Abstract
The access and recycling of the base cations are essential processes for the long-lasting functioning of forest ecosystems. While the role of soil bacterial communities has been demonstrated in mineral weathering and tree nutrition, our understanding of the link between the availability of base cations and the functioning of these communities remains limited. To fill this gap, we developed a microcosm approach to investigate how an increase in key base cations (potassium or magnesium) impacted the taxonomic and functional structures of the bacterial communities. During a 2-month period after fertilization with available potassium or magnesium, soil properties, global functions (metabolic potentials and respiration) as well as mineral weathering bioassays and 16S rRNA amplicon pyrosequencing were monitored. Our analyses showed no or small variations in the taxonomic structure, total densities and global functions between the treatments. In contrast, a decrease in the frequency and effectiveness of mineral weathering bacteria was observed in the fertilized treatments. Notably, quantitative PCR targeting specific genera known for their mineral weathering ability (i.e., Burkholderia and Collimonas) confirmed this decrease. These new results suggest that K and Mg cation availability drives the distribution of the mineral weathering bacterial communities in forest soil.
Subject terms: Microbial ecology, Element cycles
Introduction
Understanding how microorganisms adapt to variations in resource availability is a central question in ecology and evolutionary biology. For plants, different behaviours related to the competition for resources have been described, such as competitors, which adapt to rapidly consume available resources, stress tolerators, which persist in nutrient-poor environments, and ruderals, which adapt to disturbances1. These different competitive strategies as well as the related theories have been applied to the analysis of environmental microbial communities2–4. However, most of the studies dealing with this question have focused on organic nutrients (C, N, S) or iron. The important role of the composition, quality and recalcitrance of organic carbon compounds in the regulation of the composition of microbial communities has been demonstrated in several aquatic and terrestrial environments5–7. Similarly, the form of nitrogen (i.e., ammonium, nitrate, nitrite, amino acids) is known to strongly impact the composition and functioning of microbial communities8,9. As an example, N application is known to modify the abundance of nitrifiers and denitrifiers10 or methanotrophs11. For most of these substrates, the presence and increasing concentrations of compounds directly select microorganisms capable of consuming them or having a better affinity. In contrast, the absence or decreasing concentrations of certain substrates, such as iron, are known to allow the selection of competitive taxa capable of mobilizing those nutrients through the production of siderophores and the regulation of their production11–13. Comparatively, the relative impact of base cations on the soil bacterial communities at both the taxonomic and functional levels has been poorly investigated.
Base cations correspond to inorganic compounds, also termed mineral nutrients or inorganic nutrients, such as potassium (K), magnesium (Mg) or calcium (Ca), which act as nutrients, co-factors, or structural components of the cell for plants and the living biosphere14–16. These inorganic nutrients are particularly important for plant nutrition, and their depletion can cause plant nutrient deficiencies14,15. In both forest and agricultural environments, the concentrations of available K or Mg are low compared to other nutrients (e.g., C, N) but for different reasons17. In agricultural soils, those low concentrations are due to the intensive removal of the crop biomass, but the pool of base cations is usually restored by regular NPK or manure fertilizations. In temperate forest ecosystems, the low concentrations of base cations are due to the nutrient-poor soils in which forests are usually developed, the absence of amendments, and the slow replenishment of the soil fertility18. In addition, this phenomenon is accentuated in harvested forests, where trees and the cations accumulated in their biomass are exported19. In these low-input and nutrient-poor ecosystems, the available base cations are mainly derived from the dissolution of soil minerals and rocks. In this context, many of the soil microorganisms living in such conditions are likely facing nutritive stress, which probably determines their distribution and functioning.
The potential relationships between the nutrient availability and the taxonomic and functional structures of the soil bacterial communities have been reported in several studies considering soil chronosequences20,21, soil gradients22–24, soil profiles25,26, or the effect of inorganic fertilization with Ca or Mg in forest sites27,28. However, in most of these studies, the main soil factors considered were the pH and the availability of C, P or N forms and rarely the availability of base cations by themselves. Among the soil bacterial communities that may be the most impacted by variations in the availability of base cations are those involved in the dissolution of soil minerals (e.g., mineral weathering bacteria). Indeed, mineral weathering bacteria have been found in various nutrient-poor ecosystems such as deserts or forests29,30 and in different microbial habitats such as in the (mycor)-rhizosphere29,31, bulk soil26 or on the surface of minerals20,32–37. The frequency and effectiveness of mineral weathering bacteria were notably shown to vary following a mineral amendment27,28 or according to the physico-chemical properties of the mineral substrates34,37. In both cases, the conditions presenting the highest base cation availability (i.e., amended soil, highly weatherable minerals) were characterized by the lowest frequency of effective mineral weathering bacteria, suggesting a potential link between this functional group and nutrient availability. The ability to weather minerals is a functional trait observed in a wide range of bacterial genera38. It was proposed to be a functional adaptation to nutrient limiting environments39, making effective mineral weathering bacteria potentially more competitive in such conditions or stress tolerators according to the Grime definition1. However, a common feature of all these studies was the co-variation of the nutrient availability and pH, due to the in situ variations of the soil parameters40,41 and/or to the effect of the mineral amendment (i.e., liming28), limiting the conclusions. Indeed, pH is known as one of the main soil parameters determining the distribution of the soil bacterial communities24 and liming is used to rectify the acidity and fertility of the soil42,43.
In this context, our study aimed to understand how the availability of important base cations, such as potassium or magnesium, drives the taxonomic and functional distribution of the soil bacterial communities and especially the mineral weathering bacteria in forest soil. Such questioning is important in forestry, as liming is considered as an effective management practice to recover soil fertility and its use may be intensified to increase wood productivity. Consequently, it is important to know how such amendment will impact soil bacterial communities and their functioning. To do so, we considered a nutrient-poor soil (classified as Hyperdystric Cambisol) from the Montiers forest experimental site (France) characterized by a low concentration of base cations, limiting for the growth of trees for K and Mg17 and a high proportion of effective mineral weathering bacterial communities41. Calcium was not considered in our study, as it was not limiting in this soil17. Notably, the soil chemical analyses collected from this site and from other studies evidenced an enrichment of effective mineral weathering bacteria in soil conditions limited in potassium and magnesium compared to nutrient-rich conditions41. To disentangle the relative effect of the base cation availability from the effect of the pH on the soil bacterial communities, we developed a soil microcosm approach based on a fertilization with K or Mg. These nutrients were added to the soil in aqueous solution presenting the same pH as the pH of the soil considered. This experimental design allowed us to test the hypothesis that under nutrient-poor conditions (i.e., limiting in K and Mg), mineral weathering bacteria are competitive, while in nutrient-rich conditions (i.e., fertilized with K or Mg), they are not. Consequently, an increase in base cation availability is expected to directly impact mineral weathering bacteria and to reduce their frequency and effectiveness. As we did not know if the effect of the K or Mg fertilization on the soil bacterial communities was immediate, analyses were performed at different time points. During a 2-month period following K- or Mg-fertilization, the functional response of microbial communities through basal respiration (MicroResp) and carbon-substrate metabolization (Biolog EcoPlate) measurements was evaluated each 15 day-period. The soil chemical properties were measured to demonstrate the increase of available K or Mg in the soil as well as the stability of the pH. The mineral weathering potential of the bacterial communities was studied through a culture-dependent method. In addition, the taxonomic structure of bacterial communities was analysed by 16S rRNA amplicon-pyrosequencing. The same soil samples were also used for quantitative PCR analyses to quantify the total bacterial abundance as well as specific bacterial genera such as Burkholderia, Collimonas and Pseudomonas.
Results
Soil chemical analyses
Soil chemical analyses confirmed that the chemical parameters of the soil were modified according to the treatments applied (i.e., water only (Ct), water + Mg (Mg) or water + K (K) input) at each sampling time (Table 1). When considered overall, the K-fertilized treatment exhibited a significantly higher exchangeable K content (K: 0.23 ± 0.02 cmol+ kg−1) than the control and the Mg-fertilized treatments (control (Ct) and treatment fertilized with Mg: 0.12 ± 0.004 cmol+ kg−1). Similarly, the Mg-fertilized treatment exhibited a significantly higher exchangeable Mg content (Mg: 0.32 ± 0.05 cmol+ kg−1) compared to the control and the K-fertilized treatments (control (Ct): 0.07 ± 0.008 cmol+ kg−1 and treatment fertilized with K: 0.05 ± 0.007 cmol+ kg−1). Notably, the cationic exchange capacity (CEC) was not significantly modified by the fertilization treatments (P > 0.05). The other parameters remained unchanged compared to those in the control treatment, except for the exchangeable Al3+ or slight modifications at the T3 (i.e., 45 days) sampling time (Na, Mn). No significant time effect was observed for the other soil chemical characteristics (P > 0.05).
Table 1.
Chemical characteristics of the soil samples collected among the three treatments (control, K and Mg fertilized) and the three sampling times (T0, T1 and T3).
| Time | Treatment | K | Ca | Fe | Mg | Mn | Na | Al3+ | H+ | CEC | pH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cmol+ kg−1 of soil | |||||||||||
| T0 | Ct | 0.12 | 0.29 | 0.0029 | 0.077 | 0.21 | 0.067 | 2.25 | 0.14 | 3.17 | 4.65 |
| T1 | Ct | 0.12a | 0.29 | 0.0030 | 0.077a | 0.20a | 0.040 | 2.29a | 0.13 | 3.15 | 4.66 |
| +K | 0.28b | 0.16 | 0.0023 | 0.047a | 0.23b | 0.038 | 2.19b | 0.13 | 3.07 | 4.60 | |
| +Mg | 0.12a | 0.21 | 0.0024 | 0.39b | 0.23b | 0.025 | 2.11c | 0.12 | 3.20 | 4.56 | |
| Stat | <0.0001 | 0.23 | 0.29 | <0.0001 | 0.023 | 0.89 | 0.0004 | 0.4 | 0.6 | 0.06 | |
| T3 | Ct | 0.12a | 0.26 | 0.0024 | 0.07a | 0.21a | 0.003 | 2.29a | 0.12 | 3.06 | 4.64 |
| +K | 0.27b | 0.17 | 0.0024 | 0.048a | 0.23ab | 0.043 | 2.20b | 0.11 | 3.06 | 4.6 | |
| +Mg | 0.12a | 0.26 | 0.0021 | 0.45b | 0.25b | 0.043 | 1.98c | 0.10 | 3.29 | 4.59 | |
| Stat | <0.0001 | 0.49 | 0.60 | <0.0001 | 0.009 | 0.22 | <0.0001 | 0.06 | 0.13 | 0.11 | |
The quantities of exchangeable elements and the CEC are expressed in cmol of charge per kg of soil. CEC: cation exchange capacity. The data presented are mean values of 3 replicates. Different letters (a, b, c) indicated significant differences between treatments calculated for the sampling times T1 and T3 separately, according to a one-factor ANOVA analysis followed by a Tukey test (P < 0.05). Significantly higher values were indicated in bold. For legibility reasons, the standard error values are not presented.
Carbon substrate metabolization ability
When all the time points were combined and regardless the treatment, overall higher metabolization potentials were observed at T1 (i.e., 15 days after the fertilization) for carboxylic acids (T1 > T2 = T3 = T4; P = 0.003) and on the contrary lower metabolization potentials at T1 for polymers and miscellaneous substrates (T1 < T2 = T3 = T4; Ppolymers = and Pmiscellaneous = 0.0004). When the treatments were considered, a significant effect was observed for the Mg fertilization compared to the control for the polymers (P = 0.003; Mg ≥ K ≥ Ct) and the carboxylic acids (P = 0.0002; Ct ≥ K ≥ Mg).
When each time point (i.e. T1 to T4) was considered independently, our analyses revealed a significant effect of Mg fertilization on the absorbance measures (P < 0.05; Table S1). For the carboxylic acids, a significant decrease was observed in the Mg and K fertilized treatments compared to the control treatment at T2 (and a trend at T3 and T4; Table S1). For polymers, a significant increase was observed in the Mg fertilized treatment compared to the control treatment at T2, T3 (and a trend at T4; Table S1).
Respiration profiles
The MicroResp analyses revealed a decreasing respiration rate in the control treatment between T0 and the other sampling times (Fig. S1; RespT0: 2.5 µg CO2/gr of soil (Dw)/hr; RespT1: 1.77; RespT2: 1.74; RespT3: 1.64; RespT4:1.64; P < 0.0001). The same MicrResp analyses also showed both treatment and time effects according to a 2-factor (time, treatment) ANOVA (P = 0.02). When analysed globally, the addition of K and Mg induced a decrease in microbial respiration compared to that of the control treatment (RespK: 1.51 ± 0.03; RespMg: 1.40 ± 0.01 vs RespConrtol: 1.70 ± 0.03, P < 0.05), but this effect decreased with time. At T1 (i.e., 15 days), the addition of Mg caused a significant decrease compared to the control treatment (RespK: 1.70; RespMg: 1.43; RespControl: 1.77; P = 0.005), but the addition of K did not. At T2 (30 days), both K and Mg treatments allowed a significant decrease compared to the control treatment (RespK: 1.35; RespMg: 1.34; RespControl: 1.74; P < 0.0005). No significant effect of K and Mg addition was observed at T3 (i.e., 45 days) and T4 (i.e., 60 days) sampling times compared to the control, although a decreasing respiration rate was observed.
Bacterial densities
Dilutions and plate counting revealed relatively similar culturable bacterial densities between treatments and sampling times, ranging from 1.15 × 106 to 3.47 × 106 (expressed as CFU /gram of soil; Table 2).
Table 2.
Bacterial quantification and ratio analyses.
| Treatment | Culture | qPCR | (Ratio) | |||||
|---|---|---|---|---|---|---|---|---|
| Total bacteria | Total bacteria | Burkholderia | Collimonas | Pseudomonas | T/B | T/C | T/P | |
| T1-Ct | 6.50a | 7.94a | 6.06a | 6.03d | 6.03a | 76.63b | 86.80a | 81.93a |
| T1 + K | 6.38a | 7.77a | 5.49b | 5.47ab | 5.56a | 191.67a | 204.67ab | 234.97a |
| T1 + Mg | 6.11a | 7.68a | 5.52b | 5.31abc | 5.38b | 150.27ab | 249.67ab | 207.33a |
| T3-Ct | 6.42a | 7.59a | 5.74b | 5.71ad | 5.44b | 80.10b | 79.83a | 150.67a |
| T3 + K | 6.06a | 7.75a | 5.58b | 5.07bc | 5.40b | 150.00ab | 494.33ab | 224.00a |
| T3 + Mg | 6.11a | 7.74a | 5.51b | 4.93c | 5.52b | 170.67a | 717.33b | 173.00a |
Quantification done by dilution/plating or quantitative PCR are expressed as Log[cfu g−1 of soil] and Log[16S rRNA gene copies g−1 of soil], respectively. The data presented are mean values of 3 replicates. Ratio analyses (i.e., T/B: ratio Total bacteria/Burkholderia; T/P: ratio Total bacteria/Pseudomonas; T/C: ratio Total bacteria/Collimonas) have been done on the non-log transformed data. Treatments presenting different letters (a,b,c,d) are significantly different according to a one-factor ANOVA and Tukey post-hoc test (P < 0.05). Treatments are presented as follows: Ct, control; +Mg, Mg fertilized and +k, K fertilized. For legibility reasons, the standard error values are not presented.
Determination of the total bacterial densities by qPCR did not reveal any treatment or time effects, with densities ranging from 3.89 × 107 to 8.71 × 107 (expressed as 16S rRNA gene copies/gram of soil]. Quantification of Burkholderia revealed a decrease between the control and fertilized (K and Mg) treatments at T1 and T3; however, this was only significant at the T1 sampling time (Ct-T1: 1.15 × 106 vs 3.09 × 106 and 3.31 × 106 for K and Mg, respectively; P < 0.0005). Quantification of Collimonas revealed a decrease between the control and fertilized (K and Mg) treatments at T1 and T3; however, this was only significant at the T1 sampling time (Ct-T1: 1.07 × 106 vs 2.95 × 105 and 2.04 × 105 for K and Mg, respectively; P < 0.0001). Quantification of Pseudomonas revealed a decrease between the control and Mg-fertilized treatment only at the T1 sampling time (Ct-T1: 1.07 × 106 vs 2.4 × 105 for Mg; P < 0.05). The relative decrease of the abundance of Burkholderia and Collimonas according to the K and Mg fertilizations was confirmed by a global analysis of the ratios (total bacteria/Burkholderia, P = 0.003; total bacteria/Collimonas, P = 0.008), but no significant effect was observed for Pseudomonas (Table 2).
Bacterial diversity and community profiles
Analyses of the total 16S rRNA sequence dataset revealed globally high coverage according to the coverage index, ranging from 84 ± 0.01% in the control treatment at T1 (Ct_T1) to 89 ± 0% in the control and K-fertilized treatments at T2 and T4 (Table S2). Analysis of the α diversity revealed no difference between the treatments and sampling times for the Shannon indices (average: 3.82 ± 0.07) and the number of observed OTUs (average Sobs: 110 ± 3). A sampling time effect was observed for the Chao and Simpson indices with a higher diversity in the T1 (Simpson: 36.80 ± 3.72; Chao: 226.40 ± 13.06) than in the T3 (Simpson: 16.66 ± 1.91; Chao: 176.66 ± 9.30) sampling time (P = 0.0009 and P = 0.0144, respectively). Concerning the community structure, no treatment and sampling time effect was observed according to PermANOVA analyses performed on the OTU table (P > 0.05).
Taxonomic composition of bacterial communities
A total of 13 phyla (55% of the sequences), 25 classes (53%), 41 orders (52%), 49 families (46%) and 53 genera (14%) were identified in our 16S rRNA sequence dataset. The first three phyla of the dataset represented more than 50% of the total sequences and were affiliated with Acidobacteria (30%), Proteobacteria (17%; Alphaproteobacteria: 9%, Gammaproteobacteria: 4%, Betaproteobacteria: 3% and Deltaproteobacteria: 1%; Fig. S2) and Actinobacteria (3.5%). The other phyla, Saccharibacteria, Chlamydiae, Bacteroidetes, Gemmatimonadetes, Verrucomicrobia, Nitrospirae, Elusimicrobia, Firmicutes and Armatimonadetes, represented less than 1% of the total dataset each. The top 10 most represented genera in the 16S rRNA sequences were Burkholderia (1.4%), Acidothermus (1.3%), Collimonas (0.3%), Rhizomicrobium (0.3%), Variibacter (0.2%), Streptacidiphilus (0.1%), Cupriavidus (0.1%), Dokdonella (0.1%), Nitrospira (0.08%) and Roseiarcus (0.07%).
When each sampling time was considered independently, significant differences were observed between the different treatments (Table S3). Betaproteobacteria were significantly more abundant in the control treatment at T2 (i.e., 30 days) than in the K-fertilized treatment. This difference was partly explained by a higher proportion of sequences assigned to Burkholderiales in the control treatment at T1 and T2 compared to the K-fertilized treatment. However, this effect was not observed at the genus level. At the OTU level, significant differences were observed among treatments with i) the OTU0001 enriched in K-fertilized treatment compared to the control treatment (unclassified bacteria) and ii) the OTU0002 enriched in the control treatment compared to the K-fertilized treatment (unclassified bacteria).
Mineral weathering potential of bacterial strains
The functional screenings of the bacterial strains isolated from the different treatments highlighted a global negative impact of fertilization (Mg or K) on the relative distribution and effectiveness of the mineral weathering (MW) bacteria (Figs 1 and 2; P < 0.002).
Figure 1.
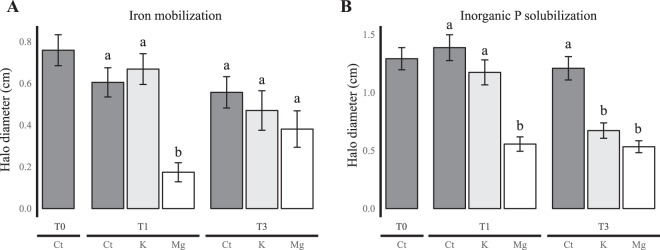
Relative mineral weathering effectiveness of the bacteria according to the treatment (control, K fertilized and Mg fertilized) and the sampling time (T0, T1 and T3). (A) Relative effectiveness at mobilizing iron and (B) Relative effectiveness at solubilizing inorganic phosphorus. For each bar, the data presented are mean +/− Standard deviation. The error bars indicate standard deviations. Lowercase letters (a or b) indicate significant differences between treatments in one sampling time. Statistics were obtained according to a one- factor ANOVA followed by a Tukey test (P < 0.05). A total of 412 and 357 bacterial strains have been tested on the TCP and CAS media, respectively. Each strain was tested with 3 replicates.
Figure 2.
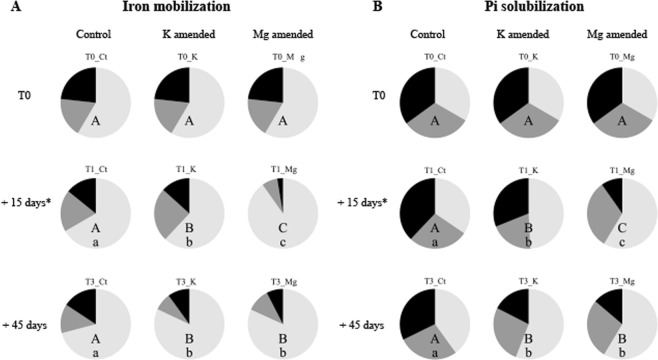
Distribution in classes of mineral weathering effectiveness of the bacteria depending on the treatment (control, K fertilized and Mg fertilized) and on the sampling time (T0, T1 and T3). (A) Iron mobilizing ability and (B) Inorganic phosphorus solubilizing ability. For each bioassay, determination of the class of effectiveness was done depending on the median (M) of the discoloration zones of the positive strains (black: strong activity, discoloration zone > M; grey: moderate activity, discoloration zone < M; white: no degradation activity). Capital letters (A,B or C) indicate significant differences between the sampling times, within one treatment. Lowercase letters (a,b of c) indicate significant differences between the treatments, within one sampling time. Statistics were obtained according to a Chi2 test (P < 0.05). A total of 412 and 357 bacterial strains have been tested on the TCP and CAS media, respectively. Each strain was tested with 3 replicates.
Notably, the Mg-fertilization treatment had the strongest effect on the ability to mobilize iron (CAS; Fig. 1A) and to solubilize inorganic phosphorus (TCP; Fig. 1B). Indeed, in this treatment, the average MW effectiveness of the bacteria passed from T0 (T0_TCP = 1.29 ± 0.10 and T0_CAS = 0.76 ± 0.07 cm of haloes) to a significantly lower average MW effectiveness at T1 (T1_TCP = 0.56 ± 0.06 and T1_CAS = 0.17 ± 0.05 cm of haloes) (P < 0.0001). The same trend was observed at T3 but was only significant for the ability to solubilize inorganic phosphorus. A significant effect of Mg fertilization was also observed on the relative distribution of the effective MW bacteria. This frequency significantly decreased from T0 (T0_TCP: T0 = 35% and T0_CAS = 23%) to T1 (TCP_T1+Mg = 10% and CAS_T1+Mg = 3%; both P < 0.0001, Fig. 2). The same trend was observed at T3.
For the K-fertilized treatment, a decrease in MW effectiveness was also observed, but later than the effect observed in the Mg-fertilized treatment (Fig. 1). Indeed, a significant reduction of the MW effectiveness occurred only between T1 and T3 [(T1_CAS = 0.67 ± 0.07 cm and T3_CAS = 0.47 ± 0.09 cm (Fig. 1A); P = 0.037; T1_TCP = 1.17 ± 0.11 cm and T3_TCP = 0.67 ± 0.07 cm of haloes (Fig. 1B); P < 0.0001)]. A significant effect of K fertilization was also observed on the relative distribution of the effective MW bacteria. This effect was observed between each sampling time for the ability to solubilize inorganic phosphorous (T0_TCP = 35%, T1_TCP = 31% and T3_TCP = 18%) and to mobilize iron (T0_CAS = 23%, T1_CAS = 13% and T3_CAS = 10%) (P < 0.0001; Fig. 2). Notably, such a time effect was not observed on the MW effectiveness and the frequency of mineral weathering bacteria in the control treatment (P > 0.05).
Taxonomic affiliation of the bacterial strains and link with function
The taxonomic identification of the bacterial strains by the partial sequencing of the 16S rRNA gene showed that they belonged to the Proteobacteria [45%; Betaproteobacteria (28%), Gammaproteobacteria (5%) and Alphaproteobacteria (1%)], Firmicutes (34%) and Actinobacteria (21%)] phyla. The dominant genera identified were Bacillus (25%), Collimonas (18%) and Arthrobacter (18%), Paenibacillus (16%) and Burkholderia (8%).
Regarding the distribution of these genera according to the treatments applied in our study, our analyses revealed a significant effect of both fertilization treatment and sampling time on the taxonomic structure of the culturable representatives. Notably, the control treatment was dominated by bacteria belonging to Proteobacteria regardless of the sampling time considered and particularly by the Collimonas genus (T0: 28% of the collection). In contrast, the proportions of Proteobacteria and Collimonas strongly decreased with time in both fertilized treatments (after 45 days (T3): Collimonas = 3%; Chi2: P < 0.0001). In parallel, the proportion of bacterial strains affiliated with Firmicutes, particularly to the Bacillus genus, increased in the fertilized treatments at T3 (37%; P = 0.004), while it remained lower in the control treatment regardless of the sampling time (15%).
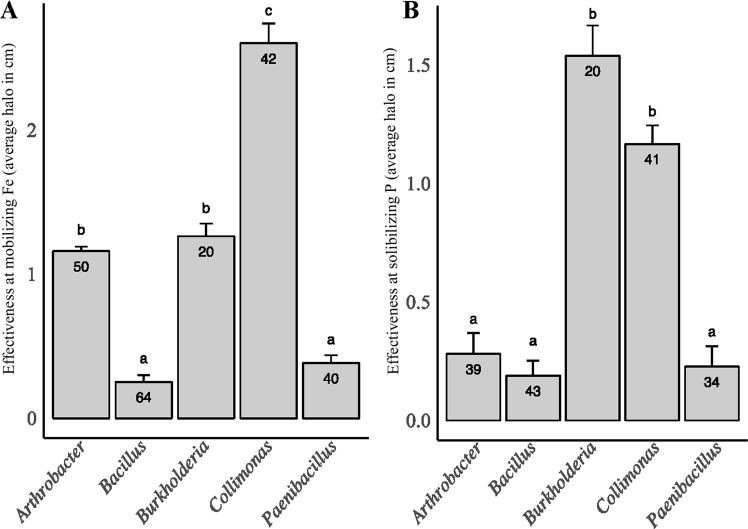
Analysis of the potential relationship between taxonomy and functional ability revealed that the members of the genera Burkholderia and Collimonas exhibited the strongest MW effectiveness regardless of the bioassay considered (i.e., mobilization of iron and solubilization of inorganic phosphorus) compared to the other genera (Fig. 3). The members of the genus Arthrobacter were relatively effective at mobilizing iron. Notably, ca. 80% of the representatives of these two genera belonged to the bacterial groups that were very effective at mobilizing iron. For the ability to solubilize inorganic phosphorous, 80% of the Collimonas strains belonged to the most effective class, while Burkholderia (80%), Arthrobacter (96%) and the remaining Collimonas strains exhibited intermediate effectiveness. In contrast, members of the Bacillus and Paenibacillus genera exhibited very low mineral weathering effectiveness in both bioassays. Notably, our analyses also revealed no significant difference in effectiveness within a genus between treatments or sampling times.
Figure 3.
Relationship between mineral weathering effectiveness and taxonomic affiliation of the bacteria. For each bacterial genus represented by a minimum of 10 isolates, the relative effectiveness to: (A) mobilize iron and (B) solubilize inorganic phosphorus is presented, whatever the origin of the bacterial isolates. For each bar, the data presented are mean +/− Standard deviation. The error bars indicate standard deviations. Lowercase letters (a–c or d) indicate significant differences between genus effectiveness. Statistics were obtained according to a one-way ANOVA followed by a Tukey test (P < 0.05). The number of bacterial isolates tested per genus is presented into the bars.
Comparison of the overlapping portion of the 16S gene sequences between the 454-pyrosequences and Sanger sequences of the bacterial strains revealed a relatively good overlap between the two datasets (Table S4). A total of 87% of the bacterial strains from the collection exhibited more than 97% sequence similarity with the pyrosequences, while only 3.5% of the pyrosequences exhibited high similarity with the Sanger sequences from the bacterial collection. Notably, many Sanger sequences of our bacterial collection presented more than 97% sequence similarity with highly represented OTUs (i.e., OTU14, Burkholderia; OTU19, Rhodanobacter; OTU55, Collimonas) or poorly represented OTUs (OTU776 and OTU829, Paenibacillus; OTU866, Rhizobium).
Discussion
Using a combination of culture-dependent and -independent methods associated with a microcosm approach, we tested the hypothesis that an increase in the concentration of a single base cation in a soil naturally depleted in this element can modify the taxonomic and functional structures of the soil bacterial communities. The resource manipulation we applied in our study to nutrient-poor soil was determined to mimic the nutrient-rich conditions occurring in the nutrient-rich soil of the Montiers toposequence41. In this sense, our fertilization reached the quantity of available K (i.e., 0.28 cmol+ kg−1 of soil) and half of the quantity of available Mg (i.e., 0.81 cmol+ kg−1 of soil) measured in the nutrient-rich soil of the Montiers toposequence41. Consequently, the fertilization applied was low and corresponded to an increase of 2 and 5 times the available K and Mg in the manipulated nutrient-poor soil. This approach allowed us to highlight how the diversity, composition and function of the bacterial communities were impacted by an increase in the availability of base cations. To our knowledge, this study represents the first experimental demonstration of the effect of the availability of base cations and of its short-term effect on the taxonomic and functional structure of soil bacterial communities. Notably, our culture-dependent approach coupled with qPCR highlighted that the modification of the functional structure observed, which corresponded to a decrease of the frequency of effective mineral weathering bacteria, was related to a decrease in specific taxa.
Compared to other edaphic parameters, such as pH or variations of the concentration of organic compounds, few studies have demonstrated a potential impact of the availability of base cations, such as Mg, K or Ca, on the soil bacterial communities40,44,45. The role of these nutrients has mainly been investigated in the context of fertilization (i.e., NPK or dolomite amendments45,46) or as potential toxic elements47. Consequently, little is known about their relative effects on soil microorganisms. This discrepancy is partly because most of the environmental studies mainly focused on the C, N or P cycles, as many soil parameters are linked, co-vary and have low concentrations of the base cations in comparison with the amounts of C and N. Indeed, base cations such as Mg, K or Ca usually present concentrations 100 to 1000 times lower than C. However, these cations are essential nutritive elements whose depletion strongly affects plant growth, such as in forest ecosystems developed on nutrient-poor soils16,48–50. These cations originated from the recycling of the cations contained in dead organic matter or from the weathering of soil minerals. In the soil considered in our study, which was sampled from the organic-mineral horizon, the base cations were mainly derived from the weathering of soil minerals17. In both organic matter decomposition and mineral weathering processes, microorganisms have been identified as key actors51. Consequently, an increase or depletion of the concentrations of base cations should directly impact their distribution and function.
The functional analyses performed during the 2-month incubation period in our study showed the impact of resource manipulation on the functional structure of the soil microbial communities. Notably, global respiration and catabolic activities appeared to be affected, but only at a low level. Indeed, both activities appeared slightly decreased in the fertilized treatments with a higher effect of Mg, suggesting a functional switch or a potential toxic effect of the cations added to the microbial communities involved in OM decomposition47. However, no effect was observable on the catabolic diversity (i.e., Biolog, Shannon index values) nor on the total bacterial quantifications performed both by dilution/plating and qPCR. These findings indicate that fertilization with K or Mg has a negligible effect on the global development of the soil bacterial communities compared to those in the control treatment, excluding the toxicity hypothesis. The few differences observed between the treatments may be related to the two functions measured (i.e., respiration and catabolic potentials). In contrast, the mineral weathering ability of the soil bacterial communities measured using bioassays highlighted a strong decrease of this functional group in the amended treatments, with a higher effect of Mg than K compared to the control treatment. Together, these results suggest an adaptation of the soil microbial communities to an increase in nutritive resources. While the mineral weathering ability seems to be advantageous for the bacteria with this functional trait in nutrient-poor conditions, it is not advantageous in nutrient-rich conditions, and this group is outcompeted.
The global analysis performed on the structure of the soil bacterial communities based on 16S rRNA amplicon pyrosequencing data did not reveal significant variations between the different treatments. Noticeably, the community composition and structure obtained in our microcosm-based study (i.e., dominance of Acidobacteria and Proteobacteria and high proportion of unclassified bacteria) appeared very similar to those obtained from the same soil, but from a field experiment in a previous study40. A BLASTN analysis revealed that most of the OTUs generated in our study, including the unclassified_bacteria, presented a high sequence homology (99 to 100% identity) with OTU sequences generated by Colin et al.40 (SRA based comparison). The absence of significant variations according to the treatments applied in our study suggests a low effect of the cation input on the taxonomic structure and composition of the soil microbiome. However, a detailed analysis revealed an increase in the relative abundance of pyrosequences assigned to Actinobacteria (corresponding mainly to Arthrobacter) and a decrease in the relative abundance of pyrosequences assigned to Betaproteobacteria (Burkholderiales) in the fertilized treatments compared to the control. Notably, the relative abundance of 16S rRNA pyrosequences assigned to Burkholderia (de novo Paraburkholderia and Caballeronia) and Collimonas, two genera known for their mineral weathering effectiveness27,52,53 decreased in the fertilized treatments (+K, +Mg) compared to that in the control. Effective mineral weathering bacteria affiliated to other genera, such as Arthrobacter, have been previously described20,26 but presented a low mineral weathering potential in our study. The decrease of Burkholderiales was confirmed by the genus-specific qPCR analyses, which showed a significant decrease of Burkholderia and Collimonas in the fertilized samples only. This effect was not observed for the qPCR analyses targeting Pseudomonas. Together, our new results highlight that K or Mg fertilization minimally affects the total density and structure of the soil bacterial communities, while specific bacterial genera such as Arthrobacter, Burkholderia and Collimonas are strongly affected.
Representatives of the Burkholderiales order are known to represent an important proportion of the taxa assigned from culture-dependent and culture-independent analyses in acidic and nutrient-poor forest soils. Burkholderia and Collimonas strains have been isolated, especially in the (mycor-)rhizosphere54 and in the mineralosphere34,37. Notably, in the mineralosphere, these taxa appeared significantly enriched on the surfaces of the less-weatherable minerals34, suggesting a link between these taxa and the mineral chemistry and/or the nutrient availability. Interestingly, Kelly et al.55, analysing the active part of the microbiome, also revealed a variation of the proportion of Burkholderiales in the presence of minerals and according to the physico-chemical properties of the mineral. In our study, the relative abundance of representatives of the Burkholderia and Collimonas genera decreased in the fertilized treatments compared to those in the control, suggesting that these genera are non-adapted or less competitive to an increase in nutrient availability. Culturable representatives of these genera are heterotrophs and have been proposed to be classified as copiotrophs56,57. Copiotrophs are organisms adapted to live in nutrient-rich environments, particularly rich in carbon. Their physiological requirements are usually superior to oligotrophs organisms which on the contrary live in nutrient-poor environments and require limited resources to grow56,57. However, our findings highlight that some of these Burkholderiales-related taxa (i.e., Burkholderia and Collimonas) have oligotrophic behaviour based on their ability to inhabit nutrient-poor environments and their effectiveness in mineral weathering37,53,58,59. Moreover, Lepleux et al.37 demonstrated that phylogenetically related Burkholderia strains isolated from the same soil but from different habitats (bulk soil vs mineralosphere) harboured different metabolic potentials and mineral weathering potentials. Indeed, the Burkholderia strains isolated from the mineral surfaces consumed few carbon substrates with a low effectiveness compared to those of the bulk soil, while they belonged to the same species. Representatives of the Collimonas genus have been mainly reported in nutrient-poor environments (soil, dune, tundra)53,58,60. Consequently, those results fit very well with Ho et al.2, who proposed that different functional strategies may exist in the same bacterial order, family or even genus.
Conclusion
Through the resource manipulation performed, this work demonstrated for the first time that an increase of base cation (i.e., K and Mg) concentration in the soil more strongly affects the functional structure of the soil bacterial communities than the taxonomic structure. This difference between the total taxonomic structure and the functional structure (i.e., mineral weathering ability) of the bacterial communities may be explained by the fact that an important proportion of the soil microbiome is driven by the availability of organic nutrients (i.e., C and N), while a smaller proportion is driven by the availability of inorganic nutrients (i.e., Ca, K and Mg). An important point in our study is that the soil pH was not modified by the resource manipulation, making a real analysis of the effect of cation availability on the soil microbiome possible. Notably, both the culture-dependent and qPCR analyses revealed no effect of fertilization on the total densities of the bacterial communities during our experiment but a significant decrease in the densities of the Burkholderia and Collimonas populations. Interestingly, the Burkholderia and Collimonas strains isolated in our study harboured a greater effectiveness to weather minerals, confirming previous findings obtained for these two taxa. Together, our results show that under nutrient-poor conditions (i.e., non-fertilized soil), these two taxa were dominant, while they were less competitive in nutrient-rich (+K or +Mg) conditions. This finding fits very well with the definition by Grime, which considers organisms capable of developing in limiting conditions as stress tolerators. Based on this definition, our results and previous findings obtained for Burkholderia and Collimonas strains isolated in different geographical locations in forest soils, we proposed to consider them as oligotrophic and effective mineral weathering taxa. These results also bring new perspectives to soil microbial ecology and land use, as the effect of fertilization with base cations on the functional diversity has been rarely considered.
Materials and Methods
Site description and soil sampling
In January 2017, soil samples were collected from the Montiers forest experimental site located in the Meuse department (northeastern France). This long-term observatory (LTO) is part of AnaEE-France (Analyses et Expérimentations sur les Ecosystèmes - France). It is co-managed by the ANDRA [French national radioactive waste management agency; Permanent Environment Observatory (OPE)] and the INRA (UMR1138 Unit; http://www.nancy.inra.fr/en/Outils-et-Ressources/montiers-ecosystem-research). The site is dominated by a homogeneous land cover of European beech trees of the even-aged stand (Fagus sylvatica L.; 50 years old in 2010) and is characterized by a Jurassic limestone (Tithonian) layer overlaid by acid detrital sediments from the lower Cretaceous (Valanginian). The site is characterized by a soil toposequence covering different soil types: from high-nutritive Cambisol Calcaric to nutrient-poor Cambisol Hyperdystric, with an intermediate Eutric Cambisol17,41,44,61. In this study, sampling was performed in Cambisol Hyperdystric soil, which is the most nutrient-poor soil type of the toposequence. The soil was sampled from the same plot after removing leaf litter. The 0–5 cm soil layer was separated, and only the 5–20-cm soil horizon was transported back to the lab. This soil horizon corresponds to the organo-mineral horizon of the soil profile and to the horizon where most of the fine roots of the trees are located. Soil samples were collected using sterile tools to avoid contamination and were returned to the laboratory immediately. All the soil samples were then mixed to homogenize and sieved by a 4 mm mesh. Then, they were stored at 4 °C overnight before the beginning of the experiment.
Microcosm experimental design
A microcosm experiment was conducted to test the relative impact of an increase in nutrient availability on the functional and taxonomic structures of the soil bacterial communities. To do so, we considered two important base cations (magnesium and potassium). These nutrients were selected as previous experiments performed on the Montiers site noted the potential role of magnesium and potassium as limiting base cations for the growth of trees in the Cambisol Hyperdystric soil compared to the Cambisol Calcaric soil17 and because these two nutrients are important cations for tree growth in nutrient-poor and acidic forests41. Moreover, according to the fertility norms established for forest soils62, the fertility in K is low in Cambisol Hyperdystric soil, and the fertility in Mg is moderate and at the limit of the norm17. The calcium was not considered in our study, as it is not limiting at the site considered as it is developed on limestone rich in calcium. The microcosms were prepared the day following the field sampling so that soil was fresh. The soil was separated into three soil subsamples to allow the application of the different treatments in the soil: (i) control (water), (ii) water + magnesium, and (iii) water + potassium. The concentrations of magnesium and potassium used to fertilize the Cambisol Hyperdystric soil were chosen to reach the same concentrations of available Mg and K in the microcosms as the concentrations of these elements measured in the most nutrient-rich soil type of the toposequence of Montiers (i.e., the Calcaric Cambisol; ref.41 [Table 1]). The different treatments considered in our microcosm experiments were (i) K fertilization (KCl, 158.7 mg kg−1 dw (dry weight) soil), (ii) Mg fertilization (MgCl2, 701.6 mg kg−1 dw soil), and (iii) distilled water (control). Distilled water was used to prepare the different solutions. Each solution was adjusted to the same pH (4.6), which was the pH of the Cambisol Hyperdystric soil sampled, to avoid any effects due to the pH of the solutions added to the microcosms. Treatments were applied in one pulse at the beginning of the experiment. The moisture of the microcosms was adjusted according to the field soil moisture (35% of the water holding capacity (WHC)), and the soil water content was adjusted throughout the experiment with sterile distilled water. After the addition of the control (water), K and Mg solutions, each of the soil subsamples was mixed and divided into 20 microcosms for the control treatment (including 4 T0 microcosms), 16 microcosms for the K treatment and 16 microcosms for the Mg treatment, giving a total of 52 independent microcosms. The microcosms used consisted of small pots (40 mL, sterile) filled with 27 g of Cambisol Hyperdystric soil (fresh weight) fertilized with the different solutions described above. The microcosms were incubated in a dark experimental chamber at 21 °C for 60 days.
Sample collection
The experiments were run over 60 days, and soil samples were collected at different time points: i) just after the application of the treatment (T0), ii) after 15 days (T1), iii) after 30 days (T2), iv) after 45 (T3) and v) after 60 days (T4). At each sampling time, four microcosms from each treatment were selected to perform the different analyses described below. For each microcosm, the soil collected (27 g) was homogenized before any analysis. A total of 1.5 g of the soil was used to determine the global respiration using the MicroResp system. A total of 5 g of the soil was suspended in 25 mL of sterile distilled water and vortexed twice for 1 min. This soil suspension was diluted at 1/20 to determine the global metabolic potential on Biolog EcoPlates. In parallel, a volume of 0.5 mL of the undiluted soil suspension was used to perform culture-dependent bacterial analyses. A total of 1 g of the soil was conserved at -20 °C for DNA-based analyses. Finally, 5 g of the soil was dried at 35 °C and sieved at 2 mm for chemical analyses.
Soil chemical analyses
The cation exchange capacity (CEC) was determined using the cobaltihexamine method63, which is based on the titration of the cobaltihexamine chloride soil extract at 472 nm compared to a reference 0.05 N cobaltihexamine chloride extract. The pH was determined by the water method using a soil/water ratio of 1:5 (w/v). Exchangeable cations (Ca, Mg, Na, K, Fe, Mn and Al) and H+ were extracted using cobaltihexamine and determined by inductively coupled plasma spectrometry-atomic emission spectrometry (ICP-AES) for cations and by potentiometric measurement using 0.05 M KOH for protons and aluminium64. The chemical analyses were performed on the four replicates of each treatment (potassium, magnesium or water) at the T0, T1 and T3 sampling times.
Substrate metabolization assay
The ability of microbial communities to metabolize various carbon substrates was assessed at each sampling time and for each treatment through the EcoPlates (Biolog®; Hayward, CA95545 U.S.A), which contain 31 different carbon substrates (pyruvic acid methyl ester;tween 40; tween 80; alpha-cyclodextrin; glycogen; D-cellubiose; alpha-D-Lactose; beta-methyl-D-glucoside; D-xylose; i-Erythritol; D-Mannitol; N-Acetyl-D-Glucosamine; D-Glucosaminidic acid; Glucose 1 phosphate; Glycerol phosphate; D-galactonique acid gamma-lactone; D-galacturonic acid; 2-Hydroxy-benzoic acid; 4-Hydroxy-benzoic acid; Hydroxybutiric acid; Itaconic acid; ketobutyric acid; malic acid; arginine; asparagine; phenylalanine; serine; threonine; glutamic acid; phenylethylamine; putrescine). A volume of 150 µL of the 1/20 diluted soil suspension described above was loaded in each well of a Biolog microplate and incubated at 25 °C for 48 h. For the soil considered 48 h of incubation was enough, saturation was obtained after this period. Colour development was measured at 595 nm with an automatic microplate reader (Bio-Rad, model iMark). The absorbance data were corrected by subtracting the blank, and negative absorbance values were set to zero. The data were expressed as average well color development (AWCD)65. The measures were performed on three replicates biological.
Respiration profiles
The basal soil microbial respiration was measured by the MicroResp method66 at each sampling time and for each treatment. The measurements were performed on the four replicates. For each replicate, 4 deep-wells of the MicroResp microplate were filled with 0.3 g of soil. Simultaneously, a CO2-detection microplate was filled with a pH indicator gel (3% agar, 2.5 mM NaHCO3, 150 mM KCl, and 12.5 µg mL−1 of cresol red). The deep-well microplate was placed face-to-face with the CO2-detection microplate, and both were firmly sealed together with a silicone rubber gasket following the manufacturer’s instructions64. After 6 h of incubation in the dark at 25 °C, the pH change was visible through a colour shift of the indicator gel and was measured at 570 nm with an automatic microplate reader (BioRad, model iMark). The rate of CO2 (µg CO2/gr of dw soil/hr) produced was calculated according to Campbell et al.66.
Bacterial collection
For each treatment, bacterial collections were performed considering three replicates. For each replicate, 5 grams of soil was sampled and suspended in 25 mL of sterile distilled water. A volume of 0.2 mL of the pure soil suspension was serially diluted in sterile distilled water and spread onto a 1/10 diluted tryptic soy agar (TSA) medium (tryptic soy broth from Difco, 3 g L−1 and agar, 15 g L−1) supplemented with 100 µg L−1 cycloheximide (Sigma; final concentration). The Petri dishes were incubated at 25 °C for 5 days. Colonies were counted after the incubation period and expressed as colony forming units (CFUs) g−1 of soil (dry weight). For each of the three biological replicates and considering the same dilution for all the treatments, all the colonies were collected. In our study, it corresponded to ca 20 colonies per replicate for a total of 60 bacterial strains per treatment and sampling time. The bacterial isolates were purified by three successive platings on 1/10 diluted TSA and then cryopreserved at −80 °C in 20% glycerol. A total of 420 bacterial strains were isolated. All the bacterial isolates used in this study were then cultivated on 1/10 diluted TSA at 25 °C for 72 h.
Mineral weathering potential of bacterial isolates
The mineral weathering potential, meaning the ability to mobilize inorganic nutrients of each bacterial isolate, was assessed through two commonly used bioassays measuring the ability: i) to mobilize iron and ii) to solubilize inorganic phosphorus28,38,59. The ability to mobilize iron was tested through the detection of siderophore production on the chrome azurol S (CAS) medium, following the protocol of Frey-Klett et al.67. This medium is composed of one litre of 800 mL of Solution 1 (34.36 g of Pipe Na2 Buffer (Sigma), 0.3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 0.246 g MgSO4∙7H2O, 0.0147 g CaCl2∙2H2O and 15 g agar in 800 mL H2O; pH 6.8), 100 mL of Solution 2 (4 g glucose in 100 mL H2O) and 100 mL of Solution 3 (Mix of 0.0905 g chrome azurol S in 75 mL, 0.0024 g FeCl3 in 15 mL, 0.1640 g hexadecyltrimethylammonium bromide in 60 mL H2O). Solutions 2 and 3 were added after autoclaving at a temperature of ca. 70 °C. The ability to solubilize inorganic phosphorous was tested using a 1/10 diluted tri-calcium phosphate (TCP) bioassay, according to Lepleux et al.28. This medium is composed of one litre of distilled water of 0.5 g NH4Cl, 0.1 g NaCl, 0.1 g MgSO4∙7H2O, 1 g glucose, 0.4 g Ca3(PO4)2 and 15 g agar (pH 6.5).
For each bioassay, bacterial isolates were grown for 72 h in 10 mL of liquid lysogeny broth (LB) medium at 25 °C. Two millilitres of each bacterial culture were then collected, washed three times in sterile water and suspended in 2 mL sterile water to obtain a calibrated inoculum suspension at λ 595 nm = 0.50 (ca. 107 cell mL−1). After vortexing, 10 µL of this suspension was then used in triplicate for the CAS and TCP assays. After incubation at 25 °C for 7 days, the diameters of colonies and the diameter of the discoloration zone were measured to determine the ability of each bacterial isolate to mobilize iron (CAS) and to solubilize inorganic phosphorous (TCP). The diameter of the discoloration zones was used to determine the relative efficacy of each bacterial isolate in each bioassay and their distribution in the three classes of efficacy based on the size of the discoloration zone on the different media (no mobilization activity, low activity and strong activity). For each medium, the determination of the class of efficacy was performed depending on the median of the discoloration zones of the positive strains (TCP = 1.4 cm; CAS = 1.8 cm) as follows: positive bacterial isolates with discoloration zones lower than the median were classified as bacterial isolates of low activity, whereas bacterial isolates with discoloration zones higher than the median were classified as bacterial isolates of strong activity. A total of 412 and 357 bacterial strains grew on TCP and CAS media, respectively.
Molecular identification of bacterial isolates
The 16S rRNA gene amplification of the bacterial isolates was performed using the universal set of primers pA [(5′-AGAGTTTGATCCTGGCTCAG-3′) and 907r (5′-CCGTCAATTCMTTTGAGTTT- 3′)68,69]. Polymerase chain reactions were performed in a total reaction volume of 50 µL containing 20 µL PCR Master mix (5-PRIME), 2 µL primers (10 µM) and 4 µL of inoculum suspension. The same inoculum suspension used for the functional assays described above was used for molecular characterization. The following temperature cycle was used: initial denaturation for 4 min at 94 °C, followed by 30 cycles of 30 s denaturation at 94 °C, 1 min annealing at 53 °C and 1 min 30 s extension at 72 °C, and a final extension for 10 min at 72 °C. PCR controls with no template or extraction control as a template were negative. PCR products were purified using the QIAquick purification kit (Qiagen, Valencia, CA) as recommended by the manufacturer. The sequencing was performed by MWG Eurofin Operon using the primers described above. The partial 16S rRNA gene sequences generated were compared with those of the GenBank databases using the BLAST programme70. They were aligned with reference 16S rRNA gene sequences using SeaView (version 4.5.471). Bootstrap values were generated using 1,000 replicates. Trees were generated using Dendroscope 3 software (version 3.4.472).
DNA extraction and quantitative PCR
For each treatment (K, Mg fertilized and control) and sampling time, total DNA was extracted from 250 mg of soil. Extractions were performed in three replicates using the ‘PowerSoilTM DNA Isolation Kit’ (MoBio Laboratories, Carlsbad, CA, USA) as recommended by the manufacturer. Concentrations of DNA were measured using a Nanodrop-1000 spectrometer (NanoDrop Technologies, Wilmington, DE, USA). The same quantity of total DNA was used to quantify the total bacterial 16S as well as the bacterial genera (Burkholderia, Collimonas and Pseudomonas) using the 16S rRNA gene-specific primers [10 μM each; 968 F/1401 R (total bacteria73), Burk3/BurkR (Burkholderia74), Eddy3For/Eddy3Rev/Sophie (Collimonas58) and PSf/Psr (Pseudomonas75), respectively]. For T0, T1 and T3, absolute quantifications were performed using serial dilutions of standard plasmids containing total or genus-specific bacterial 16S rDNA inserts (from 109 to 102 gene copies/μl) and the SsoAdvanced Universal SYBR Probes Supermix (classical qPCR for quantification of total bacteria, Burkholderia and Pseudomonas) and Probes Supermix (TaqMan qPCR for quantification of Collimonas) from Bio-Rad. PCR reaction were performed in a final volume of 20 µl containing 10 µl of the mix (SsoAdvanced Universal SYBR Probes Supermix or Probes Supermix), 1 µl of the total DNA, 2 µl of the primers (and of the Taqman probe when necessary) and milliQ water. The different bacterial genera and total bacterial quantifications were performed using the following cycle parameters: 1 cycle of 98 °C for 3 min followed by 40 cycles of 98 °C for 15 s, AT (annealing temperature) °C for 30 s (AT: 56 °C for total bacterial; 64 °C for Burkholderia; 63 °C for Pseudomonas; 66 °C for Collimonas). For each qPCR run using SYBR technology, a melting curve was performed at the end. The population sizes of total bacteria, Burkholderia, Collimonas and Pseudomonas, are expressed as log[number of 16S rRNA gene copies per gram dry weight soil].
PCR and pyrosequencing analyses
The 16S rRNA gene amplicon libraries were generated in one step using the primers 799 f and 1115r76,77 containing the specific Roche 454-pyrosequencing adaptors and 5 bases barcodes, according to Colin et al.34. PCRs contained 1X PCR Master mix (5 PRIME®), 500 nM 799 f primer, 500 nM 1115r primer and 8 ng of DNA in a final volume of 50 µL. Amplifications were performed using the following cycle parameters: 95 °C for 5 min (initial denaturation), followed by 30 cycles of 95 °C for 30 s, 57 °C for 35 s, and 72 °C for 30 s with a final extension step at 72 °C for 10 min. Triplicate PCR products were generated for each replicate and were then checked by gel electrophoresis, pooled and purified using the QIAquick purification kit (Qiagen, Valencia, CA, USA) as recommended by the manufacturer. The concentration of each purified PCR product was then measured using a Nanodrop-1000 spectrometer (NanoDrop Technologies, Wilmington, DE, USA), and an equimolar mix of the 16S rRNA gene amplicons was used for pyrosequencing on a 454 GS Junior system (Roche; Ecogenomic platform). All the treatments were sequenced, except for the Mg amended condition at T2 and T4. A total of 42,425 raw reads were generated. These 16S rRNA reads were then filtered for length (>300 bp), quality score (mean, ≥25), number of ambiguous bases (=0), and length of homopolymer runs (<8) using the trim.seqs script in Mothur v.1.30.2. These treatments generated a total of 31,216 quality sequences78. Chimeric sequences were detected using the chimera.uchime command and were removed from further analysis. To avoid any biases associated with different numbers of sequences in each of the samples, a random subsample of 448 sequences (corresponding to the smaller set of sequences after Mothur processing) from each sample was used for all downstream analyses, leading to a total of 14,784 sequences. High-quality sequences were aligned and clustered into 873 operational taxonomic units (OTUs), defined with 3% dissimilarity level and including 373 singletons. Taxonomy was assigned to each OTU by aligning sequences against the SILVA alignment database (version 128) with a bootstrap value of 80 for taxonomic assignment. The 16S rRNA gene sequences assigned as ‘unclassified bacteria’ with Silva were also verified using the BLAST programme70 against the NCBI database. Based on the OTU assignment, library richness and diversity estimates (Chao1, Shannon and inverse Simpson) were calculated in Mothur. In addition, a comparison of the 16S rRNA pyrosequences and Sanger sequences (generated from the culturable representative strains) was performed using the BLAST programme70. Alignment was performed with NCBI-BLAST + an e-value cut-off of 1 e-5 and a 100% overlap on the common portion (120 bases). Similar analyses have been performed with the 16S rRNA pyrosequences generated in a previous study on the same soil40.
Statistical analyses
All statistical analyses were performed using R 3.2.3 software79. The effect of the treatment and sampling time on the Biolog-substrate utilization profiles and on the respiration rates was determined by analysis of variance (one-factor ANOVA, P < 0.05, followed by a Tukey test). Similarly, ANOVA was conducted to determine the relative efficacy of the bacterial isolates to weather minerals (P < 0.05, followed by a Tukey test). Finally, the hypothesis of an equal distribution of the bacterial isolates according to their efficacy class in the functional assays was tested by a Chi2 test (P < 0.05) using the NCStats package80. Differences in the mineral weathering efficacy of the bacterial strains affiliated with a specific genus were assessed following the same procedure, but only for bacterial genera represented by more than 10 bacterial isolates. Estimation of the taxonomic diversity and richness based on culturable-independent data was carried out with Mothur, followed by statistical analysis (ANOVA followed by a Tukey test, P < 0.05). Permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis distances with 999 permutations were performed on the 16S rRNA pyrosequence data using the vegan package81. To assess structural differences between bacterial communities of treatments and/or sampling times, relative abundance values at all taxonomic levels (phylum, class, order, family, genus, OTU) were compared for taxa or OTUs presenting a minimum of 100 sequences. These values were transformed using the arcsine square root to achieve a normal distribution. A one-factor ANOVA was then conducted on those data, followed by a Tukey test with a threshold level of P < 0.05. A p adjustment was done using the false-Discovery-Procedure of Benjamini and Hochberg82 to determine the p value threshold.
Nucleotide sequence accession numbers
The sequences determined in this study have been deposited in the GenBank database for the Sanger sequences (MH918162-MH918556) and in the SRA trace archive for the pyrosequences (SRP013944; SRR7812734- SRR7812719).
Supplementary information
Acknowledgements
This work was funded by the Laboratory of Excellence Arbre (ANR-11-LABX-0002-01) in the frame of the multidisciplinary project ‘INABACT’. O. Nicolitch is a PhD student, and M. Feucherolles is a Master 2 student supported by a fellowship from the Laboratory of Excellence Arbre (ANR-11-LABX- 0002-01). The UMR1136 and UR1138 are supported by the French Agency through the Laboratory of Excellence Arbre (ANR-11-LABX-0002-01). The authors thank ANDRA and ONF. The English language was reviewed by American Journal Experts (ref. 874E-0EB1-A18F-F1C7-994A). The authors also thank Dr. P. Frey-Klett, P. Oger, A. Cébron, Y. Colin, L. Auer and members of the INABACT consortium for helpful discussions during the project.
Author Contributions
The present study was coordinated by S.U. and M.-P.T. The experiments were performed by O.N. and M.F., with the help of L.F. and J.-L.C. The data processing was done by O.N., S.U. and M.-P.T. The manuscript was written by S.U. and O.N. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50730-y.
References
- 1.Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. The American Naturalist, 1977;111:1169–1194. doi: 10.1086/283244. [DOI] [Google Scholar]
- 2.Ho A, Di Lonardo DP, Bodelier PL. Revisiting life strategy concepts in environmental microbial ecology. FEMS microbiology ecology, 2017;93:fix006. doi: 10.1093/femsec/fix006. [DOI] [PubMed] [Google Scholar]
- 3.Prosser JI, et al. The role of ecological theory in microbial ecology. Nature Rev. Microbiol. 2007;5:384. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 4.Wood JL, Tang C, Franks AE. Competitive Traits Are More Important than Stress-Tolerance Traits in a Cadmium-Contaminated Rhizosphere: A Role for Trait Theory in Microbial Ecology. Frontiers in Microbiol. 2018;9:121. doi: 10.3389/fmicb.2018.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langenheder S, Prosser JI. Resource availability influences the diversity of a functional group of heterotrophic soil bacteria. Environ. Microbiol. 2008;10:2245–2256. doi: 10.1111/j.1462-2920.2008.01647.x. [DOI] [PubMed] [Google Scholar]
- 6.Rasche F, et al. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. The ISME J. 2011;5:389. doi: 10.1038/ismej.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wawrik B, Kerkhof L, Kukor J, Zylstra G. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl. and Environ. Microbial. 2005;71:6776–6783. doi: 10.1128/AEM.71.11.6776-6783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enwall K, Philippot L, Hallin S. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. and Environ. Microbial. 2005;71:8335–8343. doi: 10.1128/AEM.71.12.8335-8343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Global Change Biol. 2012;18:1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x. [DOI] [Google Scholar]
- 10.Hallin S, Jones CM, Schloter M, Philippot L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. The ISME J. 2009;3:597. doi: 10.1038/ismej.2008.128. [DOI] [PubMed] [Google Scholar]
- 11.Shrestha M, Shrestha PM, Frenzel P, Conrad R. Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. The ISME J. 2010;4:1545. doi: 10.1038/ismej.2010.89. [DOI] [PubMed] [Google Scholar]
- 12.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J. of Biol. Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 13.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbio. Rev. 2003;272:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 14.Clarkson DT, Hanson JB. The mineral nutrition of higher plants. Ann. Rev. of Plant Physiol. 1980;31:239–298. doi: 10.1146/annurev.pp.31.060180.001323. [DOI] [Google Scholar]
- 15.Marschner, H. Mineral nutrition of higher plants. Academic Press, London, second edition (1995).
- 16.Tripler CE, Kaushal SS, Likens GE, Todd WM. Patterns in potassium dynamics in forest ecosystems. Ecol. Lett. 2006;9:451–466. doi: 10.1111/j.1461-0248.2006.00891.x. [DOI] [PubMed] [Google Scholar]
- 17.Calvaruso C, Kirchen G, Saint-André L, Redon PO, Turpault MP. Relationship between soil nutritive resources and the growth and mineral nutrition of a beech (Fagus sylvatica) stand along a soil sequence. Catena. 2017;155:156–169. doi: 10.1016/j.catena.2017.03.013. [DOI] [Google Scholar]
- 18.Augusto L, Ranger J, Binkley D, Rothe A. Impact of several common tree species of European temperate forests on soil fertility. Annals of Forest Sci. 2002;59:233–253. doi: 10.1051/forest:2002020. [DOI] [Google Scholar]
- 19.Akselsson, C. et al. Impact of harvest intensity on long-term base cation budgets in Swedish forest soils. Water, Air, & Soil Pollution: Focus 7, 201–210 (2007).
- 20.Frey B, et al. Weathering-associated bacteria from the Damma glacier forefield: physiological capabilities and impact on granite dissolution. Appl. and Environ. Microbial. 2010;76:4788–4796. doi: 10.1128/AEM.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uroz S, Tech JJ, Sawaya NA, Frey-Klett P, Leveau JHJ. Structure and function of bacterial communities in ageing soils: insights from the Mendocino ecological staircase. Soil Biol. and Biochem. 2014;69:265–274. doi: 10.1016/j.soilbio.2013.11.002. [DOI] [Google Scholar]
- 22.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proceed. of the Nation. Acad. of Sci. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. and Biochem. 2008;40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 24.Rousk J, et al. Soil bacteria and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Cheng C, He L, Huang Z, Sheng X. Characterization of depth related changes in bacterial communities involved in mineral weathering along a mineral-rich soil profile. Geomicrobiol. J. 2014;31:431–444. doi: 10.1080/01490451.2013.848248. [DOI] [Google Scholar]
- 26.Huang J, et al. Depth related changes in community structure of culturable mineral weathering bacteria and in weathering patterns caused by them along two contrasting soil profiles. Appl. and Environ. Microbial. 2014;80:29e42. doi: 10.1128/AEM.02335-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uroz S, Turpault MP, Van Scholl L, Palin B, Frey-Klett P. Long term impact of mineral amendment on the distribution of the mineral weathering associated bacterial communities from the beech Scleroderma citrinum ectomycorrhizosphere. Soil Biol. and Biochem. 2011;43:2275–2282. doi: 10.1016/j.soilbio.2011.07.010. [DOI] [Google Scholar]
- 28.Lepleux C, et al. short-term mineral amendment impacts the mineral weathering bacterial communities in an acidic forest soil. Res. in Microbiol. 2013;164:729–739. doi: 10.1016/j.resmic.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Puente ME, Bashan Y, Li CY, Lebsky VK. Microbial populations and activities in the rhizoplane of rock-weathering desert plants. I. Root colonization and weathering of igneous rocks. Plant Biol. 2004;6:629–642. doi: 10.1055/s-2004-821100. [DOI] [PubMed] [Google Scholar]
- 30.Uroz S, et al. Bacterial weathering and its contribution to nutrient cycling in temperate forest ecosystems. Res. in Microbiol. 2011;162:820–831. doi: 10.1016/j.resmic.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Calvaruso C, et al. Influence of forest trees on the distribution of mineral weathering-associated bacterial communities of the Scleroderma citrinum mycorrhizosphere. Appl. and Environ. Microbial. 2010;76:4780–4787. doi: 10.1128/AEM.03040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carson JK, et al. Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol. Ecol. 2009;67:381–388. doi: 10.1111/j.1574-6941.2008.00645.x. [DOI] [PubMed] [Google Scholar]
- 33.Herrera A, et al. Bacterial colonization and weathering of terrestrial obsidian in Iceland. Geomicrobiol. J. 2008;25:25–37. doi: 10.1080/01490450701828982. [DOI] [Google Scholar]
- 34.Colin, Y. et al. Mineral type and tree species determine the functional and taxonomic structure of forest soil bacterial communities. Appl. and Environ. Microbiol, 1–23, 10.1128/AEM.02684-16 (2010) [DOI] [PMC free article] [PubMed]
- 35.Minyard ML, et al. Bacterial associations with weathering minerals at the regolith-bedrock interface, Luquillo Experimental Forest, Puerto Rico. Geomicrobiol. J. 2012;29:792–803. doi: 10.1080/01490451.2011.619640. [DOI] [Google Scholar]
- 36.Olsson-Francis K, et al. A study of the microbial community at the interface between granite bedrock and soil using a culture-independent and culture-dependent approach. Advances in Microbiol. 2016;6:233–245. doi: 10.4236/aim.2016.63023. [DOI] [Google Scholar]
- 37.Lepleux, C. et al. Abundance of Beta-Proteobacteria on mineral surfaces correlates with mineral weathering in forest soils. Appl. and Environ. Microbiol, AEM-00996 (2012). [DOI] [PMC free article] [PubMed]
- 38.Uroz S, et al. Mineral weathering by bacteria: ecology, actors and mechanisms. Trends in Microbiol. 2009;17:378–87.. doi: 10.1016/j.tim.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Uroz S, et al. The mineralosphere concept: mineralogical control of the distribution and function of mineral-associated bacterial communities. Trends in Microbial. 2015;23:751–762. doi: 10.1016/j.tim.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Colin Y, et al. Taxonomic and functional shifts in the beech rhizosphere microbiome across a natural soil toposequence. Scientific reports, 2017;7:9604. doi: 10.1038/s41598-017-07639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolitch O, Colin Y, Turpault M-P, Uroz S. Soil type determines the distribution of nutrient mobilizing bacterial communities in the rhizosphere of beech trees. Soil Biol. and Biochem. 2016;103:429–445. doi: 10.1016/j.soilbio.2016.09.018. [DOI] [Google Scholar]
- 42.Court M, et al. Long-term effects of forest liming on mineral soil, organic layer and foliage chemistry: Insights from multiple beech experimental sites in Northern France. Forest Ecol. and Manag. 2018;409:872–889. doi: 10.1016/j.foreco.2017.12.007. [DOI] [Google Scholar]
- 43.Rosberg I, Frank J, Stuanes AO. Effect of liming and fertilization on tree growth and nutrient cycling in a Scots pine ecosystem in Norway. For. Ecol. Manag. 2006;237:191–207. doi: 10.1016/j.foreco.2006.09.045. [DOI] [Google Scholar]
- 44.Jeanbille, M. et al. Soil parameters drive the structure, diversity and metabolic potentials of the bacterial communities across temperate beech forest soil sequences. Microb. Ecol, 10.1007/s00248-015-0669-5 (2015). [DOI] [PubMed]
- 45.Pan Y, et al. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol. Ecol. 2014;90:195–205. doi: 10.1111/1574-6941.12384. [DOI] [PubMed] [Google Scholar]
- 46.Marschner P, Kandeler E, Marschner B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol. and Biochem. 2003;35:453–461. doi: 10.1016/S0038-0717(02)00297-3. [DOI] [Google Scholar]
- 47.Rath, K. M., Maheshwari, A., Bengtson, P. & Rousk, J. Comparative toxicity of salts to microbial processes in soil. Appl. and Environ. Microbiol. AEM-04052 (2016). [DOI] [PMC free article] [PubMed]
- 48.Huntington TG. The potential for calcium depletion in forest ecosystems of southeastern United States: review and analysis. Global Biogeochem. Cycles. 2000;14:623–638. doi: 10.1029/1999GB001193. [DOI] [Google Scholar]
- 49.Sardans J, Peñuelas J. Potassium: a neglected nutrient in global change. Global Ecol. and Biogeography. 2015;24:261–275. doi: 10.1111/geb.12259. [DOI] [Google Scholar]
- 50.Laing W, et al. Physiological impacts of Mg deficiency in Pinus radiata: growth and photosynthesis. New Phytol. 2000;146:47–57. doi: 10.1046/j.1469-8137.2000.00616.x. [DOI] [Google Scholar]
- 51.Courty PE, et al. The role of ectomycorrhizal communities in forest ecosystem processes: new perspectives and emerging concepts. Soil Biol. and Biochem. 2010;42:679–698. doi: 10.1016/j.soilbio.2009.12.006. [DOI] [Google Scholar]
- 52.Uroz S, Oger P. Caballeronia mineralivorans sp. nov., isolated from oak-Scleroderma citrinum mycorrhizosphere. System. and Appl. Microbiol., 2017;40:345–351. doi: 10.1016/j.syapm.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Leveau JH, Uroz S, De Boer W. The bacterial genus Collimonas: mycophagy, weathering and other adaptive solutions to life in oligotrophic soil environments. Environ. Microbiol. 2010;12:281–292. doi: 10.1111/j.1462-2920.2009.02010.x. [DOI] [PubMed] [Google Scholar]
- 54.Uroz, S., Oger, P., Morin, E. & Frey-Klett, P. Distinct ectomycorrhizospheres share similar bacterial communities as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. and Environ. Microbiol, AEM-06742 (2012). [DOI] [PMC free article] [PubMed]
- 55.Kelly LC, Colin Y, Turpault MP, Uroz S. Mineral type and solution chemistry affect the structure and composition of actively growing bacterial communities as revealed by bromodeoxyuridine immunocapture and 16S rRNA pyrosequencing. Microb. Ecol. 2016;72:428–442. doi: 10.1007/s00248-016-0774-0. [DOI] [PubMed] [Google Scholar]
- 56.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 57.Goldfarb KC, et al. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Frontiers in Microbial. 2011;2:94. doi: 10.3389/fmicb.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Höppener-Ogawa S, Leveau JH, Smant W, van Veen JA, de Boer W. Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems. Appl. and Environ. Microbiol. 2007;73:4191–4197. doi: 10.1128/AEM.00387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uroz S, et al. 2009b. Efficient mineral weathering is a distinctive functional trait of the bacterial genus Collimonas. Soil Biol. and Biochem. 2009;41:2178–2186. doi: 10.1016/j.soilbio.2009.07.031. [DOI] [Google Scholar]
- 60.Männistö MK, Häggblom MM. Characterization of psychrotolerant heterotrophic bacteria from Finnish Lapland. System. and Appl. Microbiol. 2006;29:229–243. doi: 10.1016/j.syapm.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 61.IUSS Working Group Wrb. World Reference Base for Soil Resources 2014, update 2015 International soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports No. 106, 192 (2015).
- 62.Bonneau, M. Fertilisation des forêts dans les pays tempérés. ENGREF, Nancy (1995).
- 63.Orsini L, Rémy JC. The use of the chloride of cobaltihexamine for the simultaneous determination of the exchange capacity and exchangeable bases in soils. Sci. Sol. 1976;4:269–275. [Google Scholar]
- 64.Rouiller J, Guillet B, Bruchert B. Cations acides échangeables et acidité de surface, approche analytique et incidences pédogénétiques. (In French.) Sci. Sol. 1980;2:161–175. [Google Scholar]
- 65.Garland JL, Mills AL. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campbell CD, et al. A rapid microtiter plate method to measure carbon dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl. and Environ. Microbiol. 2003;69:3593–3599. doi: 10.1128/AEM.69.6.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frey-Klett P, et al. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol. 2005;165:317–328. doi: 10.1111/j.1469-8137.2004.01212.x. [DOI] [PubMed] [Google Scholar]
- 68.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lane, D. J. 16S/23S rRNA sequencing. Nucleic Acid Techniques in Bacterial Systematics 125–175 (1991).
- 70.Altschul SF, et al. Gapped BLAST and PS I-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. and Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 72.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 73.Felske A, Akkermans AD, De Vos WM. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. and Environ. Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salles JF, De Souza FA, van Elsas JD. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. and Environ. Microbiol. 2002;68:1595–1603. doi: 10.1128/AEM.68.4.1595-1603.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Widmer F, et al. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Applied and Environ. Microbiol. 1998;64:2545–2553. doi: 10.1128/aem.64.7.2545-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chelius MK, Triplett EW. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb. Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 77.Reysenbach, A. L. & Pace, N. R. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction. Archaea: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor 101–107 (1995).
- 78.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. and Environ.Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team R: A language and environment for statistical computing (2016).
- 80.Ogle, D. H. NCStats: Helper Functions for Statistics at Northland College (2016).
- 81.Oksanen, J. et al. Vegan: Community Ecology Package (2017).
- 82.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological), 57, 289–300 (1995).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.