false-discovery rate < 0.05. The raw data was deposited in the NCBI Sequence Read Archive (SRP136155) and NCBI Gene Expression Omnibus (GSE112122). Gene ontology and enriched pathways were analyzed using WebGestalt and Reactome23,24.
Polymerase chain reaction
Total cellular RNA was isolated using RiboExTM solution (cat. No. 301-001, GeneAll®, Seoul, South Korea). The isolated RNA integrity and amount was examined using a Nanodrop2000 (Thermo Scientific, USA). The absorbance ratios at 260/280 and 260/230 nm were evaluated. Subsequently, RNA (1 μg) was converted to complimentary DNA using an ImProm-IITM Reverse Transcription System (cat. No. A3800, Promega). One microliter of complimentary DNA was employed for real-time polymerase chain reaction using a FastStart Essential DNA Green Master kit (cat. No. 06402712001, Roche Diagnostics, Mannheim, Germany). The reaction was performed on a LightCycler® 96 real-time polymerase chain reaction (PCR) system (Roche Diagnostics). Relative gene expression was calculated using the 2−ΔΔCt method25. According to our preliminary data, we evaluated the expression stability of three reference genes: GAPDH, 18S RNA, and ACTB. GAPDH was chosen as reference gene. The expression value was normalized to the GAPDH expression value and the control. The oligonucleotide primers are shown in Supplementary Table 1.
Enzyme linked immunosorbent assay (ELISA)
Cell lysate was extracted using RIPA buffer. Conditioned medium was collected for measuring TGF-β1 levels using a human TGF-β1 immunoassay kit (cat. No. DB100B, R&D Systems). ELISA was performed per the manufacturer’s instructions. The optical density was measured at 450 nm. The amount of TGF-β1 was calculated using a standard curve and subsequently normalized to total protein and the control condition.
Western blot
Protein extraction was performed using RIPA buffer containing a protease inhibitor cocktail (cat. No. P8340, Sigma–Aldrich). The protein samples were electrophoresed on a 12% sodium dodecyl sulfate–polyacrylamide gel and then transferred onto nitrocellulose membranes. The membranes were incubated with primary antibody at 4 °C overnight. The following primary antibodies were used: (i) a rabbit polyclonal antibody to SP7/osterix (1000× dilution, cat. No. ab22552; Abcam), and (ii) a mouse IgG against GAPDH (2000x dilution, MAB374, MILLIPORE, Temecula, CA, USA). A biotinylated secondary antibody (1000× dilution) was subsequently incubated with the membrane. The following secondary antibodies were used: (i) biotin conjugated goat anti-rabbit antibody (cat. No. sc-2040, Santa Cruz); or (ii) biotin conjugated goat-anti mouse antibody (cat. No. B2763, LifetechnologiesTM). Subsequently, membranes were incubated with peroxidase-labeled streptavidin. The signal was visualized by chemiluminescence (SuperSignal® West Pico Chemiluminescent Substrate; cat. No. 34079; Thermo SCIENTIFIC, Rockford, IL, USA). The band density was determined using ImageJ software. Band density of target proteins was normalized to band density of GAPDH and subsequently normalized to the control.
Statistical analyses
Data presentation and statistical analyses were performed using Prism 8 (GraphPad Software, CA, USA). The data are shown as mean ± standard error of mean (SEM). SEM was calculated based on the individual number of different donors employed in each experiment. Cells from at least four different donors without pooling were employed in each experiment. For RNA sequencing and western blot analysis, cells from three different donors were employed. Each dot in the graph represents the value from the independent experiments. For two independent group comparisons, the Mann Whitney U test was employed. Kruskal Wallis followed by pairwise comparison was used to determine significant differences for three or more group comparison. Correction of multiple comparisons using statistical hypothesis testing was performed by Dunn’s test with Prism 8 software. Significance was considered at p < 0.05.
Results
ICF pretreatment-induced hPDL osteogenic differentiation
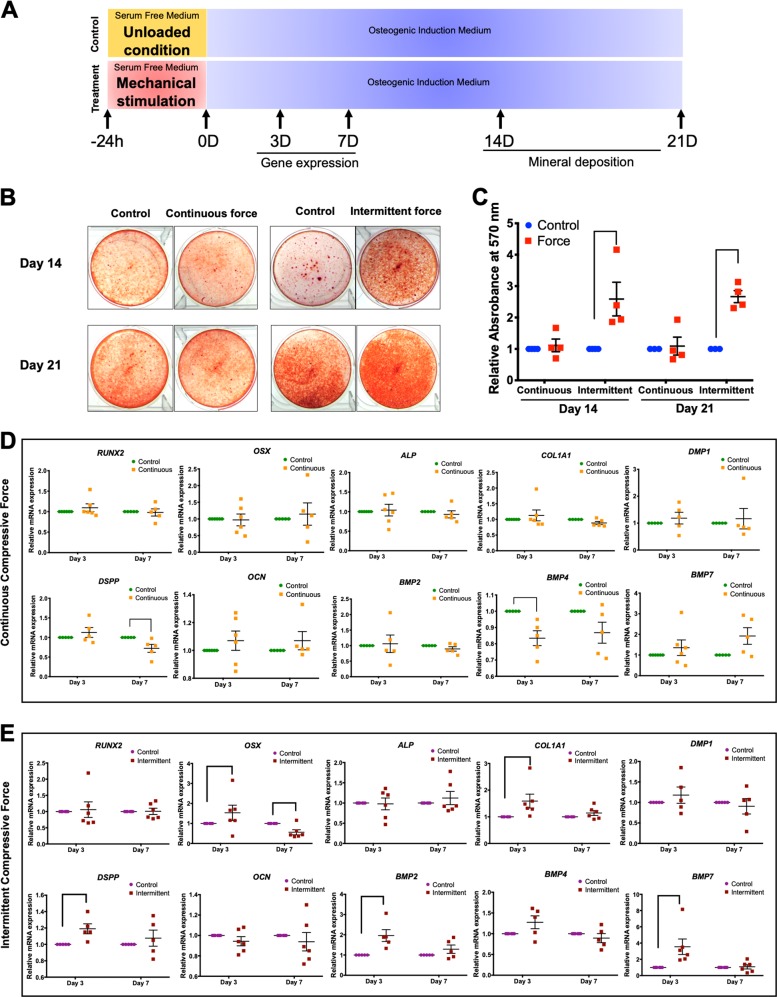
Cells were pretreated with CCF or ICF for 24 h in serum-free culture medium and subsequently maintained in osteogenic medium for 21 days (Fig. 1a). In the control condition, cells were cultured in serum-free culture medium for 24 h without mechanical loading and further maintained in osteogenic medium (Fig. 1a). The cells pretreated with ICF demonstrated markedly enhanced mineral deposition compared with the control at both day 14 and day 21 after osteogenic induction (Fig. 1b, c). However, CCF pretreatment did not significantly influence mineral deposition at either time point (Fig. 1b, c).
Fig. 1. ICF stimulated osteogenic differentiation in hPDLs.
Cells were exposed to CCF or ICF in serum-free media for 24 h and subsequently maintained in osteogenic medium (a). In the control condition, cells were cultured in serum-free culture medium for 24 h without mechanical loading and further maintained in osteogenic medium (a). Mineral deposition shown by alizarin red s staining at 14 and 21 days after osteogenic induction (b). The relative absorbance of the solubilized alizarin red dye was demonstrated (c). After pretreating cells with the CCF (d) or ICF (e), osteogenic marker gene expression was evaluated compared with the unloaded control using real-time polymerase chain reaction at 3 and 7 days after osteogenic induction. Bars indicate a significant difference between conditions
Pretreatment with CCF did not significantly influence the mRNA expression of RUNX2, OSX, ALP, COL1A1, DMP1, OCN, BMP2, or BMP7 (Fig. 1d). Reduced BMP4 and DSPP mRNA levels were observed at day 3 and 7 in osteogenic medium, respectively. hPDLs pretreated with ICF significantly upregulated OSX, COL1A1, DSPP, BMP2, and BMP7 mRNA expression at day 3 of osteogenic induction (Fig. 1e). There was no significant difference in RUNX2, ALP, DMP1, OCN, or BMP4 mRNA levels between ICF-pretreated cells and the control cells. These results imply the promotion of osteogenic marker gene expression in osteogenic inductive condition of ICF-pretreated hPDLs.
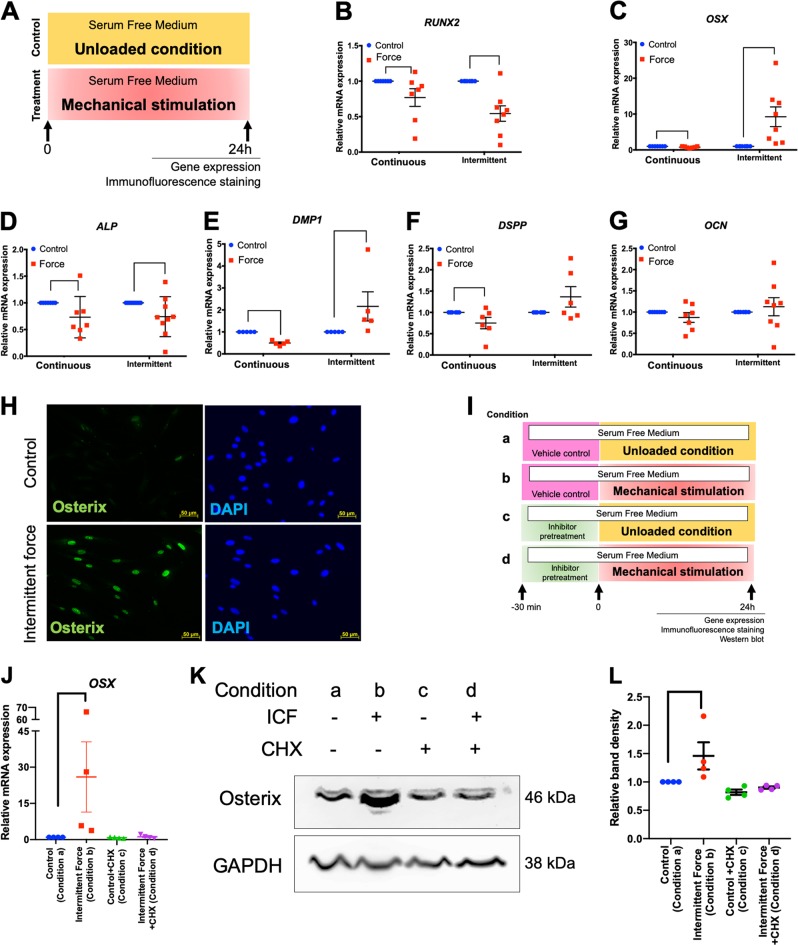
Cells were stimulated with CCF or ICF in serum-free medium for 24 h (Fig. 2a). Cells cultured in the same condition without mechanical force application were used as the control (Fig. 2a). The results demonstrated that CCF treatment significantly reduced RUNX2, OSX, ALP, DMP1, and DSPP mRNA expression in hPDLs (Fig. 2b–f). In contrast, ICF treated cells exhibited significantly increased OSX and DMP1, but reduced RUNX2 and ALP, mRNA expression (Fig. 2b–f). OCN mRNA levels were not altered by either CCF or ICF treatment at 24 h (Fig. 2g).
Fig. 2. ICF induced OSX expression in hPDLs.
Cells were treated with CCF or ICF in serum-free media for 24 h and cells cultured in the same condition without mechanical force application were used as the control (a). The mRNA expression of osteogenic marker genes was examined using real-time polymerase chain reaction (b–g). OSX protein expression was evaluated using immunofluorescence staining (h). In the inhibition experiments, cells were pretreated with inhibitor 30 min prior to ICF stimulation. Experimental conditions were illustrated (i). The OSX mRNA (j, n) were determined using real-time polymerase chain reaction. Osterix protein expression was evaluated using western blot (k, o) and the normalized band density was demonstrated (l, p). In addition, osterix protein expression was also determined by immunofluorescence staining (m, q). Bars indicate a significant difference between conditions. Scale bars indicate 50 μm
ICF-induced OSX protein expression was confirmed using immunofluorescence staining (Fig. 2h). In the chemical inhibitor experiment, the experimental condition was demonstrated in Fig. 2i. Pretreatment with cycloheximide or monensin 30 min prior to ICF stimulation abolished the force-induced OSX mRNA and protein expression in hPDLs (Fig. 2j–q).
ATP priming did not influence hPDL osteogenic differentiation
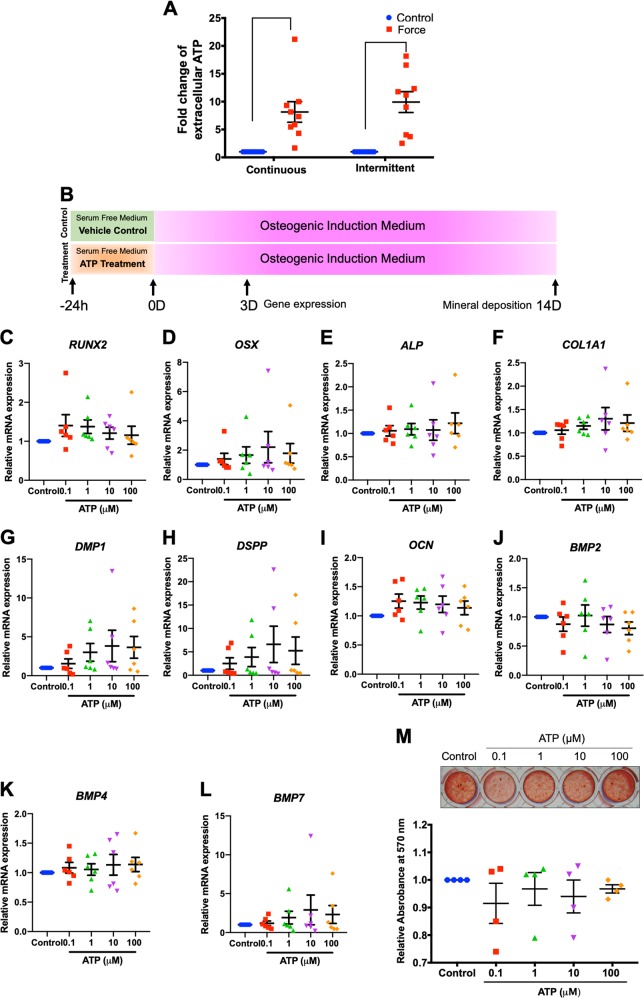
A previous report showed that compressive force stimulated adenosine triphosphate (ATP) release in hPDLs26. In addition, it has been demonstrated that low levels of ATP induced osteogenic differentiation in human osteoblast-like cells27. Hence, we hypothesized that ICF enhanced osteogenic differentiation in hPDLs via ATP release. To test this hypothesis, cells were treated with CCF or ICF for 24 h in serum-free media and the concentration of extracellular ATP was determined. The extracellular ATP concentration was significantly increased in both CCF- and ICF-treated conditions compared with the unloaded control (Fig. 3a). The fold-change of extracellular ATP of the cells treated with CCF or ICF was comparable. To evaluate if the short-term release of extracellular ATP was involved with osteogenic differentiation, cells were pretreated with ATP (0.1‒100 μM) for 24 h in serum-free medium and subsequently maintained in osteogenic medium (Fig. 3b). In control condition, cells were maintained in serum-free medium for 24 h without ATP supplementation and further maintained in osteogenic medium (Fig. 3b). The short-term exposure of 0.1‒100 μM ATP did not significantly influence osteogenic marker gene expression at day 3 of osteogenic induction (Fig. 3c–l). Correspondingly, there was no significant difference in mineral deposition between the ATP treated cells and the control as evaluated by alizarin red s staining at 14 days after osteogenic induction (Fig. 3m).
Fig. 3. ATP priming did not influence osteogenic differentiation in hPDLs.
Cells were treated with CCF or ICF in serum-free media for 24 h. Extracellular ATP was evaluated using an ATP assay in culture medium (a). Schematic diagram of the experimental plan of the ATP priming was illustrated (b). Cells were exposed to ATP for 24 h in serum-free medium. Thereafter, the culture medium was changed to osteogenic medium. Osteogenic marker gene expression was determined using real-time polymerase chain reaction at day 3 after osteogenic induction (c–l). Mineral deposition was examined using alizarin red s staining at day 14 (m). The normalized absorbance of the solubilized dye. Bars indicate a significant difference between conditions
Differential gene-expression profiling of CCF- and ICF-treated hPDLs
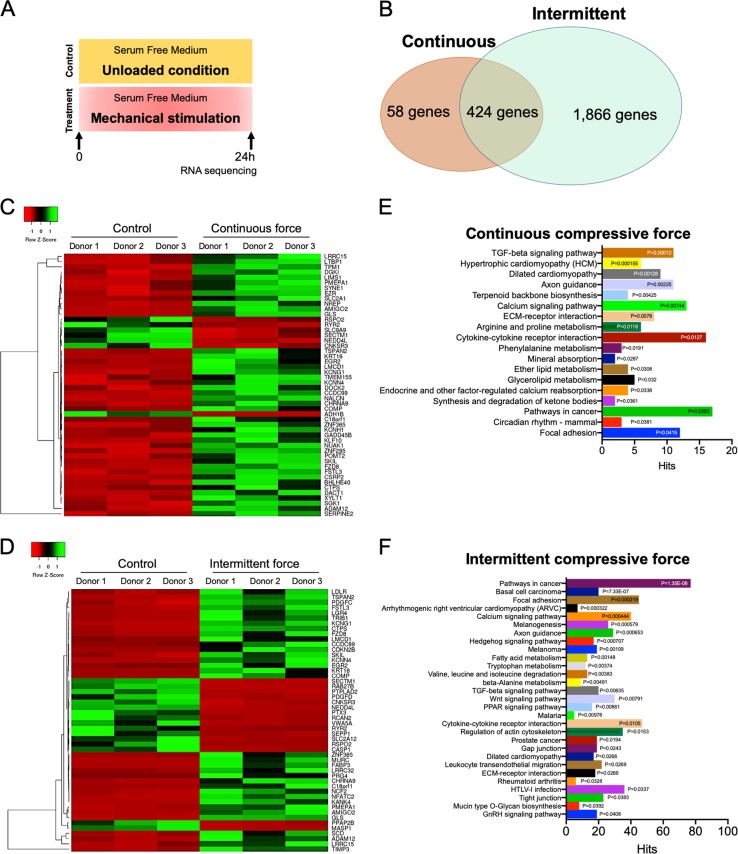
To further identify pathway(s) regulating ICF-induced osteogenic differentiation, hPDLs were serum starved and subsequently exposed to CCF or ICF for 24 h in serum-free medium. Cells maintained in serum-free medium without mechanical loading were employed as the control (Fig. 4a). Total RNA was collected and subjected to RNA sequencing analysis for differential mRNA expression profiling. The control cultures were maintained in the same condition without mechanical stimulation. The results demonstrated that CCF- and ICF-treated hPDLs exhibited 482 and 2290 differentially regulated genes, respectively. Among these differentially expressed genes, 424 genes were found in both hPDLs loaded with CCF or ICF (Fig. 4b). Heatmaps of the top 50 differentially expressed genes in CCF- or ICF-treated hPDLs are illustrated in Fig. 4c, d. The top 20 significantly upregulated and downregulated genes in CCF- or ICF-treated hPDLs are listed in Supplementary Tables 2 and 3.
Fig. 4. Gene expression profiling of mechanical force treated hPDLs.
Cells were treated with CCF or ICF in serum-free medium for 24 h. Cells maintained in serum-free medium without mechanical loading were employed as the control (a). The gene expression profile compared with the unloaded control was determined using RNA sequencing and bioinformatic analyses. Common differentially expressed genes in the CCF- or ICF-treated hPDLs are shown (b). Heat maps demonstrating the top 50 differentially expressed genes in the CCF- (c) or ICF- (d) treated cells compared with the unloaded control. Pathway enrichment of all differentially expressed genes using KEGG database revealed the differentially regulated pathways after CCF (e) and ICF (f) treatment
The enriched KEGG pathways in CCF- or ICF-treated hPDLs are presented in Fig. 4e, f as well as Supplementary Tables 4 and 5. In Fig. 4e, f, all differentially expressed genes were included for enrichment analyses. To investigate further in detail, the differentially expressed genes were categorized into upregulated and downregulated genes. Subsequently, KEGG pathway enrichment analysis was separately performed for upregulated and downregulated genes (Supplementary Tables 4 and 5). Among the upregulated enriched pathways, the upregulated genes in the CCF-treated cells were found in the ECM-receptor interaction and transforming growth factor β (TGF-β) signaling pathway genes (Supplementary Table 4). Moreover, CCF treatment downregulated genes in the calcium signaling pathway and cytokine–cytokine receptor pathway. In ICF-treated hPDLs, focal adhesion, regulation of actin cytoskeleton, TGF-β signaling pathway, and cytokine–cytokine receptor pathway genes were found among the upregulated enriched pathways (Supplementary Table 5). The downregulated genes in ICF-treated hPDLs were involved in the calcium signaling pathway. Focal adhesion and TGF-β signaling pathway genes were upregulated, while the calcium signaling pathway was downregulated in both CCF- and ICF-treated hPDLs (Supplementary Tables 4 and 5). Interestingly, the Wnt signaling pathway genes were upregulated by ICF-treatment, but not by CCF stimulation (Supplementary Tables 4 and 5).
CCF and ICF differentially regulated ECM-receptor interaction, focal adhesion, and TGF-β signaling pathway genes in hPDLs
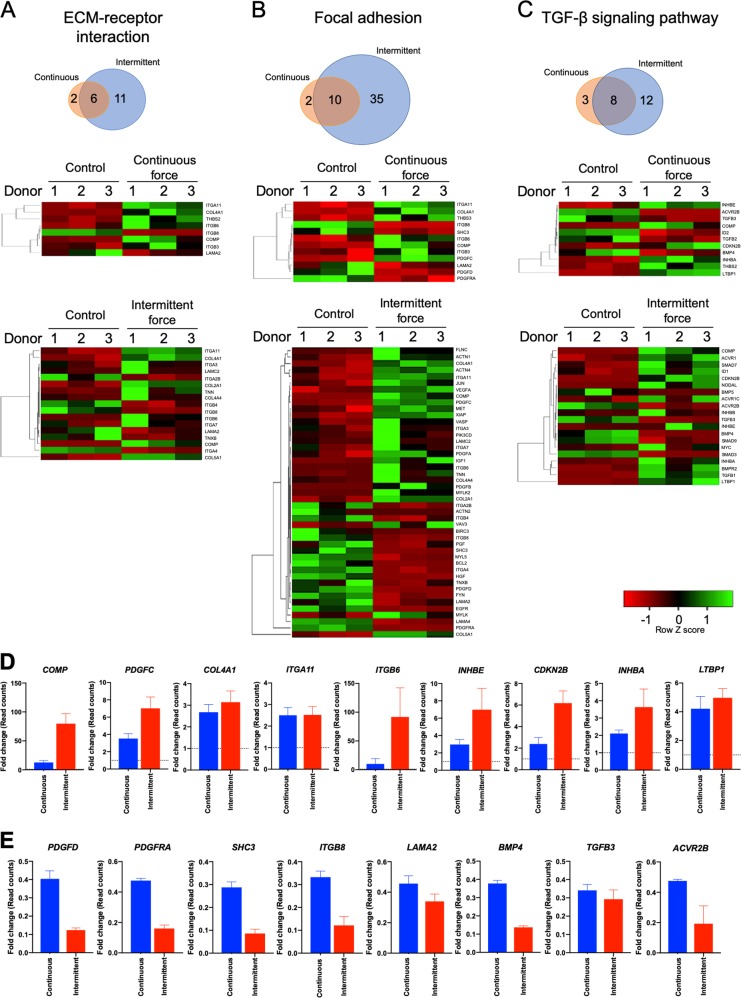
CCF differentially regulated the expression of 8, 12, and 11 genes, while ICF regulated 17, 45, and 20 genes in ECM-receptor interaction, focal adhesion, and TGF-β signaling pathways, respectively. Six, ten, and eight commonly regulated genes in both CCF- and ICF-treated cells were noted in ECM-receptor interaction, focal adhesion, and TGF-β signaling pathways, respectively (Fig. 5a–c).
Fig. 5. Mechanical force regulated ECM-receptor interaction, focal adhesion, and TGF-β signaling pathways in hPDLs.
Bioinformatic analysis demonstrated the common differentially expressed genes and heat maps in ECM-receptor interaction (a), focal adhesion (b), and TGF-β signaling (c) pathways. The significantly upregulated (d) and downregulated (e) genes were selected. The fold-change of the raw read counts from the RNA sequencing data were presented. Dotted lines indicate the normalized expression value of the unloaded control
In the TGF-β signaling pathway, CCF induced the mRNA expression of CDKN2B, INHBE, THBS2, LTBP1, COMP, and INHBA, but suppressed ID2, BMP4, ACVR2B, TGFB2, and TGFB3 expression (Fig. 5c). ICF regulated the mRNA expression of 20 genes involved in TGF-β signaling. The upregulated genes are TGFB1, CDKN2B, LTBP1, ID1, ACVR1, BMPR2, INHBB, ACVR1C, MYC, SMAD7, INHBE, COMP, INHBA, and NODAL, while the downregulated genes are BMP5, ACVR2B, TGFB3, BMP4, SMAD9, and SMAD3.
The common genes regulated by both CCF and ICF in ECM-receptor interaction, focal adhesion and TGF-β signaling pathways were illustrated in Fig. 5d, e. The fold-change of the raw read count from RNA sequencing was plotted to compare the expression of the common genes between both force types. ICF induced a dramatic fold-change in COMP, PDGFC, ITGB6, INHBE, CDKN2B, INHBA, PDGFD, PDGFRA, SHC3, ITGB8, LAMA2, BMP4, and ACVR2B expression compared with CCF (Fig. 5d, e). The expression of COL4A1, ITGA11, LTBP1, and TGFB3 mRNA was comparable between cells treated with CCF or ICF.
ICF-regulated TGF-β1 expression in hPDLs
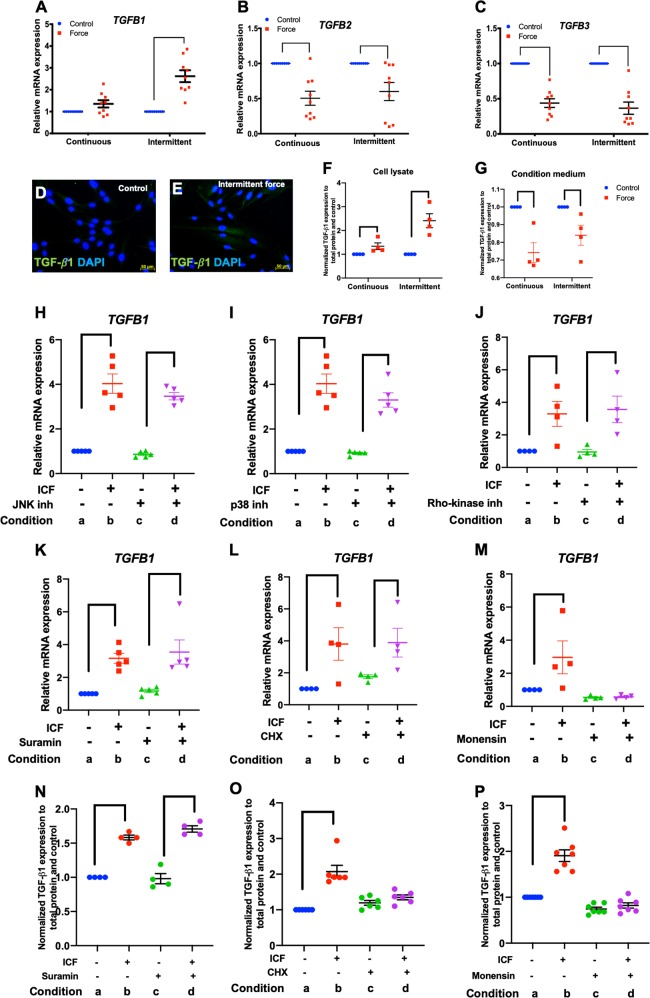
Real-time PCR was performed on selected genes in the TGF-β signaling pathway to validate the RNA sequencing data. Cells were treated with CCF or ICF in serum-free medium for 24 h. Cells cultured in the same condition without mechanical force application were used as the control. Experimental design was illustrated as in Fig. 2a. The results demonstrated that TGFB1 was differentially regulated by CCF and ICF (Fig. 6a). ICF significantly increased TGFB1 expression, while CCF did not significantly alter TGFB1 mRNA levels compared with the unloaded control (Fig. 6a). Both CCF and ICF significantly inhibited TGFB2 and TGFB3 expression (Fig. 6b, c), confirming the results from the RNA sequencing experiment. Immunofluorescence staining revealed that ICF markedly induced TGF-β1 protein expression compared with the unloaded control (Fig. 6d, e). The TGF-β1 protein levels in cell lysates and condition medium were also examined (Fig. 6f, g). ICF markedly induced TGF-β1 expression compared with CCF in cell lysates. However, the TGF-β1 expression was decreased in the conditioned medium from the compressive force treatment condition.
Fig. 6. ICF induced TGF-β1 expression.
Cells were treated with CCF or ICF in serum-free media for 24 h. Cells cultured in the same condition without mechanical force application were used as the control. Experimental conditions were illustrated in Fig. 2a. The mRNA expression of TGFB1 (a), TGFB2 (b), and TGFB3 (c) was evaluated using real-time polymerase chain reaction. TGF-β1 protein expression was examined using immunofluorescence staining (d, e) and enzyme linked immunosorbent assay (f, g). To investigate the regulatory pathways, cells were pretreated chemical inhibitor 30 min prior to ICF treatment. Experimental conditions were illustrated in Fig. 2i. Chemical inhibitors were JNK inhibitor (h), p38 inhibitor (i), Rho-kinase inhibitor (j), suramin (k, n), cycloheximide (l, o), or monensin (m, p). TGFB1 mRNA and protein levels were examined using real-time polymerase chain reaction and enzyme linked immunosorbent, respectively. Bars indicate a significant difference between conditions. ICF intermittent compressive force treatment, CHX cycloheximide. Scale bars indicate 50 μm
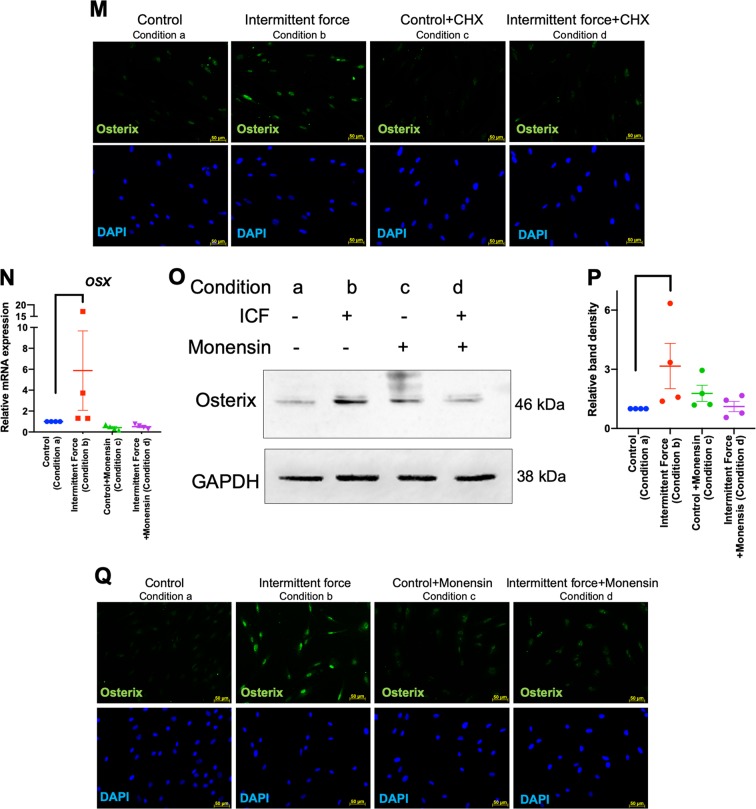
We further investigated the regulatory mechanism by which ICF promoted TGFB1 mRNA expression in hPDLs. Cells were pretreated with chemical inhibitors for 30 min before being exposed to ICF. The results demonstrated that pretreatment with a JNK inhibitor, p38 inhibitor, Rho-kinase inhibitor, cycloheximide, and suramin did not affect the ICF-induced TGFB1 mRNA expression (Fig. 6h–l). Interestingly, monensin abolished the effect of ICF on TGFB1 expression (Fig. 6m). Similar to mRNA expression, suramin pretreatment did not alter ICF-induced TGF-β1 protein levels (Fig. 6n). However, cycloheximide and monensin pretreatment inhibited the effect of ICF on TGF-β1 expression (Fig. 6o, p).
TGF-β1 participated in the ICF-induced hPDL osteogenic induction
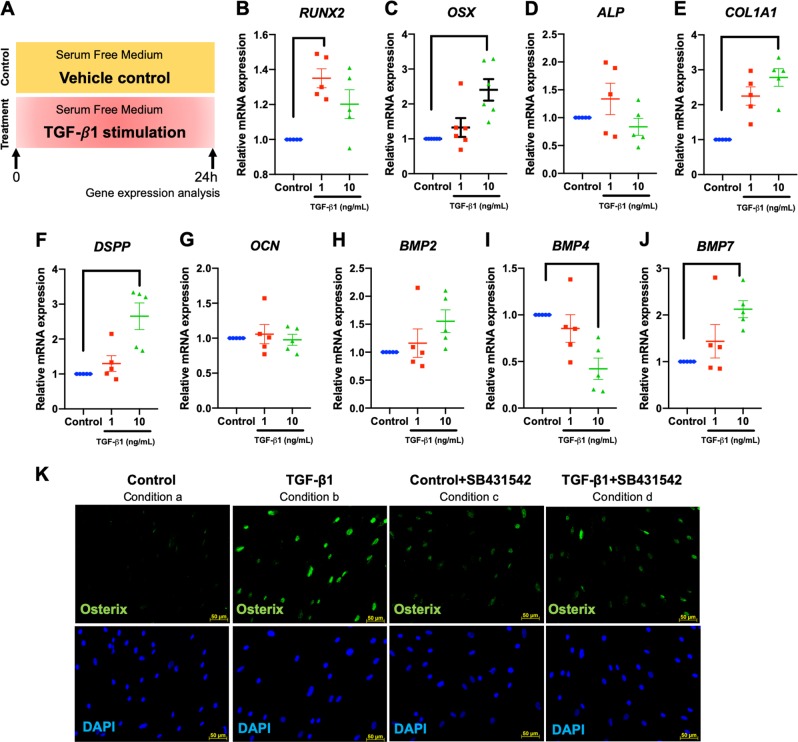
To determine the influence of TGF-β1 on hPDL osteogenic differentiation, the expression of osteogenic marker genes was evaluated after cells were exposed to various concentrations of recombinant human TGF-β1 in serum-free medium for 24 h. In the control condition, cells were cultured in serum-free condition and the vehicle control was added in the medium. Schematic diagram of the experimental plan was illustrated (Fig. 7a). TGF-β1 increased the expression of RUNX2, OSX, COL1A1, DSPP, and BMP7 mRNA levels, while attenuating BMP4 mRNA expression (Fig. 7b–j). The increased expression of OSX protein was markedly observed after cells were exposed to TGF-β1 (10 ng/mL) and this effect was attenuated by pretreatment with a TGF-β receptor inhibitor (SB431542) (Fig. 7k), suggesting the involvement of TGF-β signaling in OSX expression in hPDLs. Experimental conditions of inhibitor experiment were illustrated in Fig. 2i.
Fig. 7. Recombinant human TGF-β1 promoted osteogenic marker gene expression.
Cells were treated with 1 and 10 ng/mL of recombinant human TGF-β1 in serum-free culture medium for 24 h. In the control condition, cells were cultured in serum-free condition and the vehicle control was added in the medium. Schematic diagram of the experimental plan was illustrated (a). Osteogenic marker gene expression was determined using real-time polymerase chain reaction (b–j). OSX protein expression was evaluated using immunofluorescence staining (k). In some conditions, cells were pretreated with SB431542 30 min prior to TGF-β1 exposure. Experimental conditions were illustrated in Fig. 2i. Bars indicate a significant difference between conditions. Scale bars indicate 50 μm
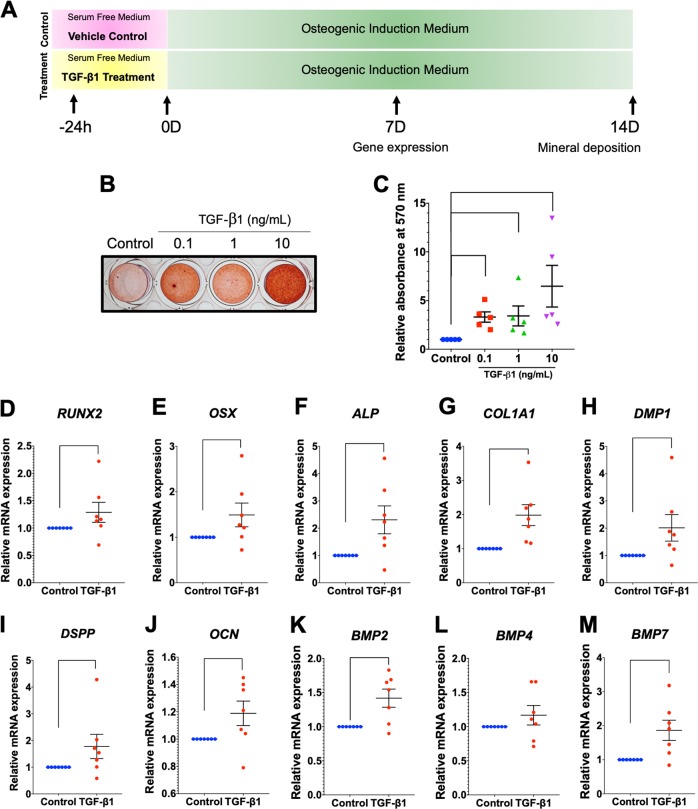
Rather than using ICF pretreatment, hPDLs were exposed to recombinant human TGF-β1 for 24 h in serum-free medium and subsequently maintained in osteogenic medium for 14 days (Fig. 8a). In control condition, cells were treated with vehicle control in serum-free condition and further maintained in osteogenic medium (Fig. 8a). TGF-β1 pretreatment dramatically induced mineral deposition in a dose-dependent manner (Fig. 8b, c). A significant difference in mineralization was observed at all concentrations of TGF-β1 pretreatment compared with the control (Fig. 8c). In addition, pretreatment with 10 ng/mL TGF-β1 significantly upregulated RUNX2, OSX, ALP, COL1A1, DMP1, DSPP, OCN, BMP2, and BMP7 mRNA levels after 7 days in osteogenic medium (Fig. 8d–m).
Fig. 8. Priming with recombinant human TGF-β1 promoted osteogenic differentiation in hPDLs.
Schematic diagram of the experimental plan of TGF-β1 priming and subsequent osteogenic induction (a). Mineralization was examined using alizarin red s staining at day 14 after osteogenic induction (b). The normalized absorbance of alizarin red dye (c). Osteogenic marker gene expression was determined using real-time polymerase chain reaction at day 7 after osteogenic induction (d–m). Bars indicate a statistically significant difference between conditions
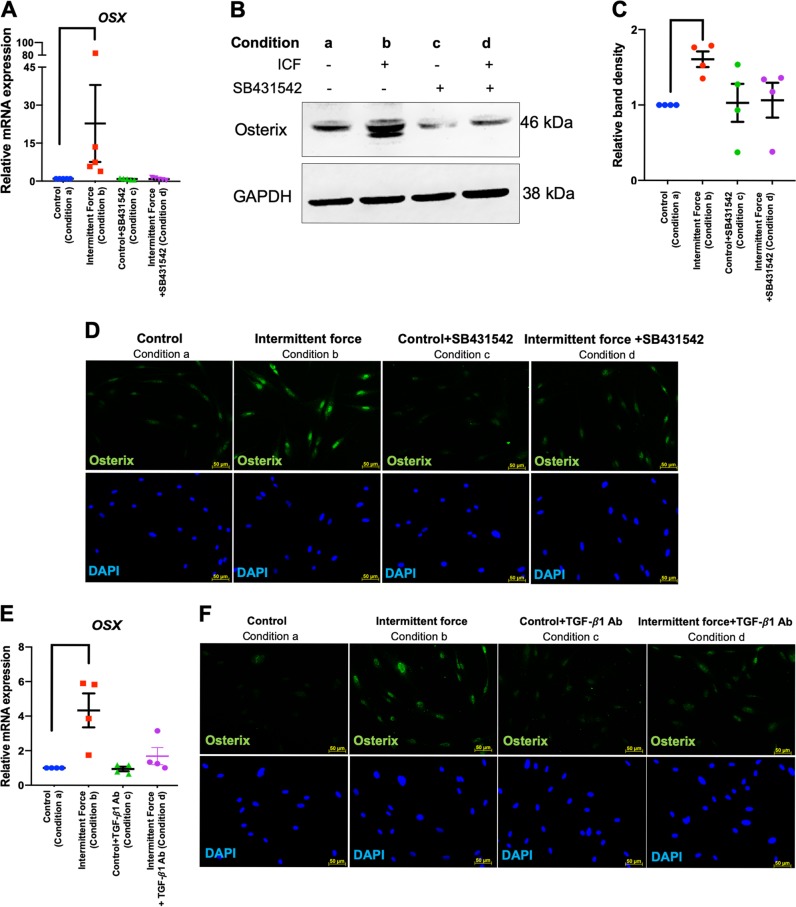
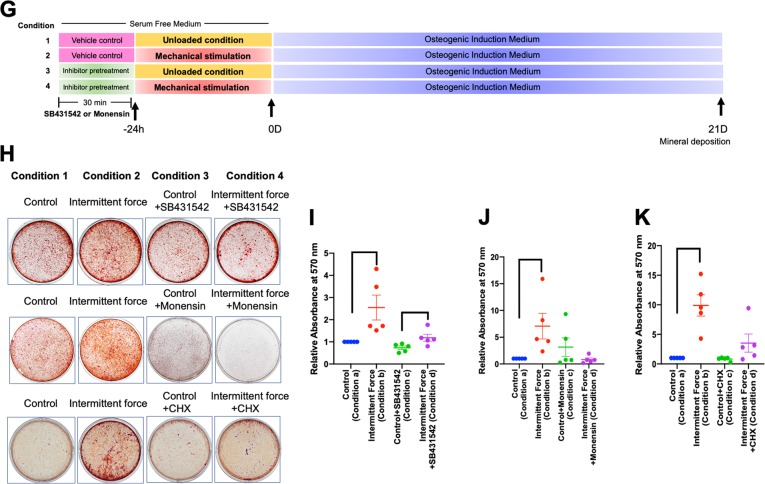
The role of TGF-β signaling in the ICF-induced osteogenic differentiation in hPDLs was determined. Pretreatment with SB431542 or neutralizing antibodies against TGF-β1 attenuated the ICF-induced OSX mRNA and protein expression at 24 h in serum-free medium (Fig. 9a–f). Cells were pretreated with SB431542 30 min prior to ICF treatment in serum-free medium for 24 h and subsequently the cells were maintained in osteogenic medium for 21 days (Fig. 9g). SB431542 pretreatment prior to force stimulation attenuated the mineral deposition compared with the ICF-treated group (Fig. 9h, i).
Fig. 9. TGF-β1 participated in the ICF-induced osteogenic differentiation in hPDLs.
Cells were pretreated with SB431542 or TGF-β1 neutralizing antibody for 30 min prior to ICF stimulation for 24 h in serum-free culture medium. Experimental conditions were illustrated in Fig. 2i. OSX mRNA expression was evaluated using real-time polymerase chain reaction (a, e). OSX protein expression was demonstrated using western blot (b) and immunofluorescence staining (d, f). The normalized band density was illustrated (c). Schematic diagram of the experimental plan for evaluating the influence of TGF-β1 on mineralization in hPDLs was demonstrated (g). Cells were exposed with SB431542 or monensin or cycloheximide for 30 min prior to ICF stimulation in serum-free culture medium for 24 h and subsequently maintained in osteogenic medium. Mineral deposition was examined using alizarin red s staining at day 21 after osteogenic induction (h). The relative absorbance of the solubilized alizarin red dye was demonstrated (i–k). Bars indicate a significant difference between conditions. Scale bars indicated 50 μm
As mentioned above, we demonstrated that monensin inhibited the effect of ICF on TGFB1 mRNA expression. We then further demonstrated that monensin pretreatment prior to ICF stimulation also inhibited mineral deposition (Fig. 9h, j). Moreover, cycloheximide was used to investigate whether the TGF-β1-induced OSX expression participates in the regulation of hPDL differentiation. Cells were pretreated with cycloheximide for 30 min prior to ICF stimulation for 24 h and subsequently maintained in osteogenic medium. Cycloheximide pretreatment attenuated the ICF pretreatment-stimulated mineral deposition (Fig. 9h, k).
Discussion
The present study demonstrated that pretreatment with ICF, but not CCF, promoted hPDL osteogenic differentiation and mineral deposition. We also found that the TGF-β signaling pathway participated in the ICF-induced osteogenic differentiation, and that ICF significantly increased TGFB1 expression. Moreover, inhibiting the TGF-β pathway attenuated the ICF-induced OSX expression. Lastly, pretreatment with a TGF-β receptor inhibitor prior to ICF abolished the ICF pretreatment-induced mineralization. These data implicate the TGF-β signaling pathway in the promotion of osteogenic differentiation by mechanical force pretreatment.
Mechanical stimulation, such as shear, tensile, centrifugation, and vibration, has been shown to regulate osteogenic differentiation28–32. Cyclic tensile strain significantly enhanced RUNX2 and OSX expression in human periodontal ligament stem cells (hPDLSCs)33. Cyclic stretching upregulated ALP enzymatic activity and osteogenic marker gene expression through the YAP, ROCK, and myosin remodeling pathways32. In addition, Wnt signaling also contributed in the hydrostatic force-induced ALP enzymatic activity as well as RUNX2 and OSX mRNA expression in hPDLSCs31. Thus, these data indicate that different types of force application may utilize different pathways to control osteogenic differentiation in hPDLs. The effect of CCF and ICF on osteogenic differentiation potency of hPDLs remains unknown. The present study illustrated that ICF treatment in serum-free culture medium for 24 h led to significantly increased OSX expression. Pretreatment with ICF prior to osteogenic induction promoted OSX, COL1A1, DSPP, BMP2, and BMP7 mRNA expression as well as mineral deposition by hPDLs. Conversely, CCF treatment inhibited OSX expression and CCF pretreatment did not markedly influence osteogenic differentiation. This information confirms that different force applications differentially regulate hPDL differentiation.
Mechanical force is known to stimulate ATP release from hPDLs. It has been shown that centrifugation-mediated force application resulted in ATP release from hPDLs34. Our results also showed that both ICF and CCF dramatically stimulated ATP release from hPDLs. Mechanistically, mechanical force stimulated ATP release via connexin43 and the intracellular calcium signaling pathway26. It has been reported that ATP supplementation in osteogenic medium enhanced osteogenic marker gene expression and mineralization in several cell types27,35–37. In the present study, short-term ATP treatment prior to osteogenic induction did not influence osteogenic marker gene expression or mineralization by hPDLs. Hence, short-term treatment with ATP may not promote osteogenic differentiation in this cell type. However, continuous long-term ATP supplementation in osteogenic medium may affect the osteogenic ability of hPDLs. Therefore, further investigation is required on this aspect.
Mechanical stimulation regulates a global change in the gene-expression profile in hPDLs. Although several studies investigated mRNA profiles, the comparison of gene expression in hPDLs subjected to different force types is lacking10,38,39. Uniaxial cyclic strain application induced genes related to cell cycle, apoptosis, and proliferation in hPDLs40. In contrast, static compressive force regulated genes related to the ECM, inflammatory cytokines, and cell growth10. These results indicate that different force types differentially regulate global gene expression patterns in hPDLs. However, a comparison among/between different force types has not yet been reported. In addition, the influence of ICF on the global gene expression in hPDLs remains unknown. In the present study, the mRNA expression profile was compared between CCF and ICF stimulation in hPDLs. The overall results suggest that ICF exhibits more influence on hPDL gene expression compared with CCF treatment. For the same genes, the effect of ICF on mRNA expression levels is more robust compared with CCF treatment. This direct comparison confirms the distinct biological effects of specific force types on hPDL behavior.
ICF differentially regulated more genes in TGF-β signaling pathway in hPDLs compared with CCF treatment. Rat periodontal tissues exhibited high Tgf-β1 expression compared with dental pulp tissues and alveolar bone41. Furthermore, rodent periodontal ligament cells expressed higher mRNA levels of Tgfb1 and Tgfb3 than that of Tgfb242,43. A study in rat periodontal ligament cells revealed that during osteogenic differentiation in vitro, Tgfb1 mRNA levels increased in a time-dependent manner43. Conversely, Tgfb3 was downregulated43. These observations imply a role for TGF-β1 in osteogenic differentiation in PDLs. It has been demonstrated that the TGF-β signaling pathway negatively regulated early osteogenic commitment in a murine periodontal ligament cell line (MPDL22), as treatment with a TGF-β receptor kinase inhibitor enhanced BMP-2-induced mineralization42. However, inhibiting TGF-β signaling alone in osteogenic medium without BMP-2 stimulation did not affect mineral deposition by this murine cell line42. Treating hPDLs twice with TGF-β1 attenuated ALP activity and osteogenic marker gene expression via the reduction of IGF-1 and pAkt44,45. In contrast, a single TGF-β1 treatment enhanced RUNX2, ALP, and IGF1 mRNA expression and ALP enzymatic activity45,46. Corresponding with the present study, TGF-β1 pretreatment for 24 h in serum-free medium before osteogenic induction promoted osteogenic differentiation in hPDLs as shown by a significant increase in mineralization and osteogenic marker gene expression. Considering all of this evidence together, TGF-β signaling has complex regulatory effects. This signaling negatively and positively controls osteogenic differentiation in hPDLs depending on the duration and differentiation stage during TGF-β1 exposure.
The present study demonstrated that 45 and 12 genes in the focal adhesion pathway were differentially expressed by ICF and CCF compressive treatment, respectively. A previous report demonstrated that tensile force stimulated the formation of actin stress fibers in hPDLs47. Mechanical force regulated the expression of molecules related to bone resorption (OPG, RANKL, and M-CSF) and inflammatory-related mediator (TNF-α, PGE2, and COX2) expression in hPDLs through the integrin-focal adhesion kinase (FAK) pathway48,49. Moreover, inhibiting FAK attenuated the tensile force-induced ALP enzymatic activity and OCN expression47. Although the present study illustrated the role of TGF-β signaling pathway in ICF-pretreatment-stimulated osteogenic differentiation in hPDLs, the influence of the focal adhesion pathway cannot be excluded. The involvement/interaction of TGF-β and focal adhesion pathways in ICF-induced osteogenic gene expression and mineralization should be further determined.
Mechanical stretching regulated genes in the ECM and adhesion molecule pathways9. Our study demonstrated that ICF induced a more than 100-fold increase in ITGB6 mRNA expression compared with the unloaded condition, implying the potential participation of ICF in controlling hPDL behavior. In a mouse dental papilla mesenchymal cell line, integrin β6 bound to dentin sialoprotein and subsequently induced cell attachment, migration, and Dmp1 and Dspp mRNA expression50, suggesting that integrin β6 is involved in the regulation of dental mesenchymal cell behavior. TGF-β1 positively regulated ITGB6 expression in oral cancer cells and gingival keratinocytes that may participate in cancer invasion and periodontal inflammation, respectively51,52. Our RNA sequencing data revealed increased ITGB6 mRNA levels concurrent with the upregulation of TGFB1 mRNA in ICF treatment. Hence, the interaction of these two molecules and their contributions to ICF pretreatment-induced osteogenic differentiation in hPDLs should be further evaluated.
In the present study, ICF significantly upregulated OSX and DMP1. We hypothesize that OSX might be a key regulatory factor in ICF-pretreatment-induced osteogenic differentiation. Osterix is an essential transcription factor regulating bone formation and can be controlled via a Runx2-independent pathway53,54. Osterix modulates the expression of various osteogenic marker genes55. Osx conditional knockout mice exhibited decreased Dmp1 and Dspp expression, which inhibited odontoblast differentiation56. The conditional deletion of Osx in dental mesenchyme resulted in reduced Dmp1 expression and cellular cementum formation57. Thus, OSX was chosen for investigation in the inhibitor experiments. However, the direct regulation of OSX on downstream osteogenic marker gene expression and mineral deposition under ICF stimulation in hPDLs requires further investigation.
ICF-induced OSX expression in hPDLs and this effect was inhibited by pretreatment with cycloheximide or monensin. Cycloheximide is a protein translation inhibitor. The attenuation of ICF-induced OSX expression by cycloheximide could imply the involvement of intermediate molecule(s). ICF also promoted the expression of TGFB1 mRNA and protein. However, cycloheximide failed to inhibit the ICF-induced TGFB1 mRNA, but not TGF-β1 protein, expression indicating an effect of ICF on TGF-β1 expression. The expression of OSX and TGFB1 under ICF was abolished by pretreating the cells with monensin, an inhibitor of intracellular protein transportation and secretion from the Golgi complex, implying similar regulatory pathway(s) of OSX and TGFB1 expression under ICF. Inhibiting TGF-β signaling using a chemical inhibitor (SB431542) or a TGF-β1 neutralizing antibody impeded ICF-induced OSX expression. Adding exogenous TGF-β1 also stimulated OSX expression. These results imply that TGF-β1 would be the upstream regulator of OSX. Further, exogenous TGF-β1 pretreatment enhanced osteogenic marker gene expression and mineralization. Correspondingly, SB431542 pretreatment prior to ICF stimulation attenuated the force-induced mineralization. Taking all this evidence together (Fig. 10), we summarize that ICF promoted the secretion of molecule(s) from Golgi complex and subsequently stimulates TGF-β1 mRNA and protein expression. Further, TGF-β1 could act in autocrine or paracrine manner to promote OSX expression, leading to the enhancement of osteogenic differentiation in hPDLs. ICF also promote ATP release but this mechanism does not involve in ICF-induced osteogenic differentiation in hPDLs.
Fig. 10. Schematic diagram of the potential regulation of OSX expression under ICF in hPDLs.
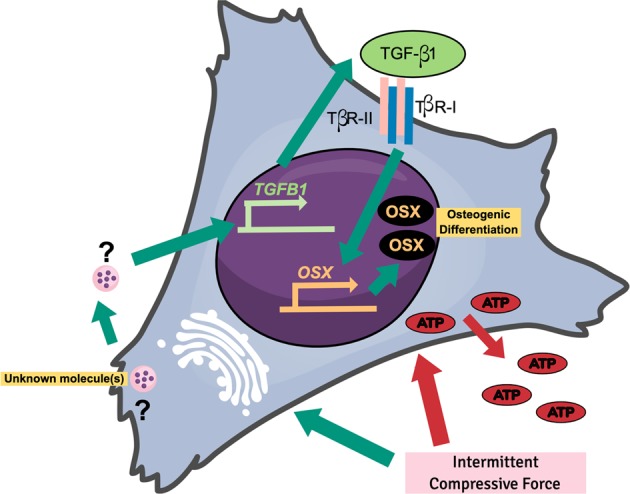
ICF induces the release of molecule(s) from the golgi apparatus and subsequently enhances TGFB1 expression. Further, TGF-β1 is released and binds to TGF-β receptors, leading to the induction of OSX expression
Supplementary information
Acknowledgements
This study was funded by Chulalongkorn University to T.O. [Grant number CU_GR_62_02_32_01] and Thailand Research Fund to P.P. (RTA6180001). The Center of Excellence for Regenerative Dentistry was supported by the Chulalongkorn Academic Advancement into Its 2nd Century Project. J.M. was supported by the Japan Society for the Promotion of Science under JSPS-RONPAKU Fellowship (FY2018), Japan. The authors thank Dr. Kevin Tompkins for language editing.
Authors’ contributions
J.M. contributed to experimental design, data acquisition, and analysis. C.S. contributed to gene-expression experiment and data analysis. N.L. and C.N.L. contributed to setting up and calibrating the computerized mechanical force loading apparatus and data analysis. P.P. and H.E. contributed to experimental design, and data interpretation. T.O. contributed to study conception and experimental design, data analysis and interpretation, and manuscript preparation. All authors critically revised the paper and gave final approve for publication.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Kaartinen
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hiroshi Egusa, Email: egu@dent.tohoku.ac.jp.
Thanaphum Osathanon, Phone: +66-2-218-8885, Email: thanaphum.o@chula.ac.th.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1992-4).
References
- 1.Tomokiyo, A. et. al. Detection, characterization, and clinical application of mesenchymsl stem cells in periodontal ligament tissue. Stem. Cell. Int.2018, 5450768 (2018). [DOI] [PMC free article] [PubMed]
- 2.Iwata Takanori, Yamato Masayuki, Ishikawa Isao, Ando Tomohiro, Okano Teruo. Tissue Engineering in Periodontal Tissue. The Anatomical Record. 2013;297(1):16–25. doi: 10.1002/ar.22812. [DOI] [PubMed] [Google Scholar]
- 3.Smith PC, Martinez C, Martinez J, McCulloch CA. Role of fibroblast populations in periodontal wound healing and tissue remodeling. Front. Physiol. 2019;10:270. doi: 10.3389/fphys.2019.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason AG, Scott BJ, van der Glas HW, Linden RW, Cadden SW. Remote noxious stimuli modulate jaw reflexes evoked by activation of periodontal ligament mechanoreceptors in man. Exp. Physiol. 2002;87:699–706. doi: 10.1113/eph8702452. [DOI] [PubMed] [Google Scholar]
- 5.Turker KS, Sowman PF, Tuncer M, Tucker KJ, Brinkworth RS. The role of periodontal mechanoreceptors in mastication. Arch. Oral. Biol. 2007;52:361–364. doi: 10.1016/j.archoralbio.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Chen FM, Jin Y. Periodontal tissue engineering and regeneration: current approaches and expanding opportunities. Tissue Eng. Part B Rev. 2010;16:219–255. doi: 10.1089/ten.teb.2009.0562. [DOI] [PubMed] [Google Scholar]
- 7.Cho MI, Garant PR. Development and general structure of the periodontium. Periodontol 2000. 2000;24:9–27. doi: 10.1034/j.1600-0757.2000.2240102.x. [DOI] [PubMed] [Google Scholar]
- 8.Poiate IA, de Vasconcellos AB, de Santana RB, Poiate E. Three-dimensional stress distribution in the human periodontal ligament in masticatory, parafunctional, and trauma loads: finite element analysis. J. Periodontol. 2009;80:1859–1867. doi: 10.1902/jop.2009.090220. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, Zhao D, Wu Y, Xu C, Zhang F. Cyclic stretch induced gene expression of extracellular matrix and adhesion molecules in human periodontal ligament cells. Arch. Oral Biol. 2015;60:447–455. doi: 10.1016/j.archoralbio.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Lee YH, et al. Differential gene expression of periodontal ligament cells after loading of static compressive force. J. Periodontol. 2007;78:446–452. doi: 10.1902/jop.2007.060240. [DOI] [PubMed] [Google Scholar]
- 11.Manokawinchoke, J., Sumrejkanchanakij, P., Pavasant, P. & Osathanon, T. Notch signaling participates in TGF-β-induced SOST expression under intermittent compressive stress. J. Cell. Physiol232, 2221–2230 (2017). [DOI] [PubMed]
- 12.Cuoghi, O. A., Aiello, C. A., Consolaro, A., Tondelli, P. M. & Mendonca, M. R. Resorption of roots of different dimension induced by different types of forces. Braz. Oral Res.28, 1–7 (2014). [DOI] [PubMed]
- 13.Kumasako-Haga T, Konoo T, Yamaguchi K, Hayashi H. Effect of 8-hour intermittent orthodontic force on osteoclasts and root resorption. Am. J. Orthod. Dentofac. Orthop. 2009;135:278 e1–278 e8. doi: 10.1016/j.ajodo.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Mowafy MI, Zaher AR. Anchorage loss during canine retraction using intermittent versus continuous force distractions; a split mouth randomized clinical trial. Prog. Orthod. 2012;13:117–125. doi: 10.1016/j.pio.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Manokawinchoke J, Sumrejkanchanakij P, Subbalekha K, Pavasant P, Osathanon T. Jagged1 inhibits osteoprotegerin expression by human periodontal ligament cells. J. Periodontal Res. 2016;51:789–799. doi: 10.1111/jre.12357. [DOI] [PubMed] [Google Scholar]
- 16.Manokawinchoke J, et al. Mechanical force-induced TGFB1 increases expression of SOST/POSTN by hPDL cells. J. Dent. Res. 2015;94:983–989. doi: 10.1177/0022034515581372. [DOI] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. (2018).
- 19.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Luckprom P, Kanjanamekanant K, Pavasant P. Role of connexin43 hemichannels in mechanical stress-induced ATP release in human periodontal ligament cells. J. Periodontal Res. 2011;46:607–615. doi: 10.1111/j.1600-0765.2011.01379.x. [DOI] [PubMed] [Google Scholar]
- 27.Cutarelli A, et al. Adenosine Triphosphate stimulates differentiation and mineralization in human osteoblast-like Saos-2 cells. Dev. Growth Differ. 2016;58:400–408. doi: 10.1111/dgd.12288. [DOI] [PubMed] [Google Scholar]
- 28.Tang M, et al. Fluid shear stress stimulates osteogenic differentiation of human periodontal ligament cells via the extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase signaling pathways. J. Periodontol. 2014;85:1806–1813. doi: 10.1902/jop.2014.140244. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, et al. Expression of Osterix in mechanical stress-induced osteogenic differentiation of periodontal ligament cells in vitro. Eur. J. Oral. Sci. 2008;116:199–206. doi: 10.1111/j.1600-0722.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Influence of different intensities of vibration on proliferation and differentiation of human periodontal ligament stem cells. Arch. Med. Sci. 2015;11:638–646. doi: 10.5114/aoms.2015.52370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, et al. Mechanical stress regulates osteogenic differentiation and RANKL/OPG ratio in periodontal ligament stem cells by the Wnt/beta-catenin pathway. Biochim. Biophys. Acta. 2016;1860:2211–2219. doi: 10.1016/j.bbagen.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Wang BK, Chang ML, Wan ZQ, Han GL. Cyclic stretch enhances osteogenic differentiation of human periodontal ligament cells via YAP activation. Biomed. Res. Int. 2018;2018:2174824. doi: 10.1155/2018/2174824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang N, et al. Up-regulated osteogenic transcription factors during early response of human periodontal ligament stem cells to cyclic tensile strain. Arch. Med. Sci. 2012;8:422–430. doi: 10.5114/aoms.2012.28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito M, et al. Gravity loading induces adenosine triphosphate release and phosphorylation of extracellular signal-regulated kinases in human periodontal ligament cells. J. Investig. Clin. Dent. 2014;5:266–274. doi: 10.1111/jicd.12049. [DOI] [PubMed] [Google Scholar]
- 35.Techatharatip O, Nowwarote N, Taebunpakul S, Pavasant P. Biphasic effect of ATP on in vitro mineralization of dental pulp cells. J. Cell Biochem. 2018;119:488–498. doi: 10.1002/jcb.26206. [DOI] [PubMed] [Google Scholar]
- 36.Sindhavajiva PR, Sastravaha P, Arksornnukit M, Pavasant P. Intermittent compressive force induces human mandibular-derived osteoblast differentiation via WNT/beta-catenin signaling. J. Cell Biochem. 2018;119:3474–3485. doi: 10.1002/jcb.26519. [DOI] [PubMed] [Google Scholar]
- 37.Ayala-Pena VB, Scolaro LA, Santillan GE. ATP and UTP stimulate bone morphogenetic protein-2,-4 and -5 gene expression and mineralization by rat primary osteoblasts involving PI3K/AKT pathway. Exp. Cell Res. 2013;319:2028–2036. doi: 10.1016/j.yexcr.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Ku SJ, et al. Static tensional forces increase osteogenic gene expression in three-dimensional periodontal ligament cell culture. BMB Rep. 2009;42:427–432. doi: 10.5483/BMBRep.2009.42.7.427. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Han M, Li S, Wang L, Xu Y. Cyclic tensile stress during physiological occlusal force enhances osteogenic differentiation of human periodontal ligament cells via ERK1/2-Elk1 MAPK pathway. DNA Cell Biol. 2013;32:488–497. doi: 10.1089/dna.2013.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, et al. Analysis of gene expression profile of periodontal ligament cells subjected to cyclic compressive force. DNA Cell Biol. 2011;30:865–873. doi: 10.1089/dna.2010.1139. [DOI] [PubMed] [Google Scholar]
- 41.Fujii S, et al. Effects of TGF-beta1 on the proliferation and differentiation of human periodontal ligament cells and a human periodontal ligament stem/progenitor cell line. Cell Tissue Res. 2010;342:233–242. doi: 10.1007/s00441-010-1037-x. [DOI] [PubMed] [Google Scholar]
- 42.Kawahara T, et al. TGF-Beta negatively regulates the BMP2-dependent early commitment of periodontal ligament cells into hard tissue forming cells. PLoS ONE. 2015;10:e0125590. doi: 10.1371/journal.pone.0125590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chien HH, Lin WL, Cho MI. Expression of TGF-beta isoforms and their receptors during mineralized nodule formation by rat periodontal ligament cells in vitro. J. Periodontal Res. 1999;34:301–309. doi: 10.1111/j.1600-0765.1999.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki E, et al. Akt activation is required for TGF-beta1-induced osteoblast differentiation of MC3T3-E1 pre-osteoblasts. PLoS ONE. 2014;9:e112566. doi: 10.1371/journal.pone.0112566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochiai H, et al. Inhibition of insulin-like growth factor-1 (IGF-1) expression by prolonged transforming growth factor-beta1 (TGF-beta1) administration suppresses osteoblast differentiation. J. Biol. Chem. 2012;287:22654–22661. doi: 10.1074/jbc.M111.279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markopoulou CE, et al. Effect of rhTGF-beta1 combined with bone grafts on human periodontal cell differentiation. Growth Factors. 2011;29:14–20. doi: 10.3109/08977194.2010.533663. [DOI] [PubMed] [Google Scholar]
- 47.Chen YJ, et al. Activation of focal adhesion kinase induces extracellular signal-regulated kinase-mediated osteogenesis in tensile force-subjected periodontal ligament fibroblasts but not in osteoblasts. J. Bone Min. Metab. 2014;32:671–682. doi: 10.1007/s00774-013-0549-3. [DOI] [PubMed] [Google Scholar]
- 48.Kim SJ, Park KH, Park YG, Lee SW, Kang YG. Compressive stress induced the up-regulation of M-CSF, RANKL, TNF-alpha expression and the down-regulation of OPG expression in PDL cells via the integrin-FAK pathway. Arch. Oral. Biol. 2013;58:707–716. doi: 10.1016/j.archoralbio.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Kang YG, Nam JH, Kim KH, Lee KS. FAK pathway regulates PGE(2) production in compressed periodontal ligament cells. J. Dent. Res. 2010;89:1444–1449. doi: 10.1177/0022034510378521. [DOI] [PubMed] [Google Scholar]
- 50.Wan C, et al. The dentin sialoprotein (DSP) domain regulates dental mesenchymal cell differentiation through a novel surface receptor. Sci. Rep. 2016;6:29666. doi: 10.1038/srep29666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghannad F, et al. Absence of alphavbeta6 integrin is linked to initiation and progression of periodontal disease. Am. J. Pathol. 2008;172:1271–1286. doi: 10.2353/ajpath.2008.071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu M, et al. Epigenetic regulation of integrin beta6 transcription induced by TGF-beta1 in human oral squamous cell carcinoma cells. J. Cell Biochem. 2018;119:4193–4204. doi: 10.1002/jcb.26642. [DOI] [PubMed] [Google Scholar]
- 53.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 54.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 55.Sinha KM, Zhou X. Genetic and molecular control of osterix in skeletal formation. J. Cell Biochem. 2013;114:975–984. doi: 10.1002/jcb.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, et al. Essential role of osterix for tooth root but not crown dentin formation. J. Bone Min. Res. 2015;30:742–746. doi: 10.1002/jbmr.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Z, et al. Genetic evidence for the vital function of Osterix in cementogenesis. J. Bone Min. Res. 2012;27:1080–1092. doi: 10.1002/jbmr.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.