Abstract
目的
建立基于肠道菌群预测摄入胆碱食物后体内氧化三甲胺(TMAO)代谢水平变化趋势的模型,为个体精准膳食提供指导,同时为潜在的心脑血管疾病高危人群筛选提供参考。
方法
采集18位健康志愿者的空腹血清样本及食用等量胆碱食品8 h后的血清样本,根据前后血清TMAO水平升降趋势,将志愿者分为升高组和降低组。采集粪便样品,对粪便样本DNA的16 SrRNA基因V4可变区进行测序分析,比较两组志愿者的肠道菌群数据;根据肠道菌群及TMAO的数据,采用机器学习随机森林方法建立预测模型,并对模型进行检验。
结果
升高组与降低组在反映肠道菌群β多样性的主坐标分析(PCOA)图展示明显的区分,两组肠道菌群物种多样性存在差异;建立的预测模型曲线下面积为86.39%,95%置信区间为(72.7%,100%);验证集合代入模型显示降低组志愿者的TMAO变化水平被模型判定为降低的可能性明显高于升高组志愿者。
结论
根据肠道菌群可以建立预测TMAO水平变化模型,模型预测效果良好。
Keywords: 肠道菌群, 氧化三甲胺, 机器学习, 模型
Abstract
Objective
To establish a machine learning model based on gut microbiota for predicting the level of trimethylamine N-oxide (TMAO) metabolism in vivo after choline intake to provide guidance of individualized precision diet and evidence for screening population at high risks of cardiovascular disease.
Methods
We quantified plasma levels of TMAO in 18 healthy volunteers before and 8 h after a choline challenge (ingestion of two boiled eggs). The volunteers were divided into two groups with increased or decreased TMAO level following choline challenge. Fresh fecal samples were collected before taking fasting blood samples for amplifying 16S rRNA V4 tags, and the PCR products were sequenced using the platform of Illumina HiSeq 2000. The differences in gut microbiata between subjects with increased and decreased plasma TMAO were analyzed using QIIME. Based on the gut microbiota data and TMAO levels in the two groups, the prediction model was established using the machine learning random forest algorithm, and the validity of the model was tested using a verified dataset.
Results
An obvious difference was found in beta diversity of the gut microbota between the subjects with increased and decreased plasma TMAO level following choline challenge. The area under the curve (AUC) of the model was 86.39% (95% CI: 72.7%-100%). Using the verified dataset, the model showed a much higher probability for correctly predicting TMAO variation following choline challenge.
Conclusion
The model is feasible and reliable for predicting the level of TMAO metabolism in vivo based on gut microbiota.
Keywords: gut microbiota, trimethylamine-N-oxide, machine learning, model
越来越多证据表明氧化三甲胺(TMAO) 在心血管疾病的发生发展起重要作用。血液中TMAO的升高可导致动脉粥样硬化、血栓等心脑血管疾病发生风险增大[1-2]。食用高卵磷脂、胆碱等食物可引起体内TMAO水平的显著升高,血液中的TMAO通过上调巨噬细胞B族清道夫受体——CD36,进而促进胆固醇的沉积和斑块的生成[3-4]。肠道菌群作为一个有机体不仅在食物消化代谢中发挥作用,同时参与到机体代谢、免疫等过程中,进而影响人类的疾病与健康,而TMAO的产生同样也±赖于肠道菌群对食物的分解代谢[5-7]。随着研究的深入,发现即使进食等量的胆碱食物,TMAO在机体产生及代谢效率也存在一定差异,这可能与个体遗传多态性、肠道菌群的组成复杂等因素相关[8-10]。目前对TMAO的研究主要停留在TMAO在疾病发生发展中的作用,或者不同食物、药物等因素引起体内TMAO及肠道菌群改变等层面[2, 11-12],而较少从肠道菌群出发,探究肠道菌群对体内TMAO变化的影响,进而对体内TMAO变化进行预测。体内TMAO的预测不仅可以为个体健康精准膳食提供指导,甚至可能为心脑血管疾病的早期预防诊断提供参考[13-14]。为了探讨肠道菌群预测TMAO变化的可行性,我们根据研究对象摄入等量的胆碱食物后体内TMAO水平升降变化趋势进行分组,一方面探究两组志愿者肠道菌群的区别与联系;另一方面通过机器学习的方法构建基于肠道菌群预测胆碱饮食后TMAO变化趋势的模型,从而利用模型筛选出对胆碱饮食对体内TMAO影响较大的人群,对个体进行精准膳食指导,避免心脑血管事件的发生,最终达到降低心脑血管疾病患病风险的目标。
1. 资料和方法
1.1. 研究对象
所有研究对象均为健康志愿者,共18人,其中男8人,女10人 (非孕妇、非月经期),年龄24~39岁。纳入标准:无既往疾病史(包括心血管、肾脏、肺部、内分泌疾病)、无感染性疾病,1个月内没有服用过抗生素。
1.2. 实验方法
实验前1晚至第2次抽血期间,志愿者避免食用含红肉、蛋类和海鲜类食物,正常作息。实验当天清晨,志愿者采集空腹静脉血样5 mL及1~2个新鲜粪便样品后,空腹食用2个水煮鸡蛋(约含500 mg卵磷脂,中国人均推荐日摄入胆碱量为500 mg)。8 h后,再次采集静脉血样5 mL。
1.3. 标本采集
采集的静脉血5 mL,常温3000 r·min-1离心10 min,取上清保存在1.5 mL无菌EP管,编号后-40 ℃保存待测。新鲜粪便样品通过受检者自行用无菌粪便采样管采集,编号后-40 ℃保存待测。
1.4. TMAO检测
参照王珊[15]的检测方法,以d9-TMAO作为同位素内标进行定量,甲酸、甲醇作为流动A、B相,得到质谱图,峰面积经计算机转化为色谱数据得出待测血清TMAO浓度。1.5肠道菌群检测与分析
1.5. 肠道菌群检测与分析
1.5.1. DNA的提取
按照磁珠法DNA提取试剂盒(深圳易瑞)说明书步骤提取粪便中的总DNA,并将DNA产物置于-20 ℃保存待用。
1.5.2. PCR扩增
以16S rDNA V4区作为靶标对粪便总DNA进行PCR扩增,选用V4可变区上游引物为:V4F:5'-GTGTGCCAGCMGCCGCGGTAA-3',下游引物为V4R:5'-CCGGACTACHVGGGTWTCTAAT-3'。成功扩增后的PCR产物保存在4 ℃冰箱待测。
1.5.3. 测序及分析
PCR产物纯化后经Illumina HiSeq2000测序仪进行测序(华大基因)。采用双末端测序(BIPES)方法[16]及QIIME分析流程[17]对测序信息数据进行提取及处理,经UCHIME筛选去除嵌合体后进行可操作单元(Operational Taxonomic Units, OTU)[18]聚类、分类学分析。由Emperor[19]软件绘制主坐标分析图(PCOA Map)后,对两组数据在PCOA图PC坐标轴的投影距离进行比较,后期进行线性判别耦合效应分析(linear discriminant analysis coupled with effect sizemeasurements, LEfSe)[20]。
1.6. 肠道菌群预测
随机森林[21]算法是一种综合能力强、预测能力高的新机器学习算法,能有效地预测多达几千个变量在模型的作用。它的主要思想是通过将样品分成X份,以其中一份作为验证集合,其余X-1份样品作为训练集合进行训练,得到对应的子模型;然后±次进行N次训练,最后综合N个子模型得出最终预测模型及其性能指标。本实验采用机器学习随机森林算法的分类模型对所有研究对象的肠道菌群数据进行分类、聚类及回归分析,随机选取部分志愿者肠道菌群及分组信息数据作为训练集,随机分为N-1份进行N-1次训练和交叉验证,最终得出预测模型及最优的受试者的工作特征(ROC)的曲线下面积(AUC)。随后将验证集合代入模型,结合caret R压缩包计算错误率及预测贡献值[22]。
1.7. 统计分析
采用SPSS20.0软件对数据进行统计分析。各分组血清TMAO数据采用配对t检验,其他数据均符合正态性检验及方差齐性检验的进行方差分析检验,不符合条件者进行独立样本非参数检验。
2. 结果
2.1. 食用鸡蛋前后TMAO水平的变化
血清TMAO检测结果显示:18名志愿者食用鸡蛋前血清TMAO浓度为(2.12±1.26)μmol/L,食用鸡蛋8 h后血清TMAO浓度为(2.50±1.63)μmol/L,食用鸡蛋前后血清TMAO浓度对比没有统计学差异。对志愿者食用鸡蛋前后血清TMAO浓度的升降趋势进行分析,发现食用鸡蛋8 h后,8人的血清TMAO水平降低,其余10人的TMAO水平升高。根据TMAO升降趋势将研究对象分为TMAO升高组(n=10)和TMAO降低组(n=8)(图 1)。降低组志愿者空腹血清TMAO平均浓度为(2.90±1.46)μmol/L,8 h后平均浓度为(2.09±1.10)μmol/L,前后对比有统计学差异(P<0.05)。升高组志愿者空腹血清TMAO平均浓度为(1.50±0.61)μmol/L,8 h后为(2.82±1.96)μmol/L,前后对比有统计学差异(P<0.05)。
1.
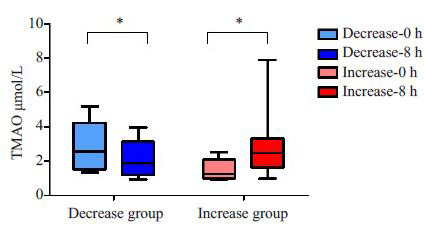
升高组与降低组TMAO变化
plasma TMAO changes in the increase group and decrease group. *P < 0.05.
2.2. TMAO变化水平与肠道菌群关系
2.2.1. 肠道菌群α多样性
肠道菌群α多样性用于反映组内肠道菌群丰富度差异,常用指标包括Chao 1,PDwhole tree,Shannon index等。Chao1用于估计菌落中OTU的数目;而PD whole tree用于比较进化关系上组间种属的多样性;Shannon index则用于计算群落中的所有物种数及平均度。由于肠道菌群α多样性数据不服从正态分布,故采用Mann-Whitney检验,统计结果显示以上指标P值均大于0.05,升高组与降低组间肠道菌群丰富度没有显著差异(表 1)。
1.
升高组与降低组肠道菌群α多样性
α diversity of intestinal microbiota between increase group and decrease group
| α Diversity | Group | n | Mann-Whitney U | Willcoxon W | Z | P |
| Chao 1 | Decrease Group | 8 | 36 | 72 | -0.355 | 0.722 |
| Increase Group | 10 | |||||
| PD whole tree | Decrease Group | 8 | 34 | 89 | -0.533 | 0.594 |
| Increase Group | 10 | |||||
| Shannon | Decrease Group | 8 | 31 | 86 | -0.800 | 0.424 |
| Increase Group | 10 | |||||
| Observed species | Decrease Group | 8 | 39 | 94 | -0.089 | 0.929 |
| Increase Group | 10 |
2.2.2. 肠道菌落构成
从属水平比较两组间整体肠道菌落结构构成(图 2),发现两组间肠道菌落构成未见明显差异,主要菌属均为Bacteroides(拟杆菌属)、Prevotella(普氏菌属)、Lachnospiraceae(毛螺菌科)等,比例分布较为相似。
2.
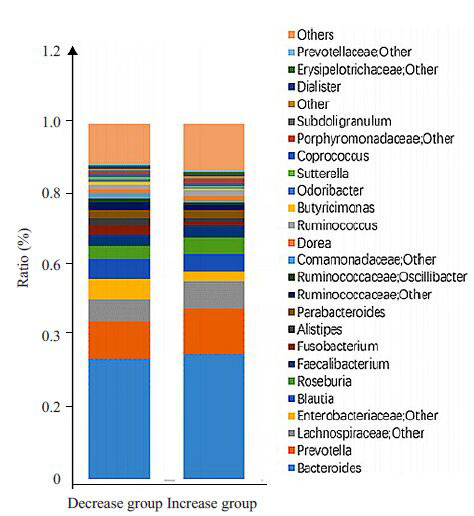
肠道菌落构成
Intestinal colony composition and proportion in the two groups.
2.2.3. 肠道菌群β多样性
肠道菌群β多样性用于反映组别间物种多样性的差异。由图 3可知,在abundant jaccard,bray curtis等PCOA图中均可直观地区分升高组和降低组。升高组与降低组在bray curtis、euclidean、pearson emperor、weighted unifrac的PC3轴;unweightedunifrac的PC5轴以及abundant jaccard的PC8轴上的投影距离具有统计学差异(P<0.05)。
3.
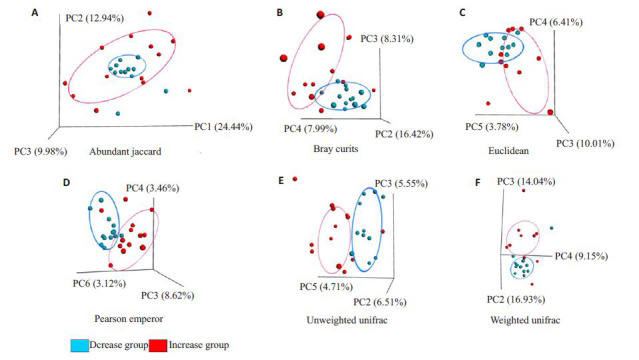
肠道菌群β多样性PCOA图
β diversity of intestinal microbiota on PCOAmap in the two groups. A: Abundant jaccard; B: Bray curtis; C: Euclidean; D: Pearson emperor; E: Unweighted unifrac; F: Weighted unifrac.
2.2.4. 差异菌属
LEfSe分析可以根据分类学对研究对象组与组之间进行线性判别分析,找出组间差异性显著的群落或物种。由图 4可知两组间差异性显著的菌属主要为Enterobacteriaceae(肠杆菌科)、Rikenellaceae(理研菌科)、Alistipes(另之菌属)、Comamonadaceae(丛毛单胞菌科)等菌属,以上菌属在降低组肠道菌群中所占比例高于升高组,均有统计学差异(P<0.05)。
4.
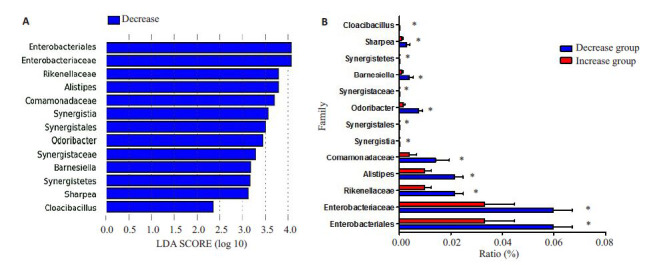
升高组与降低组肠道菌群差异性菌属
Differential gut bacterial species between the two groups. A: LEfSe analyze; B: Colony ration in intestinal microbiota. *P < 0.05.
2.3. 肠道菌群预测TMAO变化水平
机器学习建模结果(图 5A)显示:预测模型最优受试者工作特征曲线下面积为86.39%,95%置信区间为(72.7%,100%)。根据已建立的模型,将验证集合中的TMAO下降组代入模型验证,结果显示TMAO下降组的个体被预测为TMAO下降的可能性为0.59,被预测为TMAO升高的可能性为0.39(图 5B),预测效果符合预期。
5.
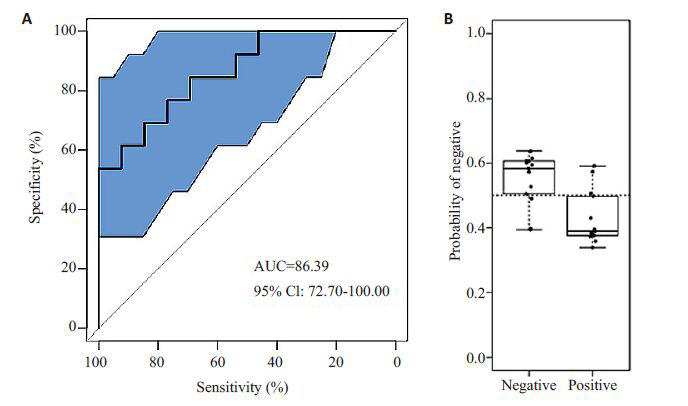
机器学习预测模型及预测效果检验
Machine learning model (A) for predicting TMAO changes and the result of verification (B).
3. 讨论
本次所有志愿者两次血清TMAO浓度为(2.31±1.45)μmol/L,与美国克利夫兰临床中心人群队列[23]正常人的TMAO水平相比(2.3~5.7 μmol/L),处于正常范围内的较低的水平,但与我国正常人群TMAO水平相近[15]。升高组及降低组志愿者进食胆碱食物前后血清TMAO浓度对比均具有统计学差异,说明两组研究对象具有较好的代表性。通过比较两组志愿者肠道菌群发现,两组志愿者肠道菌群α多样性指标均没有明显差异,菌落构成直方图亦未见明显差异,表明两组志愿者的肠道菌群丰富度比较接近。但是通过反映β多样性指标的PCOA图发现,两组志愿者的肠道菌群物种多样性具有明显差异,同时在LEfSe分析中亦可找出两组间肠道菌群中差异显著的菌属,提示肠道菌群物种多样性的差异与胆碱饮食后TMAO水平的变化趋势有一定的联系。对特征菌属进行分析,TMAO降低组特征菌以Enterobacteriaceae(肠杆菌科)、Rikenellaceae(理研菌科)、Alistipes(另之菌属)、Comamonadaceae(丛毛单胞菌科)等菌属为主,均为肠道细菌中常见菌属,并未发现文献中报道与TMAO生成或分解功能相关的特征菌属的显著变化[24]。已有文献报道降低组特征菌属中的理研菌科在肥胖患者和非酒精性脂肪肝患者肠道菌群中占有比例降低,这一结果提示血清TMAO变化可能与体内代谢状态相关[25]。
为了验证肠道菌群物种多样性差异与胆碱饮食后TMAO水平的变化趋势的关联性及强度,我们采用机器学习的方法,将个体的TMAO水平、肠道菌群种类构成及丰度度等数据纳入随机森林模型,对所有数据进行分类聚类、回归和分析,建立利用肠道菌群16SrDNA的V4片段测序数据预测胆碱饮食TMAO水平变化趋势模型。模型结果显示根据肠道菌群数据预测个体胆碱饮食后TMAO水平变化预测效果为86.39%,95%置信区间为(72.7%,100%),提示模型预测效果及可信度较高,肠道菌群与胆碱饮食后TMAO变化趋势有较高的关联性。将降低组验证集合代入构建的模型中,验证的结果显示降低组志愿者的TMAO变化水平被模型判定为下降的可能性高于升高组志愿者,证明模型预测可靠。
本次TMAO预测模型构建纳入对象为18名健康志愿者,未来应进一步扩大人群进行验证,纳入更多详细的临床资料或社区人群健康档案资料,以提高预测的准确率;同时,已有研究发现肠道菌群中的某些功能菌与体内TMAO水平密切相关[26-27],例如:肠道菌群中,厚壁菌门(Firmicutes)比例的增加可能与体内TMAO升高有关,而甲烷短杆菌属(Methanobacterium)则可以降低体内TMAO水平,这也提示TMAO预测模型的下一步改进应给予某些与TMAO代谢相关的功能菌更多的比重。
2015年以色列人群队列实验基于不同个体食用同一种食物后血糖变化差异显著的事实,首次利用肠道菌群建立餐后血糖预测模型,为“精准膳食“理念的实践提供参考[28]。介于个体间TMAO代谢的差异及肠道菌群在TMAO生成中的关键作用[29],本研究建立的TMAO预测模型,提示肠道菌群具备预测TMAO的潜力。可以设想,除了本研究提及的TMAO预测外,与菌群相关的功能或疾病均有可能建立相应的预测模型。近期,粪菌移植[30]成为新的临床研究热点,该技术被尝试应用于多种疾病的预防与治疗,包括假膜性肠炎、自身免疫性疾病、炎症性肠炎(IBD)等疾病[31]。粪菌移植被誉为“一种特殊的器官移植”[32],供体肠道菌群的优劣直接影响移植后宿主的健康。本研究建立预测模型提示,可以通过菌群模型为供体的筛选提供技术参考,剔除“不健康的供体”,降低粪菌移植后风险。
因此,基于肠道菌群及机器学习建立的TMAO预测模型可有效地预测食用胆碱食物后体内TMAO的变化趋势,可用于精准膳食指导及粪菌移植供体筛选等领域,为降低人群心脑血管疾病患病风险提供参考。
Biography
路浚齐,在读硕士研究生,E-mail: goodlujunqi@163.com
Funding Statement
国家自然科学基金(81671171);广东省自然科学基金(2014A030313353);广州市科技计划项目(201510010078)
Supported by National Natural Science Foundation of China (81671171)
Contributor Information
路 浚齐 (Junqi LU), Email: goodlujunqi@163.com.
周 宏伟 (Hongwei ZHOU), Email: biodegradation@gmail.com.
References
- 1.Martinez-del Campo A, Romano KA, Rey FE, et al. The plot thickens: Diet microbe interactions May modulate thrombosis risk. Cell Metab. 2016;23(4):573–5. doi: 10.1016/j.cmet.2016.03.017. [Martinez-del Campo A, Romano KA, Rey FE, et al. The plot thickens: Diet microbe interactions May modulate thrombosis risk [J]. Cell Metab, 2016, 23(4): 573-5.] [DOI] [PubMed] [Google Scholar]
- 2.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84. doi: 10.1056/NEJMoa1109400. [Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk[J]. N Engl J Med, 2013, 368(17): 1575-84.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease[J]. Nature, 2011, 472(7341): 57-63.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Febbraio M, Podrez EA, Smith JD, et al. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105(8):1049–56. doi: 10.1172/JCI9259. [Febbraio M, Podrez EA, Smith JD, et al. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice[J]. J Clin Invest, 2000, 105(8): 1049-56.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22(1):283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine[J]. Annu Rev Nutr, 2002, 22(1): 283-307.] [DOI] [PubMed] [Google Scholar]
- 6.Diehl G, Longman RS, Zhang J, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494(7435):116–20. doi: 10.1038/nature11809. [Diehl G, Longman RS, Zhang J, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells [J]. Nature, 2013, 494(7435): 116-20.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. https://www.researchgate.net/profile/Martina_Wallace/publication/229152782_Gut_microbiota_composition_correlates_with_diet_and_health_in_the_elderly/links/0deec534ddb665eeaa000000.pdf?inViewer=0&pdfJsDownload=0&origin=publication_detail. Nature. 2012;488(7410):178. doi: 10.1038/nature11319. [Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly[J]. Nature, 2012, 488 (7410): 178.] [DOI] [PubMed] [Google Scholar]
- 8.Miller CA, Corbin KD, da Costa K, et al. Effect of egg ingestion on trimethylamine-N-oxide production in humans: a randomized, controlled, dose-response study. Am J Clin Nutr. 2014;100(3):778–86. doi: 10.3945/ajcn.114.087692. [Miller CA, Corbin KD, da Costa K, et al. Effect of egg ingestion on trimethylamine-N-oxide production in humans: a randomized, controlled, dose-response study[J]. Am J Clin Nutr, 2014, 100(3): 778-86.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho CE, Taesuwan S, Malysheva OV, et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition:a randomized controlled trial. Mol Nutr Food Res. 2016;61(1) doi: 10.1002/mnfr.201600324. [Cho CE, Taesuwan S, Malysheva OV, et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition:a randomized controlled trial[J]. Mol Nutr Food Res, 2016, 61(1). doi: 10.1002/mnfr.201600324.] [DOI] [PubMed] [Google Scholar]
- 10.Tomoya Y. Intestinal immunity and gut microbiota in atheerogenesis. J Atheroscler Thro. 2016;23(12):1334–44. doi: 10.5551/jat.35634. [Tomoya Y. Intestinal immunity and gut microbiota in atheerogenesis [J]. J Atheroscler Thro, 2016, 23(12): 1334-44.] [DOI] [Google Scholar]
- 11.Spence JD. Dietary cholesterol and egg yolk should be avoided by patients at risk of vascular disease. https://www.researchgate.net/publication/230654642_Egg_yolk_consumption_and_carotid_plaque. J Trans Int Med. 2016;4(1):20–4. doi: 10.1515/jtim-2016-0005. [Spence JD. Dietary cholesterol and egg yolk should be avoided by patients at risk of vascular disease[J]. J Trans Int Med, 2016, 4(1): 20-4.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang WH. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl Res. 2017;179:108–15. doi: 10.1016/j.trsl.2016.07.007. [Tang WH, Microbiome, trimethylamine N-oxide, and cardiometabolic disease[J]. Transl Res, 2017, 179: 108-15.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. https://www.researchgate.net/profile/Judith_Aron-Wisnewsky/publication/285216793_The_gut_microbiome_diet_and_links_to_cardiometabolic_and_chronic_disorders/links/56faacad08ae3c0f264c01da.pdf?inViewer=0&pdfJsDownload=0&origin=publication_detail. Nat Rev Nephrol. 2016;12(3):169–81. doi: 10.1038/nrneph.2015.191. [Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders[J]. Nat Rev Nephrol, 2016, 12(3): 169-81.] [DOI] [PubMed] [Google Scholar]
- 14.Robinson-Cohen C, Newitt R, Shen DD, et al. Association of FMO3 variants and trimethylamine N-Oxide concentration, disease progression, and mortality in CKD patients. https://iths.pure.elsevier.com/en/publications/association-of-fmo3-variants-and-trimethylamine-n-oxide-concentra. PLoS One. 2016;11(8):e161074. doi: 10.1371/journal.pone.0161074. [Robinson-Cohen C, Newitt R, Shen DD, et al. Association of FMO3 variants and trimethylamine N-Oxide concentration, disease progression, and mortality in CKD patients[J]. PLoS One, 2016, 11 (8): e161074.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.王 珊, 夏 耿红, 何 彦, et al. 氧化三甲胺分布特征及其与肠道菌群的关联性. http://www.j-smu.com/oa/DArticle.aspx?type=view&id=201604455. 南方医科大学学报. 2016;36(4):455–60. [王珊, 夏耿红, 何彦, 等.氧化三甲胺分布特征及其与肠道菌群的关联性[J].南方医科大学学报, 2016, 36(4): 455-60.] [PubMed] [Google Scholar]
- 16.Zhou HW, Li DF, Tam NF, et al. BIPES, a cost-effective highthroughput method for assessing microbial diversity. ISME J. 2011;5(4):741–9. doi: 10.1038/ismej.2010.160. [Zhou HW, Li DF, Tam NF, et al. BIPES, a cost-effective highthroughput method for assessing microbial diversity[J]. ISME J, 2011, 5(4): 741-9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.盛 华芳, 周 宏伟. 微生微生物组学大数据分析方法、挑战与机遇. http://www.j-smu.com/oa/darticle.aspx?type=view&id=201507931. 南方医科大学学报. 2015;(7):931–4. [盛华芳, 周宏伟.微生微生物组学大数据分析方法、挑战与机遇[J].南方医科大学学报, 2015(7): 931-4.] [PubMed] [Google Scholar]
- 18.Rideout JR, He Y, Navas-Molina JA, et al. Subsampled openreference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [Rideout JR, He Y, Navas-Molina JA, et al. Subsampled openreference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences[J]. PeerJ, 2014, 2: e545.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez-Baeza Y, Pirrung M, Gonzalez AA. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2(1):16. doi: 10.1186/2047-217X-2-16. [Vazquez-Baeza Y, Pirrung M, Gonzalez AA. EMPeror: a tool for visualizing high-throughput microbial community data [J]. Gigascience, 2013, 2(1): 16.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang X, Wang X, Yang SG, et al. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using High-Throughput sequencing. https://www.researchgate.net/publication/308340751_Evaluation_of_the_Microbial_Diversity_in_Amyotrophic_Lateral_Sclerosis_Using_High-Throughput_Sequencing. Front Microbiol. 2016;7:149. doi: 10.3389/fmicb.2016.01479. [Fang X, Wang X, Yang SG, et al. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using High-Throughput sequencing[J]. Front Microbiol, 2016, 7: 149.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [Breiman L. Random forests[J]. Mach Learn, 2001, 45(1): 5-32.] [DOI] [Google Scholar]
- 22.黄宜娥. 孕期生殖道微生物组与不良妊娠结局的关联[D]. 广州: 南方医科大学, 2015.
- 23.Tang W., Wang ZE, Fan YY, et al. Prognostic value of elevated levels of intestinal Microbe-Generated metabolite Trimethylamine-N-Oxide in patients with heart failure refining the gut hypothesis. J Am Coll Cardiol. 2014;64(18):1908–14. doi: 10.1016/j.jacc.2014.02.617. [Tang W., Wang ZE, Fan YY, et al. Prognostic value of elevated levels of intestinal Microbe-Generated metabolite Trimethylamine-N-Oxide in patients with heart failure refining the gut hypothesis [J]. J Am Coll Cardiol, 2014, 64(18): 1908-14.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163(7):1585–95. doi: 10.1016/j.cell.2015.11.055. [Wang Z, Roberts AB, Buffa JA, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis[J]. Cell, 2015, 163(7): 1585-95.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delchierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric NAFLD and obese patients unveiled by an integrated meta-omics based approach. https://www.researchgate.net/profile/Giorgio_Capuani/publication/299518730_Gut_microbiota_profiling_of_pediatric_NAFLD_and_obese_patients_unveiled_by_an_integrated_meta-omics_based_approach/links/57430b8e08ae298602ee6e04.pdf?origin=publication_detail. Hepatology. 2016;65(2):451–64. doi: 10.1002/hep.28572. [Delchierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric NAFLD and obese patients unveiled by an integrated meta-omics based approach[J]. Hepatology, 2016, 65(2): 451-64.] [DOI] [PubMed] [Google Scholar]
- 26.Brugère JF, Borrel G, Gaci N, et al. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes. 2014;5(1):5–10. doi: 10.4161/gmic.26749. [Brugère JF, Borrel G, Gaci N, et al. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease[J]. Gut Microbes, 2014, 5(1): 5-10.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XS, Obeid S, Klingenberg R, et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes:a prognostic marker for incident cardiovascular events beyond traditional risk factors. http://eurheartj.oxfordjournals.org/content/early/2017/01/10/eurheartj.ehw582. Eur Heart J. 2017;pii:ehw582. doi: 10.1093/eurheartj/ehw582. [Li XS, Obeid S, Klingenberg R, et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes:a prognostic marker for incident cardiovascular events beyond traditional risk factors[J]. Eur Heart J, 2017, pii: ehw582.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeevi D, Korem T, Zmora N, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. doi: 10.1016/j.cell.2015.11.001. [Zeevi D, Korem T, Zmora N, et al. Personalized nutrition by prediction of glycemic responses[J]. Cell, 2015, 163(5): 1079-94.] [DOI] [PubMed] [Google Scholar]
- 29.Tang W., Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124(10):4204–11. doi: 10.1172/JCI72331. [Tang W., Hazen SL. The contributory role of gut microbiota in cardiovascular disease[J]. J Clin Invest, 2014, 124(10): 4204-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly CP. Fecal microbiota transplantation-an old therapy comes of age. N Engl J Med. 2013;368(5):474–5. doi: 10.1056/NEJMe1214816. [Kelly CP. Fecal microbiota transplantation-an old therapy comes of age[J]. N Engl J Med, 2013, 368(5): 474-5.] [DOI] [PubMed] [Google Scholar]
- 31.Brandt LJ, Aroniadis OC. An overview of fecal microbiota transplantation: techniques, indications, and outcomes. Gastrointest Endosc. 2013;78(2):240–9. doi: 10.1016/j.gie.2013.03.1329. [Brandt LJ, Aroniadis OC. An overview of fecal microbiota transplantation: techniques, indications, and outcomes[J]. Gastrointest Endosc, 2013, 78(2): 240-9.] [DOI] [PubMed] [Google Scholar]
- 32.郭 文涛, 董 丽娜, 王 俊平, et al. 粪菌移植的临床应用新进展. http://www.cnki.com.cn/Article/CJFDTOTAL-XXHB201430008.htm. 世界华人消化杂志. 2014;(30):4593–8. [郭文涛, 董丽娜, 王俊平, 等.粪菌移植的临床应用新进展[J].世界华人消化杂志, 2014(30): 4593-8.] [Google Scholar]


